Abstract
Deregulated Wnt/β-catenin signaling underlies the pathogenesis of a broad range of human cancers, yet the development of targeted therapies to disrupt the aberrant transcription has proven difficult because the pathway incorporates large protein interaction surfaces and regulates many homeostatic functions. Therefore, we have directed our efforts toward blocking the interaction with BCL9, a co-activator for β-catenin-mediated transcription that is highly expressed in tumors but not in the cells of origin. BCL9 drives β-catenin signaling through direct binding mediated by its α-helical homology domain-2. We developed a Stabilized Alpha-Helix of BCL9 (SAH-BCL9), which we show targets β-catenin, dissociates native β-catenin/BCL9 complexes, selectively suppresses Wnt transcription, and exhibits mechanism-based anti-tumor effects. SAH-BCL9 also suppresses tumor growth, angiogenesis, invasion, and metastasis in mouse xenograft models of Colo-320 colorectal carcinoma and INA-6 multiple myeloma. By inhibiting the BCL9/β-catenin interaction and selectively suppressing oncogenic Wnt transcription, SAH-BCL9 may serve as a novel prototype therapy for cancers driven by deregulated Wnt signaling.
Keywords: Wnt signaling, transcriptional activity, cancer, BCL9, B9L, stabilized alpha-helices, stapled peptides, metastasis, angiogenesis
The canonical Wnt pathway is a receptor-mediated signal transduction network required for normal embryonic development and adult tissue homeostasis. Its activity hinges on the expression, localization, and activity of β-catenin(1–4). In the absence of Wnt ligands, β-catenin binds to adenomatous polyposis coli (APC), glycogen synthase kinase 3β (GSK3β), and Axin to form a destruction complex that phosphorylates β-catenin, which targets it for proteosomal degradation(2, 3). Binding of Wnt ligands to the Frizzled and Low-density lipoprotein receptors (LRP5 and LRP6) inhibits the activity of the APC/GSK3β/Axin complex, enabling non-phosphorylated β-catenin to undergo nuclear translocation to control transcription(1). Nuclear β-catenin associates with the lymphoid enhancer factor/T-cell factor (LEF/TCF) family of transcription factors to induce the expression of cell proliferation, migration, and survival genes such as c-MYC(5) and CyclinD1(6). This transcription pathway is turned off when Wnt receptors are not occupied, but can be activated by a variety of loss-of-function mutations in APC and Axin as well as by activating mutations in β-catenin itself. These mutations enable β-catenin to escape destruction, persist in the nucleus, and drive oncogenic transcription.
Several co-activators for Wnt/β-catenin transcription have been identified and include Pygopus (PYGO), B-cell lymphoma 9 (BCL9)(7–11), and its homolog B-cell lymphoma 9-like (B9L), among others(12, 13). The formation of a quaternary complex consisting of TCF, β-catenin, BCL9 (or B9L), and PYGO enhances β-catenin-dependent Wnt transcriptional activity(7–12). In colorectal cancer (CRC), mutations in APC and β-catenin drive carcinogenesis(1). In multiple myeloma (MM), where the canonical Wnt pathway is constitutively active and promotes MM cell proliferation (14–17), APC and β-catenin mutations have not been reported(18). Instead, the mechanism of pathologic Wnt signaling in MM involves post-transcriptional regulation of β-catenin(14) and increased levels of BCL9, implicating this β-catenin co-factor as a bona fide oncogene(19).
The human BCL9 gene was first identified by cloning the t(1;14)(q21;q32) translocation from a patient with precursor B-cell acute lymphoblastic leukemia (ALL)(20). Amplifications of chromosome 1q21 containing the BCL9 locus are observed in a broad range of human cancer types(21) and have been associated with tumor progression, decreased survival, and poor clinical outcome(22). A mutation in the BCL9 gene was linked to oncogenesis in mice subjected to retroviral mutagenesis(23). In addition, B9L promotes intestinal cancer progression in transgenic mice(24) and upregulation of B9L expression coincides with aberrant activation of Wnt signaling in human CRC and breast cancers(25–27). BCL9 overexpression occurs in a subset of human tumors (Fig. S1, A and B), and BCL9-mediated enhancement of β-catenin transcriptional activity increases cell proliferation, migration, invasion, and the metastatic potential of tumor cells(19). Importantly, BCL9 is absent from the normal cellular counterparts from which tumors originate(19). Indeed, the lack of detectable phenotypic alterations in the GI tracts of mice with conditional deletion of BCL9 and B9L(28) suggests that BCL9/BL9 proteins do not play an essential homeostatic role in mammalian Wnt signaling, although BCL9/B9L regulates a subset of Wnt target genes that control epithelial mesenchymal transition (EMT) and stem cell-like behavior(28). Collectively, these data indicate that targeting the BCL9/B9L component of aberrantly activated Wnt signaling in cancer may attenuate clinically challenging aspects of tumorigenesis including tumor invasion, metastasis, and resistance to therapy, while leaving normal tissues relatively undisturbed.
We previously demonstrated the therapeutic potential of disrupting the oncogenic activity of BCL9 by showing that shRNA-induced downregulation of BCL9 in vivo suppressed the expression of Wnt targets c-MYC, CyclinD1, CD44, and VEGF. This treatment correspondingly increased the survival of mice with xenograft CRC or MM tumors by reducing tumor load, metastasis, and the host angiogenesis response(19). Here, we sought to translate this biological proof-of-concept into a novel pharmacologic strategy for inhibiting oncogenic Wnt signaling through targeted disruption of the BCL9/β-catenin complex.
RESULTS
Targeting β-catenin with stabilized α-helices of BCL9 (SAH-BCL9)
The crystal structure of the β-catenin/BCL9/TCF-4 complex revealed that the BCL9 binding site on β-catenin is distinct from those of other binding partners in that the α-helical HD2 domain of BCL9 (residues 351–374) binds to a surface groove formed by α-helices 2 and 3 of the armadillo repeat 1 of β-catenin(29) (Fig. 1A). Alanine mutagenesis of key residues at the BCL9 binding interface, such as H358A or R359A, blocks the ability of BCL9 to bind β-catenin, abrogating transactivation(29). Harnessing this natural peptidic motif, which effectively binds β-catenin in vitro(30, 31), we applied hydrocarbon stapling(32, 33) to generate cell-permeable α-helical peptides of the BCL9 HD2 domain for in vitro and in vivo studies. Non-natural amino acids with olefinic side chains were substituted at (i, i+4) positions followed by ruthenium-catalyzed olefin metathesis to yield SAH-BCL9 peptides A through C (Fig. 1B). Circular dichroism (CD) analysis confirmed that hydrocarbon stapling consistently enhanced peptide α-helicity compared to the corresponding unmodified peptide (BCL9HD2) (Fig. 1C). In addition, Colo320 cancer cells could take up fluorescent (FITC) hydrocarbonstapled derivatives, FITC-SAH-BCL9A-C, but not the unmodified BCL9HD2 peptide (Fig. 1D).
Fig. 1.
