Abstract
Sex chromosomes are the Achilles heel of male meiosis in mammals.1 Mis-segregation of the X and Y chromosomes leads to sex chromosome aneuploidies, with clinical outcomes such as infertility and Klinefelter syndrome.2 Successful meiotic divisions require that all chromosomes find their homologous partner and achieve recombination and pairing. Sex chromosomes in males of many species have only a small region of homology (the pseudoautosomal region, PAR) that enables pairing.3 Until recently, little was known about the dynamics of recombination and pairing within mammalian X and Y PARs. Here, we review our recent findings on PAR behavior in mouse meiosis.4 We uncovered unexpected differences between autosomal chromosomes and the X-Y chromosome pair, namely that PAR recombination and pairing occurs later, and is under different genetic control. These findings imply that spermatocytes have evolved distinct strategies that ensure successful X-Y recombination and chromosome segregation.
Keywords: Meiosis, recombination, sex chromosomes, chromosome pairing
In higher eukaryotes, meiotic cell divisions produce sperm and eggs. Haploid DNA content in germ cells is achieved by one round of DNA replication, followed by two successive rounds of cell division. To successfully segregate chromosomes to daughter cells in the first meiotic cell division, homologous chromosomes (homologs) must find each other and stably pair. In most organisms studied to date, including mammals, homolog recognition and pairing is achieved by DNA recombination interactions that are initiated by developmentally programmed DNA double-stranded breaks (DSBs).5
Evolutionary geneticists have long appreciated the role of meiotic recombination in reshuffling genetic material from one generation to the next (see e.g. ref. 6). Gene mapping efforts are possible thanks only to this feature of meiosis. What has received less attention in this field is the fact that meiotic recombination also has an essential function in chromosome pairing7–9 and in establishing a physical connection (crossover) between homologs,10, 11 which in turn is critical for chromosome segregation. To initiate recombination, mouse spermatocytes typically make 200–250 DSBs per nucleus.12, 13 As meiosis progresses, most of these DSBs are repaired without the exchange of flanking chromosome arms (non-crossover) while a subset matures into crossovers involving a reciprocal exchange between homologs14 (see also Cole et al., this volume). Recombination failure leads to inviable gametes.7, 9
Once a DSB is made, the proper partner — the unbroken template for homologous recombination — must be located and engaged in a stable pairing interaction. Not all chromosomes, however, are on a level playing field with regard to pairing. Autosomal chromosomes are homologous along their entire length (~60–200 Mb, depending on the chromosome) and are hence able to pair from end to end. The X and Y chromosomes, on the other hand, are non-homologous save for a short segment called the pseudoautosomal region (PAR).3 X-Y chromosome homology search and pairing therefore can only be mediated by DSBs in the PAR, conceivably making X-Y segregation particularly difficult. Indeed, X-Y aneuploidy is a common paternally inherited chromosome disorder.2 In most cells, however, X-Y chromosome segregation is completed successfully, raising the question how the challenging task of recombination between heteromorphic sex chromosomes is achieved. We applied sophisticated cytological methods to investigate DSB formation and pairing of the PARs in mice, and review here our recent findings on mechanisms that secure faithful X-Y recombination.4
Unusual higher-order chromatin structure in the PAR
Ensuring that at least one PAR DSB is formed presents the first potential hurdle on the path to successful X-Y recombination: in mice, one meiotic DSB forms every 10 Mb on average,15 while the PAR spans just 0.7 Mb.16 How do spermatocytes promote a DSB frequency 10- to 20-fold higher than average in this tiny region? Insight into this question came from consideration of higher-order chromosome structure. Meiotic DSB formation takes place in the context of highly organized chromatin;17 replicated sister chromatids emanate from chromosome axes as chromatin loops (Figure 1). Blat et al.18 proposed a model in which chromatin loops are accessible to the DSB machinery, with each loop therefore presenting an opportunity for DSB formation. Thus, DNA packaged into many small loops may be more prone to DSB formation than if the same kilobase-amount of DNA was packaged into fewer and larger loops. We therefore speculated that one way to render the PAR more conducive to DSB formation could be packaging into larger numbers of smaller loops.
Figure 1.
