Abstract
Import of Hansenula polymorpha alcohol oxidase (AO) into peroxisomes is dependent on the PTS1 receptor, HpPex5p. The PTS1 of AO (-LARF) is sufficient to direct reporter proteins to peroxisomes. To study AO sorting in more detail, strains producing mutant AO proteins were constructed. AO containing a mutation in the FAD binding fold was mislocalized to the cytosol. This indicates that the PTS1 of AO is not sufficient for import of AO. AO protein in which the PTS1 was destroyed (-LARA) was normally sorted to peroxisomes. Moreover, C-terminal deletions of up to 16 amino acids did not significantly affect AO import, indicating that the PTS1 was not necessary for targeting. Consistent with these observations we found that AO import occurred independent from the C-terminal TPR-domain of HpPex5p, known to bind PTS1 peptides. Synthesis of the N-terminal domain (amino acids 1-272) of HpPex5p in pex5 cells restored AO import, whereas other PTS1 proteins were mislocalized to the cytosol. These data indicate that AO is imported via a novel HpPex5p-dependent protein translocation pathway, which does not require the PTS1 of AO and the C-terminal TPR domains of HpPex5p, but involves FAD binding and the N-terminus of HpPex5p.
INTRODUCTION
Growth of the yeast Hansenula polymorpha on methanol as sole carbon and energy source is associated with the massive development of peroxisomes in the cells (Veenhuis and Harder, 1987). These organelles are crucial for growth because they harbor the key enzymes of methanol metabolism, namely alcohol oxidase (AO), dihydroxyacetone synthase (DHAS), and catalase.
One topic of research in the laboratory includes the biosynthetic pathway of AO, focused on the mechanisms of the sorting and assembly/activation of the enzyme. AO is a homo-octameric flavoenzyme of 600 kDa; each of the eight identical subunits contains one noncovalently bound flavin molecule (FAD) as a cofactor. In wild-type (WT) H. polymorpha cells AO enzyme activity is confined to peroxisomes (van der Klei et al., 1990).
AO monomers are synthesized in the cytosol and subsequently posttranslationally imported into the target organelle, where assembly into the active octamer is presumed to take place. AO may represent one of the exceptions to the rule that peroxisomal matrix proteins are imported in their oligomerized state (Stewart et al., 2001; (Faber et al., 2002). In riboflavin auxotrophic mutants of H. polymorpha that produce reduced levels of FAD, AO import into peroxisomes is strongly inhibited (Evers et al., 1994, 1996). Therefore, binding of the cofactor FAD to AO monomers may be essential to allow import. Recently, evidence was presented that FAD-binding requires the function of pyruvate carboxylase protein (HpPyc1p), a cytosolic protein essential for import of AO into peroxisomes (Ozimek et al., 2003).
Here, we study sorting and import of AO in H. polymorpha peroxisomes in more detail. Strains were constructed in which the WT AOX gene was replaced by mutant ones. The analysis of these strains revealed that FAD binding is essential for targeting of AO, whereas the extreme C-terminus of AO, which contains the putative PTS1, was not required. Also, the C-terminal TPR domain of HpPex5p, known to bind PTS1 tripeptides, was dispensable for AO sorting. The details of this work are included in this article.
MATERIALS AND METHODS
Organisms and Growth
The H. polymorpha strains used in this study are H. polymorpha NCYC 495 (leu1.1; Gleeson and Sudbery, 1988), pex3::URA3 (leu1.1.pex3; Baerends et al., 1996), pex5::URA3 (leu1.1.pex5; Salomons et al., 2000a), and HF295, a DHAS deletion strain (leu1.1.ΔDHAS; van Dijk R. et al., 2001). Yeast cells were grown at 37°C in selective minimal media containing 0.67% yeast nitrogen base without amino acids supplemented with 0.5% glucose (YND) or 0.5% methanol (YNM) or in mineral media (van Dijken et al., 1976) containing 0.25% ammonium sulfate as nitrogen source supplemented with 0.5% glucose, 0.5% methanol, or 0.5% methanol + 0.1% glycerol as carbon sources. When required leucine or uracil were added to a final concentration of 30 mg/l. Escherichia coli DH5α (Sambrook et al., 1989) was grown on LB medium supplemented with the appropriate antibiotics.
Molecular Techniques
Standard recombinant DNA techniques were carried out essentially as described by Sambrook et al. (1989). Transformation of H. polymorpha was performed by electroporation (Faber et al., 1994b). Restriction enzymes and biochemicals were obtained from Roche (Almere, The Netherlands) and used as detailed by the manufacturer. The plasmids used in this study are listed in Table 1.
Table 1.
Plasmids used in this study
| Plasmid | Relevant properties | Reference |
|---|---|---|
| pAOX | E. coli plasmid containing PAOXAOX | Distel et al. (1987) |
| pHIPX1 | Shuttle vector containing PAOX and S. cerevisiae | Faber et al. (1994a) |
| LEU2 flanked by promoter and terminator regions of HpAOX; allows gene replacement of WT AOX by mutant versions | ||
| pHIPX4 | Shuttle vector containing the AO promoter (PAOX) | Gietl et al. (1994) |
| pHIPX4-PEX5 | pHIPX4 containing the PEX5 gene under control of PAOX | van der Klei et al. (1995) |
| pHIPX4-PEX5-ZEO | pHIPX4-PEX5 containing ZEO resistance gene | This study |
| pHIPX4-N-PEX5 | pHIPX4-PEX5-ZEO encoding C-terminal truncated HpPex5p instead of full length HpPex5p | This study |
| pHIPX5-PYC | pHIPX5 containing the PYC gene under control of the amine oxidase promoter (PAMO) | Ozimek et al. (2003) |
| pHIPX5-PYC-ZEO | pHIPX5-PYC containing ZEO resistance gene | This study |
| pHIPX1-AOXΔN (N = 1, 4, 10, 16, 22) | pHIPX1 encoding different truncated AOX genes | This study |
| pHIPX1-AOX-LARA | pHIPX1 encoding AO-LARA | This study |
| pHIPX4-AOX-G15A | pHIPX4 encoding AO-G15A | This study |
Construction of AOX Mutants
AO mutant strains were constructed by replacing the WT AOX gene by mutant ones using targeted integration. The mutant AOX genes were created by PCR using the universal primer and the primers listed in Table 2 using plasmid pAOX (Distel et al., 1987) as template. The obtained DNA fragments were inserted in pHIPX1 (Faber et al., 1994a). The resulting plasmids were designated pHIPX1-AOXΔ1, pHIPX1-AOXΔ4, pHIPX1-AOXΔ10, pHIPX1-AOXΔ16, and pHIPX1-AOXΔ22 and encode AO proteins lacking 1, 4, 10, 16, or 22 C-terminal residues. pHIPX1-AOX-LARA encodes AO containing a C-terminal A instead of F. The plasmids were linearized by BamHI and transformed to WT NCYC 495. pHIPX1-AOXΔ22 was also transformed to pex3::URA3 and leu1.1 ΔDHAS. Correct integration in the AOX locus was analyzed by Southern blot analysis. The resulting strains were designated WT-AOΔ1, AOΔ4, AOΔ10, AOΔ16, AOΔ22, AO-LARA (Figure 1), pex3- AOΔ22, and ΔDHAS-AOΔ22.
