Abstract
Bioorthogonal ligation methods that allow the selective conjugation of fluorophores or biotin to proteins and small molecule probes that contain inert chemical handles are an important component of many chemical proteomic strategies. Here, we present a new catch-and-release enrichment strategy that utilizes a hexylchloride group as a bioorthogonal chemical handle. Proteins and small molecules that contain a hexylchloride tag can be efficiently captured by an immobilized version of the self-labeling protein HaloTag. Furthermore, by using a HaloTag fusion protein that contains a protease cleavage site, captured proteins can be selectively eluted under mild conditions. We demonstrate the utility of the hexylchloride-based catch-and-release strategy by enriching protein kinases that are covalently and non-covalently bound to ATP-binding site-directed probes from mammalian cell lysates. Our catch-and-release system creates new possibilities for profiling enzyme families and for the identification of the cellular targets of bioactive small molecules.
Introduction
Derivatized small molecule probes are valuable reagents for studying biology. Support-bound small molecule ligands have facilitated the enrichment of specific classes of low abundance proteins, such as protein kinases.1–4 Furthermore, immobilized analogs of small molecules that show interesting properties in phenotypic screens are useful for identifying the intracellular targets of bioactive molecules.5–7 Fluorophore- and biotin-modified derivatives of small molecule probes that covalently modify the active sites of their binding partners have served as effective tools for profiling the activities of various enzyme families. These activity-based protein profiling (ABPP) probes have allowed the discovery of enzymatic activities that are misregulated in various disease models and for the selectivity profiling of inhibitors in physiologically relevant contexts.8
The development of a number of robust bioorthogonal reactions has revolutionized the design and use of small molecule probes. These reactions allow the use of small molecule probes that contain an inert chemical handle that minimally perturbs their solubility, cell permeability, and binding properties. Examples of bioorthogonal reactions that have been successfully used for conjugation include Diels-Alder cycloadditions,9–10 nucleophile additions to carbonyl groups,11 Michael additions,12 thiol-ene reactions,13 Staudinger ligations,14 and alkyne-azide cycloaddition reactions.15 Bioorthogonal reactions, in particular cycloaddition reactions utilizing alkyne and azide tags, have found widespread use in chemical proteomic studies. For example, azide and alkyne tags have been incorporated into ABPP probes and used to examine large families of enzymes.16–18
Many chemical proteomic studies rely on selectively enriching covalently or non-covalently bound proteins for subsequent identification and quantification. For small molecule probes that contain a bioorthogonal chemical handle, this is usually accomplished through selective conjugation to biotin, followed by the enrichment of probe-bound proteins with an immobilized protein (avidin or streptavidin) that recognizes biotin. While this two-step enrichment procedure has been successfully used in a number of proteomic applications, there are several drawbacks to its implementation. The bioorthogonal reactions used to conjugate biotin are not always quantitative and in some cases can lead to irreversible protein aggregation and precipitation from solution.19–20 In addition, endogenously biotinylated proteins and proteins that bind non-specifically to the affinity matrix can lead to an increase in the complexity of the sample being analyzed.21 Furthermore, the harsh elution conditions required to elute captured proteins do not allow differentiation of specifically versus non-specifically bound proteins. While a number of biotin analogs that contain releasable linkers have been developed to overcome this limitation,22 the use of these reagents adds an additional non-quantitative handling step to proteomic analyses. Therefore, new bioorthogonal tags that circumvent the use of biotin-streptavidin are needed.
Here, we present a new catch-and-release strategy that utilizes a hexylchloride group as a bioorthogonal chemical handle. The hexylchloride tag is unique because it allows chemoselective and direct conjugation to a self-labeling protein through a covalent bond. By incorporating a hexylchloride tag into a small molecule probe of interest, probe-bound proteins can be enriched with an immobilized version of HaloTag, which is an engineered form of Rhodococcus dehalogenase that undergoes a self-labeling reaction with alkylchlorides (Supplementary Figure 1).23 Furthermore, by using a HaloTag fusion protein that contains a protease cleavage site, captured proteins can be selectively released under mild conditions. To demonstrate the overall utility of this strategy, we show that our hexylchloride/HaloTag catch-and-release system can be used to enrich proteins that are either covalently or non-covalently bound to kinase-directed probes.
RESULTS AND DISCUSSION
Design of a hexylchloride-based catch-and-release system
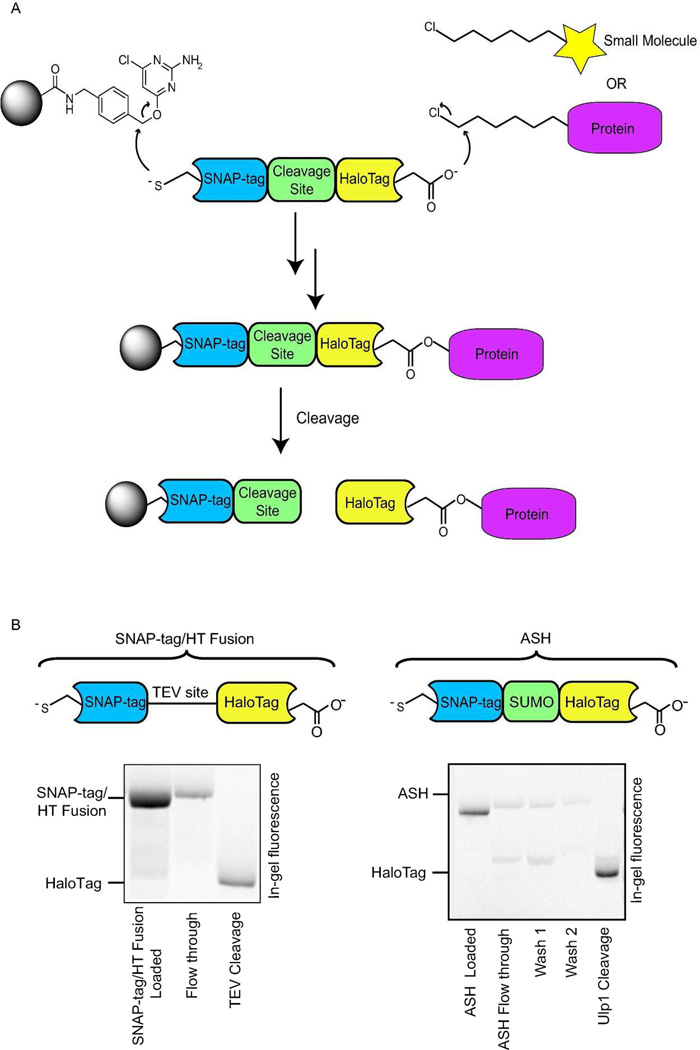