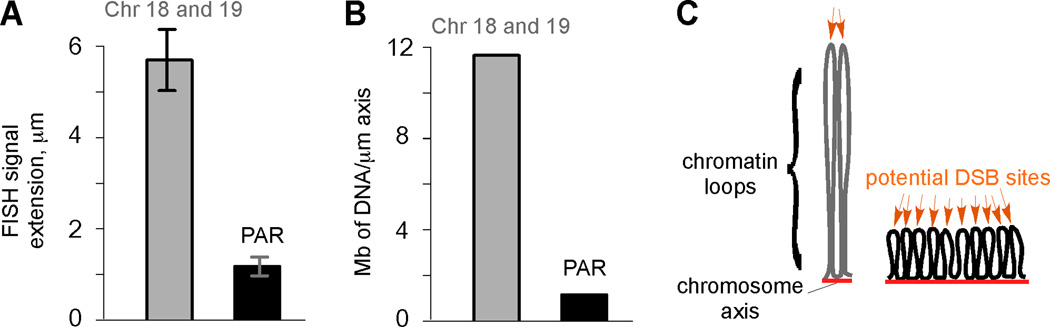
Higher-order structure of the PAR compared to autosomes. A) Measurements of the maximal distance of FISH signals from chromosome axes (average for subtelomeric chromosome 18 and 19 probes, gray bar; PAR probe, black bar) in cells at pachynema (mean ± SD). B) DNA content (Mb) per µm of chromosome axis in the subtelomeric regions of chromosomes 18 and 19, and the PAR. C) Model of how organization of DNA into chromatin loops can influence DSB frequency (after ref. 18). Only one homolog (each consisting of a pair of sister chromatids) is shown, with chromatin loops tethered to chromosome axes (red lines). Each chromatin loop is envisioned to provide an opportunity for meiotic DSB formation (orange arrows). DNA organized into more and smaller loops per Mb (right) presents the DSB machinery with more opportunities for break formation than if the same length of DNA is organized into fewer and larger loops (left).
To gain insight into the higher-order structure of the PAR, we measured how far a fluorescent in situ hybridization (FISH) probe signal extended from chromosome axes, compared to probes on small autosomes, in nuclei isolated from spermatocytes in early meiosis . Autosomal probes extended on average >5-fold further than the PAR probe (Figure 1A).4 This observation, combined with the fact that PAR chromosome axes are disproportionately long considering their DNA content (Figure 1B) indicates that PAR chromatin loops are several-fold smaller than loops in autosomal genomic regions (Figure 1C), potentially providing the meiotic DSB machinery with increased opportunity for cutting. Hence, distinct chromatin packaging may be the first level of control that helps promote high-frequency DSB formation in the PAR.
Most DSBs in PARs form later than on autosomes
We asked whether PAR DSBs form with the same timing as “bulk” (nucleus-wide) DSBs. To answer this question, we examined meiotic chromosome preparations of nuclei at various stages of meiotic progression. Stages are defined by the extent of chromosome axis development, assessed by the synaptonemal complex protein SYCP3, used as a cytological marker.4 We first performed immunofluorescence (IF) against SYCP3 (to visualize meiotic chromosome axes) and RAD51 (which marks sites of DSB repair), to quantify nucleus-wide DSBs (Figure 2Ai). IF was followed by PAR FISH (Figure 2Aii) to examine whether PARs displayed a RAD51 focus (Figure 2Aiii). In control mice (blue circles in Figure 2B), nucleus-wide RAD51 focus numbers peaked in early/mid-zygonema. Surprisingly, in the same mice, PAR RAD51 foci were not observed until late zygonema in most nuclei (blue squares in Figure 2B). This analysis indicated that many cells do not experience a PAR DSB until late zygonema, whereas nucleus-wide DSBs reach a maximum earlier, in early/mid-zygonema (Figure 2B, compare blue solid line with blue dashed line). For data and in-depth discussion of RAD51 foci observed on the X PAR versus the Y PAR, see ref. 4.
Figure 2.