Table 2.
Primers used in this study
| Name | Sequence 5′-3′ |
|---|---|
| AOX-LARA | CCCAGGCTTAGGCCTTAGGCTCTGGCAAGTCCGGTC |
| AOXΔ1 | CCCAAGCTTGAATTCTTATCTGGCAAGTCCGGTCTCC |
| AOXΔ4 | CCCAAGCTTATCCGGTCTCCTCGTAAGTTCC |
| AOXΔ10 | CCCAGGCTTGCTAGCTTATCCGAGTCTGAAGTTTGGAATCG |
| AOXΔ16 | CCCAGGCTTACTAGTTAAATCGTCATGTCCAGGTCGG |
| AOXΔ22 | CCCAAGCTTCTAGATTAGGAGCCTGAGTAGCCAAGATC |
| AOX G15A | CGATATCATTGTTGTTGGTGGCGCCTCCACCGGCTGC |
| AOX-downstream | CCCCTGCAGTTAGAATCTGGCAAGTCCGGTCTCC |
| N-PEX5-downstream | AGTCATCGTACGCAGATTGTTACGGAACTGATTATTC |
Figure 1.
C-terminal amino acid sequences of AO from WT H. polymorpha and the mutated and truncated AO proteins used in this study. The introduced cloning sites in the corresponding DNA fragment are indicated as well.
AO protein with a mutation in the FAD binding fold was created by changing G at position 15 into A. The mutation was introduced into the AOX gene using primers AOX G15A and AOX-downstream (Table 2). The PCR product was cloned into plasmid pHIPX4 (Gietl et al., 1994). The resulting plasmid, designated pHIPX4-AOX G15A, was linearized and integrated into the AOX locus of WT NCYC 495 as detailed above. The resulting strain was designated AO-G15A.
HpPex5p and HpPyc1p Overproduction Strains
An H. polymorpha strain producing AOΔ22 (Figure 1) and overproducing HpPex5p was obtained by placing the PEX5 gene under control of the strong AOX promoter. To this purpose a fragment containing the ZEO gene, obtained from pHIPZ4-NIa (Faber et al., 2001) upon digestion with SacI/KpnI, was inserted into plasmid pHIPX4-PEX5 (van der Klei et al., 1995). The resulting plasmid pHIPX4-PEX5-ZEO was linearized by NotI and transformed to WT-AOΔ22. Correct integration in the AOX promoter was analyzed by Southern blotting.
A strain overproducing HpPyc1p was created by inserting a ZEO containing fragment, obtained upon digestion of pHIPZ4-NIa with SpeI/ScaI, into pHIPX5-PYC (Ozimek et al., 2003) digested with SpeI/HpaI. The resulting plasmid pHIPX5-PYC-ZEO was linearized by SpeI and transformed to WT-AOΔ22. Transformants were tested for correct integration in the AMO promoter by Southern blotting.
Construction of a Truncated H. polymorpha PEX5 Gene
A strain, producing the first N-terminal 272 amino acids of HpPex5p in a PEX5 deletion strain (pex5), was constructed by insertion of a DNA fragment consisting of the first 816 base pairs of PEX5 (1-816PEX5) in pex5::URA3 (leu1.1pex5) by targeted integration. For this purpose the region encoding the N-terminal HpPex5p-domain was created by PCR using the universal primer and primer N-PEX5-downstream (Table 2) and inserted into plasmid pHIPX4 via NotI/SphI. The resulting plasmid pHIPX4-N-PEX5 was linearized by ApaI and transformed to pex5::URA3 (leu1.1pex5). Transformants were tested for correct integration in the PEX5 promoter by Southern blot analysis. The resulting strain was designated pex5(1-272)Pex5p.
Protein Purification
AO was isolated from WT H. polymorpha, WT-AOΔ16, and WT-AOΔ22 as detailed previously (van der Klei et al., 1990). A HpPex5p-His6 fusion protein was overproduced in E. coli and purified by affinity chromatography using Ni-NTA-resin (Qiagen, Hilden, Germany) as described previously (Wang et al., 2003).
Biochemical Methods
Crude extracts of H. polymorpha cells were prepared as described previously (van der Klei et al., 1991c). AO activity was measured according to Verduyn et al. (1984). Protein concentrations were determined using the Bio-Rad Protein Assay system (Bio-Rad GmbH, Munich, Germany) using BSA as a standard.
Sucrose-gradient centrifugation for the separation of monomeric and octameric AO was performed according to Goodman et al. (1984).
AO protein was immunoprecipitated from crude extracts using specific anti-AO antibodies and protein A-Sepharose beads essentially as described before (van der Klei et al., 1989). The crude extracts were prepared from cells producing WT AO or AOΔ16 precultivated on glucose medium and shifted for 5 h to methanol-containing medium. Immunoprecipitates were analyzed by Western blotting using anti-HpPex5p or anti-AO antibodies.
SDS-PAGE was carried out according to Laemmli (1970). Western blotting was performed as described by Kyhse-Andersen (1984). The blots were decorated using specific antibodies against H. polymorpha AO. Levels of AO protein were determined by densitometric scanning of the blots.
HpPex5p-AO Binding Experiments
Purified native octameric AO was dissociated into monomers by incubation in 80% glycerol (final AO protein concentration 0.1 μM) for 20 min at 23°C (Evers et al., 1995). To study interactions of HpPex5p with WT AO or the C-terminally truncated form AOΔ16, AO protein incubated in 80% glycerol was diluted 10 times in 50 mM potassium phosphate buffer, pH 6.0, containing 0.3 μM purified HpPex5p (Evers et al., 1995). The samples were kept on ice for 15 min before being subjected to nondenaturating gel electrophoresis using 4-10% polyacrylamide gradient gels (Musgrove et al., 1987). Subsequently, Western blots were prepared from the gels and decorated with anti-AO antibodies.
Protein concentrations of purified AO and HpPex5p were determined from the absorbance at 280 nm using the corresponding molar extinction coefficient of AO (ε280 = 93500 M-1 cm-1) and HpPex5p (ε280 = 58000 M-1 cm-1).
FAD content of purified AO
Purified AO protein (WT and truncated forms) was precipitated with 12.5% TCA, a procedure that caused the release of noncovalently bound FAD (van der Klei et al., 1989). After centrifugation (20 min, 20,000 × g, 4°C) fluorescence intensities were determined in the supernatants (excitation at 450 nm, emission at 521 nm) using a FluoroMax-3 spectrofluorimeter (Jobin Yvon, Edison, NJ). Solutions of purified FAD were used as standard.
UV illumination of native gels was used to qualitatively determine the presence of FAD in various AO proteins.
