Abstract
Cells re-use signaling proteins in multiple pathways, raising the potential for improper crosstalk. Scaffold proteins are thought to insulate against such miscommunication by sequestering proteins into distinct physical complexes. We show that the scaffold protein Ste5, which organizes the yeast mating MAP kinase pathway, does not use sequestration to prevent misactivation of the mating response. Instead, Ste5 appears to use a conformation mechanism: under basal conditions, an intramolecular interaction of the PH domain with the VWA domain blocks its the ability to co-activate the mating-specific MAPK, Fus3. Pheromone-induced membrane binding of Ste5 triggers release of this autoinhibition. Thus, in addition to serving as a conduit guiding kinase communication, Ste5 directly receives input information to decide if and when signal can be transmitted to mating output.
Cells use a complex network of signaling proteins to respond to diverse signals and stresses. Execution of proper decisions is complicated by the fact that individual cells contain many closely related signaling proteins (1). In fact, the same proteins are often reused in multiple signaling pathways (2, 3). The resulting interlinked networks could lead to inappropriate crosstalk between signaling pathways.
Scaffold proteins, which physically assemble components of a signaling pathway (4–6), provide a possible solution to this problem. By binding and organizing pathway components into complexes, scaffold proteins promote efficient signaling along a particular pathway. Scaffold proteins may also insulate against improper communication by physically sequestering signaling proteins into distinct pools (7–15). However, to prevent shared proteins from exchanging between pools, a scaffold must bind its partners with dissociation rates that are slow compared to the timescale for signaling. Direct evidence for this prevailing view of scaffold-based insulation is limited.
A prototypical scaffold protein is Ste5, which coordinates the yeast mating mitogen-activated protein kinase (MAPK) response by binding to all three components of the MAPK cascade and serving as a required co-activator of the mating-specific MAPK, Fus3 (16, 17). The Ste5 scaffold is thought to insulate the mating response from other MAPK pathways in yeast, such as the starvation response, which uses the identical MAPK kinase (MAPKK), Ste7, and MAPKK kinase (MAPKKK), Ste11, proteins, but activates a distinct starvation-specific MAPK Kss1 to produce an invasive growth response (Fig. 1A) (2, 17). How the common MAPKK, Ste7, when activated by a specific input, is directed to the correct downstream MAPK is only partially understood. With mating input, both Fus3 and Kss1 are activated (binding to the Ste5 scaffold does not prevent the MAPKK Ste7 from activating Kss1) (16, 18). However, activation of Kss1 by mating input does not lead to crosstalk because activated Fus3 overrides the Kss1-induced starvation response by phosphorylating and downregulating a starvation-specific transcription factor (19, 20). Thus, proper starvation response hinges upon preventing Fus3 misactivation by starvation inputs, which would both launch the mating program and directly inhibit the starvation response.
Fig. 1. Exchange of the Ste7 MAPKK from the Ste5 scaffold protein.
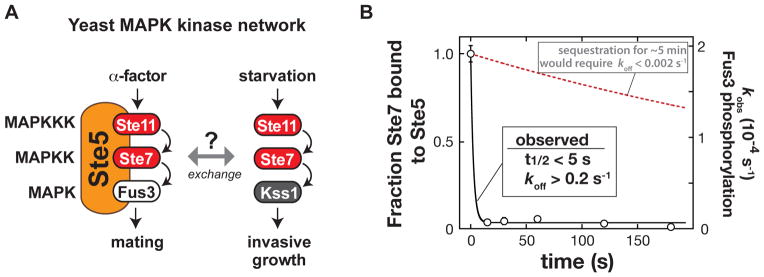
(A) Shared components of the yeast mating and invasive growth pathways yield physiologically distinct input-output responses.
(B) Dissociation rate of the MAPKK Ste7 from the Ste5 scaffold protein measured with purified recombinant Ste5, the MAPKKK Ste11, the MAPK Fus3, and a constitutively active form of the MAPKK Ste7 (Ste7EE, bearing phosphomimic mutations in the Ste7 activation loop (16)). To a preassembled Ste5-Ste11-Ste7-Fus3 complex, an excess of a Ste7 binding domain (a minimal Ste7 binding domain from Ste5 [residues 759–810]) was added to capture Ste7 as it dissociated from Ste5 (Fig. S1). At various times, ATP was added, and the initial rate of Fus3 phosphorylation was measured (the amount of Ste5-Ste7-Fus3 complex remaining at each timepoint). Error bars are standard deviations. The observed koff of 0.2 s−1 is a lower limit – dissociation occurred on a timescale faster than could be measured with mixing by hand.
