Abstract
3-Hydroxy-3-methyl-glutaryl-CoA reductase (HMGR) is a key enzyme in the sterol biosynthesis pathway, but its subcellular distribution in the Trypanosomatidae family is somewhat controversial. Trypanosoma cruzi and Leishmania HMGRs are closely related in their catalytic domains to bacterial and eukaryotic enzymes described but lack an amino-terminal domain responsible for the attachment to the endoplasmic reticulum. In the present study, digitonin-titration experiments together with immunoelectron microscopy were used to establish the intracellular localization of HMGR in these pathogens. Results obtained with wild-type cells and transfectants overexpressing the enzyme established that HMGR in both T. cruzi and Leishmania major is localized primarily in the mitochondrion and that elimination of the mitochondrial targeting sequence in Leishmania leads to protein accumulation in the cytosolic compartment. Furthermore, T. cruzi HMGR is efficiently targeted to the mitochondrion in yeast cells. Thus, when the gene encoding T. cruzi HMGR was expressed in a hmg1 hmg2 mutant of Saccharomyces cerevisiae, the mevalonate auxotrophy of mutant cells was relieved, and immunoelectron analysis showed that the parasite enzyme exhibits a mitochondrial localization, suggesting a conservation between the targeting signals of both organisms.
INTRODUCTION
Trypanosoma cruzi and Leishmania species are the etiological agents of Chagas' disease and leishmaniases, respectively, and both exhibit complex life cycles. After invasion of mammalian cells, parasites differentiate into intracellular amastigotes, which multiply and are able to infect new cells or gain access to a new vector (De Souza, 1984). Despite considerable work, neither a vaccine to prevent these diseases nor satisfactory drugs are available. Current treatments are either expensive, have severe side effects, or are ineffective in many cases (Croft et al., 1997). Several promising target molecules for the treatment of this disease are enzymes involved in isoprenoid metabolism also called the mevalonate pathway (Urbina, 1997). The mevalonate pathway provides precursors for the diverse spectrum of isoprenoid compounds (Sacchettini and Poulter, 1997; Edwards and Ericsson, 1999), some of which have been shown to be essential for the growth and development of eukaryotic cells (Rao, 1995). The pathway starts with the synthesis of mevalonate catalyzed by 3-hydroxy-3-methyl-glutaryl-CoA reductase (HMGR), a key enzyme that is subject to several regulatory mechanisms (Goldstein and Brown, 1990; Stermer et al., 1994; Hampton et al., 1996). Potent HMGR inhibitors with Ki values in the nanomolar range are available (Endo and Hasumi, 1992). In trypanosomatids, the importance of isoprenoids for cell viability and proliferation has been proved, and the combination of inhibitors that act at different points of the pathway seems to be a useful strategy against T. cruzi and Leishmania infections (Gebre-Hiwot and Frommel, 1993; Maldonado et al., 1993; Urbina, 1997).
Although eukaryotic HMGR enzymes are very conserved in sequence and subcellular distribution, they differ in size, membrane topology, quaternary structure, and mechanisms of regulation. HMGR and the mevalonate pathway have been studied in detail in eukaryotes, yet the information available regarding this enzyme in trypanosomatids is limited. Thus, the localization of the enzyme remains obscure and controversial; HMGR from Trypanosoma brucei was reported to be microsomal (Coppens et al., 1995) and mitochondrial (Heise and Opperdoes, 2000), whereas the T. cruzi enzyme was found to be glycosomal (Concepción et al., 1998).
We have previously isolated the gene encoding HMGR from T. cruzi and Leishmania major and demonstrated that they lack the membrane N-terminal domain characteristic of eukaryotic HMGRs and responsible for the attachment to endoplasmic reticulum (ER), thus determining a soluble form of the enzyme (Peña-Díaz et al., 1997; Montalvetti et al., 2000). Comparison of several trypanosomatid mitochondrial proteins revealed that T. cruzi (Tc)HMGR and Leishmania (Lm)HMGR exhibit a short, putative mitochondrial signal. All evidence so far indicates that the mechanism of mitochondrial protein import is conserved among eukaryotes. Nuclear-encoded mitochondrial matrix proteins from trypanosomatids have cleaved N-terminal peptides. Sequence comparisons suggested that two groups of mitochondrial targeting signals could function in trypanosomes. One group consists of sequences 15-20 amino acids in length that are similar to signals known in yeast and other organisms. The other group of sequences is exceptionally short (7-9 amino acids) and also function in yeast albeit rather inefficiently (Häusler et al., 1997).
In an effort to resolve the controversy regarding cellular localization of HMGR in the Trypanosomatidae, permeabilization analysis, and immunoelectron microscopy have been used to study in detail the subcellular distribution of the enzyme, showing it to be mitochondrial and that amino terminal sequences are responsible for targeting of the enzyme to the mitochondrial matrix. Furthermore, expression of TcHMGR in the yeast double mutant hmg1 hmg2 allows cells to grow in the absence of mevalonate, and TcHMGR was efficiently targeted to the mitochondrion, hence showing conserved import mechanisms among both organisms (Basson et al., 1987).
MATERIALS AND METHODS
Materials
Restriction enzymes, T4 DNA ligase, Taq polymerase, aprotinin, leupeptin, reverse transcriptase M-MuLV, and the RNase inhibitor were from Roche Diagnostics (Mannheim, Germany). [α-32P]ATP was from ICN Pharmaceuticals (Irvine, CA). d,l-[3-14C]3-hydroxy-3-methylglutaryl-CoA was from Amersham Biosciences (Piscataway, NJ), and R,S-[5-3H(N)]mevalonolactone was from DuPont (Wilmington, DE). Benzamidine, bovine serum albumin (BSA), dithiothreitol, d,l-3-hydroxy-3-methylglutaryl-CoA, d,l-mevalonic acid lactone, 1,10-phenanthroline, phenylmethylsulfonyl fluoride, trypsin inhibitor, sodium phosphate, and mevalonic acid were from Sigma-Aldrich (St. Louis, MO). The mRNA purification kit was from Pharmacia (Peapack, NJ). Oligonucleotides CLC1 and CLC2 were hybrid constructions that anneal with the 5′ and 3′region, respectively, of S. cerevisiae HMG2 (uppercase) and a part of kanMX4 from Ashbya gossypii (lowercase) and were purchased from ISOGEN. The sequences were as follows: CLC1 (5′-TGAAGAGCCAAAGATACCAACTGAATAGTGTCTGAAAACGGAACcgtacg ctgcaggtcgacg-3′) and CLC2 (5′-TCTTTGGTTAAAACAGTTGTTGCACCACCACCAGC ATTGATGGCTatcgatgaattcgagctcgtt-3′). Other oligonucleotides were synthesized at Analytical Services (Instituto deParasitología y Biomedicina “López Neyra”, Granada, Spain). BamHI-ATG (5′-CGCGGATCCCCATGTTTCGTAGGGCAATTCT-3′) and HindIII-TGA (5′-CGCTAAGCTTTCACTTCTTCTTGCGATTCAG-3′) were designed to amplify the coding region of the TcHMGR gene, Neo-ATG (5′-CTCGAAGCTTATGGGATCGGCCATTGAACA-3′) and Neo-TGA (5′-CTGGAATTCAGCGGGGAAAAAACAAAAAACACGC-3′) were used to amplify the coding region of the aminoglycoside phosphotransferase gene (NEO), and 5′rDNA (5′-GATATTTGCGCACCCACCTT-3′) and 3′rDNA (5′-GCGACAGACAATTCACGCAC-3′) were used for amplifying an rDNA region. For overexpression of truncated forms of LmHMGR, the oligonucleotides LmHMGR1i (5′-GGAATTGGATCCATGGAGAGCTGGGCATCC-3′), LmHMGR2i (5′-GGAATTGGATCCATGTCCGATACAGAG-3′), and LmHMGR3f (5′-ATCGCAAGCTTTTACGGAGTCGGAGGC-3′) were used.