Synthesis, characterization, and β-catenin binding of SAH-BCL9 peptides. (A) The α-helical HD2 domain of BCL9 (orange), which directly engages a surface groove of β-catenin (gray), provided the template for structural stabilization by hydrocarbon stapling. Structure adapted from Sampietro et al. (29) (PDB ID 2GL7). (B) SAH-BCL9 sequences and design, with location of hydrocarbon staples shown in purple. (C) CD analysis of α-helical stabilization of SAH-BCL9 peptides and the unmodified peptide template. [θ], Ellipticity (deg. cm2 dmol−1). (D) Immunoprecipitation of wild type (HD2) and SAH-BCL9 (A–C) peptides and β-catenin from lysates of Colo320 cells treated with FITC-labeled peptides using anti-FITC and anti-β-catenin antibodies. TCL, total cellular lysate. (E) CD analysis of H358D and R359E reverse polarity mutants of FITC-SAH-BCL9B showing similar high percent α-helicity to FITC-SAH-BCL9B. (F) Immunoprecipitation of SAH-BCL9B, H358D and R359E mutant peptides, and β-catenin from lysates of Colo320 cells treated with FITC-labeled peptides using anti-FITC and anti-β-catenin antibodies.
To assess the relative capacity of FITC-SAH-BCL9A-C peptides to bind β-catenin in these treated cells, we performed both anti-FITC and anti-β-catenin immunoprecipitation analyses, which identified FITC-SAH-BCL9B as the most effective β-catenin-targeting peptide (Fig. 1D). Live cell microscopy (Fig. S2A) demonstrated the intracellular distribution of FITC-SAH-BCL9B, including nuclear localization, which was confirmed by cellular fractionation (Fig. S2B). Of note, FITC-SAH-BCL9 peptides demonstrated superior cellular penetrance and intracellular accumulation compared to a TAT fusion construct of BCL9 HD2 (Fig. S2C–E), and manifested a striking 26 to 35-fold enhancement in proteolytic resistance (Fig. S2F), highlighting the beneficial properties conferred by hydrocarbon stapling (34).
With a lead stapled peptide in hand, we then developed two negative control constructs for our studies, FITC-SAH-BCL9B(H358D) and FITC-SAH-BCL9B(R359E), which contain single reverse polarity point mutations of key binding interface residues. Both mutants displayed similar α-helical enhancement (Fig. 1E) and cellular uptake compared to FITC-SAH-BCL9B (Fig. 1F), yet showed impaired β-catenin interaction by co-immunoprecipitation analysis (Fig. 1F), with the R359E construct being less effective. Thus, we selected the SAH-BCL9B peptide and its R359E mutant (hereafter SAH-BCL9MUT) for functional testing in Wnt-dependent CRC and MM cell lines(19, 35).
Selective dissociation of BCL9/β-catenin complexes by SAH-BCL9B
Consistent with its reduced capacity to immunoprecipitate native β-catenin (Fig. 1F), FITC-SAH-BCL9MUT was 5 times less effective in direct binding to recombinant β-catenin protein, as measured by ELISA assay (Fig. 2A and Fig. S3A, left). We next conducted in vitro and in situ binding analyses to test the capacity of SAH-BCL9B to disrupt preformed BCL9/β-catenin complexes, the activity required for Wnt signaling blockade. First, we generated recombinant GST-β-catenin and His-BCL9 proteins (Fig. S3A, right) and demonstrated that SAH-BCL9B could dissociate the complex in a dose dependent manner, with an IC50 of 135 nM, whereas FITC-SAH-BCL9MUT was 6 times less effective (Fig. 2B). Consistent with equivalent cellular uptake by an energy-dependent endocytic mechanism(33, 36, 37), SAH-BCL9B and SAH-BCL9MUT displayed parallel temperature- and dose- dependent penetrance of Colo 320 and MM1S cell lines (Fig. 2C and Fig. S3B). We then performed a series of co-immunoprecipitation analyses to determine whether treating intact cells with SAH-BCL9B could disrupt the native interactions of β-catenin with BCL9 and its close homolog B9L(12), which contains an identical HD2 domain. FITC-SAH-BCL9B, but not FITC-SAH-BCL9MUT, caused dose-dependent disruption of BCL9/β-catenin and B9L/β-catenin complexes (Fig. 2D). Correspondingly, FITC-SAH-BCL9B, but not FITC-SAH-BCL9MUT, co-immunoprecipitated with β-catenin in a dose-dependent manner, linking β-catenin targeting by FITC-SAH-BCL9B with dissociation of the native protein complexes. Mindful of the documented toxicities associated with agents that disrupt β-catenin's protein interactions, we confirmed that FITC-SAH-BCL9B had no effect on β-catenin's homeostatic interaction with E-cadherin (Fig. S3C, top), consistent with the distinct, non-overlapping location of the BCL9/β-catenin binding site. We further documented the target-based selectivity of FITC-SAH-BCL9B by anti-FITC immunoprecipitation, which co-precipitated β-catenin but not other unrelated cellular proteins such as IκBα and actin (Fig. S3C, bottom).
Fig. 2.
SAH-BCL9B disruption of β-catenin-BCL9/B9L complexes. (A) Differential binding affinities of SAH-BCL9B and SAH-BCL9MUT for recombinant β-catenin. (B) SAH-BCL9B dissociation of recombinant β-catenin/BCL9 complexes as demonstrated by GST-pull-down assay. Similar results were obtained in three independent experiments. Equal amounts of pre-formed His-tagged-BCL9/GST-tagged-β-catenin protein complexes bound to agarose beads were incubated in the absence (lane 1) or presence of increasing amounts (lanes 2–7) of acetylated SAH-BCL9B or SAH-BCL9MUT peptides. After 2 hour incubation at room temperature, agarose beads were serially washed, spun down by centrifugation, denatured, and loaded on acrylamide gels. After immunoblotting, β-catenin and BCL9 were detected with anti-FLAG or anti-His antibodies respectively (top). ImageJ software was used to measure the ratio of the density of BCL9 and β-catenin bands in relation to the concentration of acetylated SAH-BCL9B or SAH-BCL9MUT peptides (bottom). An arbitrary value of 1.0 was assigned to the BCL9/β-catenin density ratio in the absence of peptides (lane 1). (C) Cellular uptake of FITC-SAH-BCL9B and SAH-BCL9MUT by Colo320 cells. (D) Effect of peptides on the native association of β-catenin with BCL9 in Colo320 cells (upper) and with B9L in DLD1 cells (bottom). TCL, total cellular lysate.
SAH-BCL9B inhibition of Wnt transcriptional activity
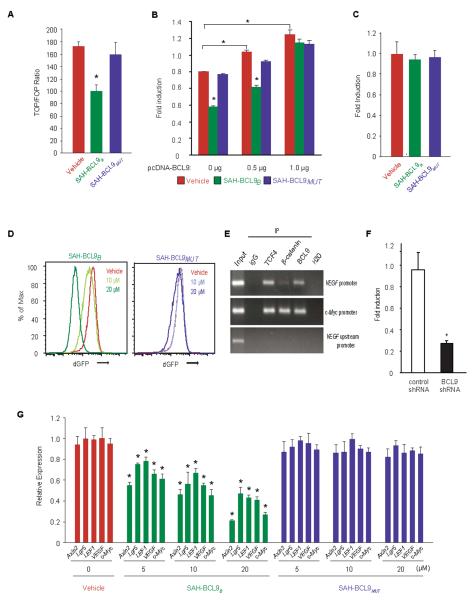
To examine the functional consequences of disrupting BCL9/β-catenin complexes, we evaluated the effects of SAH-BCL9B and SAH-BCL9MUT in a Wnt-specific TCF reporter gene transcriptional assay (TOP/FOP-FLASH)(11, 19). Whereas SAH-BCL9B treatment reduced reporter activity by nearly 50%, vehicle and SAH-BCL9MUT had no effect (Fig. 3A). Importantly, SAH-BCL9B's specificity-of-action was demonstrated by transfecting HCT116 cells with increasing amounts of a vector encoding BCL9 cDNA sequences, which abrogated the peptide's inhibitory effect (Fig. 3B), and by its complete inactivity in an NFκB reporter gene transcriptional assay (Fig. 3C). In a second Wnt-specific reporter assay that monitors destabilized GFP (dGFP) under the transcriptional control of TCF regulatory sequences, SAH-BCL9B, but not vehicle or SAH-BCL9MUT, dose-dependently blocked dGFP expression (Fig. 3D).