Electron Microscopy and Immunocytochemistry
Whole cells were fixed and prepared for electron microscopy and immunocytochemistry as described (Waterham et al., 1994). Immunolabeling was performed on ultrathin sections of Unicryl-embedded cells using specific polyclonal antibodies against various H. polymorpha proteins and gold-conjugated goat anti-rabbit antibodies (Waterham et al., 1994).
RESULTS
A Point Mutation (G15A) in the Putative FAD-binding Site Prevents AO Import and Assembly
Recently, Ozimek et al. (2003) suggested that cytosolic binding of FAD to newly synthesized AO monomers, a process mediated by pyruvate carboxylase (HpPyc1p), is essential for targeting of the protein to peroxisomes. To determine the importance of FAD-binding for AO import in a direct way, an H. polymorpha strain (AO-G15A) was constructed in which the WT AOX gene was replaced by a mutant one encoding AO protein containing an amino acid substitution in the FAD binding fold (Gly15 into Ala).
Growth experiments revealed that H. polymorpha AOG15A cells were unable to grow in media containing methanol as sole carbon and energy source and lacked AO enzyme activity. Western blotting experiments, however, indicated that AO protein was normally produced, but contained no FAD judged from fluorescence analysis of native gels prepared from crude extracts of AO-G15A cells (Figure 2A). In control experiments using crude extracts of WT cells, FAD fluorescence was evident at the position in the gel where octameric AO protein was located (Figure 2A). Western blot analysis of the native gels also revealed that AO protein produced in H. polymorpha AO-G15A was not oligomerized but present as monomers (Figure 2B).
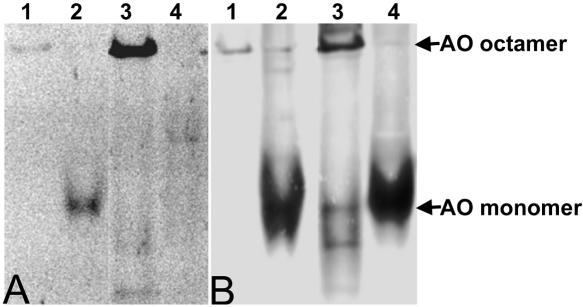
Figure 2.
Analysis of oligomerization and FAD binding in AOG15A. (A) A native gel exposed to UV light to visualize FAD fluorescence. (B) A Western blot of a similar native gel decorated with anti-AO antibodies. Purified WT octameric AO (20 μg protein; lane 1) or an equal amount of WT AO protein dissociated into monomers (incubated in 80% glycerol; lane 2) were used as controls. Lanes 3 and 4: 200 μg protein of crude extracts prepared from WT (lane 3) and AO-G15A (lane 4) cells. (A) Octameric (lane 1 and 3) and monomeric AO (lane 2) obtained by dissociation of native WT AO contained FAD, as evident from the fluorescent bands. Monomeric AO present in crude extracts of strain AO-G15A lacked FAD (lane 4). (B) Western blotting revealed that in crude extracts of AO-G15A (lane 4) only monomers were observed, whereas in crude extracts of WT cells (lane 3) octameric AO also is found. The locations of AO octamers and monomers are indicated by arrows and were determined using purified WT octameric (lane 1) or monomeric (lane 2) AO as controls. Because of less efficient blotting of AO octamers compared with monomers, the signal of monomeric AO is relatively strong.
In ultrathin sections of cells of H. polymorpha AO-G15A, anti-AO labeling was confined to the cytosol, indicating that AO protein was mislocalized to the cytosol (Figure 3) under conditions that the PTS1 import machinery was normally functioning.
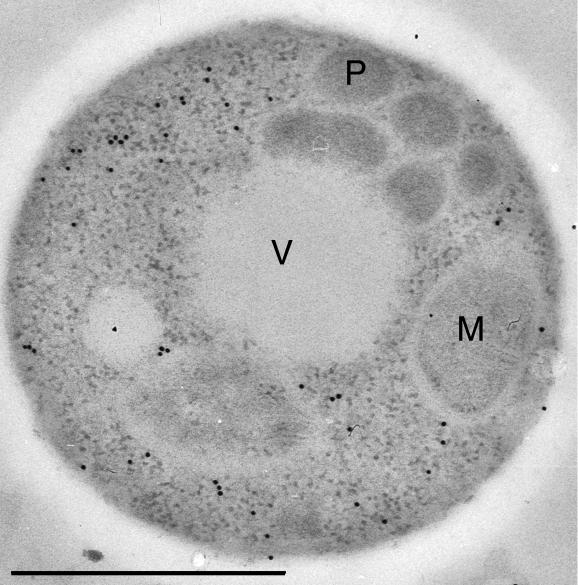
Figure 3.
AO-G15A protein is mislocalized to the cytosol. Methanol-induced AO-G15A cells were analyzed immunocytochemically using anti-AO antibodies. Labeling was confined to the cytosol and not detected on peroxisomal profiles. M, mitochondrion; P, peroxisome; V, vacuole. Aldehyde fixation. Bar, 0.5 μm.
Short C-terminal Deletions Do Not Affect AO Protein Sorting and Enzyme Activity
AO-G15A is not imported into peroxisomes despite the presence of a functional PTS1 (-LARF; Waterham et al., 1997; Salomons et al., 2000b) at the extreme C-terminus of the protein. Therefore, we analyzed the significance of the C-terminus for AO sorting in more detail. Six mutants were constructed in which either the putative PTS1 was inactivated (the extreme C-terminal F changed into A) or deleted (deletions of 1, 4, 10, 16, or 22 C-terminal amino acids, respectively; see Figure 1). All strains, except AOΔ22 (for details see below), grew normally in media containing methanol as sole carbon source at WT rates and displayed normal specific AO activities in crude extracts (approximately 4 U/mg protein). Also, immunolabeling experiments revealed that these mutant AO proteins were normally imported into peroxisomes (Figure 4). Sucrose gradients prepared of crude extracts from these cells indicated that these mutant AO proteins were predominantly in the octameric state (shown for WT and AOΔ16; Figure 5). Also, the specific activity of purified WT and mutant AOs (except for AOΔ22) were similar and amounted to ∼20 U/mg protein.
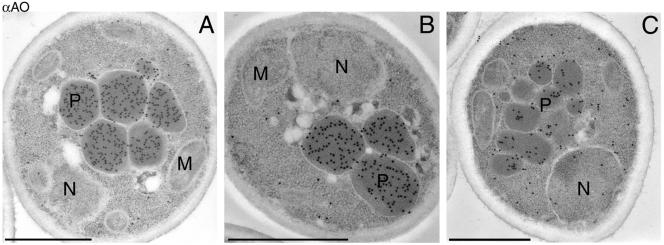
Figure 4.
Immunocytochemical localization of the mutant AO proteins AO-LARA (A), AOΔ16 (B), and AOΔ22 (C). To illustrate (partial) AO import, sections of cells were selected in which several peroxisomes were present. The distribution of the labeling demonstrates that AO-LARA and AOΔ16 protein is confined to peroxisomes, whereas AOΔ22 protein is present both in peroxisomes and in the cytosol. Aldehyde fixation. M, mitochondrion; N, nucleus; P, peroxisome. Bar, 0.5 μm.