For Ste5 to act as a sequestration-based insulator would require exchange rates for the scaffold-bound shared kinases (Ste11 or Ste7) to be slow relative to the timescale of signaling. Otherwise, shared kinases activated by non-mating inputs would be able to exchange onto the Ste5 scaffold protein and activate the mating response. We measured the dissociation of purified Ste7 from Ste5 to have a t1/2 of <5 seconds (Fig. 1B), faster by several orders of magnitude than the typical ~5 minute timescale of MAPK signaling pathways, and far faster than the timescale of days on which the yeast starvation response operates. Thus, physical sequestration is unlikely to be the primary mechanism that prevents activation of the mating MAPK Fus3 by non-mating inputs.
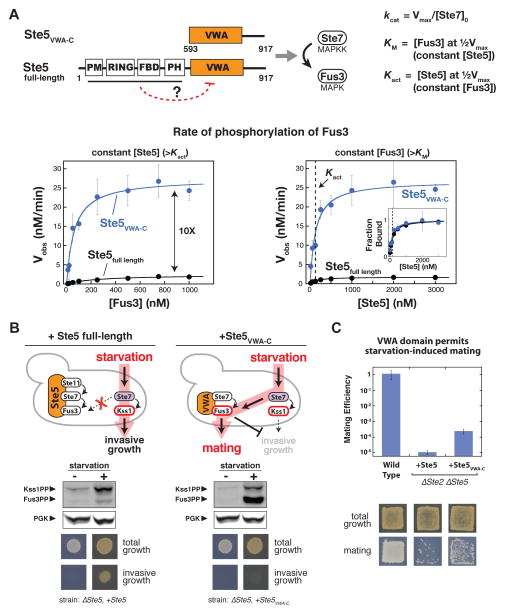
An alternative model for insulation is that, in the absence of mating input, Ste5 adopts an inactive conformation that blocks its ability to co-catalyze Fus3 phosphorylation (17). To test this possibility, we measured rates of Fus3 phosphorylation by the MAPKK Ste7 with full-length Ste5 and with a minimal Ste5 fragment (Ste5VWA-C, containing a von-Willebrand Type-A (VWA) domain that is required for Fus3 co-activation together with active MAPKK Ste7 (16)). Under maximal rate (kcat) conditions (saturating concentrations of all components), the rate of Fus3 phosphorylation with full-length Ste5 was one-tenth that in the presence of Ste5VWA-C (Fig. 2A). Assembly of the Ste5-Ste7-Fus3 complex was similar with either Ste5 construct (Fig. 2A), suggesting that the dominant contribution to the difference in activity is not a binding effect (disruption of kinase complex assembly) but rather a disruption of the catalytic co-activator function of the VWA domain.
Fig. 2. Autoinhibition of the Ste5 scaffold protein insulates the MAPK Fus3 from activation by incorrect inputs.
(A) Full-length Ste5 (residues 1–917) and Ste5VWA-C (residues 593–917) used for in vitro kinetic assays for phosphorylation of Fus3 by Ste7EE, Michaelis-Menten plot of Vobs vs. [Fus3], and plot of Vobs vs. [Ste5]. Kact corresponds to the midpoint of the Vobs vs. [Ste5] plot and represents the dissociation constant for Ste5-Ste7. Error bars are standard deviations. It was unnecessary to measure binding affinity for Fus3 assembly into the ternary complex because the Ste5 VWA domain does not bind with any detectable affinity to Fus3 (Fus3 is recruited to this ternary catalytic complex via binding to Ste7; the interaction between the Ste5 VWA domain and Fus3 is a transient catalytic interaction (16)). See fig. S2-S4 and table S1 for values of fitted kinetic constants.