Fig. 3.
SAH-BCL9B inhibits Wnt transcriptional activity. (A) Colo320 cells were transfected with TOP-FLASH, incubated with vehicle or SAH-BCL9 peptides (10 μM), and assayed for luciferase activity, which was normalized to Renilla luciferase control. Error bars are mean +/− s.d. for assays performed in triplicate. *p < 0.01. (B) HCT116 cells that express low levels of BCL9 were transfected with TOP-FLASH and the indicated amounts of pcDNA-BCL9, treated with vehicle or SAH-BCL9 peptides (5 μM), and subjected to dual luciferase assay, performed at 24h. *p < 0.01. (C) Effect of SAH-BCL9B on NFκB transcriptional activity in HCT116 cells. (D) Effect of SAH-BCL9 peptides on dGFP expression in Colo320 cells lentivirally-transduced with a reporter containing TCF regulatory sequences (7xTdG). (E) ChIP of Colo320 cells using anti-TCF-4, β-catenin, and BCL9 antibodies. Negative controls included ChIP with IgG and the use of primers to a non-specific, upstream region of the VEGF promoter. (F) Colo320 cells lentivirally-transduced with control or BCL9 shRNA vectors were transfected with VEGF promoter-luciferase reporter plasmids. Reporter activity was assayed using the dual luciferase assay system and results normalized to Renilla values for each sample. *, p< 0.001. (G) qRT-PCR analysis of Wnt target gene expression in response to SAH-BCL9B treatment of Colo320 cells. Error bars are mean +/− s.d. for assays performed in quadruplicate. * p < 0.01.
We next employed quantitative PCR (qRT-PCR) analysis to measure the effects of vehicle, SAH-BCL9B, and SAH-BCL9MUT on the expression of Wnt/β-catenin target genes, including VEGF (Fig. 3E and Fig. 3F), in Colo320 (Fig. 3G) and MM1S (Fig. 4A) cell lines. Treatment with SAH-BCL9B, but not vehicle or SAH-BCL9MUT, reduced the mRNA levels of VEGF, c-MYC, LGR5, LEF1, and AXIN2(5, 28, 38–40). Actin, which is not a non-Wnt pathway target gene, was used as a reference and was not altered by SAH-BCL9B treatment. Interestingly, LGR5, which is regarded as a Wnt target gene in CRC cells, was reduced in Colo320 cells but not in MM1S cells, consistent with the notion that the expression of Wnt target genes may vary according to the physiologic and pathologic state of the cells, and also differ across discrete cellular subtypes.
Fig. 4.
SAH-BCL9B selectively blocks the intestinal Wnt/TCF signature. (A) qRT-PCR analysis revealed repression of Wnt target genes in response to SAH-BCL9B treatment of MM1S cells at 10 μM. Error bars are mean +/− s.d. for assays performed in quadruplicate. * p < 0.01. (A) Quantitative comparison of genes down-regulated by SAH-BCL9B and dominant-negative TCF1/TCF4 expression in DLD1 cells across adenoma (top) and carcinoma (bottom) signatures. (C) Heat map representation of the 50 most down-regulated genes (p<0.001) of the leading edge - the genes contributing most to the correlation between SAH-BCL9B and dominant-negative TCF1/TCF4 - for the adenoma (left) and carcinoma (right) signatures. (D) qRT-PCR validation of key Wnt target genes in DLD1 cells treated with SAH-BCL9B. (E) Affimetrix gene expression profiling analysis of VEGF-A in DLD1 cells.
To further investigate the specificity of SAH-BCL9B in blocking Wnt transcriptional activity, we performed comparative genome-wide expression analyses of Wnt target genes in the DLD1 colon cancer cell line, for which a Wnt transcription pathway signature has been described(35) (Fig. 4B–E). We first generated triplicate data sets from SAH-BCL9B- and vehicle-treated DLD1 samples using Affimetrix oligonucleotides microarrays and compared the resultant profiles with published gene expression data from DLD1 cells bearing inducible dominant-negative forms of TCF1 and TCF4(35). Gene set enrichment analysis (GSEA) revealed a statistically significant correlation between the genes down-regulated by SAH-BCL9B and by the dominant-negative forms of TCF1 and TCF4 in both adenoma (family-wise error (FWER) p-value <0.001; false discovery rate (FDR) q-value <0.001) and carcinoma (FWER and FDR <0.01) (Fig. 4B), highlighting the specificity of SAH-BCL9B in blocking Wnt transcriptional activity. Axin2, a robust and specific Wnt target gene(38) was among the most down-regulated genes by SAH-BCL9B treatment, as were other Wnt targets involved in cell metastasis (CD44, CLDN2), cell proliferation (CyclinA2, CDK4), and EMT (FOXQ1) (Fig. 4C), findings that were then validated by qRT-PCR (Fig. 4D). VEGF-A was also among the genes downregulated in cells treated with SAH-BCL9B (Fig. 4E), linking the β-catenin/BCL9 complex to tumor-induced angiogenesis.
Targeted disruption of BCL9/β-catenin inhibits cell proliferation, angiogenesis, and migration
To examine the phenotypic consequences of pharmacologic disruption of the β-catenin/BCL9/B9L complex, we conducted cellular proliferation, angiogenesis, and migration assays based on our previous findings that these key physiologic processes are regulated by β-catenin/BCL9 in CRC and MM cells(14–17, 19). A consistent pattern emerged whereby SAH-BCL9B, but not vehicle or SAH-BCL9MUT, significantly reduced the proliferation of CRC and MM cell lines, and primary tumor cells (Fig. 5A–D and Fig. S4A). All of these SAH-BCL9B-susceptible cancer cells express BCL9 (Fig. S4B–D), whereas the LS174T cell line that does not express BCL9 showed no response, consistent with an inhibitory effect of SAH-BCL9B only in the context of BCL9 expression. As further cellular controls for SAH-BCL9B's specificity-of-action, we tested HCT116 cells and its two derivative cell lines, HCT116DO20 and HCT116KO58, whose proliferative capacity does not depend on Wnt/β-catenin activity(41). Notably, these cell lines showed no sensitivity to SAH-BCL9B (Fig. 5A). To test whether the anti-proliferative effect of SAH-BCL9B could synergize with other agents commonly used to treat MM or CRC, we conducted combination treatment studies. Indeed, the cytotoxic effects of 5-flurouracil on CRC cells and of doxorubicin on MM cells were enhanced by SAH-BCL9B, but not by vehicle or the mutant peptide (Fig. S4E).
Fig. 5.