Figure 5.
Separation of octameric and monomeric AO protein by sucrose density centrifugation. Crude extracts (A) prepared from methanol-induced cells producing WT AO, AOΔ16, or AOΔ22 protein or purified, octameric AO (B) isolated from WT cells or cells producing AOΔ22 were subjected to sucrose density centrifugation. The fractions obtained were analyzed by Western blotting using anti-AO antibodies. In crude extracts of WT cells and cells producing AOΔ16, AO protein was predominantly octameric (peak fraction in lane 3), whereas crude extracts prepared from cells producing AOΔ22 contained both AO monomers (peak fraction lane 7) and octamers. Purified, octameric WT AO obtained by gel filtration chromatography remained octameric upon sucrose density centrifugation (B), whereas AO octamers purified from cells synthesizing AOΔ22 partly had dissociated into monomers. Equal volumes of the fractions obtained from each gradient were loaded per lane. From the gradients prepared from AOΔ22 cells larger volumes of each fraction were loaded per lane compared with WT and AOΔ16, to allow detection of the strongly reduced amounts of AO protein in these cells.
Cells that produced AOΔ22 showed an aberrant behavior. The initial growth of such cells on methanol was normal, but growth ceased at an OD corresponding to the midexponential growth stage in WT cells (unpublished data). These cells displayed >90% reduction in specific AO activities (0.3 U/mg protein) relative to WT cells. Analysis of Western blots prepared from crude extracts of these cells, indicated that this reduction in AO activity was associated with a comparable reduction in AO protein levels (unpublished data).
Immunocytochemically, AOΔ22 showed a dual location in both peroxisomes and the cytosol (Figure 4). Also, sucrose gradient analysis indicated that ∼70% of the total AOΔ22 protein was in the monomeric state (Figure 5A). Because AOΔ16 was predominantly octameric, this indicates that deletion of six additional amino acids in AOΔ22 severely affected the oligomerization and stability of the protein. This view was consistent with the observation that the specific activity of purified AOΔ22 protein had decreased to 9 U/mg protein. Also, the FAD content of AOΔ22 was significantly lower than WT and AOΔ16 (Table 3), suggesting that the capacity to bind or retain FAD is also affected by the deletion of 22 C-terminal amino acids.
Table 3.
FAD content of purified AO proteins
| WT AO | AOΔ16 | AOΔ22 | |
|---|---|---|---|
| Fluorescence intensities | 1 | 1.13 | 0.20 |
FAD was extracted from equal amounts of purified octamers of WT AO, AOΔ16, and AOΔ22 and quantified by fluorescence spectroscopy. The fluorescence intensity of WT AO was arbitrarily set to 1.
Possibly, the strong reduction in AO protein levels in AOΔ22 cells was due to instability and degradation of not properly imported or assembled AO protein in vivo. To analyze this, we took advantage of the fact that in H. polymorpha PEX3 deletion (pex3) cells, AO is normally synthesized, assembled, and stable in the cytosol but is not subject to selective degradation upon a shift of cells to glucose-containing media, as is peroxisomal AO in WT cells (van der Klei et al., 1991b). Because glucose fully represses AO synthesis, the analysis of the fate of cytosolic AOΔ22 relative to WT AO protein in a pex3 background allowed to determine the stability of both proteins in vivo upon a shift to glucose media. The results, depicted in Figure 6, indicated that WT AO protein showed the expected behavior and remained unaffected in methanol-induced pex3 cells, exposed to excess glucose. However, the level of mutant AOΔ22 protein rapidly diminished in identical cells to <10% of the original amounts in a few hours time interval, indicative of a decreased stability of this protein in vivo. Because ∼70% of the total AO protein was octameric in methanol-induced AOΔ22 cells (Figure 5A), these data imply that both monomeric and octameric AOΔ22 proteins display reduced stability relative to WT AO. Indeed, when purified octameric AOΔ22, obtained by gel filtration chromatography, was subsequently subjected to sucrose density centrifugation, both monomers and octamers were found (Figure 5B), indicating that part of the AOΔ22 octamers was dissociated into monomers during the procedure. In a control experiment using WT AO, only AO octamers were found, indicating that these oligomers are more stable (Figure 5B).
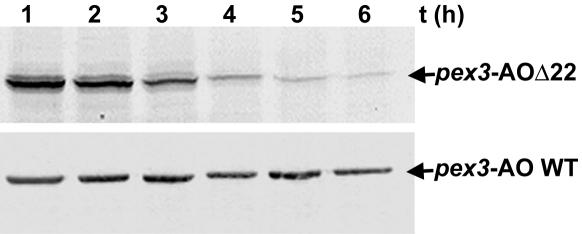
Figure 6.
AOΔ22 is less stable than WT AO in vivo. Methanol-induced pex3 cells, producing WT AO or AOΔ22 protein, were shifted to glucose media to fully repress AO synthesis. Samples were taken at the indicated time points after the shift and analyzed by Western blotting using anti-AO antibodies. The level of AOΔ22 protein decreased with time, whereas WT AO levels remained constant. Equal portions of the cultures were loaded per lane.
Import of Truncated AO Is Dependent on HpPex5p and Is Not Due to Piggybacking with DHAS
Next, we addressed whether the import of C-terminal truncated AO species was still dependent of HpPex5p. To this end, AOΔ22 was produced in a PEX5 deletion strain (pex5) and localized by immunocytochemistry. Inspection of ultrathin sections of glycerol/methanol-grown cells revealed that mutant AOΔ22 protein was confined to the cytosol (unpublished data). Hence, import of AOΔ22 (and also the other C-terminal truncated AO species) is still dependent of HpPex5p function.
The observed import of mutant AO protein lacking the carboxyterminal PTS1 raised the question whether this import could be due to piggyback import together with another high abundant PTS1 matrix protein, DHAS. This aspect was investigated by producing AOΔ22 protein in a DHAS deletion strain. The distribution pattern of AOΔ22 protein in the DHAS deletion strain was indistinguishable from that in a WT background (Figure 7, compare Figure 4C), thus suggesting that AOΔ22 import was not due to piggyback import together with DHAS. Also, overproduction of HpPex5p or HpPyc1p, a protein shown to be essential for FAD binding and AO import (Ozimek et al., 2003), did not affect the dual subcellular AOΔ22 distribution (unpublished data), suggesting that the key components of the AO sorting machinery were not limiting for import.
Figure 7.
AOΔ22 is not imported via piggybacking together with DHAS. Immunocytochemical localization of AOΔ22 protein synthesized in a H. polymorpha DHAS deletion strain. AO specific labeling is found on peroxisomal profiles and in the cytosol. Aldehyde fixation. M, mitochondrion; P, peroxisome; N, nucleus. Bar, 0.5 μm.