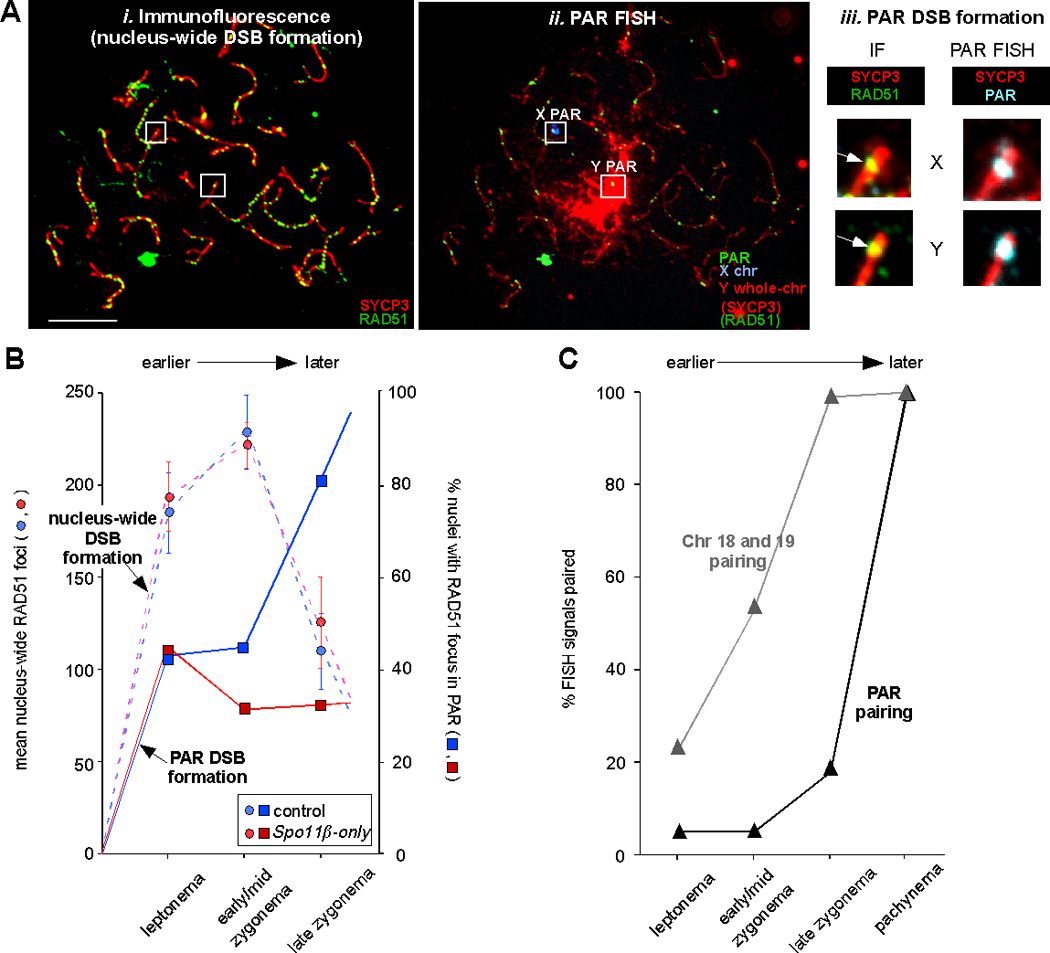
Many PAR DSBs form later than nucleus-wide DSBs, and are under different genetic control. A) Assay for nucleus-wide and PAR DSB formation. i-ii) Example of immunofluorescent (IF) and FISH staining of a spermatocyte in late zygonema. Nuclei were first stained with antibodies against RAD51 and SYCP3 and photographed (i), then FISH was performed using PAR, X and Y probes (as cartooned above the FISH image) and the same nuclei were photographed again (ii).4 Under the FISH conditions used in these experiments, some IF signal remains visible. The presence or absence of a RAD51 focus in the X and Y PAR was scored by comparing the IF image to an IF+FISH image overlay (iii). In this example, both the X and Y PAR (inside white squares in i-ii) display a RAD51 focus (white arrows in iii). Scale bar, 10 µm. B) Mean nucleus-wide RAD51 focus numbers (bars ± 95% confidence interval of the mean; left Y axis) and percentage of nuclei with a PAR RAD51 focus on the X and/or Y PAR (right Y axis) as prophase I progresses from leptonema to late zygonema, in mice of the indicated genotypes (see text). C) Percentage of nuclei with paired FISH signals for autosomal and PAR probes as prophase I progresses from leptonema to pachynema.
PAR DSB formation is under distinct genetic control
The temporally distinct (delayed) DSB formation of the PARs raised the possibility of differential genetic control. The protein responsible for meiotic DSB formation, SPO11,19 has two major isoforms in mammals, SPO11α and SPO11β.20–22 Interestingly, the two isoforms appear with differential timing: SPO11β predominates in early meiotic cells when most DSBs form, whereas SPO11α is more abundant later.4, 22 It therefore seemed possible that the late isoform, SPO11α, may play a role in coordinating later-forming DSBs in general, or PAR DSBs in particular. To elucidate the genetic requirements for nucleus-wide and PAR DSB formation, we analyzed RAD51 foci in a transgenic mouse model carrying only the Spo11β isoform (Spo11β-only mice).4
Nucleus-wide (“global”) RAD51 focus numbers in Spo11β-only mice were indistinguishable from those in control mice (Figure 2B, compare red dashed line with blue dashed line), suggesting that SPO11β by itself is sufficient for normal overall DSB levels. In contrast, the percentage of cells with PAR RAD51 foci was significantly lower at late zygonema, the stage when most PARs in control mice display a RAD51 focus (Figure 2B, compare red solid line with blue solid line). Late PAR DSBs therefore are genetically separable from both nucleus-wide DSBs and early PAR DSBs. The absence of SPO11α apparently affects only late-forming PAR DSBs. The cellular consequences are discussed in the next section.