SAH-BCL9B inhibits proliferation of cultured and primary colon cancer and multiple myeloma cells and blocks angiogenesis and cell migration. (A and B) Effect of SAH-BCL9B treatment on the growth of CRC cell lines (A) and primary tumors (CRCPT) (B) that express BCL9 and depend on Wnt signaling, as measured by [3H]-thymidine uptake at 24 h. (C and D) Effect of SAH-BCL9B, on the growth of MM cell lines (C) and primary tumors (D) (MMPT). *p < 0.01. Error bars are mean +/− s.d. for experiments performed in triplicate. (E) Effect of SAH-BCL9B on VEGF secretion by Colo320 and MM1S cells as measured by ELISA. * p < 0.001. (F) Effect of SAH-BCL9B on capillary tube formation. HUVEC cells were cultured with supernatants collected from Colo320 or MM1S cells, incubated with vehicle or SAH-BCL9B peptides, and the number of tubes (black arrows) formed per high power field analyzed by microscopy at 5 h. *p <0.01 (n=3). (G) Effect of SAH-BCL9B on migration of Colo320 cells, as monitored using Matrigel Boyden chambers. Vehicle and SAH-BCL9MUT had no effect. *p<0.01. Error bars are mean +/− s.d. for experiments performed in triplicate. SAH-BCL9 dosing, 10 μM.
We next investigated the effect of SAH-BCL9B in non-cancer cells to further probe specificity-of-action. We screened normal peripheral blood mononuclear cells (PBMCs) and skin fibroblasts (WS1 and BJ) for BCL9 and β-catenin expression by western analysis (Fig. S5A). PBMCs lacking BCL9 and β-catenin showed no anti-proliferative response to SAH-BCL9B or SAH-BCL9MUT upon induction with Concanavalin A (ConA) (Fig. S5B), highlighting the absence of peptide activity in non-cancer cells lacking Wnt/β-catenin signaling. In contrast, BCL9 and β-catenin were detected at high levels in normal skin fibroblasts and because these cells are susceptible to transduction with a Wnt reporter vector, we evaluated the effect of SAH-BCL9B on Wnt transcriptional activity in WS1 cells as compared to Colo320 cells. Whereas SAH-BCL9B treatment suppressed Wnt reporter activity in Colo320 cells, no such effect was observed in WS1 cells (Fig. S5C). These striking results prompted us to investigate the mechanism underlying the differential activity of SAH-BCL9B in WS1 vs. Colo320 cells. Interestingly, cellular fractionation (Fig. S5D) and confocal microscopy (Fig. S5E) analyses documented only a cytosolic localization for BCL9 and β-catenin in WS1 cells, whereas Colo320 cells also manifested an abundant nuclear colocalization of the proteins, which is required for activation of Wnt transcriptional activity and thus modulation by SAH-BCL9B. These PBMC and fibroblast data highlight that the anti-proliferative activity of SAH-BCL9B specifically depends on the expression of BCL9 and β-catenin, and their colocalization in the nuclear compartment.
Having previously documented a role for BCL9 in VEGF-A production and tumor-associated angiogenesis(19), we next examined the effect of SAH-BCL9B on tumor cell-induced angiogenesis by treating CRC (Colo320) and MM (MM1S) cells with vehicle or SAH-BCL9B peptides and then quantified VEGF-A in the media. Consistent with the qRT-PCR analysis, SAH-BCL9B, but not vehicle or the mutant peptide, reduced the level of secreted VEGF-A (Fig. 5E). In an in vitro angiogenesis assay, human umbilical vein endothelial cells (HUVEC) were cultured with supernatants from treated Colo320 or MM1S cells and then scored for capillary tube-like formations by microscopy. HUVEC cells exposed to the supernatant from SAH-BCL9B-treated cells showed reduced capillary tube formation compared to the vehicle- and SAH-BCL9MUT-treated controls (Fig. 5F). SAH-BCL9B also decreased the adhesive and invasive potential of Colo320 cells, as reflected by a significant reduction in the capacity of SAH-BCL9B-treated cells to pass through the extracellular matrix of Matrigel-coated invasion chambers (Fig. 5G). Taken together, these data demonstrate that SAH-BCL9B specifically disrupts physiologic processes regulated by the BCL9/β-catenin transcriptional complex.
Anti-tumor activity of SAH-BCL9B in mouse xenograft models of colon cancer and multiple myeloma
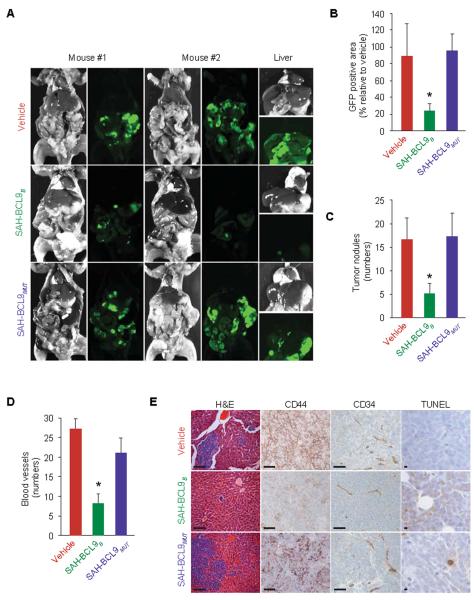
To explore the therapeutic potential of targeting the interactions between BCL9/B9L and β-catenin, we examined the capacity of SAH-BCL9B to suppress tumor growth in vivo using established mouse xenograft models of CRC and MM(14, 19). GFP-expressing Colo320 cells (1×106) were injected into the peritoneum of NOD/SCID mice. Two days after cellular injection, mice (n=6) were treated with vehicle (2.5% DMSO in a solution of 5% dextrose in water), SAH-BCL9B, or SAH-BCL9MUT peptides (20 mg/kg/day). On day 40, mice were sacrificed and evaluated for tumor burden and metastasis by whole-body imaging and histologic examination of harvested GFP-positive tissues. Tissue fluorescence was markedly reduced (p<0.01) in mice treated with SAH-BCL9B compared to vehicle and SAH-BCL9MUT-treated animals (Fig. 6, A and B). These data are consistent with an overall reduction in the number and size of metastatic tumor nodules observed in livers of SAH-BCL9B-treated mice (Fig. 6, C and E). Tumor tissue from SAH-BCL9B-treated mice also showed decreased tumor cell CD44 immunoreactivity, a reduction in the number of intratumoral blood vessels, and less intense capillary CD34 immunoreactivity (Fig. 6, D and E), suggesting that SAH-BCL9B-mediated suppression of tumor growth and metastasis may derive at least in part from reduction of cell migration and angiogenesis. We also detected an increase in apoptotic tumor cells in animals treated with SAH-BCL9B compared to vehicle or SAH-BCL9MUT-treated mice, as evaluated by TUNEL staining (Fig. 6E and Fig. S6A). No histologic changes in normal tissues were observed in any the treatment group upon necropsy (Fig. S6B).
Fig. 6.
SAH-BCL9B inhibits tumor growth, angiogenesis, and metastasis in a mouse xenograft model of CRC. (A) Whole body imaging of NOD/SCID mice bearing intraperitoneal GFP-positive Colo320 cells treated with vehicle or SAH-BCL9B peptides. (B) Tumor burden and liver metastases in SAH-BCL9B-treated mice. *p < 0.01. (C) Total number of intraparenchymal nodules for each experimental group (n=6), as quantified by examining liver sections at 5 mm intervals, in SAH-BCL9B-treated mice. Error bars are mean +/− s.d. *p < 0.01. (D) Angiogenesis, as evaluated by tumor blood vessel quantitation and anti-CD34 immunostaining in SAH-BCL9B-treated mice. Error bars are mean +/− s.d. *p < 0.0001 (n=6). (E) Histologic analysis of the liver and intraperitoneal tumors for tumor invasion, CD34- and CD44-, and TUNEL-positivity in SAH-BCL9B-treated mice. Scale bar: 50μm.