(B) Fus3 misactivation in response to starvation in yeast cells expressing Ste5VWA-C (see Methods for growth conditions). Fus3 and Kss1 phosphorylation was monitored with an antibody to phosphorylated MAPKs by protein immunoblotting, and phosphoglycerate kinase (PGK) is shown as a loading control for yeast lysates (see fig. S5-S7 for quantitative analysis). The invasive growth response was assayed with yeast cells grown on solid agar plates (see Methods). Similar results were obtained with constitutively active alleles of Ste11 and Ste7 (Fig. S8).
(C) Ste5VWA-C misdirects signaling to Fus3 and allows cells to mate in response to starvation. Mating efficiencies were determined using a quantitative mating assay, and patch assays were done as described (see Methods). Error bars are standard deviations.
To test the possibility that this activity difference contributes to insulation of the mating pathway in vivo, we introduced the fully active Ste5VWA-C fragment into yeast. In these cells, starvation led to substantial activation of the mating MAPK Fus3, whereas cells with full-length Ste5 predominantly activated the starvation MAPK Kss1 (Fig. 2B). Thus the minimal Ste5VWA-C fragment appears to promote Fus3 activation when the MAPKK Ste7 is activated, regardless of whether cells have received the mating signal or not. Because Fus3 activation inhibits the invasive growth response at the transcriptional level (19, 20), misactivation of Fus3 by cells expressing Ste5VWA-C overrides the invasive growth phenotype (Fig. 2C). Further, under starvation conditions, Ste5VWA-C restores a partial mating phenotype in cells that lack the mating receptor, Ste2 (Fig. 2D); full rescue of mating likely requires mating pathway components upstream of the kinase cascade that are not activated by starvation (21). Thus when Fus3 activation is promiscuous, cells misinterpret the starvation condition as a signal to initiate the mating program.
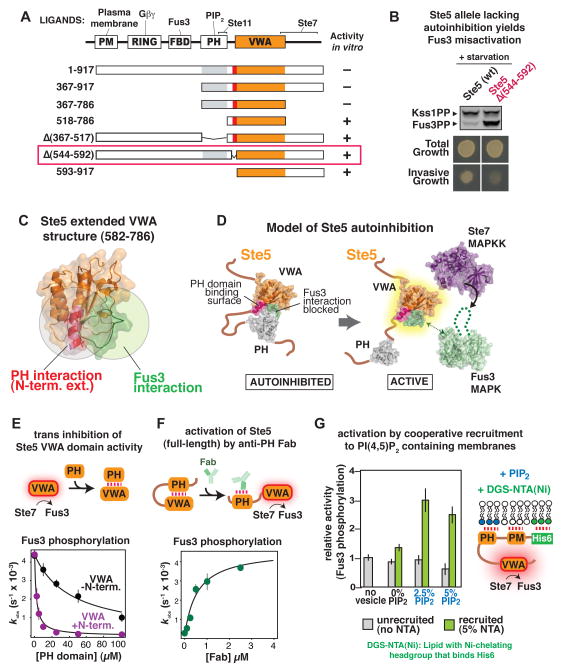
By deletion analysis, we identified two regions of Ste5 essential for autoinhibition of the VWA domain in vitro: the PH domain, which binds to phosphoinositol 4,5-bisphosphate (PIP2) to facilitate membrane binding (22) and binds to the MAPKKK Ste11 (23), and an N-terminal extension to the VWA domain (residues 544–592), within the linker that connects the PH and VWA domains (Fig. 3A and fig. S9). When Ste5 was replaced by Ste5Δ (544–592) in vivo, activation of Fus3 in response to mating pheromone was normal (Fig. S7), but Fus3 was misactivated in response to starvation (Fig. 3B). Because all kinase binding sites are intact in Ste5Δ (544–592), this result supports the idea that physical sequestration of kinases by Ste5 is not sufficient for pathway insulation.
Fig. 3. Mechanism of Ste5 autoinhibition and activation by mating-specific input.
(A) Diagram of truncation mapping of Ste5 (red is residues 544–592). The minimal autoinhibited fragment (367–786) contains all elements necessary to assemble the three-tiered MAPK cascade (Fus3 is recruited to the VWA domain by Ste7 (16)). See fig. S9 for additional constructs.