Our strategy for designing a hexylchloride-based catch-and-release system relies on the selective and rapid reaction between alkylchloride-labeled molecules and an immobilized version of the self-labeling protein HaloTag. In order to exploit this bioorthogonal reaction for proteomic studies, HaloTag must be able to be immobilized on a solid support without loss of catalytic activity. Furthermore, a method for the selective release of captured proteins is required. Towards this end, we envisioned generating a fusion protein that contains HaloTag linked through a protease cleavage site to a domain that allows immobilization to a solid support (Figure 1a). The self-labeling protein SNAP-tag (also referred to as AGT), which is a mutant of O6–alkylguanine-DNA alkyltransferase that undergoes an efficient self-labeling reaction with benzylguanine derivatives, was chosen as the immobilization domain.24–26 SNAP-tag is an attractive immobilization option because it has already been demonstrated that fusion proteins containing this self-labeling domain can be displayed from surfaces derivatized with benzylguanine analogues. Furthermore, it has been shown that both enzymes maintain their catalytic activity in the context of a SNAP-tag/HaloTag fusion protein.27 We next investigated the use of TEV protease, which is used in the purification of TAP-tagged proteins,28 to selectively release immobilized HaloTag. To test the efficiency of this protease in our catch-and-release system, a fluorescently-labeled SNAP-tag/HaloTag fusion protein that contains a TEV cleavage site was generated and tested (Figure 1b). As expected, the fluorescently-labeled SNAP-tag/HaloTag fusion protein was efficiently captured (>90%) by an agarose resin displaying the benzylguanine derivative chloropyrimidine (CLP) (Figure 1b). After immobilization of the fusion protein and washing of the resin, TEV protease’s efficiency in releasing the HaloTag portion of the SNAP-tag/HaloTag fusion protein was determined. Disappointingly, the ability of this protease to cleave the TEV site of this immobilized fusion protein was modest. Even at a 1:2 ratio of TEV protease to immobilized HaloTag protein, a low cleavage efficiency (~20%) was observed (Figure 1b). Therefore, the TEV cleavage site of the fusion protein was replaced with a SUMO (small ubiquitin-like modifier) tag, which when fused to proteins has been shown to increase their solubility and expression levels.29 Furthermore, the SUMO Protease Ulp1 is able to cleave SUMO-tagged fusion proteins directly after the C-terminal glycine residue in SUMO with a high catalytic efficiency. This is most likely because Ulp1 recognizes the tertiary structure of SUMO, rather than a short peptide motif like the one recognized by TEV protease. A fluorescently-labeled SNAP-tag(AGT)/SUMO/HaloTag (ASH) fusion protein was generated to test the immobilization and release of this construct. Like the fusion protein containing a TEV cleavage site, >90% of fluorescently-labeled ASH was captured by the resin. Gratifyingly, the SUMO Protease Ulp1 was able to rapidly and efficiently release the HaloTag portion of the immobilized fusion construct. A 1:80 (w/w) ratio of Ulp1:immobilized ASH allowed >85% of fluorescently-labeled HaloTag to be released from the resin (Figure 1b).
Figure 1.
The hexylchloride-based catch-and-release strategy. a) Overall strategy for the catch-and-release of hexylchloride-labeled small molecules probes and proteins. A protein that contains SNAP-tag fused to HaloTag through a selectively cleavable site serves as a linker between a solid support and captured small molecule probes or proteins. The selective reaction between SNAP-tag and a chloropyrimidine (CLP)-derivatized resin allows immobilization of the bi-functional fusion protein. The support-bound HaloTag enzyme is then able to capture hexylchloride-labeled probes or proteins. Captured proteins are selectively released with a protease that cleaves the linker between SNAP-tag and HaloTag. b) Comparison of the release efficiencies for SNAP-tag/HaloTag fusions that contain either a TEV or SUMO protease cleavage site. A fluorescently-labeled SNAP-tag/HaloTag fusion protein is captured with a CLP-derivatized resin and the fluorophore-modified HaloTag protein is released with a protease. Quantitation of the fluorescence intensities of the flow through and elution fractions allows the efficiency of the capture and release steps to be determined.
To demonstrate the stringency of the washes that can be performed with the immobilized ASH constructs, this fusion protein was immobilized on chloropyrimidine resin and then subjected to various buffers, detergents, and salts. Following these washes, HaloTag was cleaved from the resin with Ulp1 to determine the overall cleavage efficiency. The ASH catch-and-release system was found to be compatible with wash buffers containing 0.1% Tween, 0.1% NP-40, 0.5 mM EDTA, 1 M NaCl, and at pH 5–9 (Supplementary Table 1). Subjecting immobilized ASH to high concentrations of detergents (> 1%), 8M urea, 6M guanidine, or SDS (> 0.1%) resulted in a dramatic decrease in the cleavage efficiency of Ulp1. This is most likely due to the partial unfolding of SUMO under these conditions.
Catch-and-Release of Hexylchloride-Labeled Protein in Cell Lysate
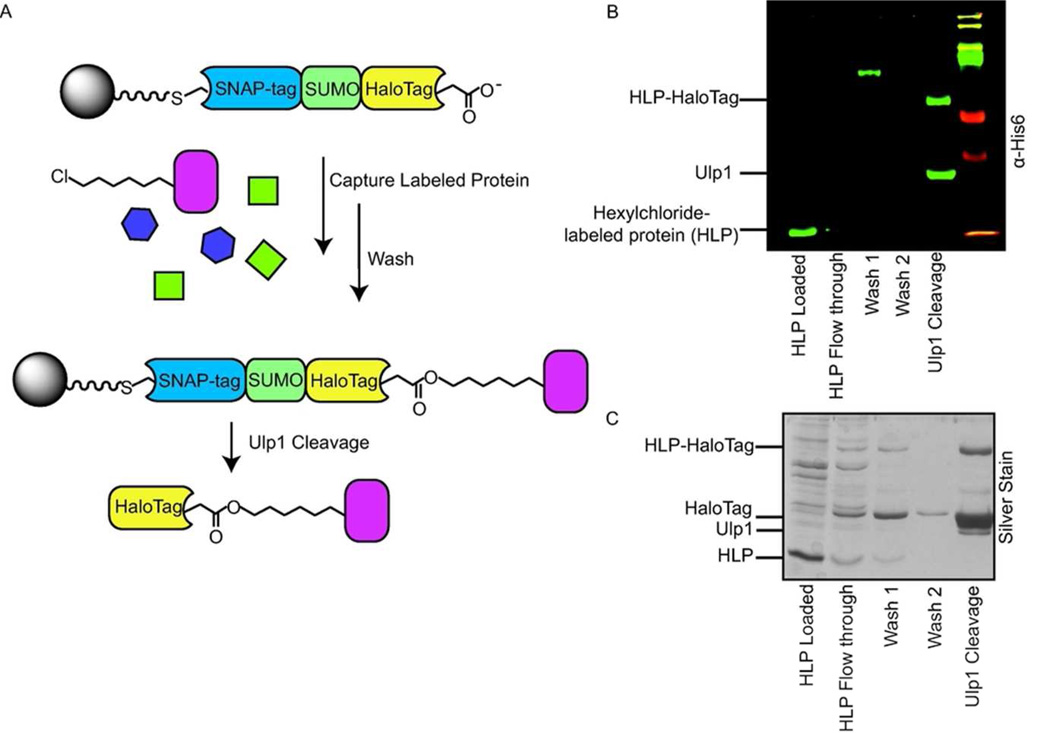
Encouraged by the high cleavage efficiency of HaloTag from resin by Ulp1, we investigated the ability of the fusion protein ASH to capture proteins labeled with a hexylchloride tag. To test this, His6-SNAP-tag was covalently labeled with the hexylchloride derivative 2 (Supplementary Figure 2), resulting in a protein containing a single hexylchloride moiety. This hexylchloride-labeled protein (HLP) was then added to mammalian cell lysate and incubated with resin displaying ASH. After a series of washes, HaloTag and the HLP/HaloTag conjugate were cleaved from the resin using Ulp1 (Figure 2a). Western blot analysis showed that >90% of the hexylchloride-labeled protein was captured by pre-immobilized HaloTag (Figure 2b). Gratifyingly, >90% of the captured HLP was released from the resin as an HLP/HaloTag conjugate following incubation with Ulp1. Only three proteins (Ulp1, unmodified HaloTag, and the HLP/HaloTag conjugate) could be detected in a silver stain analysis of the elution fraction, demonstrating the selectivity of the Ulp1 release strategy (Figure 2c). After optimizing the catch-and-release strategy with a pre-immobilized ASH protein, we next explored whether it would be possible to first capture hexylchloride-labeled proteins with soluble ASH in a cell lysate and then to subsequently capture this covalent complex with beads displaying a SNAP-tag substrate. Gratifyingly, this strategy provided a comparable catch-and-release efficiency to the pre-immobilization strategy (Supplementary Figure 3).