Strains and Culture Methods
The epimastigote form of T. cruzi Y was cultured in filter-sterilized LIT medium with 10% (vol/vol) heat-inactivated fetal calf serum (Invitrogen, Carlsbad, CA) in tissue culture flasks at 28°C. Leishmania major 252 wild-type cells were grown in M199 medium with 10% (vol/vol) heat-inactivated fetal calf serum under the same conditions as T. cruzi cells.
Yeast strains were grown at 30°C in rich medium containing 2% glucose (YPD) and minimal medium containing 2% glucose. Amino acids and mevalonate supplements were added, when needed, at the following concentrations: 20 μg/ml adenine, histidine, and tryptophan; 30 μg/ml leucine; and 20 mM mevalonate. Solid medium contained 2% agar.
Overexpression of HMGR in T. cruzi Y Cells
The vector pRIBOTEX was kindly provided by Roberto Hernandez (Martínez-Calvillo et al., 19973). Insertion of the coding sequence for TcHMGR flanked by the restriction sites BamHI and HindIII into the polylinker of pRIBOTEX created the expression vector pRIBOTEX1-HMGR. The oligonucleotides for the amplification of the insert were the sense BamHI-ATG and the antisense HindIII-TGA. Conditions for electroporation of the cells were those reported by Martínez-Calvillo et al., 1997. A control of transfection by using void pRIBOTEX was run in parallel. Poly (A)+ RNA was obtained and the splice acceptor site was assessed by reverse transcription-polymerase chain (PCR) reaction and sequencing as described previously (Peña-Díaz et al., 1997).
Overexpression of HMGR in Leishmania major Cells
The coding region of L. major HMGR was cloned in the BamHI site of the vector pSP72αNEOα (Papadopoulou et al., 1994) to generate pSP72hmgr. Truncated versions of the LMHMGR gene were amplified by PCR and cloned in the BamHI and HindIII sites of pSP72αneoα to give pSP72hmgr1 and pSP72hmgr2, which lack the first 42 and 57 base pairs, respectively, of the LMHMGR gene. Conditions of transfection and selection of positive clones were as reported previously (Papadopoulou et al., 1994).
Contour-clamped Homogeneous Electric Field (CHEF) Electrophoresis
Low-melting-point agarose blocks were prepared as described previously (Garvey and Santi, 1986; Ellenberg and Beverley, 1989). Chromosomes were separated on a 1% agarose gel in 0.5 M Tris borate-EDTA by using a CHEF electrophoresis system (Pharmacia). The parameters used were frequencies of 75 s for 28 h, frequencies of 100 s for 18 h, frequencies of 200 s for 18 h, and frequencies of 250 s for 0 h at 120 V. Molecular masses of the chromosomal DNA bands were determined by comparison with DNA standards from S. cerevisiae strain YNN295 (Bio-Rad, Hercules, CA). The resulting gel was transferred to a Hybond-N nylon filter (Amersham Biosciences) and subjected to Southern blot analysis by using as probes the TcHMGR and LMHMGR genes, NEO gene, and an rDNA fragment.
Yeast Strain Construction
Plasmid pAN10HMGTc carrying TcHMGR under the control of the ADH1 promoter of S. cerevisiae was constructed as follows: an NdeI-BamHI segment containing the complete TcHMGR coding sequence was obtained from pETHMGR. HindIII linkers were added, and the resulting fragment was cloned in the HindIII site of pAN10 (Navas et al., 1993). The plasmid carrying the TcHMGR gene in the adequate orientation was termed pAN10HMGTc.
All strains used in this study were derived from W303 (ade2 try1 leu2 his3 ura3). Two yeast strains with disruptions in HMG1 and HMG2, respectively, were constructed as follows. Plasmid pJR429 (Basson et al., 1986) was digested with SphI, and the 2.9-kb fragment containing HMG1 was inserted in the SphI site of pUC18. The resulting construct was digested with BglII that eliminates a 1.8-kb fragment internal to the HMG1 gene. This fragment was replaced by a 1.76-kb fragment from plasmid YDpH carrying the HIS3 gene (Berben et al., 1991). The 2.86-kb SphI-SphI fragment of this construct was used to disrupt the chromosomal copy from S. cerevisiae. Correct integration was verified by Southern blot analysis.
HMG2 was disrupted with the gene kanMX4 from A. gossypii that confers resistance to geneticine (Wach et al., 1994). Oligonucleotides CLC1 and CLC2 hybridize, respectively, with regions 5′ and 3′ of HMG2 and kanMX4. A PCR reaction was performed with these oligonucleotides by using plasmid pFA6akanMX4 as template (Wach et al., 1994), and the product of the reaction was used to disrupt the chromosomal copy of HMG2 in S. cerevisiae. Southern blot analysis of the transformants showed that the integration occurred at the correct place. To obtain the double hmg1 hmg2 mutant, the strains bearing the hmg1 and hmg2 disruptions were crossed and the diploid sporulated. Sporulation was poor, and the spores that ought to have carried the hmg1::HIS3 hmg2::KanMX4 interruptions did not germinate well even in media supplemented with mevalonate. Therefore, the diploid hmg1 and hmg2 was transformed with pAN10HMGTc and sporulated. All spores germinated and those bearing the double interruption were identified by the disruption markers. Results of plasmid loss experiments showed the gene of T. cruzi complements the lack of the yeast HMGR genes. A tetratype was selected among the tetrads and after plasmid loss, a spore with the double disruption (strain CFL9) was retained.
Measurement of HMG-CoA Reductase Activity
The activity of the enzyme was determined by a radiometric assay as described previously (Peña-Díaz et al., 1997). Yeast cells were grown to mid-log phase (15 × 106 cells/ml), harvested, and disrupted using glass beads (Daum et al., 1982). Supernatants obtained after centrifugation at 1500 × g were used for determining enzyme activity. Strains W303 and CFL9 were resuspended in sorbitol-HEPES buffer (0.6 M sorbitol, 20 mM HEPES-KOH, pH 7.4) containing 50 μg/ml aprotinin, 20 μg/ml leupeptin, 10 mM 1,10-phenanthroline, 1 mM benzamidine, 50 μg/ml trypsin inhibitor, and 50 μM phenylmethylsulfonyl fluoride as protease inhibitors. One unit of enzyme activity was expressed as nanomoles of [14C]HMG-CoA converted to [14C]mevalonate min-1 mg-1 protein. Protein was determined by the method of Bradford (Bradford, 1976).