Truncated AO Binds to HpPex5p
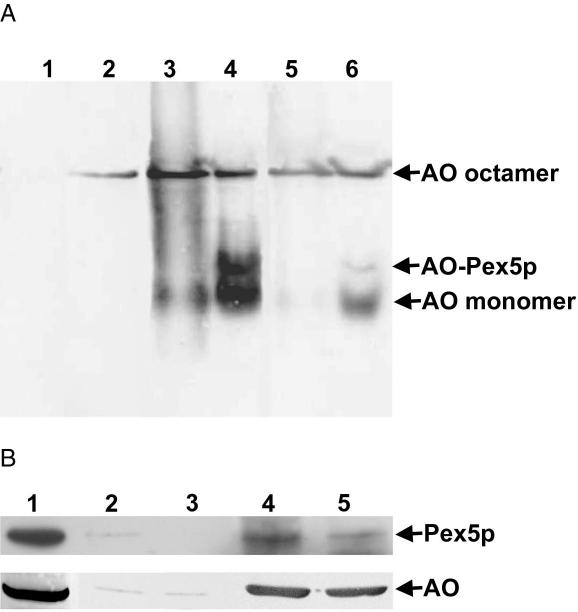
To study whether the C-terminal truncated AO forms were capable of binding to HpPex5p, in vitro binding studies were performed using purified HpPex5p, WT AO, and AOΔ16 protein. We previously showed that HpPex5p solely binds AO monomers, but not octamers (Faber et al., 2002). Therefore purified octameric WT AO and AOΔ16 proteins were first dissociated into monomers by incubation in 80% glycerol (compare Figure 2, lanes 2). This procedure does not result in removal of FAD from the monomers and therefore is associated with reoligomerization of the protein after dilution of the glycerol (Evers et al., 1995; Boteva et al., 1999; see also Figure 8A, lane 3). To analyze HpPex5p binding, the AO/glycerol solutions were diluted in buffer containing purified HpPex5p. On native gel electrophoresis and Western blotting, an additional anti-AO cross-reacting band was observed, located in between the bands of AO monomers and octamers (Figure 8A, lane 4). This band was absent in samples of purified HpPex5p (Figure 8A, lane 1) and thus is not due to cross-reactivity of the AO antiserum with a contaminating protein in the purified HpPex5p sample. Therefore it most likely represents a complex containing HpPex5p and AO. Using truncated AOΔ16 the additional band was also observed, indicating that HpPex5p binds this truncated AO protein as well (Figure 8A, lane 6). These results were confirmed by coimmunoprecipitation experiments using crude extracts of WT and AOΔ16 cells. As shown in Figure 8B, HpPex5p coprecipitated with both forms of AO in immunoprecipitation experiments using specific antibodies against AO.
Figure 8.
HpPex5p binds AOΔ16 protein. (A) Demonstration of in vitro association of purified HpPex5p with purified WT AO or AOΔ16 protein analyzed by native gel electrophoresis followed by Western blotting using anti-AO antibodies. Purified HpPex5p (lane 1), octameric WT AO (lane 2), and octameric AOΔ16 (lane 5) are used as controls. WT octameric AO was dissociated into monomers by incubation in 80% glycerol (see Figure 2, lane 2). On dilution in buffer, the reduction in glycerol concentration causes that monomeric AO reassociates into octamers (Evers et al., 1995). This explains the presence of two bands, octameric AO and monomeric AO, on the Western blot (lane 3). On dilution of the AO sample incubated in 80% glycerol with buffer containing purified HpPex5p, an additional band above the monomeric AO band appeared (lane 4). After the same procedure performed with AOΔ16 protein, this additional band was detected as well (lane 6), indicating that also AOΔ16 protein associates with HpPex5p. (B) Coimmunoprecipitation experiments using crude extracts of cells producing WT AO or AOΔ16 and specific anti-AO antibodies and protein A-Sepharose beads. Immunoprecipitates were analyzed by Western blotting using anti-HpPex5p or anti-AO antibodies as indicated. Lane 1: total crude extract of WT cells to show the position of AO and HpPex5p on the Western blots. Lanes 2 and 3: control experiments using protein A-Sepharose beads without anti-AO antiserum; lanes 4 and 5: immune-precipitation experiment using protein A-Sepharose beads and anti-AO antiserum. Lane 2 and 4 WT cells. Lanes 3 and 5: cells producing AOΔ16. In control experiments (lanes 2 and 3) minor amounts of HpPex5p and AO protein were detected most likely because of nonspecific binding to the protein A-Sepharose beads. In the presence of anti-AO antiserum high amounts of AO protein were precipitated from both extracts. These precipitates also contained HpPex5p (lanes 4 and 5).
Import of WT AO and AOΔ16 Is Mediated by the N-terminal Domain of HpPex5p
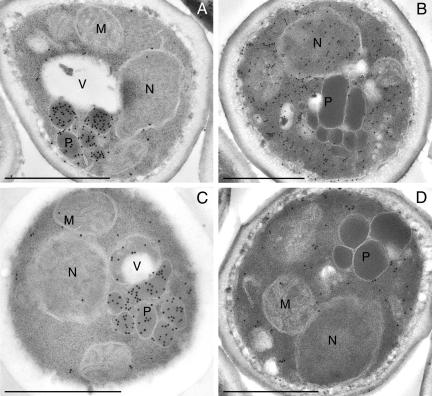
The finding that import of AO occurred independent of the PTS1 suggested that it may require other domains of HpPex5p than the C-terminus where the PTS1 binding TPR repeats are located. To investigate this, we constructed a truncated PEX5 gene consisting of nucleotides 1-816, encoding amino acids 1-272 of HpPex5p, and expressed this gene under control of the PEX5 promoter in a pex5 deletion strain. Glycerol/methanol grown cells of this strain contained normal specific AO activities (approximately 4U/mg protein in crude cell extracts). The location of AO was analyzed immunocytochemically. The data, shown in Figure 9, show that anti-AO-dependent labeling was predominantly localized on peroxisomal profiles, indicating that the protein was localized in these organelles (Figure 9A). Essentially similar results were obtained in a strain in which also the WT AOX gene was replaced by AOXΔ16 (Figure 9C). For the other PTS1 proteins, DHAS and catalase, specific labeling was confined to the areas of cytosol, indicating that these proteins were mislocalized in these strains (Figure 9, B and D).
Figure 9.
AO is imported into peroxisomes in cells producing C-terminally truncated HpPex5p. Immunocytochemical localization of AO (A and C), DHAS (B), and catalase (D) in a PEX5 deletion strain producing the N-terminal domain of HpPex5p (residues 1-272) that lacks the TPR motifs. WT AO (A) and AOΔ16 (C) proteins were normally located in peroxisomes, whereas other PTS1 proteins (DHAS; B), or catalase (D) were mislocalized to the cytosol. Aldehyde fixation. M, mitochondrion; N, nucleus; P, peroxisome; V, vacuole. Bar, 0.5 μm.