Figure 2.
Selective catch-and-release of a hexylchloride-labeled protein in HeLa lysate. a) Schematic for the selective catch-and-release of hexylchloride-labeled protein (HLPs). ASH is immobilized on resin and then incubated with mammalian lysate supplemented with a protein, HLP, which contains a single hexylchloride tag (shown in pink). HaloTag selectively captures the HLP and the SUMO protease Ulp1 releases HaloTag (and any proteins that HaloTag has captured) from the beads. b) Western Blot analysis (anti-His6) of the catch-and-release experiment described in (a). c) Silver Stain analysis of the catch-and-release experiment described in (a).
While performing our catch-and-release experiments in cell lysate, we noticed that a small percentage of HaloTag was cleaved from the resin during the wash steps. We felt that this was likely due to the presence of endogenous SUMO proteases. To prevent the premature cleavage of HaloTag from the resin by native mammalian SUMO proteases, various protease inhibitors and inhibitor cocktails were employed. Unfortunately, none of the protease inhibitors tested was able to prevent premature cleavage of the ASH protein (data not shown). Therefore, we explored the use of an orthogonal SUMO/SUMO protease pair.30 Mutation of two amino acid residues in the SUMO domain prevents recognition by native SUMO proteases. This SUMO variant, called SUMO*, can be cleaved by an engineered form of Ulp1, Ulp1*. Like Ulp1, Ulp1* cleaves the C-terminal glycine residue of SUMO*. Catch-and-release experiments in mammalian lysate were repeated with a fluorescently-labeled variant of ASH, ASH*, that contains SUMO*. As expected, endogenous SUMO proteases present in mammalian lysate do not cleave immobilized ASH*. However, Ulp1*’s efficiency in releasing HaloTag from resin (> 85%) is similar to that of Ulp1’s ability to cleave ASH (Supplementary Figure 4). Therefore, the Ulp1*/ASH* pair was used in all subsequent experiments in mammalian cell lysate.
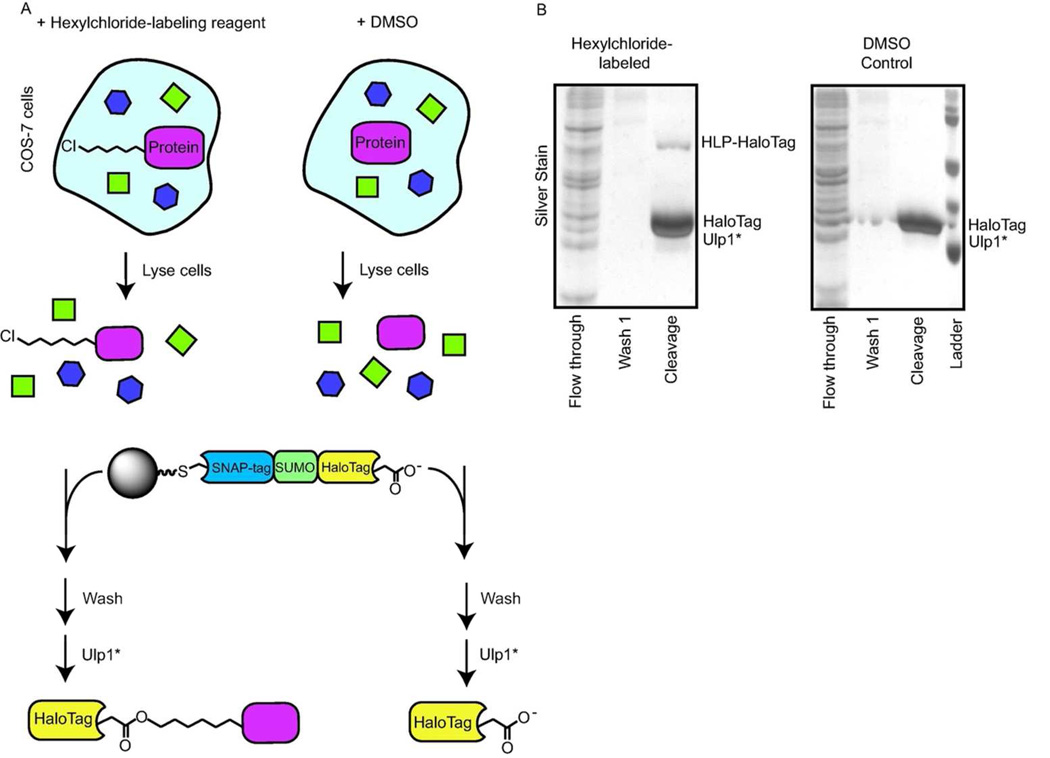
We next investigated whether this system could be used to enrich a protein, HLP, that had been labeled with a hexylchloride tag in live cells (Figure 3). COS-7 cells were transfected with a protein, His6-SNAP-tag, which can be labeled with a single hexylchloride tag in cells. Transfected cells were then incubated with the hexylchloride-labeling reagent 2. In parallel, transfected control cells were incubated with TMRstar, a rhodamine derivative that labels the model protein but does not contain a hexylchloride tag, instead of hexylchloride-labeling reagent 2. Both sets of cells were then washed, lysed, and subjected to our optimized catch-and-release conditions with the ASH*/Ulp1* pair (Figure 3). Similar to the lysate pull-downs described above, only three proteins (HaloTag, Ulp1*, and the HLP/HaloTag conjugate) were detected in the elution from the catch-and-release experiments performed with cells that contain HLP (Figure 3b). As expected, no band corresponding to the molecular weight of HLP/HaloTag was observed in the elution from the control experiment. Therefore, the ASH*/Ulp1* pair is capable of selectively enriching hexylchloride-labeled proteins in the presence of a full complement of cellular components.