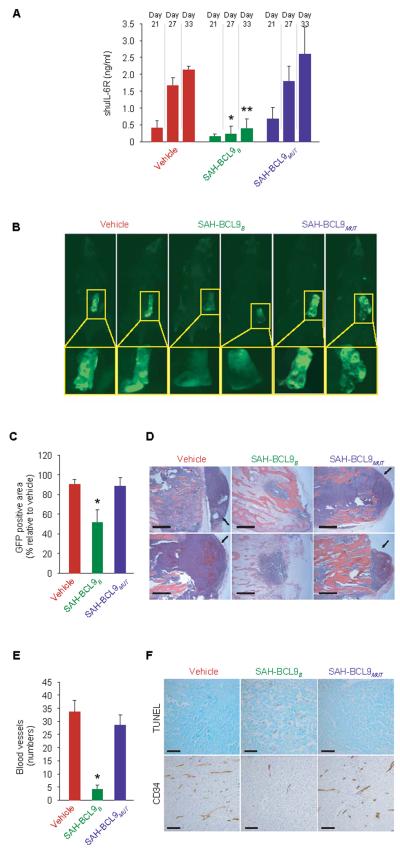
In a second in vivo model, we examined the effect of SAH-BCL9B treatment on the growth of INA-6 MM cells within a human bone graft implanted in the flank of SCID-hu mice(42). INA-6 cells (5×106) - which express both BCL9 and β-catenin (Fig. S4C), are dependent on Wnt transcriptional activity for proliferation in vitro (fig. S7A), and are inhibited by SAH-BCL9B (Fig. 5C) - were labeled with GFP and injected into bone grafts four weeks after implantation. Two days later, cohorts of mice (n=5 each) were treated by local injection with vehicle (2.5% DMSO in a solution of 5% dextrose in water), SAH-BCL9B, or SAH-BCL9MUT peptides (5 mg/kg/day) for a total of 10 doses administered every other day. To monitor tumor burden, we measured the serum level of soluble human interleukin-6 receptor (shuIL-6R), which is first detectable 3–4 weeks after INA-6 tumor engraftment(42). Whereas mice treated with vehicle or SAH-BCL9MUT showed a progressive increase in shuIL-6R levels, reflective of tumor growth, SAH-BCL9B-treated mice maintained low to undetectable levels throughout the evaluation period (Fig. 7A). Mice were sacrificed 33 days after INA-6 cell injection and evaluated for MM tumor burden by fluorescence imaging, histological analysis, and anti-CD34 staining. Consistent with the levels of shuIL-6R, tumor burden within the bone chip, as evaluated by tissue fluorescence, was significantly reduced (p<0.01) in SAH-BCL9B-treated mice (Fig. 7, B and C). Tumor cells in SAH-BCL9B-treated mice resided within the confines of the bone chip, whereas in vehicle- and SAH-BCL9MUT-treated mice tumor had invaded the surrounding soft tissue (Fig. 7D). As in the CRC model, local angiogenesis was suppressed in SAH-BCL9B-treated SCID-hu mice, as monitored by anti-CD34 staining and blood vessel quantitation (Fig. 7E), and INA-6 tumors likewise exhibited a notable increase in apoptotic tumor cells in animals treated with SAH-BCL9B (Fig. 7F and Fig. S7B). Thus, in two distinct mouse models of Wnt-driven cancer, SAH-BCL9B effectively suppressed tumor growth, invasion, and angiogenesis.
Fig. 7.
SAH-BCL9B inhibits tumor growth, angiogenesis, and metastasis in a mouse xenograft model of MM. SCID-hu mice (n=5) bearing human bone chips populated by GFP-positive INA-6 cells were injected locally with vehicle or SAH-BCL9B peptides. (A−C) Tumor burden evaluated by shuIL-6R serum levels at the indicated days after injection of tumor cells (A) and fluorescent whole body imaging upon sacrifice on day 33 (B and C). *p< 0.01. (D) Histologic analysis of INA-6 cells in the bone chips of SAH-BCL9B-treated mice. In vehicle- and SAH-BCL9MUT-treated mice, tumor cells migrated outside of the bone chip and invaded adjacent soft tissue (black arrows). Scale bar: 250 μm. (E and F) Intratumoral angiogenesis (E) and apoptosis (F) in SAH-BCL9B-treated mice, as monitored by anti-CD34 immunostaining and TUNEL staining, respectively. Scale bar: 50 μm.
DISCUSSION
The β-catenin transcriptional complex is a high-priority pharmacologic target because of its pathologic role in a broad range of human cancers including CRC(35) and MM(14, 16, 17, 19, 22). As β-catenin participates in a variety of homeostatic functions and engages the majority of its interaction partners through the same binding surface(43), achieving anti-cancer activity and selectivity remains a pressing challenge. For example, PKF115-584, a small molecule identified by high-throughput screening for inhibitors of the β-catenin/TCF interaction, blocked Wnt-specific transcriptional activity and reduced the growth of CRC and MM, but induced severe bone marrow hypoplasia, anemia, and generalized wasting of treated mice, likely a result of disrupting homeostatic Wnt signaling in normal hematopoietic and intestinal stem cells(16, 44, 45). Such therapeutic limitations, which were not observed in SAH-BCL9B-treated mice, may derive from small molecule disruption of β-catenin-TCF and β-catenin-E-cadherin interactions, and the attendant effects on epithelial tissue integrity(43). Other small molecules, such as ICG-001(46), IWR-1(47), XAV939(48), and pyrvinium(49), indirectly affect the Wnt pathway, among other activities, through engagement of CBP, Porcupine, Tankyrase, and CK1α respectively(50).
As an alternative strategy, direct and selective targeting of β-catenin by disruption of the BCL9/β-catenin complex (Fig. S8) is appealing because: (i) BCL9 drives pathologic β-catenin transcriptional activity; (ii) the β-catenin binding site is unique and corresponds to its homology domain 2 (HD2), which comprises a single amphipathic α-helix(29); and (iii) the complex is predominantly found in tumor tissue but not in the cells of origin(19), and eliminating BCL9/B9L-β-catenin interactions through genetic deletion of BCL9 and B9L in the murine gut had no overt phenotypic consequences, indicating that blockade of BCL9 function may not be harmful to normal cells(28). Thus, harnessing one of nature's solutions to β-catenin targeting by mimicking the BCL9 HD2 domain represents a rational approach to selective disruption of pathologic Wnt signaling.