DISCUSSION
We have studied the biosynthetic pathway of peroxisomal AO focused on sorting of the protein to its target organelle. The extreme carboxyterminal amino acids of AO contains a PTS1 (Waterham et al., 1997;Salomons et al., 2000b) and import of AO into peroxisomes requires the function of the PTS1 receptor, Pex5p (McCollum et al., 1993; van der Klei et al., 1995). However, previous studies revealed that the sorting pathway of AO displays several unusual features. First, only AO monomers can be imported into peroxisomes. Pulse-chase experiments in combination with cell fractionation studies using Candida boidinii cells, convincingly demonstrated that AO octamers never occur in the cytosol of WT cells, whereas oligomerization could be followed within the peroxisomal matrix (Waterham et al., 1993;Stewart et al., 2001). In line with these results are the observations of Faber et al. (2002), who showed that H. polymorpha Pex5p (HpPex5p) is capable of binding monomeric but not octameric AO, explaining why AO octamers that artificially accumulated in the cytosol were not imported into peroxisomes. Another peculiar finding is that deletion of the carboxyterminal four amino acids of Pichia pastoris AO did not fully block AO import, suggesting that P. pastoris AO contains a second PTS in another part of the protein (Waterham et al., 1997). Finally, several observations in H. polymorpha suggested that translocation of AO monomers requires binding of the cofactor FAD (Evers et al., 1994, 1995, 1996; Ozimek et al., 2003).
In this article we provide direct evidence that FAD binding is a prerequisite for import of H. polymorpha AO into peroxisomes, because a point mutation (AO-G15A) that prevents FAD binding fully blocks import of the protein. This finding is remarkable in view of the fact that H. polymorpha AO-G15A still contains a PTS1. The failure to import AOG15A was not due to a general defect in PTS1 protein import in the cells, because other PTS1 proteins were normally imported. This suggests that in H. polymorpha AO the PTS1 may not be recognized by HpPex5p. The reason behind this phenomenon is fully unknown.
Consistent with this finding is our observation that import of AO occurred efficiently in the absence of the PTS1 sequence. In fact up to 16 amino acids could be deleted from the C-terminus without affecting AO import or assembly. Import of the proteins also remained dependent on the function of HpPex5p and was not due to piggyback import together with DHAS, as previously reported for Saccharomyces cerevisiae Eci1p (Yang et al., 2001). Eci1p is a peroxisomal enzyme involved in beta-oxidation of unsaturated fatty acids and, like AO, is imported into peroxisomes in a Pex5p-dependent way upon deletion of the PTS1. However, in this case it has been demonstrated that in the absence of the PTS1, Eci1p was translocated as a hetero-oligomer with another PTS1 protein, Dcilp (Yang et al., 2001). In vitro binding experiments using purified HpPex5p and purified AOΔ16 protein demonstrated that HpPex5p directly interacts with AO lacking a PTS1, in line with the view that the protein is not imported by a piggyback mechanism.
The most remarkable finding in this study includes that AO sorting is not dependent on the PTS1 of AO and the C-terminal part of HpPex5p, which contains the TPR motifs known to bind PTS1 sequences (Gatto et al., 2000). Instead, only the N-terminal half of the PTS1 receptor was essential to mediate AO import. The fact that in PEX5 deletion cells production of the N-terminal half of HpPex5p solely facilitated import of AO but not of the other PTS1 proteins catalase and DHAS stresses the exceptional import pathway of AO.
Recently, Distel and coworkers (Klein et al., 2002) indicated that S. cerevisiae acyl-CoA oxidase, a protein that lacks both a PTS1 and PTS2, interacts with the N-terminus of ScPex5p and Purdue and Lazarow (2001) reported that the region of ScPex5p responsible for this interaction was mapped to residues 136-292 of ScPex5p, a domain clearly distinct from the TPR repeat region. Whether the corresponding region of HpPex5p is involved in AO binding is the topic of current investigations.
Our data suggest that a not yet identified internal PTS of AO (a putative PTS3) is recognized by HpPex5p. This PTS3 apparently becomes functional upon FAD binding, explaining why in the absence of FAD binding, import of AO is fully blocked (Evers et al., 1994;Ozimek et al., 2003; this article). This also suggests that the PTS3 of AO represents a structural rather than an amino acid sequence motif.
The importance of FAD binding for AO import may explain why AOΔ22 was only partially imported. Most likely deletion of 22 carboxyterminal residues interferes with the tertiary structure of AO (Boteva et al., 1999) and affects FAD binding and subsequent octamerization. This is indicated by the reduced FAD content of purified AOΔ22 protein and the increased amounts of monomeric AO found in crude extracts of WT-AOΔ22 cells. Also, the stability of AOΔ22 protein was strongly reduced in vivo. Because our in vivo stability test revealed that AOΔ22 protein disappeared almost completely during incubation of cells for 6 h, apparently both AOΔ22 monomers and octamers are degraded. Whether degradation of octamers is preceded by dissociation of octamers into monomers and release of FAD is not known. Also, whether the putative peroxisomal protease (Stewart et al., 2002) contributes to degradation of AOΔ22 protein inside peroxisomes of WT-AOΔ22 cells remained elusive.
At present several peroxisomal matrix proteins are known to be imported in their oligomeric enzymatically active state, which implies that folding and cofactor binding of these proteins occurs in the cytosol. For Yarrowia lipolytica acyl-CoA oxidase, Titorenko et al. (2002) showed that assembly of pentameric complexes is obligatory for import into the peroxisome. Most likely the proteins that facilitate folding, assembly, and cofactor binding to these peroxisomal enzymes do not occur in the organelles but are located to the cytosol. Examples are Y. lipolytica Pex20p, involved in thiolase assembly (Titorenko et al., 1998) and HpPyc1p, a cytosolic protein that is necessary for FAD binding to AO (Ozimek et al., 2003). This pathway of protein translocation has characteristics of the Tat-system for secretion of cofactor containing enzymes in bacteria. These enzymes are not secreted by the Sec system, because the Sec translocon can only accommodate unfolded proteins. Because bacteria cannot control cofactor binding after the protein has been secreted, this process occurs in the cytosol before export (Palmer and Berks, 2003). An example of a Tat protein is glucose-fructose oxidoreductase, which requires a twin arginine (Tat) motif in the signal peptide for secretion. Interestingly, mutations that affect binding of the cofactor severely affected secretion (Halbig et al., 1999). For another Tat-protein evidence was obtained that a specific chaperone may exist that keeps the protein in a conformation competent to bind the cofactor and at the same time shelters the signal peptide to prevent interaction with the Tat-translocase (Sargent et al., 2002). It is tempting to speculate that cytosolic HpPyc1p, which has been suggested to facilitate FAD binding to AO, fulfils a comparable function in the AO sorting pathway (Ozimek et al., 2003).