Figure 3.
Catch-and-release of an HLP from mammalian cells. a) Schematic for the catch-and-release of an HLP from mammalian cells. COS-7 cells were transfected with a model protein capable of being singly-labeled with a hexylchloride tag. Transfected cells were either incubated with a hexylchloride-tagging reagent or a control compound. After washing, cells were lysed and the lysate was incubated with immobilized ASH*. Captured proteins were released by incubating the solid support with Ulp1*. b) Silver stain analysis of the experiment described in (a).
Enrichment of Hexylchloride-Labeled Protein Kinases
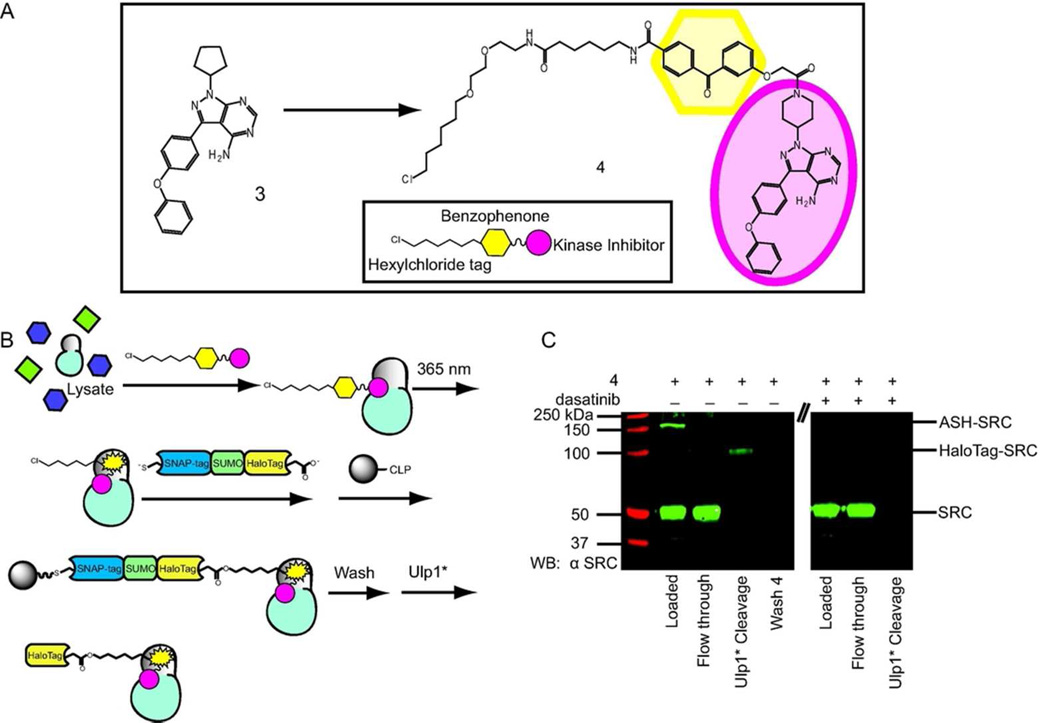
A chemical proteomic application for our catch-and-release system was next investigated. We are particularly interested in chemical probes that are able to covalently label the ATP-binding sites of protein kinases. Affinity-based probes of this type can be used for inhibitor selectivity profiling and for the identification of aberrant kinase activities. Towards exploring this application, a kinase-directed photo-crosslinker based on the ATP-competitive inhibitor 3 was generated (Figure 4). Inhibitor 3 is a potent inhibitor of the tyrosine kinase BTK and the SRC-family of protein kinases.31 A crystal structure of the catalytic domain of BTK bound to 3 shows that this inhibitor occupies the ATP-binding site of this kinase, with the pyrazolopyrimidine scaffold forming the same hydrophobic and hydrogen-bonding interactions as the adenine ring of ATP (Supplementary Figure 5). Interestingly, the ATP-binding site of BTK adopts an inactive conformation when bound to 3 in which a salt bridge between a conserved glutamate and the catalytic lysine is disrupted by the movement of helix αC in the N-terminal lobe.32,33 In order to convert inhibitor 3 into a kinase-directed probe that is able to covalently label kinase targets, it was derivatized with a benzophenone photo-crosslinker at a position that would not be expected to disrupt its interaction with the ATP-binding sites of protein kinases. A hexylchloride tag was also incorporated into the benzophenone moiety of probe 4 to allow enrichment of labeled kinase targets with our catch-and release system (Figure 4). Probe 4 was tested for its inhibitory activity in an in vitro assay with the tyrosine kinase SRC to confirm that modification of the kinase inhibitor scaffold does not adversely affect its ability to interact with protein kinases. Gratifyingly, probe 4 potently inhibits the catalytic activity of SRC (IC50 = 49 nM).
Figure 4.
Catch-and-release of proteins labeled by the kinase-directed photo-crosslinker 4. a) Chemical structure of Probe 4. Kinase inhibitor 3 was converted into a photo-crosslinker by attaching a photo-activatable benzophenone moiety and a hexylchloride tag. b) Pulldown schematic for the catch-and-release of proteins photo-labeled by probe 4. Cell lysate was incubated with probe 4, followed by irradiation with UV light. The lysate was then successively incubated with ASH* and CLP-resin. After extensive washing, captured proteins were released from the resin by incubating with Ulp1*. A parallel experiment was performed in the presence of an active site competitor (dasatinib). c) Western blot analysis (anti-SRC) of the catch-and-release of the tyrosine kinase SRC. Photo-labeled SRC is captured by ASH* and a SRC/HaloTag complex is released from the beads with Ulp1*.
The chemo-selectivity of the HaloTag labeling reaction allows for two different labeling and enrichment strategies with probe 4. In the first strategy (pre-conjugation), probe 4 is conjugated to the active site of HaloTag in the ASH* fusion protein. The ASH*-4 conjugate is then incubated with cell lysate, and subsequently irradiated with UV light to covalently label any bound targets. The SNAP-tag portion of ASH* can be used to enrich any targets that are covalently linked to the ASH*-4 conjugate. In the second strategy (post-conjugation), unconjugated probe 4 is incubated with cell lysate, and then irradiated with UV light. Labeled proteins can be captured using ASH*. With both strategies it is possible to determine the labeling efficiency of probe 4 for a specific protein target by subjecting samples to SDS-PAGE and immunoblotting with a target-specific antibody. Tethering a covalently modified target to ASH* results in a significant gel shift compared to the unlabeled protein and the percentage of crosslinked target protein can be determined with a gel-shift assay (Supplementary Figure 6).