Antibody Production and Western Blot Analysis
Polyclonal antiserum against recombinant both TcHMGR and LmHMGR was generated by immunizing rabbits with the purified protein. Monoclonal anti-TcHMGR antibodies were obtained as described previously (Coligan et al., 1995) and purified by affinity chromatography over a protein A-Sepharose column (Pharmacia) according to the recommendations of the supplier. T. cruzi and Leishmania cells were ruptured by sonication. Yeast cells were ruptured using glass beads and the extracts were clarified by centrifugation (5 min, 1500 × g). In all cases, 15 μg of protein was subjected to electrophoresis through a 12% SDS-polyacrylamide gel and blotted onto Immobilon-P membranes (Millipore, Bedford, MA) at 25 V for 30 min, by using a SemiDry Transfer Cell (Bio-Rad). Western analysis of TcHMGR was performed with monoclonal anti-TcHMGR at 1:5000 dilution by using anti-mouse IgG peroxidase conjugate (Promega, Madison, WI) as the secondary antibody. Bound antibody was visualized with the ECL system from Amersham Biosciences. LmHMGR was analyzed at a 1:25000 dilution of polyclonal anti-LmHMGR by using anti-rabbit peroxidase conjugate.
Digitonin Permeabilization
Exponential phase T. cruzi cells (6 × 107 cells/ml) were collected and washed with phosphate-buffered saline (PBS) and suspended in buffer A (0.25 M sucrose/10 mM HEPES, pH 7.4/50 mM NaCl/20 mM EDTA/2 mM EGTA/5 mM dithiothreitol) plus protease inhibitors at a concentration of 2 × 108 cells/ml. The required amount of digitonin was added, and the suspension was incubated at 28°C for 30 min and centrifuged at 13,000 × g for 2 min. The supernatant was used to assay activity of citrate synthase, pyruvate kinase, hexokinase, and HMGR as described previously (Cannata and Cazzulo, 1984; Callens et al., 1991; Peña-Díaz et al., 1997).
Immunoelectron Microscopy
Yeast cells growing in early exponential phase were harvested by centrifugation, washed several times with PBS, and fixed with 1% paraformaldehyde and 1% glutaraldehyde in cacodylate buffer (pH 6.8, 0.05 M). After 4 h at 4°C, cells were centrifuged and washed three times with cacodylate buffer and resuspended in 5 ml of 1% sodium metaperiodate. The suspension was incubated for 15 min at room temperature after which the cells were pelleted and washed once in distilled water. Subsequent steps of dehydration with ethanol, infiltration in increasing concentrations of LRWhite resin (ESB) and embedding were carried out as described previously (Wright and Rine, 1989).
Trypanosomes growing in exponential phase were collected and washed in PBS. Cells (107) were fixed in Karnowsky solution (1% glutaraldehyde, 4% paraformaldehyde, 0.05 cacodylate, pH 7.2) at 4°C for 2 h and then washed with cacodylate, centrifuged, and the pellet dehydrated in progressively increasing concentrations of ethanol. The sample was embedded in increasing concentrations of LRWhite and polymerized at -20°C under UV light exposure for 4 d.
Ultrathin sections were treated with NH4Cl for 2 h, incubated 30 min with 3% BSA in PBS (pH 7.4) and 10 min with 0.05% Tween 20, 1% BSA in PBS to block nonspecific binding sites. Labeling was performed by incubation with primary monoclonal antibodies raised against recombinant TcHMGR diluted 1:20 and a 1:75 dilution of goat anti-mouse IgG conjugated to 10-nm gold (Sigma-Aldrich) both diluted with 5% rabbit serum, 0.05% Tween 20, 1% BSA in PBS. For L. major cells, parasite sections were labeled with purified anti-LmHMGR antibody (1:80) for 1 h, washed with BSA-PBS solution, and then goat anti-rabbit IgG coupled with 10-nm gold particles (Sigma-Aldrich) as secondary antibody. Controls consisted of samples previously incubated with an excess of recombinant protein or samples without the respective primary antibody. After staining with uranyl acetate, sections were examined in a Zeiss 902 electron microscope at 80 kV (Centro deInstrumentacion Científica, Universidad deGranada, Granada, Spain). For ultrastructural studies, parasites were collected by centrifugation and fixed in a solution containing 2.5% glutaraldehyde in 0.1 M cacodylate buffer, pH 7.4, 450 mOsM; washed in cacodylate buffer; postfixed in 2% osmium tetroxide stained with 2% uranyl acetate; dehydrated in ethanol; and embedded in Epon.
RESULTS
Construction of Stable Epimastigote Cell Lines Overexpressing TcHMGR
To investigate the subcellular location of TcHMGR, a cell line overexpressing the enzyme was obtained. The TCHMGR gene was obtained by PCR with the plasmid pETHMGR (Peña-Díaz et al., 1997) as template DNA and inserted into the polylinker of the integrative vector pRIBOTEX (Martínez-Calvillo et al., 1997) by using the BamHI and HindIII cloning sites so that transcription was under the control of the rDNA promoter. Parasites were transfected with pRIBOTEX-HMGR and selected with increasing geneticine concentrations up to 2 mg ml-1. Transfectants with this construct exhibited new copies of the TCHMGR gene located in the rDNA locus as determined by CHEF and Southern blot analysis. Analysis by RT-PCR showed that the addition of the splice leader took place at an AG site in position -88 in transfectant cells and in position -37 in wild-type cells. Specific activity of TcHMGR in transfectant cells was 21.5 nmol min-1 mg-1, threefold higher than that of wild-type cells. Transfected cells did not show any special morphological alterations with regard to wild-type cells as determined by ultrastructural studies.
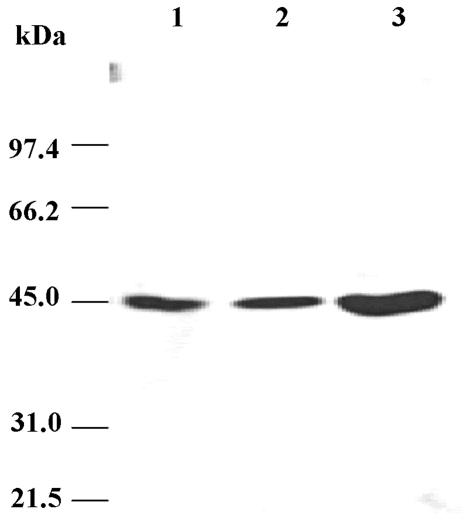
To determine the location of TcHMGR in T. cruzi cells, antibodies directed toward epitopes of the recombinant enzyme were generated in rabbits. Monoclonals were also generated and isotyped as Ig G1. In a Western blot analysis, both types of antibodies recognized a single band in wild-type parasite lysates that corresponded to a molecular mass of 46 kDa, similar to that predicted for the translated TCHMGR coding region. This band was established to be TcHMGR, and T. cruzi cells overexpressing the enzyme showed a more intense band of the same size than the wild type (Figure 1).