Interestingly, binding of the cofactor and—at least partial—folding of the AO monomer occurs in the cytosol, whereas the oligomerization process is postponed to a stage after import. Why is this mechanism used? We hypothesize that this is related to specific physiological advantages. We showed before that the presence of low amounts of active oligomeric AO in the cytosol is associated with severe energetic disadvantages and prevents growth of WT cells on methanol (van der Klei et al., 1991a). Only few yeast species have gained the capacity of methylotrophic growth. As these species are closely related, acquisition of this property may have been a rare and relatively late event in the evolution of the organisms. Because cytosolic assembly/activation of the protein had to be prevented for metabolic reasons, probably only those species could survive that invented an intermediate system: partial folding and FAD-binding before import and—most likely spontaneous (Evers et al., 1995)—assembly upon translocation in the target organelle. Possibly this adaptation has changed the interaction of the PTS1 of AO with the TPR domains of HpPex5p into a novel mode of binding involving the N-terminus of HpPex5p and an internal PTS3 that requires FAD binding.
Acknowledgments
I.J.V.D.K. holds a PIONIER fellowship (NWO). K.G. is supported by ALW/NWO and R.V.D. by STW. We thank Klaas Sjollema and Ineke Keizer-Gunnink for assistance in electron microscopy and Dong Yuan Wang for help in fluorescence measurements.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03-04-0258. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-04-0258.
References
- Baerends, R.J.S. et al. (1996). The Hansenula polymorpha PER9 gene encodes a peroxisomal membrane protein essential for peroxisome assembly and integrity. J. Biol. Chem. 271, 8887-8894. [DOI] [PubMed] [Google Scholar]
- Boteva, R., Visser, A.J., Filippi, B., Vriend, G., Veenhuis, M., and van der Klei, I.J. (1999). Conformational transitions accompanying oligomerization of yeast alcohol oxidase, a peroxisomal flavoenzyme. Biochemistry 38, 5034-5044. [DOI] [PubMed] [Google Scholar]
- Distel, B., Veenhuis, M., and Tabak, H.F. (1987). Import of alcohol oxidase into peroxisomes of Saccharomyces cerevisiae. EMBO J. 6, 3111-3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evers, M.E., Harder, W., and Veenhuis, M. (1995). In vitro dissociation and re-assembly of peroxisomal alcohol oxidases of Hansenula polymorpha and Pichia pastoris. FEBS Lett. 368, 293-296. [DOI] [PubMed] [Google Scholar]
- Evers, M.E., Titorenko, V.I., Harder, W., van der Klei, I.J., and Veenhuis, M. (1996). Flavin adenine dinucleotide binding is the crucial step in alcohol oxidase assembly in the yeast Hansenula polymorpha. Yeast 12, 917-923. [DOI] [PubMed] [Google Scholar]
- Evers, M.E., Titorenko, V.I., van der Klei, I.J., Harder, W., and Veenhuis, M. (1994). Assembly of alcohol oxidase in peroxisomes of the yeast Hansenula polymorpha requires the cofactor flavin adenine dinucleotide. Mol. Biol. Cell 5, 829-837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faber, K.N., Haima, P., Gietl, C., Harder, W., Ab, G., and Veenhuis, M. (1994a). The methylotrophic yeast Hansenula polymorpha contains an inducible import pathway for peroxisomal matrix proteins with an N-terminal targeting signal (PTS2 proteins). Proc. Natl. Acad. Sci. USA 91, 12985-12989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faber, K.N., Haima, P., Harder, W., Veenhuis, M., and Ab, G. (1994b). Highly-efficient electrotransformation of the yeast Hansenula polymorpha. Curr. Genet. 25, 305-310. [DOI] [PubMed] [Google Scholar]
- Faber, K.N., Kram, A.M., Ehrmann, M., and Veenhuis, M. (2001). A novel method to determine the topology of peroxisomal membrane proteins in vivo using the tobacco etch virus protease. J. Biol. Chem. 276, 36501-36507. [DOI] [PubMed] [Google Scholar]
- Faber, K.N., van Dijk, R., Keizer-Gunnink, I., Koek, A., van der Klei, I.J., and Veenhuis, M. (2002). Import of assembled PTS1 proteins into peroxisomes of the yeast Hansenula polymorpha: yes and no! Biochim. Biophys. Acta 1591, 157-162. [DOI] [PubMed] [Google Scholar]
- Gatto, G.J., Jr., Geisbrecht, B.V., Gould, S.J., and Berg, J.M. (2000). Peroxisomal targeting signal-1 recognition by the TPR domains of human PEX5. Nat. Struct. Biol. 7, 1091-1095. [DOI] [PubMed] [Google Scholar]
- Gietl, C., Faber, K.N., van der Klei, I.J., and Veenhuis, M. (1994). Mutational analysis of the N-terminal topogenic signal of watermelon glyoxysomal malate dehydrogenase using the heterologous host Hansenula polymorpha. Proc. Natl. Acad. Sci. USA 91, 3151-3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleeson, M.A.G., and Sudbery, P.E. (1988). Genetic analysis in the methylotrophic yeast Hansenula polymorpha. Yeast 4, 293-303. [Google Scholar]
- Goodman, J.M., Scott, C.W., Donahue, P.N., and Atherton, J.P. (1984). Alcohol oxidase assembles post-translationally into the peroxisome of Candida boidinii. J. Biol. Chem. 259, 8485-8493. [PubMed] [Google Scholar]
- Halbig, D., Wiegert, T., Blaudeck, N., Freudl, R., and Sprenger, G.A. (1999). The efficient export of NADP-containing glucose-fructose oxidoreductase to the periplasm of Zymomonas mobilis depends both on an intact twin-arginine motif in the signal peptide and on the generation of a structural export signal induced by cofactor binding. Eur. J. Biochem. 263, 543-551. [DOI] [PubMed] [Google Scholar]
- Klein, A.T., Van den Berg, M., Bottger, G., Tabak, H.F., and Distel, B. (2002). Saccharomyces cerevisiae acyl-CoA oxidase follows a novel, non-PTS1, import pathway into peroxisomes that is dependent on Pex5p. J. Biol. Chem. 277, 25011-25019. [DOI] [PubMed] [Google Scholar]
- Kyhse-Andersen, J. (1984). Electroblotting of multiple gels: a simple apparatus without buffer tank for rapid transfer of proteins from polyacrylamide to nitrocellulose. J. Biochem. Biophys. Methods 10, 203-209. [DOI] [PubMed] [Google Scholar]
- Laemmli, U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680-685. [DOI] [PubMed] [Google Scholar]