The crosslinking efficiency of both strategies was determined for probe 4 with the kinase domain of SRC (SRC KD) using a gel-shift assay. For the pre-conjugation method, the pre-assembled ASH*-4 conjugate was added to mammalian cell lysate (1 mg mL−1) supplemented with SRC KD and then irradiated with UV light for 20 minutes. Immunoblot analysis of the crosslinking reaction shows that 2.6% of SRC KD is labeled by ASH*-4 (Supplementary Figure 6). As expected, no gel-shifted SRC KD is observed when crosslinking is performed in the presence of a competitor (dasatinib, 10 µM), demonstrating that labeling is active site directed. When unconjugated probe 4 was first incubated with cell lysate (1 mg mL−1) supplemented with SRC KD and then irradiated with UV light prior to conjugation to ASH* (post-conjugation method), a higher crosslinking efficiency was observed (10% of SRC KD). Like the pre-conjugation method, no photo-labeled SRC KD was observed in the presence of an active site competitor. Due to the higher crosslinking efficiency of the post-conjugation method, all subsequent experiments were performed with unconjugated probe 4. Next, a catch-and-release experiment was performed with probe 4 in COS-7 cell lysate that had been supplemented with 50 nM SRC (Figure 4b). Lysate containing probe 4 was irradiated with UV light, and then incubated with ASH*. Western blot analysis showed that 11% of total SRC was labeled by probe 4 (Figure 4c). The irradiated lysate was then added to beads displaying CLP and the affinity matrix was then washed extensively prior to incubation with Ulp1* (Figure 4b). Analysis of the flow-through of the capture step shows that nearly all of the ASH*-conjugated SRC was captured by the resin. Furthermore, almost all of the captured SRC was released from the beads as a SRC/HaloTag conjugate upon incubation with Ulp1*. Thus, the ASH* construct can be used to efficiently catch and release proteins that have been covalently labeled with a hexylchloride-derivatized probe.
Enrichment of Non-covalently Bound Kinases
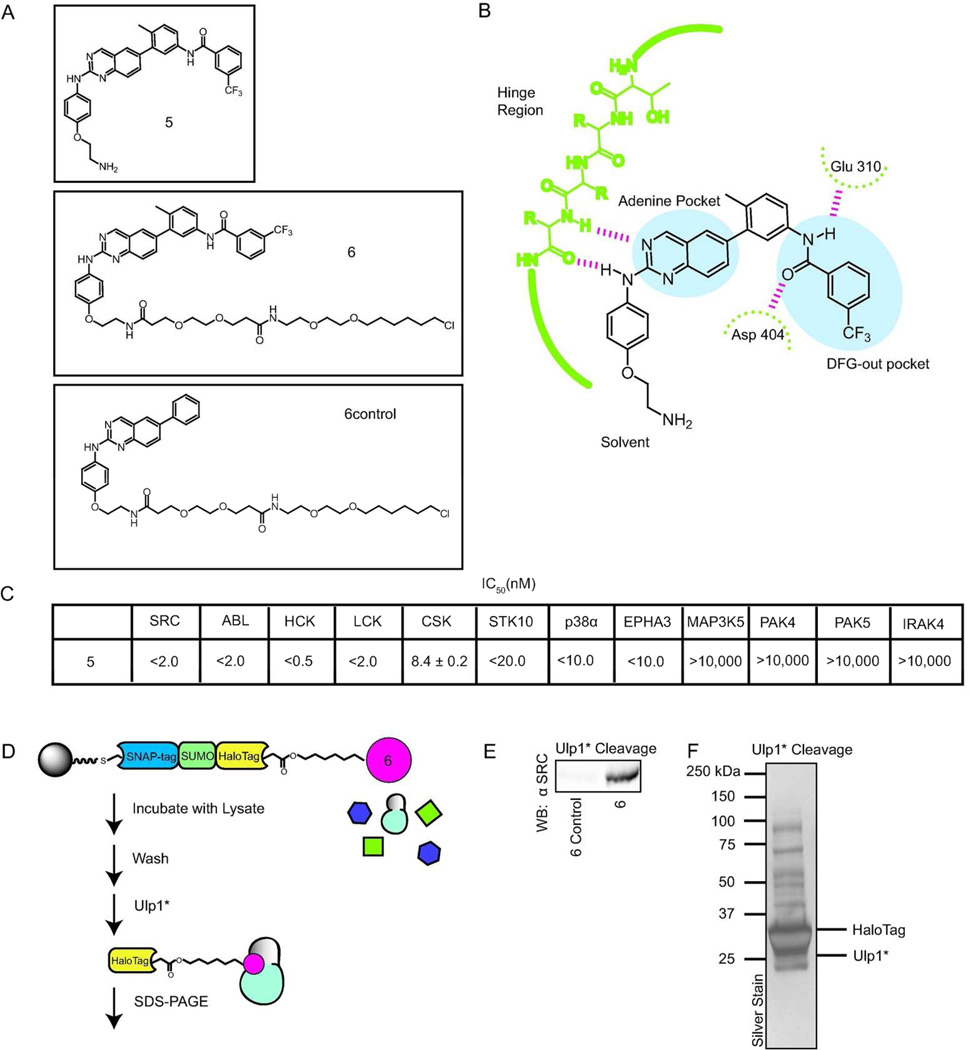
Beyond the catch and release of covalently labeled proteins, we were interested in exploring whether the ASH* system can be used to enrich proteins that are non-covalently bound to a hexylchloride-tagged probe. To test this, catch-and-release experiments were performed with hexylchloride-derivatized ligands that stabilize a specific inactive form, called the DFG-out conformation, of the ATP-binding sites of protein kinases (Figure 5a). The DFG-out inactive conformation is characterized by an almost 180° rotation of the conserved Asp-Phe-Gly (DFG) motif, relative to the active conformation, which exposes a hydrophobic pocket that is accessible to small molecule inhibitors. A number of inhibitors, commonly referred to as type II inhibitors, have been identified that stabilize kinases in the DFG-out conformation (Figure 5b).34–37 Type II ligands contain three conserved features: (1) A hetero-aromatic moiety that makes many of the same contacts as the adenine ring of ATP, (2) a hydrophobic group that occupies the pocket created by the movement of the DFG motif, and (3) a hydrogen bond donor/acceptor pair that interacts with the backbone of the DFG motif and a conserved glutamic acid in helix αC. In this study, we were interested in identifying the kinase targets of inhibitor 5, which is an analogue of a series of potent type II inhibitors that target the SRC-family kinase LCK.38 5 potently inhibits the catalytic activities of protein kinases that have been structurally characterized in the DFG-out conformation (Figure 5c).
Figure 5.