Figure 1.
Western blot analysis of the levels of TcHMGR in overexpressing mutants. Cells were ruptured by sonication and the extract was clarified by centrifugation (1500 × g for 10 min). Lane 1, supernatant from wild-type T. cruzi; lane 2, supernatant from T. cruzi cells harboring the vector pRIBOTEX; and lane 3, supernatant of T. cruzi cells harboring pRIBOTEX-TcHMGR and overexpressing the TcHMGR protein. All samples were subjected to SDS-PAGE, transferred to an Immobilon-P membrane, and incubated with a mAb generated against the recombinant T. cruzi protein.
TcHMGR in T. cruzi Undergoes Mitochondrial Targeting
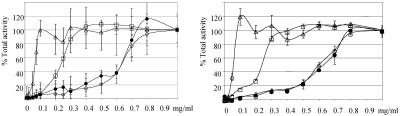
An analysis of the amino-terminal sequence of TcHMGR reveals similarity with proteins described to undergo mitochondrial targeting in trypanosomatids. These proteins carry a short signal (7-9 amino acids) for import to mitochondria (Table 1). To determine whether indeed TcHMGR is located in this compartment, a digitonin titration experiment was performed. Analysis of the proteins obtained at different detergent concentrations showed that TcHMGR from wild-type and transfected parasites was poorly recovered at digitonin concentrations <0.5 mg ml-1 but coeluted with the enzyme citrate synthase characteristic from mitochondria at higher concentrations. Elution of the mitochondrial enzymes occurred at 0.5 mg ml-1, glycosomal enzymes were released from 0.2 to 0.3 mg ml-1 digitonin, and elution was complete at 0.5 mg ml-1 (Figure 2). Aliquots obtained after permeabilization were subjected to Western blot analysis with polyclonal anti-HMGR, further confirming that TcHMGR is not released at low detergent concentrations (our unpublished data).
Table 1.
Comparison of the amino-terminal sequence of TcHMGR and LmHMGR with those of different trypanosomatid proteins that undergo mitochondrial import
| Organism | Amino terminal sequencea | Protein |
|---|---|---|
| T. cruzi | MFRRAILLGCSAAK | HMGR |
| L. major (Montalvetti et al., 2000) | MRRSLLLACSAAKGESWASM | HMGR |
| T. brucei (Else et al., 1994) | MFRRCFPIF*NPYD | Lipoamide dehydrogenase |
| C. fasciculata (Xu and Ray, 1993) | MLRRSPTLL*RVSP | p17 |
| T. cruzi (Giambiagi de Marval et al., 1993) | MFRSAARF*AGKE | hsp60 |
| L. tarentolae (Bringaud et al., 1995) | MLRATLAR*EMAP | p51, aldehyde dehydrogenase |
| L. tarentolae (Bringaud et al., 1995) | MRRLSSQLMCTAAAVRF*ASAG | p18 |
| L. major (Searle et al., 1993) | MFARRVCGTAAASAACLVRC*ASDK | mhsp70 |
Positively charged residues are in bold. Sequence homologies are underlined.
Denotes known cleavage sites.
Figure 2.
Digitonin titration of TcHMGR. Activities of pyruvate kinase (open triangles), hexokinase (open squares), citrate synthase (open circles), and 3-hydroxy-3-methyl-glutaryl-CoA reductase (closed circles) in supernatants of whole T. cruzi suspensions in solutions of increasing digitonin concentrations. Enzyme total activity is expressed as a percentage, taking as 100% the total activity obtained in supernatants after permeabilization with 0.5% Triton X-100. (A) Activities recovered from T. cruzi wild-type supernatants. (B) Activities recovered in supernatants of T. cruzi parasites transfected with pRIBOTEX-TcHMGR and overexpressing HMGR.
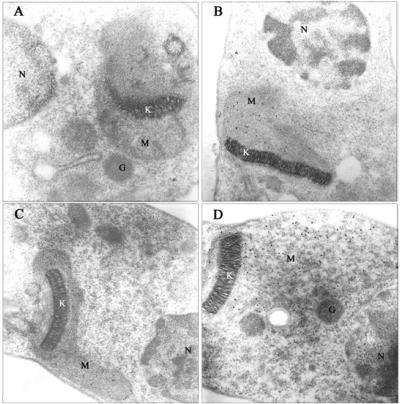
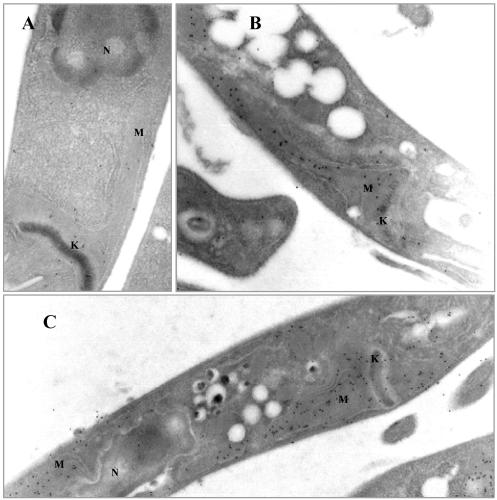
Immunoelectron microscopy was used to further corroborate mitochondrial targeting of the enzyme, and a monoclonal antibody (mAb) was used for this purpose. As shown in Figure 4A, wild-type epimastigotes showed a small, but significant number of particles inside the mitochondrion that can be clearly identified by the presence of the electrodense mitochondrial DNA network or kinetoplast. Furthermore, epimastigotes overexpressing HMGR (Figure 3, B-D) showed a profuse and specific labeling restricted to the mitochondrial matrix because the number of gold particles was notably increased relative to wild-type cells. A significant number of particles associated to glycosomes or any other internal organelle could not be evidenced. Likewise, immunogold labeling indicative of a cytoplasmic milieu was not apparent either. Particle counting revealed that >90% of labeling was related to the mitochondrion in T. cruzi cells overexpressing the enzyme. Although the possibility cannot be ruled out that HMGR is a minor constituent of other structures, clearly the major location of the enzyme was within the matrix of this organelle, and the distribution pattern was not affected by culture conditions such as increased oxygenation or different culture media (our unpublished data).
Figure 4.
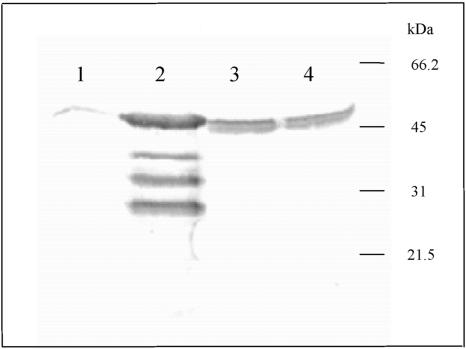
Western blot analysis of the levels of LmHMGR in overexpressing mutants. Cells were ruptured by sonication and the extract was clarified by centrifugation (1500 × g for 10 min). Lane 1, supernatant from wild-type L. major; lane 2, supernatant of L. major cells harboring pSP72hmgr and overexpressing LmHMGR; lane 3, supernatant of L. major cells harboring pSP72hmgr1; and lane 4, supernatant of L. major cells harboring the pSP72hmgr2. All samples were subjected to SDS-PAGE, transferred to an Immobilon-P membrane, and incubated with a polyclonal antibody generated against recombinant LmHMGR.