Reinforcement of peptide α-helicity by insertion of chemical restraints is proving to be an effective method for investigating and modulating protein interactions(51, 52). For example, Kawamoto et al. used click chemistry to generate triazole-stapled BCL9 peptides, which demonstrate enhanced α-helicity, β-catenin binding affinity, and proteolytic stability in vitro(31). To couple such biophysical enhancements with the capacity to penetrate intact cells so that the mechanism and therapeutic potential of BCL9 HD2-based targeting of β-catenin can be interrogated in cells and in vivo, we applied hydrocarbon stapling to generate SAH-BCL9 peptides. In doing so, we determined that SAH-BCL9B targets β-catenin in cells with high affinity and selectively disrupts native β-catenin-BCL9/B9L complexes. Pharmacologic blockade of these interactions inhibited β-catenin-dependent transcriptional activity and target gene expression, and suppressed tumor cell growth, angiogenesis, and metastasis without overt damage to normal tissues. Cells and tissues that do not overexpress BCL9 or depend on Wnt signaling for cell proliferation were unaffected by SAH-BCL9B treatment. These proof-of-principle experiments document that selective targeting of the BCL-9/β-catenin interface in cancer is a promising strategy for investigating and combating oncogenic Wnt signaling. Next steps include exploring opportunities to further optimize SAH-BCL9B based on analyses of its pharmacokinetic and pharmacodynamic properties and in vivo efficacy in a broad variety of Wnt-dependent cancer models. Indeed, the emergence of this and other peptidic(31) and small molecule(53) approaches to targeting the BCL9/β-catenin interface highlights the clinical translation potential of this pharmacologic strategy to achieve a therapeutic window for combating Wnt-driven cancer.
MATERIALS AND METHODS
Peptide Synthesis and Circular Dichroism (CD) Spectroscopy
Peptides were produced on an Apex 396 (Aapptec) automated peptide synthesizer using Rink amide AM LL resin (EMD Biosciences, 0.2 mmol/g resin), at 50mmol scale. The standard Fmoc protocol employed 2×10 min deprotections in 20% piperidine/NMP followed by a pair of consecutive methanol and dimethylformamide (DMF) washes. The incorporated non-natural amino acids were treated with 4×10 min incubations in 20% piperidine/NMP to achieve complete deprotection. Amino acid coupling was performed using 400mM stock solutions of Fmoc-protected amino acids, 0.67M 2-(6-chloro-1H-benzotriazole-1-yl)-1,1,3,3-tetramethylaminium hexafluorophosphate (HCTU), and 2M N,N-diisopropyl ethylamine (DIEA), yielding 1mL of 200mM active ester (4 equivalents). Coupling frequency and incubation times were 2×30min for standard residues, 2×45min for the olefinic non-natural amino acids, and 3×45min for the residue following a non-natural amino acid. Upon completion of automated synthesis, the amino terminus was either acetylated or capped with Fmoc-β-Ala for FITC derivatization. To generate hydrocarbon staples by olefin metathesis, the resin was charged with a 10mM solution of bis(tricyclohexylphosphine)-benzylidene ruthenium (IV) dichloride (Grubbs' first generation catalyst) in 1,2-dichloroethane and stirred for 2h twice. For FITC derivatization, Fmoc-β-Ala was deprotected with piperidine in NMP and then reacted with FITC and triethylamine in dimethylformamide overnight. The peptide was cleaved from the resin and deprotected in TFA/triisopropyl silane (TIS)/water (95%, 2.5%, 2.5%), and precipitated with diethylether/hexanes. Stapled peptides were purified by reversed-phase HPLC (Agilent) using a C18 column (Zorbax), characterized by LC/MS (mass spectra obtained using electrospray in positive ion mode), and quantified by amino acid analysis on a Beckman 6300 high-performance amino acid analyzer. Stock solutions were generated by dissolving the lyophilized powder in 100% DMSO at 1 to 10mM. SAH-BCL9 powders and DMSO stock solutions were stored at −20°C. CD analysis was conducted as described(33). The solvent was 5 mM potassium phosphate buffer, pH 7.4, and the peptide concentration was 50μM.
Immunoblotting and co-immunoprecipitation
Western blotting, performed as described(16), employed the following primary antibodies: BCL9 (6109)(19), BCL9 (ab37305, Abcam), B9L (AF4967, R&D Systems), β-catenin (CAT5-H10, Zymed), FITC (ab19224, Abcam), Actin-HRP (C-11, Santa Cruz), Caspase3 (#9662, Cell Signaling), IκBα (#9242, Cell Signaling), PARP (#9542, Cell Signaling), E-cadherin (#3195, Cell Signaling), and Lamin B (sc-6217, Santa Cruz). Horseradish peroxidase conjugated secondary antibodies were purchased from Santa Cruz and SouthernBiotech. Co-immunoprecipitation was performed as described(33). Briefly, cells were lysed in 50mM Tris, 150mM NaCl, and 1% CHAPS buffer containing protease and phosphatase inhibitors. Lysates were precleared with Protein A/G PLUS-agarose beads (Santa Cruz Biotechnologies) for 3 hours followed by overnight incubation at 4°C with the respective antibodies. Agarose A/G beads were then added for 4h, pelleted, and washed as described(33).
Recombinant proteins and GST pull-down assays
Recombinant human BCL9 (residues 243–469) cloned into pET-23a (+) vector containing carboxy- terminal hexa-histidine tag and full-length human β-catenin cloned into pGEX-4T-1 vector with an amino-terminal glutathione-S-transferase (GST) tag were expressed and purified as reported(29). For GST pull-down assays, equal amounts (1 nM) of His-tagged BCL9 and GST-tagged β-catenin bound to glutathione-Sepharose 4B beads (GE) were incubated overnight at 4°C in assay buffer (100 mM Na2PO4 [pH7.4], 100μg/mL bovine serum albumin, 0.01% Triton X-100 and 4% DMSO). Complexes of His-tagged BCL9 bound to bead-immobilized, GST-tagged β-catenin were isolated by centrifugation, resuspended in 1 mL of assay buffer, and 50 μL of slurry incubated in the presence or absence of SAH-BCL9B or SAH-BCL9MUT in 500 μl assay buffer for 2 hr at room temperature. Glutathione bead-bound proteins were washed twice by centrifugation, eluted, and resolved by gel electrophoresis. GST-β-catenin was detected by Coomassie blue staining and the presence of retained His–BCL9 was detected by immunoblot analysis (anti-His 23655, Cell Signaling) and quantified with ImageJ software (NIH). The experiment was repeated three times with similar results.
Binding affinity by ELISA
Glutathione microtiter plates (Pierce) were incubated with 50 ng recombinant GST-β-catenin in 100 μL of ELISA buffer (PBS, 1% BSA, 0.05% Tween-20) per well and rotated (200 rpm) at 37 °C for 1 hr, followed by 4-cycles of automated plate-washing with PBS, 0.05% Tween-20. Two-fold serial dilution of FITC-conjugated peptides in ELISA buffer were prepared in a separate 96-well plate and transferred (100 μL) to the β-catenin-bound plate. The experimental plate was incubated for 2 hr at 37 °C (200 rpm), subjected to automated plate washing, and then 100 μL of a 1:7500 dilution of anti-FITC-conjugated HRP in ELISA buffer was transferred to each well for an additional 1 hr incubation at 37 °C (200 rpm), followed by automated plate washing. Wells were developed by adding 50 μL of tetramethylbenzidine (TMB) solution, incubating at room temperature for 20 min, and then stopping the reaction with 50 μL of 2 M H2SO4. The absorbance at 450 nm was read on a microplate reader (Molecular Devices), the binding isotherms plotted and EC50 values determined by nonlinear regression analysis using Prism software (GraphPad). Binding assays were performed in triplicate and repeated at least twice with freshly prepared recombinant proteins.
Cell Proliferation, Viability, and Apoptosis Assays
Cell proliferation assays were performed as described(16). Cell viability was measured using the CellTiter-Glo assay (Promega) according to the manufacturer's instructions. Apoptosis was evaluated by activated caspase-3 and PARP western blotting.