Catch-and-release of protein kinases non-covalently bound to a probe that stabilizes an inactive ATP-binding site conformation. a) Structures of inhibitors 5, 6, and 6control. b) A schematic of the binding interactions that the type II inhibitor 5 makes with the ATP-binding sites of protein kinases (residue numbering for the tyrosine kinase SRC is shown). c) Inhibition data for 5 against a panel of protein kinases. d) The overall strategy for the catch-and-release of lysate proteins bound to probes 6 and 6control. HeLa lysate was added to pre-immobilized ASH* displaying 6 or 6control. After extensive washing, bound proteins were released with Ulp1*. Eluted proteins were subjected to SDS-PAGE and then identified my tandem mass spectrometry (MS/MS). e) Western blot analysis (anti-SRC) of a catch-and-release experiment performed with HeLa lysate that was enriched with the tyrosine kinase SRC. f) Silver stain analysis of the catch-and-release experiment performed with 6. Proteins that were selectively enriched with probe 6 are shown in Table 1.
Many type II inhibitors form high affinity interactions with their kinase targets and have slow association and dissociation kinetics. These features have made performing affinity chromatography with type II ligands challenging because it is difficult to elute inhibitor-bound kinases under non-denaturing conditions. For example, we previously developed a set of affinity reagents based on a general type II pyridinyl triazine inhibitor scaffold that are able to efficiently capture kinase targets, but we were unable to elute bound proteins with a soluble competitor.39,40 Support-bound kinases could only be released in the presence of a detergent, which increases the number of non-specifically captured proteins that are eluted, and complicates mass spectrometric analysis. To determine whether this is also true for affinity reagents based on inhibitor 5, we generated a support bound analogue of this ligand. Immobilized 5 was incubated with mammalian lysate supplemented with SRC kinase, washed extensively, and subjected to elution conditions of increasing stringency. Like the previous series of type II affinity reagents that we have characterized, none of the captured kinases could be eluted with a soluble competitor (100 µM of 5). Elution with 0.5% and saturated SDS released a small percentage of SRC from the resin; only boiling the affinity matrix in SDS-loading buffer resulted in the release of a majority of the captured SRC kinase (Supplementary Figure 7). Unfortunately, the latter conditions also led to the elution of a number of other proteins that were non-specifically retained by the affinity matrix. Clearly, conditions that allow selective release of probe-bound protein targets would greatly benefit the proteomic characterization of ligand 5.
Modification of inhibitor 5 with a hexylchloride tag generated probe 6. To aid in the identification of specific kinase targets during proteomic analysis, a control hexylchloride-tagged compound, 6control, was also generated. 6control contains the same core inhibitor scaffold as 6 but lacks the substituents that interact with the hydrophobic pocket created by movement of the DFG motif, which greatly reduces the affinity of most kinases for this inhibitor. With the new hexylchloride-tagged compounds in hand, we optimized our catch-and-release conditions with mammalian lysate that had been supplemented with SRC kinase. 6 and 6control were each immobilized on resin displaying ASH* and then incubated with mammalian lysate. Both resins were washed to remove any non-specifically-bound proteins and then incubated with Ulp1* (Figure 5). Western blot analyses of the elutions from both catch-and-release experiments demonstrate that immobilized 6 is able to enrich SRC kinase, while immobilized 6control cannot (Figure 5e).
Encouraged by these results, we tested the ability of ligand 6 to enrich endogenous kinases from HeLa lysate. Silver stain analysis of the eluted fractions from this pull-down experiment demonstrated that Ulp1* was able to release a number of proteins from ASH*-immobilized 6 (Figure 5f). The proteins eluted from catch-and-release experiments performed with 6 and 6control were separated by SDS-PAGE, trypsinized, and then analyzed by mass spectrometry for identification. In total, there were 13 protein kinases that were enriched by ASH*-immobilized 6 but not by ASH*-immobilized 6control (Table 1). (See Supporting Information for a complete list of proteins selectively enriched by probe 6) The majority of the selectively enriched kinases are members of the Tyrosine Kinase group. Three of the tyrosine kinases, SRC, LCK, and CSK, have been structurally characterized in the DFG-out conformation.38,41–43 In addition to SRC and LCK, three additional SRC-family kinase members, YES1, LYN, and FRK, were also enriched. While these three kinases have not been observed in the DFG-out conformation, the similar sensitivity of SRC-family members to type II inhibitors makes it highly likely that they are able to adopt this inactive form. All other enriched kinases have not been structurally characterized in the DFG-out conformation, although EPHA3 and EPHA7, which are highly homologous to EPHA2, have been observed in this inactive form.44 To verify that newly identified kinases are sensitive to the type II inhibitor displayed from immobilized 6, we performed activity assays with PTK2 and EIF2AK2 in the presence of 5 and 5control (Supplementary Table 3). Consistent with their selective enrichment by ASH*-immobilized 6, 5 is a submicromolar inhibitor of both PTK2 and EIF2AK2. Furthermore, neither kinase is inhibited by the highest concentration of 5control (10,000 nM) tested.
Table 1.
Kinases selectively enriched from HeLa lysate with ASH*-immobilized 6.
| Kinase | # of peptides identified |
Kinase Group |
|---|---|---|
| PTK2 | 3 | Tyrosine |
| EPHA2 | 2 | Tyrosine |
| FRK | 1 | Tyrosine |
| SRC | 8 | Tyrosine |
| YES1 | 2 | Tyrosine |
| LYN | 3 | Tyrosine |
| CSK | 18 | Tyrosine |
| CRKRS | 1 | CMGC |
| CDK3 | 1 | CMGC |
| ULK3 | 1 | Other |
| EIF2AK2 | 1 | Other |
| SMG1 | 1 | Atypical |
| ZAK | 1 | Tyrosine Kinase-like |
In conclusion, we have developed a new catch-and-release strategy that allows the selective enrichment of hexylchloride-labeled probes and proteins. This new enrichment strategy leverages the rapid and selective reaction between the self-labeling protein HaloTag and alkylchlorides. By using a HaloTag fusion protein that contains an immobilization domain (SNAP-tag) and a protease cleavage site, it is possible to selectively release captured proteins with a protease. We demonstrate that our strategy is able to efficiently capture and selectively release proteins that have been labeled with a hexylchloride tag. Furthermore, we show that kinases that are non-covalently bound to a hexylchloride-derivatized probe can be enriched with our catch-and-release system. The ability to selectively elute probe-bound proteins creates numerous opportunities for proteomic analysis, including the identification of the targets of small molecules that show interesting properties in phenotypic screens and the active site profiling of low abundance enzyme families.
METHODS
Fluorescence assay for determination of catch-and-release efficiency
Purified ASH or ASH* (30 µM) was labeled with 1 (45 µM) in 50 mM HEPES buffer, pH=7.5, 100 mM NaCl, and 1 mM DTT for 1 h at RT. Fluorescently-labeled protein was then rotated with CLP resin for 1.5 h at RT. After incubation, the resin was washed and then incubated with Ulp1 or Ulp1* protease. Eluted samples were separated by SDS-PAGE and scanned with a GE Typhoon FLA 9000 fluorescent scanner. The intensities of fluorescently-labeled protein bands were quantified with ImageQuant software.