(B) Effect of deletion of the N-terminal extension of the VWA domain in Ste5 (residues 544–592) to disrupt pathway insulation in vivo. Activation of Fus3 measured by protein immunoblotting, and invasive growth assayed on solid agar plates (assays conducted as in fig. 2C, see fig. S6).
(C) Crystal structure of Ste5582–786 (see Table S4 for crystallographic statistics). The N-terminal extension (582–592) is shown in pink, with surface residues necessary for trans inhibition by the PH domain (Fig. S10) shown as sticks in red. The Fus3-coactivating loop of Ste5 (743–756) is shown in green. The spatial proximity of the PH-domain interface and the site of Fus3 coactivation is illustrated by overlapping circles.
(D) Model for Ste5 autoinhibition inferred from truncation mapping data and Ste5582–786 crystal structure.
(E) Titration of the VWA domain with the PH domain results in inhibition of the Fus3 coactivator activity (kobs) of the VWA construct bearing the N-terminal extension (582–786, shown in pink), or lacking the N-terminal extension (593–786, shown in black). Error bars are standard deviations.
(F) Relief of autoinhibition in full-length Ste5 by a Fab antibody (SR13) that can bind the PH domain. Error bars are standard deviations.
(G) Recruitment of Ste5 to membranes with PIP2 stimulates coactivation of Fus3. A minimal, autoinhibited Ste5 fragment bearing a hexahistidine tag can be recruited to small unilamellar vesicles of varying lipid compositions by the DGS-NTA(Ni) lipid (see fig. S13 for exact details of the Ste5 construct used here). Error bars are standard deviations.
We determined the crystal structure of an extended VWA fragment (residues 582–786; we were unable to obtain crystals for a PH-VWA complex) (Fig. 3C); this construct includes the minimal N-terminal extension that binds the PH domain (Fig. S10). This extension forms an N-terminal α-helix lying directly adjacent to the VWA domain “coactivator loop” that contains residues essential for Fus3 coactivation (16). The spatial proximity of the autoinhibitory PH domain binding site (the N-terminal extension) and the Fus3 coactivator loop indicates that PH domain binding and Fus3 activation might be mutually exclusive, providing a molecular mechanism for Ste5 autoinhibition (Fig. 3D). Indeed, the isolated PH domain of Ste5 inhibited the Fus3 co-activator function of the VWA domain in trans (Fig. 3E). Further, a Fab antibody fragment that binds the PH domain completely relieved autoinhibition (Fig. 3F and fig. S11). Also, an allele of Ste5 (S770N) that was previously found to constitutively activate the mating pathway (24) is not autoinhibited in vitro (Fig. S12).
An early step in mating pathway activation is pheromone-induced membrane recruitment of Ste5, which requires a cooperative set of membrane interactions that includes the PH domain binding to PIP2 lipids (22). Thus binding of Ste5 to PIP2-containing membranes might disrupt the PH-VWA interaction and relieve autoinhibition. We designed a minimal, membrane-binding Ste5 construct that is autoinhibited, but PIP2-containing lipid vesicles did not bind or activate this construct in vitro (Fig. S13). Because pheromone-induced membrane recruitment of Ste5 is a cooperative process that requires several membrane-binding motifs (21), we induced association of the autoinhibited Ste5 construct to the lipid vesicles using other cooperative membrane interactions (Fig. 3G and Fig. S13). Under these conditions, PIP2 caused a 3-fold activation of Ste5 (Fig. 3G), suggesting that membrane recruitment of Ste5 and its interaction with PIP2 contributes to relief of autoinhibition of Ste5. The inability of such membrane association to completely relieve autoinhibition of Ste5 (a 10-fold effect, Fig. 2A) could result from incomplete binding to lipid vesicles in vitro (Fig. S13), or because complete activation requires additional interactions present in vivo. Ste5 oligomerization has been suggested to contribute to pathway activation (25), but we find no evidence that oligomerization plays a direct role in relief of Ste5 autoinhibition (Fig. S14).