X and Y chromosomes pair later than autosomes
Given our observation that PAR DSBs form later than autosomal DSBs, one would predict that PAR (that is, X and Y chromosome) pairing would occur later than autosomal pairing in wild-type mice. We assessed the pairing status of PARs and autosomes by asking whether cells at various stages of meiosis displayed two (i.e. unpaired) or one (i.e. paired) FISH probe signals. We discovered that the X and Y PARs indeed pair late in comparison to the autosomes we examined (Figure 2C). Contemporaneously, and independent of DSB formation, a heterochromatic domain forms called the sex body which brings the X and Y chromosomes closer together.23 This physical proximity of the X and Y at late zygonema likely is an additional (but not sufficient) factor that facilitates PAR pairing.
A further prediction is that since PAR DSBs fail to form in many Spo11β-only spermatocytes, PAR pairing should also often fail. This is exactly what we found: 69% of PARs were unpaired in pachynema in Spo11β-only mice, compared to 5% in control mice; in contrast, no change in autosomal pairing was found.4 As Spo11β-only spermatocytes progress to metaphase I, premature X-Y separation was observed in a similarly high percentage of cells. Massive apoptosis was seen in the testes at this stage and mice are largely infertile.4 The simplest explanation for these meiotic defects is that in the majority of cells, PAR DSB formation and subsequently crossover formation fails, leading to unconnected X and Y chromosomes at metaphase whose presence triggers the spindle checkpoint and an apoptotic response.
Interestingly, we found that Spo11β-only female mice are fertile, and all chromosomes (including the fully homologous X chromosomes) pair normally.4 Therefore in female meiosis, although SPO11α is expressed,4, 21 it appears to be entirely dispensable. If SPO11α’s main function indeed is to ensure efficient PAR DSB formation, the Spo11β-only female fertility phenotype also indicates that PAR DSBs are dispensable for X-X pairing. This is perhaps not surprising, since recombination in females can take place along the nearly 170 Mb length of the X chromosome, instead of being restricted to the <1-Mb PAR.
Conclusions and future directions
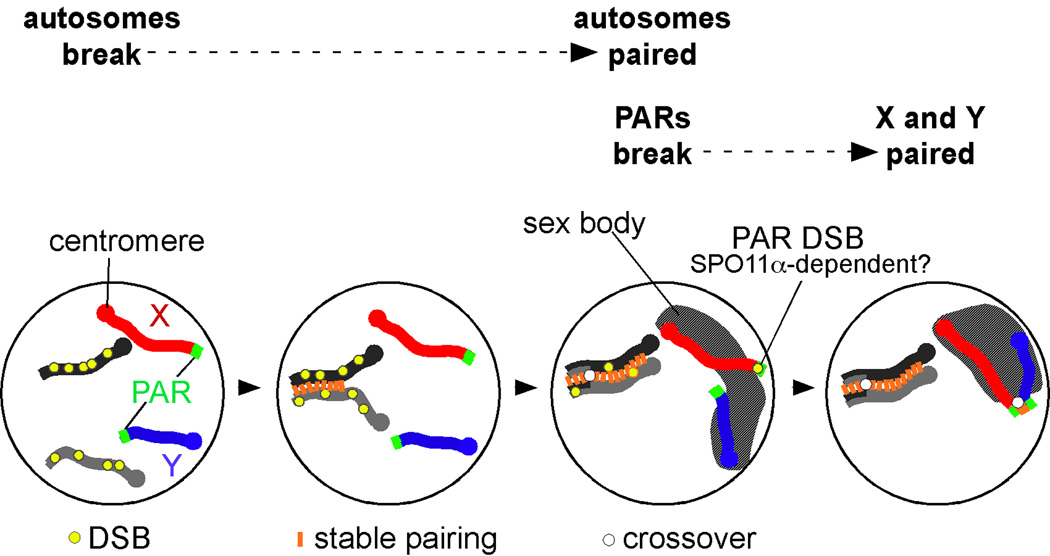
Our experiments demonstrated that PAR DSB formation, recombination and pairing are temporally and genetically distinct from that of autosomal chromosomes. A model consolidating these findings is shown in Figure 3. Autosomes undergo DSB formation earlier, and pair earlier than PARs. Not until autosomal pairing is nearly complete, do most cells form a DSB in the PAR. Sex body formation likely aids in PAR pairing by gathering the X and Y chromosomes into the same nuclear territory. In transgenic mice that lack SPO11α (the late-appearing isoform), late PAR DSBs are not formed, and PAR pairing frequently fails, leading to X-Y mis-segregation and male infertility. In a subset of cells, PAR DSBs form earlier, around the same time as nucleus-wide DSBs; these cells do not need SPO11α for pairing or crossing-over. The simplest interpretation is that SPO11α‘s main and perhaps only role is to promote late PAR DSB formation, either directly or indirectly. A Spo11α-only mouse model, by itself and crossed with Spo11 β-only mice, should shed light on this matter.