Histopathological Analysis and Immunohistochemistry
Tissue sections were processed as described (16). Sections were incubated with primary antibodies (5μg/ml) or the corresponding IgG fraction of preimmune serum overnight at 4°C in blocking solution (3% BSA/PBS). BCL9 (ab37305, Abcam), mouse CD34 (RAM34, eBiosciences), human CD34 (M7165, Dako) and human CD44H (2C5, R&D Systems) antibodies were employed. Blood vessel formation in the CRC and MM models was evaluated using anti-mouse CD34 and anti-human CD34 antibodies, respectively and the corresponding biotinylated antibodies coupled to streptavidin peroxidase (Vector). The number of blood vessels was determined by counting the mean number of independent blood vessels in 6 randomly selected fields at 50 × magnification as highlighted by CD34 staining.
Gene Expression Profiling and Statistical Analysis
RNA from triplicate SAH-BCL9B- and vehicle-treated DLD1 samples (10 μM each for 12 hours) was isolated for gene expression profiling analyses. Affymetrix Human U133 Plus 2.0 arrays were processed using the function of the affy Bioconductor package (URL http://www.R-project.org.). Gene sets were compiled from Van der Flier et al.(35) and gene set enrichment and statistical analyses performed using GSEA software (http://www.broad.mit.edu/GSEA) and a two-tailed t-test, respectively. Microarray data has been deposited in the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) and comply with MIAME annotation standards.
Angiogenesis and Invasion Assays
Angiogenesis was evaluated as previously described(19) using an in vitro angiogenesis assay kit (Millipore). For capillary tube formation analysis, HUVEC were cultured on polymerized matrix gel and exposed to supernatant media collected from Colo320 or MM1S cells treated with vehicle (0.5% DMSO), SAH-BCL9B peptides (5μM) for 24h. The number of capillary tubes formed after 5h treatment at 37°C was determined by counting 5 randomly selected fields at 40× magnification, according to manufacture's instructions. HUVEC cultured in VEGF media and VEGF-free media were used as positive and negative controls, respectively(19). Cellular invasion assays were performed using Matrigel Boyden chambers (BD Biosciences) as described (19). The reported data represent the average of three independent experiments performed in triplicate.
Xenograft Models
GFP-positive Colo320 cells were generated as previously reported (19). Cells were pelleted, resuspended in sterile 1x PBS and injected intraperitoneally (1×106 cells/mouse) into 5 week-old sublethally irradiated NOD.CB17-PrkdcSCID/J mice (Jackson Laboratory) (n=6 per cohort). Two days after cellular inoculation, mice were treated by intraperitoneal injection with vehicle (2.5% DMSO in D5W) or SAH-BCL9 peptides (20mg/kg) on alternate days for a total of 6 doses. Forty days after tumor cell injection, the mice were euthanized and GFP-positive tumor visualized using an ImageQuant LAS-4000 (GE Healthcare). Complete necropsies were performed for each experimental animal and livers were sectioned in their entirety at 5 mm intervals for quantitation of tumor metastases. Tissues were subjected to H&E staining and immunohistochemical analysis using anti-CD34 and anti-CD44 antibodies.
For the SCID-hu murine model of human MM, human fetal bone grafts measuring 1.5×0.5 cm were subcutaneously implanted into eight week-old male CB-17 SCID mice (Taconic) as previously described(42). Four weeks after bone implantation, 5×106 GFP-positive INA-6 MM cells were injected directly into each bone implant. Two days later, mice were treated with 100μl injections of vehicle (2.5% DMSO in D5W) or SAH-BCL9 peptides (5mg/kg) instilled adjacent to the bone chips on alternate days for a total of 10 doses. Mouse sera were serially monitored for shuIL-6R levels by ELISA (R&D Systems). Thirty-three days after tumor cell injection, the mice were sacrificed and analyzed for tumor burden by fluorescence imaging and histologic analysis of the bone grafts. All animal experiments were performed in accordance with approved protocols of the Dana-Farber Cancer Institute Animal Care and Use Committee.
Reporter Assays
Luciferase activity was measured using the Dual Luciferase Reporter Assay System (Promega) as previously described(3). To measure Wnt or NFκB reporter activity, Colo320 cells were transfected with TOP-FLASH, FOP-FLASH plasmid (Millipore Corporation) or NFκB luciferase reporter (Stratagene), along with an internal Renilla control plasmid (hRL-null). Transfection was accomplished using FuGENE (Roche) according to the manufacturer's protocol. The results were normalized to control Renilla activity. The reported data represent the average of three independent transfection experiments performed in triplicate.
Supplementary Material
ACKNOWLEDGMENTS
We thank Drs. Jeremy Ryan, Yu-Tsu Tai, Abdelkareem Azab, Teru Hideshima, and Victor Penta-Cruz for technical assistance. Kenneth C. Anderson and Paul Richardson for providing myeloma tumor samples (Dana-Farber Cancer Institute), and Anthony Letai for helpful discussions. Funding: D.R.C. is supported by a Claudia Adams Barr Award, the Multiple Myeloma Research Foundation, a P50CA127003 DF/HCC SPORE in gastrointestinal cancer, and a 1R01CA151391-01. This work was also supported by a Burroughs Welcome Career Award in the Biomedical Sciences and a Todd J. Schwartz Pediatric Oncology grant to L.D.W.
Footnotes
AUTHOR CONTRIBUTIONS D.R.C. designed the study, with input from G.H.B. and L.D.W. G.H.B. and L.D.W. designed, synthesized, and characterized SAH-BCL9 peptides, and conducted the in vitro binding analyses. K.T., D.Z., J.J.Z., M.M., D.C., K.S., J.R., D.H. and D.R.C. performed the cellular and in vivo experiments. R.A.S and D.H provided and flow sorted primary CR cells. M.F. and N.C.M. performed the SCID-hu experiment. A.L.K. and M.L. performed the bioinformatic analysis. W.X provided vectors and purified GST-β-catenin protein. D.R.C. and L.D.W. wrote the manuscript, which was reviewed and edited by the co-authors.
COMPETING FINANCIAL INTERESTS L.D.W. is a scientific advisory board member and consultant for Aileron Therapeutics.
DATA AND MATERIALS AVAILABILITY Oligonucleotide microarray data has been deposited in the Gene Expression Omnibus under the accession number GSE33143.