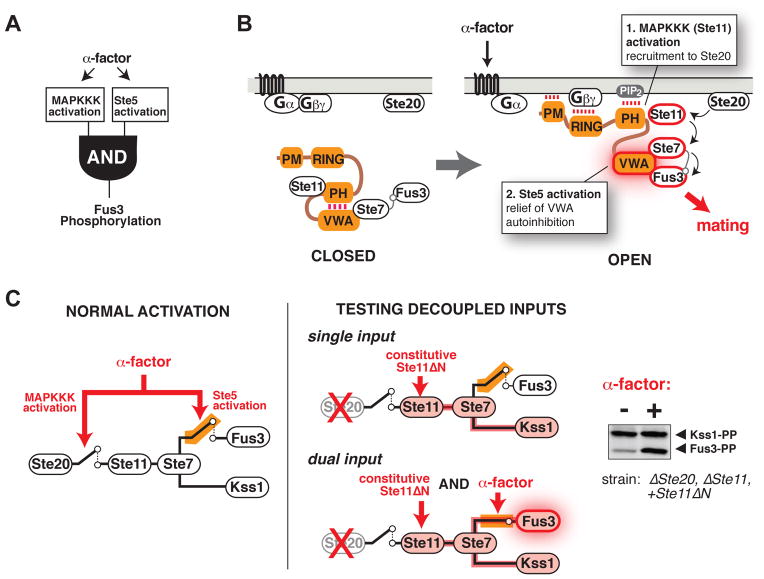
We propose that although the shared upstream kinases (MAPKKK Ste11 and MAPKK Ste7) can be activated by other inputs, only mating input activates both the kinase cascade and the Ste5 scaffold protein to permit Fus3 activation (Fig. 4A). Activation of the mating pathway recruits Ste5 to the membrane (21), thus activating the MAP kinase cascade by bringing the MAPKKK Ste11 in proximity to its upstream kinase Ste20. Membrane recruitment may also relieve autoinhibition in Ste5 when the PH domain interacts with PIP2 at the membrane (Fig. 4B).
Fig. 4. Mating input-mediated conformational activation of Ste5.
(A) Simple AND-gate model for specific mating pathway activation. Non-mating inputs that activate the shared MAPKKK do not activate Fus3.
(B) Revised molecular model for mating pathway activation mediated by the Ste5 scaffold protein. Mating pheromone (α-factor) activates a heterotrimeric G protein, leading to release of the Gβγ subunit from Gα and recruitment of Ste5 to free Gβγ at the membrane (21). Membrane recruitment triggers activation of the MAPKKK Ste11 and PH domain binding to PIP2, leading to release of the VWA domain and relief of autoinhibition.
(C) Fus3 activation in vivo when kinase cascade activation is decoupled from the mating signal (α-factor), measured by protein immunoblotting. See fig. S15 for additional Ste11 alleles and controls.
To further test this model, we decoupled the two functions of Ste5 by deleting the upstream kinase Ste20 (preventing normal activation of the MAPK cascade), and introducing a constitutively-active allele of the MAPKKK Ste11, thus rendering activation of the kinase cascade independent of the mating signal. Previous experiments of this type demonstrated that full pathway activation still requires the mating input, suggesting that the input acts on a step downstream of kinase cascade activation (24, 26). Here we take this approach one step further by using a constitutively active allele, Ste11ΔN (27), which lacks the Ste5 binding site (28), so that any observed effects of Ste5 activation are likely to arise from promoting the Ste7→Fus3 reaction rather than the Ste11→Ste7 reaction. When wild type Ste11 was replaced by Ste11ΔN in a yeast strain lacking Ste20, the MAPK Kss1 was preferentially phosphorylated, but when this strain was treated with α-factor, activation of Fus3 was observed (Fig. 4), supporting the idea that pheromone-induced membrane recruitment of Ste5 has two distinct and separable functions: to activate the MAPKKK Ste11 and to relieve autoinhibition in Ste5 to permit Fus3 activation.
Our data do not support the prevailing model that scaffold proteins primarily insulate signaling by sequestration of proteins. Instead, Ste5 appears to function as a conformational switch to gate the flow of information between two distinct signaling outcomes. This mechanism provides a potentially general means to control information flow in complex signaling networks with shared components.