Figure 3.
Model summarizing our findings on the distinct behavior of meiotic X and Y chromosomes.4 A pair of autosomal homologous chromosomes is shown as light and dark gray lines, the X and Y as red and blue lines, respectively. Most DSBs on autosomes form before PAR DSBs; as a consequence, autosomal chromosomes pair earlier than the X and Y. Only DSBs that can facilitate homolog pairing are depicted (yellow circles); not shown are the numerous DSBs that form on the non-PAR portion of the X chromosome (see ref. 4) but cannot mediate pairing. In many cells, PARs undergo DSB formation later, around the same time as the sex body (hatched area) begins to form. This chromatin domain brings the X and Y closer together and likely facilitates PAR pairing.
Heteromorphic sex chromosomes and PARs are a widespread genomic feature in the animal kingdom.3 This raises the question whether the distinct properties of PARs that we uncovered in mouse meiosis are a frequently utilized solution to the sex chromosome recombination problem. Circumstantial evidence suggests that this may be the case, at least for mammals: humans have the same SPO11 isoforms as mice,20, 22 and human X and Y chromosomes have been reported to pair later than autosomes.24 With the immuno-FISH methods established in mice, detailed cytological studies into human X-Y chromosome dynamics are now feasible for the first time.
Accurate DNA recombination, with DSBs as initiating DNA lesions, is critical for genome stability and for fertility. DSBs represent potentially toxic DNA damage that can lead to genome re-arrangements if not faithfully repaired. Yet, in each meiotic cell, >200 self-inflicted DSBs on average are made and subsequently processed in a remarkably error-free manner. Our data highlight multiple layers of exquisite control that underlie this fidelity. First, higher-order chromatin structure can affect which genomic regions are accessible to the recombination machinery. Second, the position of chromosomes in three-dimensional space within the nucleus influences DNA transactions. Third, the expression of proteins that interact with germline DNA is under stringent temporal regulation and restricts DSB formation to a limited window of opportunity. Future studies will further elucidate factors that affect genome plasticity. Nowhere is the repair of DNA damage more important than in the germline, where it has the potential to give rise to heritable changes and thereby to provide fuel for evolution.