Catch-and-release of singly-labeled hexylchloride proteins from cell lysate
ASH protein (2 µM) was added to a 200 µL mixture of singly-labeled protein (SNAP-tag) (500 nM), 50 µg mammalian cell lysate, 1X protease inhibitor cocktail (Roche complete), and 1 mM DTT in 50 mM HEPES, pH=7.5. The mixture was incubated at RT for 1 h and then incubated for 1.5 h with 60 µL (50% slurry) CLP resin. The beads were then washed twice with Cleavage Buffer (50 mM Tris, pH=8.0, 150 mM NaCl, 0.2% NP40, and 1 mM DTT) and then incubated with Ulp1 (1:80 mass ratio) in Cleavage Buffer for 2 h at 30 °C. Samples were then subjected to SDS-PAGE and immunoblot analysis (anti-His, abcam) to determine capture and release efficiency. Blots were quantified with Li-cor Odyssey software.
Catch-and-release of proteins labeled in cells by a single hexylchloride tag
HeLa cells were grown in a 12-well plate, transfected with SNAP-tag (New England BioLabs), and cultured for 24 h. Cells were then treated with 2 (10 µM) in 1 mL of serum-free media for 1 h at 37 °C. Control cells were incubated with TMRstar (10 µM) (New England Biolabs). Cells were then washed with media (3X, 10 min) and PBS (2X). Cells were then transferred to a 1.5 mL microcentrifuge tube and sonicated to lyse the cells. The lysate was then incubated with resin displaying preimmobilized ASH*. After a series of washes (50 mM Tris, pH=8.0, 300 mM NaCl, 0.1% Tween), immobilized proteins were eluted from the resin with Ulp1* (1:20 mass ratio Ulp1*:ASH*). Samples were separated on 10% SDS-PAGE gels and either silver stained (Invitrogen SilverXpress Staining Kit) or immunoblotted (anti-His, abcam). Blots were quantified with Li-cor Odyssey software.
Catch-and-release of hexylchloride-labeled protein kinases
After crosslinking using the post-conjugation technique (Supporting Information), 4×50 µL wells were combined and DTT was added to a final concentration of 2 mM. The sample was then incubated with CLP resin for 90 min. After a series of washes (50 mM Tris, pH=8.0, 300 mM NaCl, 0.1% Tween), immobilized proteins were eluted from the resin with Ulp1* (1:20 mass ratio Ulp1*:ASH*). Samples were separated on 10% SDS-PAGE gels and immunoblotted (SRC (36D10) antibody, Cell Signaling). Blots were quantified with Li-cor Odyssey software.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Y. A. Goo (University of Washington, Seattle) for performing mass spectrometry experiments. This work was supported by the National Institutes of Health (R01GM086858) and the Alfred P. Sloan and Camille and Henry Dreyfus Foundations.
Footnotes
SUPPORTING INFORMATION
Supplementary figures and tables, along with detailed material and methods are included. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Bantscheff M, Eberhard D, Abraham Y, Bastuck S, Boesche M, Hobson S, Mathieson T, Perrin J, Raida M, Rau C, Reader V, Sweetman G, Bauer A, Bouwmeester T, Hopf C, Kruse U, Neubauer G, Ramsden N, Rick J, Kuster B, Drewes G. Quantitative chemical proteomics reveals mechanisms of action of clinical ABL kinase inhibitors. Nat. Biotechnol. 2007;25:1035–1044. doi: 10.1038/nbt1328. [DOI] [PubMed] [Google Scholar]
- 2.Daub H, Olsen JV, Bairlein M, Gnad F, Oppermann FS, Korner R, Greff Z, Keri G, Stemmann O, Mann M. Kinase-selective enrichment enables quantitative phosphoproteomics of the kinome across the cell cycle. Mol. Cell. 2008;31:438–448. doi: 10.1016/j.molcel.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 3.Dulla K, Daub H, Hornberger R, Nigg EA, Korner R. Quantitative site-specific phosphorylation dynamics of human protein kinases during mitotic progression. Mol. Cell Proteomics. 2010;9:1167–1181. doi: 10.1074/mcp.M900335-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oppermann FS, Gnad F, Olsen JV, Hornberger R, Greff Z, Keri G, Mann M, Daub H. Large-scale proteomics analysis of the human kinome. Mol. Cell Proteomics. 2009;8(7):1751–1764. doi: 10.1074/mcp.M800588-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taunton J, Hassig CA, Schreiber SL. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- 6.Ong SE, Schenone M, Margolin AA, Li X, Do K, Doud MK, Mani DR, Kuai L, Wang X, Wood JL, Tolliday NJ, Koehler AN, Marcaurelle LA, Golub TR, Gould RJ, Schreiber SL, Carr SA. Identifying the proteins to which small-molecule probes and drugs bind in cells. Proc. Natl. Acad. Sci. USA. 2009;106:4617–4622. doi: 10.1073/pnas.0900191106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rix U, Superti-Furga G. Target profiling of small molecules by chemical proteomics. Nat. Chem. Biol. 2009;5:616–624. doi: 10.1038/nchembio.216. [DOI] [PubMed] [Google Scholar]
- 8.Evans MJ, Cravatt BF. Mechanism-Based Profiling of Enzyme Families. Chem. Rev. 2006;106:3279–3301. doi: 10.1021/cr050288g. [DOI] [PubMed] [Google Scholar]
- 9.Taylor MT, Blackman ML, Dmitrenko O, Fox JM. Design and synthesis of highly reactive dienophiles for the tetrazine-trans-cyclooctene ligation. J. Am. Chem. Soc. 2011;133:9646–9649. doi: 10.1021/ja201844c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blackman ML, Royzen M, Fox JM. Tetrazine ligation: fast bioconjugation based on inverse-electron-demand Diels-Alder reactivity. J. Am. Chem. Soc. 2008;130:13518–13519. doi: 10.1021/ja8053805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mahal LK, Yarema KJ, Bertozzi CR. Engineering chemical reactivity on cell surfaces through oligosaccharide biosynthesis. Science. 1997;276:1125–1128. doi: 10.1126/science.276.5315.1125. [DOI] [PubMed] [Google Scholar]
- 12.Sampathkumar S-G, Li AV, Jones MB, Sun Z, Yarema KJ. Metabolic installation of thiols into sialic acid modulates adhesion and stem cell biology. Nat. Chem. Biol. 2006;2:149–152. doi: 10.1038/nchembio770. [DOI] [PubMed] [Google Scholar]