Figure 3.
Immunoelectron microscopy localization of HMGR to the mitochondrion. T. cruzi epimastigotes were fixed in 4% paraformaldehyde and 0.1% glutaraldehyde and embedded in LRWhite resin. Thin sections were immunolabeled with anti-TcHMGR monoclonal antibodies followed by goat anti-mouse immunoglobulin conjugated to gold (10-nm probe). Magnification is 40,000×. (A) Section of wild-type T. cruzi epimastigotes showing gold labeling of the mitochondrial matrix. No label is present in glycosomes or the cytosol. (B-D) Longitudinal sections of T. cruzi epimastigote cells transfected with the expression vector pRIBOTEX-TcHMGR showing intense gold labeling of the mitochondrial matrix. G, glycosome; K, kinetoplast; M, mitochondrion.
Production of Leishmania major Cell Lines Overexpressing Full-Length and Truncated Forms of LmHMGR
The amino terminus of LmHMGR, similar to T. cruzi, also contains a putative mitochondrial targeting sequence (Table 1). To determine the intracellular location of HMGR in other members of the Trypanosomatidae family and the importance of this sequence in mitochondria internalization, we have generated a L. major cell line overexpressing full-length and two truncated versions of LmHMGR, HMGR1 and HMGR2, that lack the first 14 and 19 amino acids of the amino terminal region, respectively. For HMGR1, a methionine was introduced in the amino terminus for correct translation, whereas for HMGR2, advantage was taken of a preexisting methionine in position 20. The resulting transfectants were analyzed by Western blot for overproduction of HMGR, and enzyme activity was quantified. As shown in Figure 4, cells overexpressing HMGR present approximately a ninefold increase in protein when assessed by Western blot, whereas in cells overproducing HMGR1 and HMGR2 bands of a slightly lower molecular mass seemed to be increased approximately fivefold, as revealed by densitometric analysis with regard to wild-type cells. Likewise, when enzyme activity values were measured in extracts, cells overproducing HMGR and HMGR2 rendered increased specific activities of approximately nine- and five-fold, respectively, with regard to controls.
Mitochondrial Targeting Is Impaired in Mutants Expressing Truncated Forms of LmHMGR
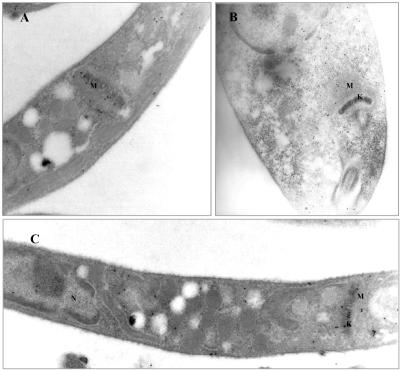
We have assessed if in Leishmania HMGR is also situated in the mitochondrion and furthermore if mitochondrial internalization is impaired in mutants overexpressing modified versions of LmHMGR were the targeting sequence has been eliminated. Immunoelectron microscopy was also accomplished for this purpose by using a rabbit polyclonal antibody raised against homogeneous purified recombinant protein. Photographs of thin sections of cells overexpressing full-length HMGR as well as wild-type cells are shown in Figure 5. Again, in the trypanosomatide L. major, the enzyme was clearly located in the mitochondrial matrix. Profuse labeling was observed in overexpressing cells (Figure 5, B and C), indicating that the intact enzyme, when overproduced, is efficiently transported across the mitochondrial membrane. However in mutants were the amino terminal sequence has been perturbed (Figure 6, A-C), HMGR seems to accumulate in the cytosol, and an intense labeling of the mitochondrial matrix is no longer evident. The small number of gold particles within the mitochondrion would correspond to the full-length enzyme encoded by the chromosomal gene copy, whereas the transgene-encoded enzyme is retained in the cytoplasm.
Figure 5.
Immunoelectron microscopy localization of HMGR in wild-type and mutant cells overexpressing full-length HMGR. L. major promastigotes were fixed in 4% paraformaldehyde and 0.1% glutaraldehyde and embedded in LRWhite resin. Thin sections were immunolabeled with anti-LmHMGR (dilution 1:70) polyclonal antibodies followed by goat anti-rabbit immunoglobulin conjugated to gold (10-nm probe). Magnification is 31,500× in A and B and 16,000× in C. (A) Section of wild-type L. major promastigotes showing gold labeling of the mitochondrial matrix. (B and C) Longitudinal sections of L. major promastigote cells transfected with the expression vector pSP72hmgr showing intense gold labeling of the mitochondrial matrix. G, glycosome; K, kinetoplast; M, mitochondrion.
Figure 6.
Immunoelectron microscopy localization of HMGR in L. major promastigotes overexpressing HMGR1 and HMGR2. Cells were fixed in 4% paraformaldehyde and 0.1% glutaraldehyde and embedded in LRWhite resin. Thin sections were immunolabeled with anti-LmHMGR (dilution 1:70) polyclonal antibodies followed by goat anti-rabbit immunoglobulin conjugated to gold (10-nm probe). Magnification is 25,000× in A and C and 31,500× in B. (A) Section of L. major promastigotes transfected with the expression vector pSP72hmgr1 expressing a form of HMGR that lacks the first 14 amino acids. (B and C) Longitudinal sections of L. major promastigote cells transfected with the expression vector pSP72hmgr2 expressing a form of HMGR that lacks the first 19 amino acids. Increased gold labeling of the cytoplasm is evidenced. G, glycosome; K, kinetoplast; M, mitochondrion.
Expression of T. cruzi 3-Hydroxy-3-methylglutaryl-CoA Reductase Supports Growth of hmg1 hmg2 S. cerevisiae Mutant Cells
Considering the sequence similarity between HMGR from T. cruzi and the soluble domain of S. cerevisiae reductase, we tested whether expression of the parasite enzyme could rescue the growth defect of a hmg1 hmg2 yeast double mutant. This mutant lacks both HMGR isozymes and cannot grow in the absence of mevalonate, the product of the reaction catalyzed by HMGR. Plasmid pAN10HMGTc, in which the ADH1 promoter drives the expression of the TcHMGR coding sequence from T. cruzi, complemented the phenotype of the double mutant and allowed it to grow in the absence of mevalonate. Extracts from wild type and the transformed double hmg1 hmg2 mutant were analyzed for expression of the HMGR protein by SDS-PAGE. Although a 46-kDa band corresponding to the T. cruzi enzyme was not evident in transformed cells, Western blot analysis with a specific mAb for T. cruzi HMGR that had no cross-reaction with the yeast enzymes revealed a specific band in the transformants (our unpublished data). Moreover, the specific activity of HMGR in the yeast transformants was one order of magnitude higher than that in wild-type type cells grown under the same conditions (5.08 and 0.48 nmol min-1 mg-1, respectively). All these results clearly demonstrate that the T. cruzi enzyme is functional in yeast.