Supplementary Material
Acknowledgments
J.G.Z. is supported by the Damon Runyon Cancer Research Foundation (DRG# 2012-09). S.M.C. is supported by a National Science Foundation Graduate Research Fellowship. S.S.S. and S.R. are supported by a grant from the Canadian Institutes for Health Research (MOPS-93725). This work was also supported by NIH grants RO1 GM55040, RO1 GM62583, PN2 EY016546, P50 GM081879 (W.A.L.) and the Howard Hughes Medical Institute (W.A.L.). We thank G. Narlikar, H. Madhani, and M. Good for helpful discussions and comments, N. Helman and S. Vidal for plasmids, H. Madhani for providing yeast strains, and C. Waddling for assistance with X-ray data collection. Coordinates have been deposited in the Protein Data Bank (4F2H).
Footnotes
This manuscript has been accepted for publication in Science. This version has not undergone final editing. Please refer to the complete version of record at http://www.sciencemag.org/. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS.
References and Notes
- 1.Taylor SS, et al. A template for the protein kinase family. Trends Biochem Sci. 1993;18:84. doi: 10.1016/0968-0004(93)80001-r. [DOI] [PubMed] [Google Scholar]
- 2.Schwartz MA, Madhani HD. Principles of MAP kinase signaling specificity in Saccharomyces cerevisiae. Annu Rev Genet. 2004;38:725. doi: 10.1146/annurev.genet.39.073003.112634. [DOI] [PubMed] [Google Scholar]
- 3.Qi MS, Elion EA. MAP kinase pathways. J Cell Sci. 2005;118:3569. doi: 10.1242/jcs.02470. [DOI] [PubMed] [Google Scholar]
- 4.Good MC, Zalatan JG, Lim WA. Scaffold proteins: Hubs for controlling the flow of cellular information. Science. 2011;332:680. doi: 10.1126/science.1198701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morrison DK, Davis RJ. Regulation of MAP kinase signaling modules by scaffold proteins in mammals. Annu Rev Cell Dev Biol. 2003;19:91. doi: 10.1146/annurev.cellbio.19.111401.091942. [DOI] [PubMed] [Google Scholar]
- 6.Shaw AS, Filbert EL. Scaffold proteins and immune-cell signalling. Nat Rev Immunol. 2009;9:47. doi: 10.1038/nri2473. [DOI] [PubMed] [Google Scholar]
- 7.Saito H. Regulation of cross-talk in yeast MAPK signaling pathways. Curr Opin Microbiol. 2010;13:677. doi: 10.1016/j.mib.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 8.Dhanasekaran DN, Kashef K, Lee CM, Xu H, Reddy EP. Scaffold proteins of MAP-kinase modules. Oncogene. 2007;26:3185. doi: 10.1038/sj.onc.1210411. [DOI] [PubMed] [Google Scholar]
- 9.Harris K, et al. Role of scaffolds in MAP kinase pathway specificity revealed by custom design of pathway-dedicated signaling proteins. Curr Biol. 2001;11:1815. [PubMed] [Google Scholar]
- 10.Burack WR, Shaw AS. Signal transduction: hanging on a scaffold. Curr Opin Cell Biol. 2000;12:211. doi: 10.1016/s0955-0674(99)00078-2. [DOI] [PubMed] [Google Scholar]
- 11.Garrington TP, Johnson GL. Organization and regulation of mitogen-activated protein kinase signaling pathways. Curr Opin Cell Biol. 1999;11:211. doi: 10.1016/s0955-0674(99)80028-3. [DOI] [PubMed] [Google Scholar]
- 12.Whitmarsh AJ, Davis RJ. Structural organization of MAP-kinase signaling modules by scaffold proteins in yeast and mammals. Trends Biochem Sci. 1998;23:481. doi: 10.1016/s0968-0004(98)01309-7. [DOI] [PubMed] [Google Scholar]
- 13.Pawson T, Scott JD. Signaling through scaffold, anchoring, and adaptor proteins. Science. 1997;278:2075. doi: 10.1126/science.278.5346.2075. [DOI] [PubMed] [Google Scholar]