References
- 1.Shi Q, et al. Single sperm typing demonstrates that reduced recombination is associated with the production of aneuploid 24, XY human sperm. Am. J. Med. Genet. 2001;99:34–38. doi: 10.1002/1096-8628(20010215)99:1<34::aid-ajmg1106>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 2.Shi Q, Martin RH. Aneuploidy in human spermatozoa: FISH analysis in men with constitutional chromosomal abnormalities, and in infertile men. Reproduction. 2001;121:655–666. doi: 10.1530/rep.0.1210655. [DOI] [PubMed] [Google Scholar]
- 3.Raudsepp T, et al. The pseudoautosomal region and sex chromosome aneuploidies in domestic species. Sex. Dev. 2012;6:72–83. doi: 10.1159/000330627. [DOI] [PubMed] [Google Scholar]
- 4.Kauppi L, et al. Distinct properties of the XY pseudoautosomal region crucial for male meiosis. Science. 2011;331:916–920. doi: 10.1126/science.1195774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zickler D, Kleckner N. Meiotic chromosomes: integrating structure and function. Annu. Rev. Genet. 1999;33:603–754. doi: 10.1146/annurev.genet.33.1.603. [DOI] [PubMed] [Google Scholar]
- 6.Barton NH, Charlesworth B. Why sex and recombination? Science. 1998;281:1986–1990. [PubMed] [Google Scholar]
- 7.Baudat F, et al. Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking Spo11. Mol. Cell. 2000;6:989–998. doi: 10.1016/s1097-2765(00)00098-8. [DOI] [PubMed] [Google Scholar]
- 8.Peoples TL, et al. Close, stable homolog juxtaposition during meiosis in budding yeast is dependent on meiotic recombination, occurs independently of synapsis, and is distinct from DSB-independent pairing contacts. Genes Dev. 2002;16:1682–1695. doi: 10.1101/gad.983802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Romanienko PJ, Camerini-Otero RD. The mouse Spo11 gene is required for meiotic chromosome synapsis. Mol. Cell. 2000;6:975–987. doi: 10.1016/s1097-2765(00)00097-6. [DOI] [PubMed] [Google Scholar]
- 10.Eaker S, et al. Meiotic prophase abnormalities and metaphase cell death in MLH1-deficient mouse spermatocytes: insights into regulation of spermatogenic progress. Dev. Biol. 2002;249:85–95. doi: 10.1006/dbio.2002.0708. [DOI] [PubMed] [Google Scholar]
- 11.Lipkin SM, et al. Meiotic arrest and aneuploidy in MLH3-deficient mice. Nat. Genet. 2002;31:385–390. doi: 10.1038/ng931. [DOI] [PubMed] [Google Scholar]
- 12.Cherry SM, et al. The Mre11 complex influences DNA repair, synapsis, and crossing over in murine meiosis. Curr. Biol. 2007;17:373–378. doi: 10.1016/j.cub.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roig I, et al. Mouse TRIP13/PCH2 is required for recombination and normal higher-order chromosome structure during meiosis. PLoS Genet. 2010;6:e1001062. doi: 10.1371/journal.pgen.1001062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cole F, Keeney S, Jasin M. Comprehensive, fine-scale dissection of homologous recombination outcomes at a hot spot in mouse meiosis. Mol Cell. 39:700–710. doi: 10.1016/j.molcel.2010.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barchi M, et al. ATM promotes the obligate XY crossover and both crossover control and chromosome axis integrity on autosomes. PLoS Genet. 2008;4:e1000076. doi: 10.1371/journal.pgen.1000076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perry J, et al. A short pseudoautosomal region in laboratory mice. Genome Res. 2001;11:1826–1832. doi: 10.1101/gr.203001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Panizza S, et al. Spo11-accessory proteins link double-strand break sites to the chromosome axis in early meiotic recombination. Cell. 2011;146:372–383. doi: 10.1016/j.cell.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 18.Blat Y, et al. Physical and functional interactions among basic chromosome organizational features govern early steps of meiotic chiasma formation. Cell. 2002;111:791–802. doi: 10.1016/s0092-8674(02)01167-4. [DOI] [PubMed] [Google Scholar]
- 19.Keeney S, Giroux CN, Kleckner N. Meiosis-specific DNA doublestrand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 1997;88:375–384. doi: 10.1016/s0092-8674(00)81876-0. [DOI] [PubMed] [Google Scholar]
- 20.Keeney S, et al. A mouse homolog of the Saccharomyces cerevisiae meiotic recombination DNA transesterase Spo11p. Genomics. 1999;61:170–182. doi: 10.1006/geno.1999.5956. [DOI] [PubMed] [Google Scholar]
- 21.Bellani MA, et al. The expression profile of the major mouse SPO11 isoforms indicates that SPO11beta introduces double strand breaks and suggests that SPO11alpha has an additional role in prophase in both spermatocytes and oocytes. Mol. Cell. Biol. 2010;30:4391–4403. doi: 10.1128/MCB.00002-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Romanienko PJ, Camerini-Otero RD. Cloning, characterization, and localization of mouse and human SPO11. Genomics. 1999;61:156–169. doi: 10.1006/geno.1999.5955. [DOI] [PubMed] [Google Scholar]
- 23.Burgoyne PS, Mahadevaiah SK, Turner JM. The consequences of asynapsis for mammalian meiosis. Nat. Rev. Genet. 2009;10:207–216. doi: 10.1038/nrg2505. [DOI] [PubMed] [Google Scholar]
- 24.Rasmussen SW, Holm PB. Human meiosis II. Chromosome pairing and recombination nodules in human spermatocytes. Carlsberg Res. Comm. 1978;43:275–327. [Google Scholar]