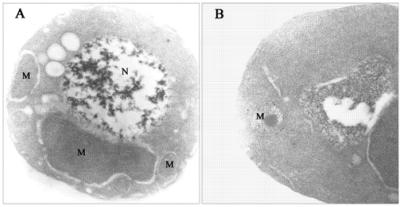
To assess the intracellular location of the overexpressed TcHMGR in S. cerevisiae, cells were analyzed by immunoelectron microscopy. Thin sections corresponding to the wild-type S. cerevisiae strain W303 and the S. cerevisiae strain CFL9 were labeled with anti-TcHMGR monoclonal antibodies (Figure 7). Gold particles were mainly observed within the mitochondrion and concentrated in the membrane region, although a certain amount of labeling remained associated to the outer mitochondrial membrane. Cells not overexpressing the protein did not show any significant labeling.
Figure 7.
Transmission electron micrographs of ultrathin section of yeast cells embedded in LRWhite resin and immunolabeled with anti-HMGR monoclonal antibodies. Cells were fixed in 1% paraformaldehyde and 1% glutaraldehyde and immunolabeled with anti-HMGR monoclonal antibodies followed by goat anti-mouse immunoglobulin conjugated to gold (10-nm probe). (A) S. cerevisiae W303 cells. (B) S. cerevisiae CFL9 cells. Magnification is 31,500× in A and 50,000× in B. N, nucleus; M, mitochondrion.
DISCUSSION
The indication of a atypical subcellular location for HMGR from T. cruzi came from the isolation of the gene and characterization of the recombinant protein (Peña-Díaz et al., 1997). An outstanding feature was the lack of the amino-terminal membrane binding domain characteristic of the other eukaryotic proteins reported to date. The enzyme was shown to exist in a soluble form constituting the sole example of a soluble HMGR in eukaryotes. The enzyme from L. major was also characterized and defined as a soluble protein (Montalvetti et al., 2000). In addition, the T. brucei gene for HMGR is currently available in the database and also lacks sequences coding for a membrane spanning domain.
In Eukaryotae, the initial steps of cholesterol biosynthesis probably occur mainly in the cytosol, and the later steps in the ER. However, several enzymes of the cholesterol biosynthetic pathway have also been reported to exist in peroxisomes (Olivier and Krisans, 2000). For example although the majority of HMGR resides in the ER, significant peroxisomal activity has also been detected in rat liver (Keller et al., 1986). In trypanosomatids, the localization of this enzyme has been subjected to controversy; thus, it has been reported to be microsomal in T. cruzi (Concepción et al., 1998) and microsomal and mitochondrial in T. brucei (Coppens et al., 1995; Heise and Opperdoes, 2000). In both cases, localization analysis was performed by cell fractionation and digitonin titration. Difficulties in the use of other conventional techniques arise from the lack of characterized enzymes from these sources and the special morphology of the mitochondria in these parasites, although protocols to isolate sealed and tRNA import competent mitochondria are available (Hauser et al., 1996). The mitochondrion in trypanosomatids is unique. There is a single mitochondrion per cell, and it extends throughout in a complex branched structure (Clayton et al., 1995; De Souza, 1999).
Overexpressing mutants were used in this study due to very low levels of HMGR in wild-type cells. The moderate levels of overexpression obtained did not occasion any apparent physiological changes in the cell. Thus, modifications in membrane proliferation such as those reported for yeast cells overexpressing HMGR were not found in overexpressing T. cruzi cells (Wright and Rine, 1989). In yeast HMGR, sequences involved in the formation of these membrane arrays have been found in one of the lumenal loops between transmembrane domains, which is absent in TcHMGR (Parrish et al., 1995). For Leishmania, LmHMGR was expressed from a transgene located in the episomal vector pSP72αNEOα developed by Papadopoulou et al. (1994), and again no profound morphological alterations seemed to occur in overexpressing mutants with regard to wild-type cells.
Monoclonal antibodies raised against the recombinant enzyme were tested in immunoelectron microscopy studies after their specificity was established. Results showed a distinct and specific mitochondrial matrix labeling in T. cruzi epimastigotes, whereas no significant labeling occurred in other cell compartments. Similar observations were obtained in Leishmania promastigote cells. The present results provide the first clear-cut demonstration of the mitochondrial location of an enzyme involved in isoprenoid biosynthesis in the Trypanosomatidae family by using both biochemical and immunological techniques. It has been argued that differential metabolic conditions may be responsible for the differences found in sensitivity to digitonin (Rodrigues et al., 2001). However, we have failed to obtain a different distribution pattern for the enzyme under different growth conditions (i.e., increased oxygenation).
T. cruzi HMGR in yeast cells devoid of the genes coding for the two HMGR isoenzymes was found to overcome the auxotrophy for mevalonate, indicating functional conservation of enzyme activity. Levels of activity found in transfected yeast cells were increased 1 order of magnitude with respect to the wild-type cells; thus, a correct translation and folding seems to occur. A hydrophobic domain found in the sequence in position 96-112 may account for a weak matrix stop-transfer signal responsible for the arrest of the protein in the mitochondria and or the intermembrane space (Hovius, 1998). On the other hand, the small amount of enzyme that remained unimported in the cytosolic compartment may have contributed toward the maintenance of viability in the absence of endogenous yeast reductase.
The reason for the mitochondrial compartimentalization of TcHMGR in T. cruzi remains unclear, because the enzyme is ER attached in mammalian cell systems. Recent results point out toward a possible metabolic role of a HMGR in mitochondria correlating leucine degradation and sterol biosynthesis. The majority of T. brucei HMGR activity (Heise and Opperdoes, 2000) has been also found associated to mitochondria. This may represent a common feature of trypanosomatids and raises the question on the origin of the necessary precursors for HMGR activity inside the mitochondrion. Reports by Ginger et al. (1999, 2000, 2001) have shown that trypanosomatids rely to different extents on the degradation of leucine as a carbon source for isoprenoid. This process, in parallel with mammalian cells, takes place in the mitochondrial matrix. Although incorporation of carbon from a leucine source in T. cruzi seems to be considerable less significant than the amount seen with Leishmania and Endotrypanum species (Ginger et al., 2001), it is uncertain to what extent results obtained in the related work are affected by the contribution of unlabeled leucine from exogenous or endogenous protein. The compartimentalization of the enzymes involved in the degradation of leucine and first steps of the mevalonate pathway would confer considerable advantages in energy economy for these organisms. The localization of HMG-CoA synthase has not been yet established in trypanosomatids, but in mammals, this enzyme has been described to have a dual localization, both cytosolic and mitochondrial (Royo et al., 1993), with the latter being involved in ketogenesis.