- 14.Marcus S, Polverino A, Barr M, Wigler M. Complexes between Ste5 and Components of the Pheromone-Responsive Mitogen-Activated Protein-Kinase Module. Proc Natl Acad Sci USA. 1994;91:7762. doi: 10.1073/pnas.91.16.7762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choi KY, Satterberg B, Lyons DM, Elion EA. Ste5 tethers multiple protein kinases in the MAP kinase cascade required for mating in S. cerevisiae. Cell. 1994;78:499. doi: 10.1016/0092-8674(94)90427-8. [DOI] [PubMed] [Google Scholar]
- 16.Good M, Tang G, Singleton J, Remenyi A, Lim WA. The Ste5 scaffold directs mating signaling by catalytically unlocking the Fus3 MAP kinase for activation. Cell. 2009;136:1085. doi: 10.1016/j.cell.2009.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flatauer LJ, Zadeh SF, Bardwell L. Mitogen-activated protein kinases with distinct requirements for Ste5 scaffolding influence signaling specificity in Saccharomyces cerevisiae. Mol Cell Biol. 2005;25:1793. doi: 10.1128/MCB.25.5.1793-1803.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elion EA, Brill JA, Fink GR. Fus3 represses Cln1 and Cln2 and in concert with Kss1 promotes signal transduction. Proc Natl Acad Sci USA. 1991;88:9392. doi: 10.1073/pnas.88.21.9392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bao MZ, Schwartz MA, Cantin GT, Yates JR, Madhani HD. Pheromone-dependent destruction of the Tec1 transcription factor is required for MAP kinase signaling specificity in yeast. Cell. 2004;119:991. doi: 10.1016/j.cell.2004.11.052. [DOI] [PubMed] [Google Scholar]
- 20.Chou S, Huang L, Liu HP. Fus3-regulated Tec1 degradation through SCF Cdc4 determines MAPK signaling specificity during mating in yeast. Cell. 2004;119:981. doi: 10.1016/j.cell.2004.11.053. [DOI] [PubMed] [Google Scholar]
- 21.Bardwell L. A walk-through of the yeast mating pheromone response pathway. Peptides. 2005;26:339. doi: 10.1016/j.peptides.2004.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garrenton LS, Young SL, Thorner J. Function of the MAPK scaffold protein, Ste5, requires a cryptic PH domain. Gene Dev. 2006;20:1946. doi: 10.1101/gad.1413706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Inouye C, Dhillon N, Durfee T, Zambryski PC, Thorner J. Mutational analysis of STE5 in the yeast Saccharomyces cerevisiae: Application of a differential interaction trap assay for examining protein-protein interactions. Genetics. 1997;147:479. doi: 10.1093/genetics/147.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lamson RE, Takahashi S, Winters MJ, Pryciak PM. Dual role for membrane localization in yeast MAP kinase cascade activation and its contribution to signaling fidelity. Curr Biol. 2006;16:618. doi: 10.1016/j.cub.2006.02.060. [DOI] [PubMed] [Google Scholar]
- 25.Inouye C, Dhillon N, Thorner J. Ste5 RING-H2 domain: Role in Ste4-promoted oligomerization for yeast pheromone signaling. Science. 1997;278:103. doi: 10.1126/science.278.5335.103. [DOI] [PubMed] [Google Scholar]
- 26.Lyons DM, Mahanty SK, Choi KY, Manandhar M, Elion EA. The SH3-domain protein Bem1 coordinates mitogen-activated protein kinase cascade activation with cell cycle control in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:4095. doi: 10.1128/mcb.16.8.4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cairns BR, Ramer SW, Kornberg RD. Order of action of components in the yeast pheromone response pathway revealed with a dominant allele of the STE11 kinase and the multiple phosphorylation of the STE7 kinase. Genes Dev. 1992;6:1305. doi: 10.1101/gad.6.7.1305. [DOI] [PubMed] [Google Scholar]
- 28.Jansen G, Buhring F, Hollenberg CP, Ramezani Rad M. Mutations in the SAM domain of STE50 differentially influence the MAPK-mediated pathways for mating, filamentous growth and osmotolerance in Saccharomyces cerevisiae. Mol Genet Genomics. 2001;265:102. doi: 10.1007/s004380000394. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.