REFERENCES
- 1.Logan CY, Nusse R. Annu Rev Cell Dev Biol. 2004;20:781. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 2.Clevers H. Cell. 2006 Nov 3;127:469. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 3.Klaus A, Birchmeier W. Nat Rev Cancer. 2008 May;8:387. doi: 10.1038/nrc2389. [DOI] [PubMed] [Google Scholar]
- 4.Valenta T, Hausmann G, Basler K. EMBO J. 2012 May 22; doi: 10.1038/emboj.2012.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.He TC, et al. Science. 1998 Sep 4;281:1509. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 6.Tetsu O, McCormick F. Nature. 1999 Apr 1;398:422. doi: 10.1038/18884. [DOI] [PubMed] [Google Scholar]
- 7.Kramps T, et al. Cell. 2002 Apr 5;109:47. doi: 10.1016/s0092-8674(02)00679-7. [DOI] [PubMed] [Google Scholar]
- 8.Townsley FM, Cliffe A, Bienz M. Nat Cell Biol. 2004 Jul;6:626. doi: 10.1038/ncb1141. [DOI] [PubMed] [Google Scholar]
- 9.Hoffmans R, Stadeli R, Basler K. Curr Biol. 2005 Jul 12;15:1207. doi: 10.1016/j.cub.2005.05.054. [DOI] [PubMed] [Google Scholar]
- 10.Stadeli R, Basler K. Mech Dev. 2005 Nov;122:1171. doi: 10.1016/j.mod.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 11.Sustmann C, Flach H, Ebert H, Eastman Q, Grosschedl R. Mol Cell Biol. 2008 May;28:3526. doi: 10.1128/MCB.01986-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brembeck FH, et al. Genes Dev. 2004 Sep 15;18:2225. doi: 10.1101/gad.317604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hecht A, Vleminckx K, Stemmler MP, van Roy F, Kemler R. EMBO J. 2000 Apr 17;19:1839. doi: 10.1093/emboj/19.8.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dutta-Simmons J, et al. Blood. 2009 Sep 24;114:2699. doi: 10.1182/blood-2008-12-194290. [DOI] [PubMed] [Google Scholar]
- 15.Qiang YW, Endo Y, Rubin JS, Rudikoff S. Oncogene. 2003 Mar 13;22:1536. doi: 10.1038/sj.onc.1206239. [DOI] [PubMed] [Google Scholar]
- 16.Sukhdeo K, et al. Proc Natl Acad Sci U S A. 2007 May 1;104:7516. doi: 10.1073/pnas.0610299104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Derksen PW, et al. Proc Natl Acad Sci U S A. 2004 Apr 20;101:6122. doi: 10.1073/pnas.0305855101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chapman MA, et al. Nature. 2011 Mar 24;471:467. doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mani M, et al. Cancer Res. 2009 Oct 1;69:7577. doi: 10.1158/0008-5472.CAN-09-0773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Willis TG, et al. Blood. 1998 Mar 15;91:1873. [PubMed] [Google Scholar]
- 21.Beroukhim R, et al. Nature. 2010 Feb 18;463:899. [Google Scholar]
- 22.Carrasco DR, et al. Cancer Cell. 2006 Apr;9:313. doi: 10.1016/j.ccr.2006.03.019. [DOI] [PubMed] [Google Scholar]
- 23.Rad R, et al. Science. 2010 Nov 19;330:1104. [Google Scholar]
- 24.Brembeck FH, et al. Gastroenterology. 2011 Oct;141:1359. doi: 10.1053/j.gastro.2011.06.039. [DOI] [PubMed] [Google Scholar]
- 25.Adachi S, et al. Cancer Res. 2004 Dec 1;64:8496. doi: 10.1158/0008-5472.CAN-04-2254. [DOI] [PubMed] [Google Scholar]
- 26.Sakamoto I, et al. Cancer Sci. 2007 Jan;98:83. doi: 10.1111/j.1349-7006.2006.00363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Toya H, et al. Cancer Sci. 2007 Apr;98:484. doi: 10.1111/j.1349-7006.2007.00430.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deka J, et al. Cancer Res. 2010 Aug 15;70:6619. doi: 10.1158/0008-5472.CAN-10-0148. [DOI] [PubMed] [Google Scholar]
- 29.Sampietro J, et al. Mol Cell. 2006 Oct 20;24:293. doi: 10.1016/j.molcel.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 30.Kawamoto SA, et al. Biochemistry. 2009 Oct 13;48:9534. doi: 10.1021/bi900770z. [DOI] [PubMed] [Google Scholar]
- 31.Kawamoto SA, et al. J Med Chem. 2011 Dec 23; [Google Scholar]
- 32.Bird GH, Bernal F, Pitter K, Walensky LD. Methods Enzymol. 2008;446:369. doi: 10.1016/S0076-6879(08)01622-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walensky LD, et al. Science. 2004 Sep 3;305:1466. doi: 10.1126/science.1099191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bird GH, et al. Proc Natl Acad Sci U S A. 2010 Aug 10;107:14093. doi: 10.1073/pnas.1002713107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van der Flier LG, et al. Gastroenterology. 2007 Feb;132:628. doi: 10.1053/j.gastro.2006.08.039. [DOI] [PubMed] [Google Scholar]
- 36.Bernal F, Tyler AF, Korsmeyer SJ, Walensky LD, Verdine GL. J Am Chem Soc. 2007 Mar 7;129:2456. doi: 10.1021/ja0693587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brooks H, Lebleu B, Vives E. Adv Drug Deliv Rev. 2005 Feb 28;57:559. doi: 10.1016/j.addr.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 38.Lustig B, et al. Mol Cell Biol. 2002 Feb;22:1184. doi: 10.1128/MCB.22.4.1184-1193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang X, Gaspard JP, Chung DC. Cancer Res. 2001 Aug 15;61:6050. [PubMed] [Google Scholar]
- 40.Zirn B, et al. Genes Chromosomes Cancer. 2006 Jun;45:565. doi: 10.1002/gcc.20319. [DOI] [PubMed] [Google Scholar]
- 41.Chan TA, Wang Z, Dang LH, Vogelstein B, Kinzler KW. Proc Natl Acad Sci U S A. 2002 Jun 11;99:8265. doi: 10.1073/pnas.082240999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tassone P, et al. Blood. 2005 Jul 15;106:713. doi: 10.1182/blood-2005-01-0373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barker N, Clevers H. Nat Rev Drug Discov. 2006 Dec;5:997. doi: 10.1038/nrd2154. [DOI] [PubMed] [Google Scholar]
- 44.Lepourcelet M, et al. Cancer Cell. 2004 Jan;5:91. doi: 10.1016/s1535-6108(03)00334-9. [DOI] [PubMed] [Google Scholar]
- 45.Reya T, Clevers H. Nature. 2005 Apr 14;434:843. doi: 10.1038/nature03319. [DOI] [PubMed] [Google Scholar]
- 46.Emami KH, et al. Proc Natl Acad Sci U S A. 2004 Aug 24;101:12682. doi: 10.1073/pnas.0404875101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen B, et al. Nat Chem Biol. 2009 Feb;5:100. doi: 10.1038/nchembio.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang SM, et al. Nature. 2009 Oct 1;461:614. doi: 10.1038/nature08356. [DOI] [PubMed] [Google Scholar]
- 49.Thorne CA, et al. Nat Chem Biol. 2010 Nov;6:829. doi: 10.1038/nchembio.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Polakis P. EMBO J. 2012 May 22; doi: 10.1038/emboj.2012.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Henchey LK, Jochim AL, Arora PS. Curr Opin Chem Biol. 2008 Dec;12:692. doi: 10.1016/j.cbpa.2008.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bird GH, Crannell WC, Walensky LD. Current Protocols in Chemical Biology. 2011 Sep 1;3:99. doi: 10.1002/9780470559277.ch110042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.de la Roche M, et al. Nat Commun. 2012;3:680. doi: 10.1038/ncomms1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shang Y, Hu X, DiRenzo J, Lazar MA, Brown M. Cell. 2000 Dec 8;103:843. doi: 10.1016/s0092-8674(00)00188-4. [DOI] [PubMed] [Google Scholar]
- 55.Clifford RL, Deacon K, Knox AJ. J Biol Chem. 2008 Dec 19;283:35337. doi: 10.1074/jbc.M803342200. [DOI] [PubMed] [Google Scholar]
- 56.Basu A, et al. Cancer Res. 2008 Jul 15;68:5689. doi: 10.1158/0008-5472.CAN-07-6603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Naldini L, Blomer U, Gage FH, Trono D, Verma IM. Proc Natl Acad Sci U S A. 1996 Oct 15;93:11382. doi: 10.1073/pnas.93.21.11382. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.