In summary, the present study clarifies the controversy raised around the localization of this enzyme in trypanosomatids. Work is in progress to characterize other enzymes of the mevalonate pathway and to determine the extent of mitochondria-associated isoprenoid biosynthesis in these organisms.
Acknowledgments
We thank Drs. Marcus A. Varnier and David Porcel for useful comments on the immunoelectron microscopy studies. This work was supported by grants from the UNDP/World Bank/World Health Organization Program for Research and Training in Tropical diseases (T24/181/30 ID 980139), the Spanish Programa Nacional deBiotecnología (BIO97-0659), the EC INCO-DC project contract no. CT980371, and the Plan Andaluz deInvestigación (Cod. CVI-199). J.P. and R.H. are fellows of the Spanish PFPI of the Ministerio deEducación y Ciencia, A.M. and C.F.L. had a fellowship from the Instituto deCooperacion Iberoamericana (Spain).
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03-10-0720. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-10-0720.
References
- Basson, M.E., Moore, R.L., O'Rear, J., and Rine, J. (1987). Identifying mutations in duplicated functions in Saccharomyces cerevisiae: recessive mutations in HMG-CoA reductase genes. Genetics 117, 645-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basson, M.E., Thorsness, M., and Rine, J. (1986). Saccharomyces cerevisiae contains two functional genes encoding 3-hydroxy-3-methylglutaryl Coenzyme A reductase. Proc. Natl. Acad. Sci. USA 83, 5563-5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berben, G., Dumont, J., Gilliquet, V., Bolle, P.A., and Hilger, F. (1991). The YDp plasmids: a uniform set of vectors bearing versatile gene disruption cassettes for Saccharomyces cerevisiae. Yeast 7, 475-477. [DOI] [PubMed] [Google Scholar]
- Bradford, M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein dye binding. Anal. Biochem. 131, 499-503. [DOI] [PubMed] [Google Scholar]
- Bringaud, F., Peris, M., Zen, K.H., and Simpson, L. (1995). Characterization of two nuclear-encoded protein components of mitochondrial ribonucleoprotein complexes from Leishmania tarentolae. Mol. Biochem. Parasitol. 71, 65-69. [DOI] [PubMed] [Google Scholar]
- Callens, M., Kuntz, D.A., and Opperdoes, F.R. (1991). Characterization of pyruvate kinase of Trypanosoma brucei and its role in the regulation of carbohydrate metabolism. Mol. Biochem. Parasitol. 47, 19-29. [DOI] [PubMed] [Google Scholar]
- Cannata, J.J.B., and Cazzulo, J.J. (1984). Glycosomal and mitochondrial malate dehydrogenases in epimastigotes of Trypanosoma cruzi. Mol. Biochem. Parasitol. 11, 37-49. [DOI] [PubMed] [Google Scholar]
- Clayton, C., Häusler, T., and Blattner, J. (1995). Protein trafficking in kinetoplastid protozoa. Microbiol. Rev. 59, 325-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coligan, J.E., Kruibeek, A.M., Margulies, D.H., Shevach, E.M., and Strober, W. (1995). Current Protocols in Immunology. New York: John Wiley & Sons.
- Concepción, J.L., González-Pacanowska, D., and Urbina, J.A. (1998). 3-Hydroxy-3-methyl-glutaryl-CoA reductase in Trypanosoma (Schizotrypanum) cruzi: subcellular localization and kinetic properties. Arch. Biochem. Biophys. 352, 114-120. [DOI] [PubMed] [Google Scholar]
- Coppens, I., Bastin, P., Levade, T., and Courtoy, P.J. (1995). Activity, pharmacological inhibition and biological regulation of 3-hydroxy-3-methylglutaryl coenzyme A reductase in Trypanosoma brucei. Mol. Biochem. Parasitol. 69, 29-40. [DOI] [PubMed] [Google Scholar]
- Croft, S.L., Urbina, J.A., and Brun, R. (1997). Chemotherapy of human Leishmaniasis and Trypanosomiasis. In: Trypanosomiasis and Leishmaniasis; Biology and Control, ed. G. Hide, J.C. Mottram, G.H. Coombs, and P.H. Holmes. Wallingford, United Kingdom: CAB International, 245-258.
- Daum, G., Bohni, P.C., and Schatz, G. (1982). Import of proteins into mitochondria. Cytochrome b2 and cytochrome c peroxidase are located in the intermembrane space of yeast mitochondria. J. Biol. Chem. 257, 13028-13033. [PubMed] [Google Scholar]
- De Souza, W. (1984). Cell biology of Trypanosoma cruzi. Int. Rev. Cytol. 86, 197-282. [DOI] [PubMed] [Google Scholar]
- De Souza, W. (1999). “A short review on the morphology of Trypanosoma cruzi: from 1909 to 1999.” Mem. Inst. Oswaldo Cruz. 94, 17-36. [DOI] [PubMed] [Google Scholar]
- Edwards, P.A., and Ericsson, J. (1999). Sterols and isoprenoids: signaling molecules derived from the cholesterol biosynthetic pathway. Annu. Rev. Biochem. 68, 157-185. [DOI] [PubMed] [Google Scholar]
- Ellenberg, T.E., and Beverley, S.M. (1989). Multiple drug resistance and conservative amplification of the H region in Leishmania major. J. Biol. Chem. 264, 15094-15103. [PubMed] [Google Scholar]
- Else, A.J., Clarke, J.F., Willis, A., Jackman, S.A., Hough, D.W., and Danson, M.J. (1994). Dihydrolipoamide dehydrogenase in the Trypanosoma subgenus, trypanozoon. Mol. Biochem. Parasitol. 64, 233-239. [DOI] [PubMed] [Google Scholar]
- Endo, A., and Hasumi, K. (1993). HMG-CoA reductase inhibitors. Nat. Prod. Rep. 10, 541-50. [DOI] [PubMed] [Google Scholar]
- Garvey, E.P., and Santi, D.V. (1986). Stable amplified DNA in drug-resistant Leishmania exists as extrachromosomal circles. Science 233, 535-540. [DOI] [PubMed] [Google Scholar]
- Gebre-Hiwot, A., and Frommel, D. (1993). The in-vitro anti-leishmanial activity of inhibitors of ergosterol biosynthesis. J. Antimicrob. Chemother. 32, 873-842. [DOI] [PubMed] [Google Scholar]
- Giambiagi de Marval, M., Gottesdiener, K., Rondinelli, E., and Van der Ploeg, L.H.T. (1993). Predicted amino acid sequence and genomic organization of Trypanosoma cruzi hsp60 genes. Mol. Biochem. Parasitol. 58, 25-31. [DOI] [PubMed] [Google Scholar]
- Ginger, M.L., Prescott, M.C., Reynolds, D.G., Chance, M.L., and Goad, L.J. (2000). Utilization of leucine and acetate as carbon sources for sterol and fatty acid biosynthesis by Old and New World Leishmania species, Endotrypanum monterogeii and Trypanosoma cruzi. Eur. J. Biochem. 267, 2555-2566. [DOI] [PubMed] [Google Scholar]