- McCollum, D., Monosov, E., and Subramani, S. (1993). The pas8 mutant of Pichia pastoris exhibits the peroxisomal protein import deficiencies of Zellweger syndrome cells—the PAS8 protein binds to the COOH-terminal tripeptide peroxisomal targeting signal, and is a member of the TPR protein family. J. Cell Biol. 121, 761-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musgrove, J.E., Johnson, R.A., and Ellis, R.J. (1987). Dissociation of the ribulosebisphosphate-carboxylase large-subunit binding protein into dissimilar subunits. Eur. J. Biochem. 163, 529-534. [DOI] [PubMed] [Google Scholar]
- Ozimek, P., van Dijk, R., Latchev, K., Gancedo, C., Wang, D.Y., van der Klei, I.J., and Veenhuis, M. (2003). Pyruvate carboxylase is an essential protein in the assembly of yeast peroxisomal oligomeric alcohol oxidase. Mol. Biol. Cell 14, 786-797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer, T., and Berks, B.C. (2003). Moving folded proteins across the bacterial cell membrane. Microbiology 149, 547-556. [DOI] [PubMed] [Google Scholar]
- Purdue, P.E., and Lazarow, P.B. (2001). Peroxisome biogenesis. Annu. Rev. Cell Dev. Biol. 17, 701-752. [DOI] [PubMed] [Google Scholar]
- Salomons, F.A., Faber, K.N., Veenhuis, M., van der Klei, I.J. (2000a). Peroxisomal remnant structures in Hansenula polymorpha pex5 cells can develop into normal peroxisomes upon induction of the PTS2 protein amine oxidase. J. Biol. Chem. 276, 4190-4198. [DOI] [PubMed] [Google Scholar]
- Salomons, F.A., Kiel, J.A.K.W., Faber, K.N., Veenhuis, M., and van der Klei, I.J. (2000b). Overproduction of Pex5p stimulates import of alcohol oxidase and dihydroxyacetone synthase in a Hansenula polymorpha pex14 null mutant. J. Biol. Chem. 275, 12603-12611. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Sargent, F., Berks, B.C., and Palmer, T. (2002). Assembly of membrane-bound respiratory complexes by the Tat protein-transport system. Arch. Microbiol. 178, 77-84. [DOI] [PubMed] [Google Scholar]
- Stewart, M.Q., Esposito, R.D., Gowani, J., and Goodman, J.M. (2001). Alcohol oxidase and dihydroxyacetone synthase, the abundant peroxisomal proteins of methylotrophic yeasts, assemble in different cellular compartments. J. Cell Sci. 114, 2863-2868. [DOI] [PubMed] [Google Scholar]
- Stewart, M.Q., van Dijk, R., Veenhuis, M., and Goodman, J.M. (2002). Monomeric alcohol oxidase is preferentially digested by a novel protease from Candida boidinii. Biochim. Biophys. Acta 1542, 160-172. [DOI] [PubMed] [Google Scholar]
- Titorenko, V.I., Nicaud, J.M., Wang, H., Chan, H., and Rachubinski, R.A. (2002). Acyl-CoA oxidase is imported as a heteropentameric, cofactor-containing complex into peroxisomes of Yarrowia lipolytica. J. Cell Biol. 156, 481-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titorenko, V.I., Smith, J.J., Szilard, R.K., and Rachubinski, R.A. (1998). Pex20p of the yeast Yarrowia lipolytica is required for the oligomerization of thiolase in the cytosol and for its targeting to the peroxisome. J. Cell Biol. 142, 403-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Klei, I.J., Bystrykh, L.V., and Harder, W. (1990). Alcohol oxidase from Hansenula polymorpha CBS 4732. Methods Enzymol. 188, 420-427. [DOI] [PubMed] [Google Scholar]
- van der Klei, I.J., Harder, W., and Veenhuis, M. (1991a). Methanol metabolism in a peroxisome-deficient mutant of Hansenula polymorpha: a physiological study. Arch. Microbiol. 156, 15-23. [DOI] [PubMed] [Google Scholar]
- van der Klei, I.J., Harder, W., and Veenhuis, M. (1991b). Selective inactivation of alcohol oxidase in two peroxisome-deficient mutants of the yeast Hansenula polymorpha. Yeast 7, 813-821. [DOI] [PubMed] [Google Scholar]
- van der Klei, I.J. et al. (1995). The Hansenula polymorpha PER3 gene is essential for the import of PTS1 proteins into the peroxisomal matrix. J. Biol. Chem. 270, 17229-17236. [DOI] [PubMed] [Google Scholar]
- van der Klei, I.J., Sulter, G.J., Harder, W., and Veenhuis, M. (1991c). Assembly of alcohol oxidase in the cytosol of a peroxisome-deficient mutant of Hansenula polymorpha—properties of the protein and architecture of the crystals. Yeast 7, 195-209.1882546 [Google Scholar]
- van der Klei, I.J., Veenhuis, M., van der Ley, I., and Harder, W. (1989). Heterologous expression of alcohol oxidase in Saccharomyces cerevisiae: properties of the enzyme and implications for microbody development. FEMS Microbiol. Lett. 48, 133-137. [DOI] [PubMed] [Google Scholar]
- van Dijk, R., Faber, K.N., Hammond, A.T., Glick, B.S., Veenhuis, M., and Kiel, J.A.K.W. (2001). Tagging Hansenula polymorpha genes by random integration of linear DNA fragments. Mol. Genet. Genom. 266, 646-656. [DOI] [PubMed] [Google Scholar]
- van Dijken, J.P., Otto, R., and Harder, W. (1976). Growth of Hansenula polymorpha in a methanol-limited chemostat. Physiological responses due to the involvement of methanol oxidase as a key enzyme in methanol metabolism. Arch. Microbiol. 111, 137-144. [DOI] [PubMed] [Google Scholar]
- Veenhuis, M., and Harder, W. (1987). Metabolic significance and biogenesis of microbodies in yeast. In: Peroxisomes in Biology and Medicine, ed. D. Fahimi, H. Sies, Berlin: Springer-Verlag, 436-457.
- Verduyn, C., Van Dijken, J.P., and Scheffers, W.A. (1984). Colorometric alcohol assays with alcohol oxidase. J. Microbiol. Methods 2, 15-25. [Google Scholar]
- Wang, D., Visser, N.V., Veenhuis, M, and Van der Klei, I.J. (2003). Physical interactions of the peroxisomal targeting signal receptor, Pex5p, studied by fluorescence correlation spectroscopy. J. Biol. Chem. 278, 43340-43345. [DOI] [PubMed] [Google Scholar]
- Waterham, H.R., Russell, K.A., Vries, Y., and Cregg, J.M. (1997). Peroxisomal targeting, import, and assembly of alcohol oxidase in Pichia pastoris. J. Cell Biol. 139, 1419-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterham, H.R., Titorenko, V.I., Haima, P., Cregg, J.M., Harder, W., and Veenhuis, M. (1994). The Hansenula polymorpha PER1 gene is essential for peroxisome biogenesis and encodes a peroxisomal matrix protein with both carboxy- and amino-terminal targeting signals. J. Cell Biol. 127, 737-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterham, H.R., Titorenko, V.I., Swaving, G.J., Harder, W., and Veenhuis, M. (1993). Peroxisomes in the methylotrophic yeast Hansenula polymorpha do not necessarily derive from pre-existing organelles. EMBO J. 12, 4785-4794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, X., Purdue, P.E., and Lazarow, P.B. (2001). Eci1p uses a PTS1 to enter peroxisomes: either its own or that of a partner, Dci1p. Eur. J. Cell Biol. 80, 126-138. [DOI] [PubMed] [Google Scholar]