- Ginger, M. L. Chance, M.L., and Goad, L.J. (1999). Elucidation of carbon sources used for the biosynthesis of fatty acids and sterol in the trypanosomatid Leishmania mexicana. Biochem. J. 342, 397-405. [PMC free article] [PubMed] [Google Scholar]
- Ginger, M.L., Chance, M.L. Sadler, I.H., and Goad, L.J. (2001). The biosynthetic incorporation of the intact leucine skeleton into sterol by the trypanosomatid Leishmania mexicana. J. Biol. Chem. 276, 11674-82. [DOI] [PubMed] [Google Scholar]
- Goldstein, J.L. and Brown, M.S. (1990). Regulation of the mevalonate pathway. Nature 343, 425-430. [DOI] [PubMed] [Google Scholar]
- Hampton, R., Dimster-Denk, D., and Rine, J. (1996). The biology of HMG-CoA reductase: the pros of contra-regulation. Trends Biochem. Sci. 21, 140-145. [PubMed] [Google Scholar]
- Hauser, R., Pypaert, M., Häusler, T., Horn, E.K., and Schneider, A. (1996). In vitro import of proteins into mitochondria of Trypanosoma brucei and Leishmania tarentolae. J. Cell Sci. 109, 517-523. [DOI] [PubMed] [Google Scholar]
- Häusler, T., Stierhof, Y.D., Blattner, J., and Clayton, C. (1997). Conservation of mitochondrial targeting sequence function in mitochondrial and hydrogenosomal proteins from the early-branching eukaryotes Crithidia, Trypanosoma, and Trichomonas. Eur. J. Cell Biol. 73, 240-251. [PubMed] [Google Scholar]
- Heise, H., and Opperdoes, F.R. (2000). Localisation of a 3-hydroxy-3-methylglutaryl-Coenzyme A reductase in the mitochondrial matrix of Trypanosoma brucei procyclics. Z. Naturforsch. 55, 473-477. [DOI] [PubMed] [Google Scholar]
- Hovius, R. (1998). Mitochondrial targeting and import. In: Protein Targeting and Translocation, ed. D. A. Phoenix, London: Portland Press Ltd., XII: 231-244.
- Keller, G.A., Pazirandeh, M., and Krisans, S. (1986). 3-hydroxy-3-methylglutaryl-coenzyme A reductase localization in rat liver peroxisomes and microsomes of control and cholestyramine-treated animals: quantitative biochemical and inmunoelectron microscopical analyses. J. Cell Biol. 103, 875-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado, R.A., Molina, J., Payares, G., and Urbina, J.A. (1993). Experimental chemotherapy with combinations of ergosterol biosynthesis inhibitors in murine models of Chagas' disease. Antimicrob. Agents Chemother. 37, 1353-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Calvillo, S., Lopez, I., and Hernandez, R. (1997). pRIBOTEX expression vector: a pTEX derivative for a rapid selection of Trypanosoma cruzi transfectants. Gene 199, 71-76. [DOI] [PubMed] [Google Scholar]
- Montalvetti, A., Pena-Diaz, J., Hurtado, R., Ruiz-Perez, L.M., and Gonzalez-Pacanowska, D. (2000). Characterization and regulation of Leishmania major 3-hydroxy-3-methylglutaryl-CoA reductase. Biochem. J. 349, 27-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navas, M.A., Cerdán, S., and Gancedo, J.M. (1993). Futile cycles in Saccharomyces cerevisiae strains expressing the gluconeogenic enzymes during growth in glucose. Proc. Natl. Acad. Sci. USA 90, 1290-1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivier, L.M., and Krisans, S.K. (2000). Peroxisomal protein targeting and identification of peroxisomal targeting signals in cholesterol biosynthesis enzymes. Biochim. Biophys. Acta 1529, 89-102. [DOI] [PubMed] [Google Scholar]
- Papadopoulou, B., Roy, G., and Ouellette, M. (1994). Autonomous replication of bacterial DNA plasmid oligomers in Leishmania. Mol. Biochem. Parasitol. 65, 39-49. [DOI] [PubMed] [Google Scholar]
- Parrish, M.L., Sengstag, C., Rine, J.D., and Wright, R.L. (1995). Identification of the sequences in HMG-CoA reductase required for karmellae assembly. Mol. Biol. Cell 6, 1535-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña-Díaz, J., Montalvetti, A., Camacho, A. Gallego, C., Ruiz-Perez, L.M., and Gonzalez-Pacanowska, D. (1997). A soluble 3-hydroxy-3-methylglutaryl-CoA reductase in the protozoan Trypanosoma cruzi. Biochem J. 324, 619-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao, K.N. (1995). The significance of the cholesterol biosynthetic pathway in cell growth and carcinogenesis. Anticancer Res. 15, 309-314. [PubMed] [Google Scholar]
- Rodrigues, C.O., Catisti, R., Uyemura, S.A., Vercesi, A.E., Lira, R., Rodriguez, C., Urbina, J.A., and Docampo, R. (2001). The sterol composition of Trypanosoma cruzi changes after growth in different culture media and results in different sensitivity to digitonin-permeabilization. J. Eukaryot. Microbiol. 48, 588-594. [DOI] [PubMed] [Google Scholar]
- Royo, T., Pedragosa, M.J., Ayte, J., Gil-Gomez, G., Vilaro, S., and Hegardt, F.G. (1993). Testis and ovary express the gene for ketogenic mitochondrial 3-hydroxy-3-methylglutaryl-CoA synthase. J. Lipid Res. 34, 867-874. [PubMed] [Google Scholar]
- Sacchettini, J.C., and Poulter, C.D. (1997). Creating isoprenoid diversity. Science 277, 1788-1789. [DOI] [PubMed] [Google Scholar]
- Searle, S., McCrossan, M.V., and Smith, D.F. (1993). Expression of a mitochondrial stress protein in the protozoan parasite Leishmania major. J. Cell Sci. 104, 1091-1000. [DOI] [PubMed] [Google Scholar]
- Stermer, B.A., Bianchini, G.M., and Korth, K.L. (1994). Regulation of HMGCoA reductase activity in plants 35, 1133-1140. [PubMed] [Google Scholar]
- Urbina, J.A. (1997). Lipid biosynthesis pathways as chemotherapeutic targets in kinetoplastid parasites. Parasitology 114, 91-99. [PubMed] [Google Scholar]
- Wach, A., Brachat, A., Pohlmann, R., and Philippsenm, P. (1994). New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10, 1793-808. [DOI] [PubMed] [Google Scholar]
- Wright, R., and Rine, J. (1989). Transmission electron microscopy and inmmunocytochemical studies of yeast: analysis of HMG-CoA reductase overproduction by electron microscopy. Methods Cell Biol. 31, 473-512. [DOI] [PubMed] [Google Scholar]
- Xu, C., and Ray, D.S. (1993). Isolation of proteins associated with kinetoplast DNA networks in vivo. Proc. Natl. Acad. Sci. 90, 1786-1789. [DOI] [PMC free article] [PubMed] [Google Scholar]