- 13.Dondoni A. The Emergence of Thiol–Ene Coupling as a Click Process for Materials and Bioorganic Chemistry. Angew. Chem. Int. Edit. 2008;47:8995–8997. doi: 10.1002/anie.200802516. [DOI] [PubMed] [Google Scholar]
- 14.Saxon E, Bertozzi CR. Cell Surface Engineering by a Modified Staudinger Reaction. Science. 2000;287:2007–2010. doi: 10.1126/science.287.5460.2007. [DOI] [PubMed] [Google Scholar]
- 15.Best MD. Click chemistry and bioorthogonal reactions: unprecedented selectivity in the labeling of biological molecules. Biochemistry. 2009;48:6571–6584. doi: 10.1021/bi9007726. [DOI] [PubMed] [Google Scholar]
- 16.Cravatt BF, Wright AT, Kozarich JW. Activity-based protein profiling: from enzyme chemistry to proteomic chemistry. Annu. Rev. Biochem. 2008;77:383–414. doi: 10.1146/annurev.biochem.75.101304.124125. [DOI] [PubMed] [Google Scholar]
- 17.Salisbury CM, Cravatt BF. Activity-based probes for proteomic profiling of histone deacetylase complexes. Proc. Natl. Acad. Sci. USA. 2007;104:1171–1176. doi: 10.1073/pnas.0608659104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saghatelian A, Jessani N, Joseph A, Humphrey M, Cravatt BF. Activity-based probes for the proteomic profiling of metalloproteases. Proc. Natl. Acad. Sci. USA. 2004;101:10000–10005. doi: 10.1073/pnas.0402784101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Besanceney-Webler C, Jiang H, Zheng T, Feng L, Soriano Del Amo D, Wang W, Klivansky LM, Marlow FL, Liu Y, Wu P. Increasing the efficacy of bioorthogonal click reactions for bioconjugation: a comparative study. Angew. Chem. Int. Edit. 2011;50:8051–8056. doi: 10.1002/anie.201101817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Speers AE, Adam GC, Cravatt BF. Activity-Based Protein Profiling in Vivo Using a CoppeRI)-Catalyzed Azide-Alkyne [3 + 2] Cycloaddition. J. Am. Chem. Soc. 2003;125:4686–4687. doi: 10.1021/ja034490h. [DOI] [PubMed] [Google Scholar]
- 21.Praul CA, Brubaker KD, Leach RM, Gay CV. Detection of Endogenous Biotin-Containing Proteins in Bone and Cartilage Cells with Streptavidin Systems. Biochem. Bioph. Res. Co. 1998;247:312–314. doi: 10.1006/bbrc.1998.8757. [DOI] [PubMed] [Google Scholar]
- 22.Yang Y-Y, Grammel M, Raghavan AS, Charron G, Hang HC. Comparative Analysis of Cleavable Azobenzene-Based Affinity Tags for Bioorthogonal Chemical Proteomics. Chem. Biol. 2010;17:1212–1222. doi: 10.1016/j.chembiol.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Los GV, Encell LP, McDougall MG, Hartzell DD, Karassina N, Zimprich C, Wood MG, Learish R, Ohana RF, Urh M, Simpson D, Mendez J, Zimmerman K, Otto P, Vidugiris G, Zhu J, Darzins A, Klaubert DH, Bulleit RF, Wood KV. HaloTag: A Novel Protein Labeling Technology for Cell Imaging and Protein Analysis. ACS Chem. Biol. 2008;3:373–382. doi: 10.1021/cb800025k. [DOI] [PubMed] [Google Scholar]
- 24.Gronemeyer T, Chidley C, Juillerat A, Heinis C, Johnsson K. Directed evolution of O6- alkylguanine-DNA alkyltransferase for applications in protein labeling. Protein Eng. Des. Sel. 2006;19:309–316. doi: 10.1093/protein/gzl014. [DOI] [PubMed] [Google Scholar]
- 25.Keppler A, Kindermann M, Gendreizig S, Pick H, Vogel H, Johnsson K. Labeling of fusion proteins of O6-alkylguanine-DNA alkyltransferase with small molecules in vivo and in vitro. Methods. 2004;32:437–444. doi: 10.1016/j.ymeth.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 26.Keppler A, Gendreizig S, Gronemeyer T, Pick H, Vogel H, Johnsson K. A general method for the covalent labeling of fusion proteins with small molecules in vivo. Nat. Biotechnol. 2003;21:86–89. doi: 10.1038/nbt765. [DOI] [PubMed] [Google Scholar]
- 27.Chidley C, Mosiewicz K, Johnsson K. A Designed Protein for the Specific and Covalent Heteroconjugation of Biomolecules. Bioconjugate Chem. 2008;19:1753–1756. doi: 10.1021/bc800268j. [DOI] [PubMed] [Google Scholar]
- 28.Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 29.Butt TR, Edavettal SC, Hall JP, Mattern MR. SUMO fusion technology for difficult-to-express proteins. Protein Expres. Purif. 2005;43:1–9. doi: 10.1016/j.pep.2005.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu L, Spurrier J, Butt TR, Strickler JE. Enhanced protein expression in the baculovirus/insect cell system using engineered SUMO fusions. Protein Expres. Purif. 2008;62:21–28. doi: 10.1016/j.pep.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marcotte DJ, Liu Y-T, Arduini RM, Hession CA, Miatkowski K, Wildes CP, Cullen PF, Hong V, Hopkins BT, Mertsching E, Jenkins TJ, Romanowski MJ, Baker DP, Silvian LF. Structures of human Bruton's tyrosine kinase in active and inactive conformations suggest a mechanism of activation for TEC family kinases. Protein Sci. 2010;19:429–439. doi: 10.1002/pro.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sicheri F, Moarefi I, Kuriyan J. Crystal structure of the Src family tyrosine kinase Hck. Nature. 1997;385:602–609. doi: 10.1038/385602a0. [DOI] [PubMed] [Google Scholar]
- 33.Xu W, Harrison SC, Eck MJ. Three-dimensional structure of the tyrosine kinase c-Src. Nature. 1997;385:595–602. doi: 10.1038/385595a0. [DOI] [PubMed] [Google Scholar]
- 34.Pargellis C, Tong L, Churchill L, Cirillo PF, Gilmore T, Graham AG, Grob PM, Hickey ER, Moss N, Pav S, Regan J. Inhibition of p38 MAP kinase by utilizing a novel allosteric binding site. Nat. Struct. Biol. 2002;9:268–272. doi: 10.1038/nsb770. [DOI] [PubMed] [Google Scholar]
- 35.Liu Y, Gray NS. Rational design of inhibitors that bind to inactive kinase conformations. Nat. Chem. Biol. 2006;2:358–364. doi: 10.1038/nchembio799. [DOI] [PubMed] [Google Scholar]
- 36.Wan PT, Garnett MJ, Roe SM, Lee S, Niculescu-Duvaz D, Good VM, Jones CM, Marshall CJ, Springer CJ, Barford D, Marais R. Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell. 2004;116:855–867. doi: 10.1016/s0092-8674(04)00215-6. [DOI] [PubMed] [Google Scholar]
- 37.Schindler T, Bornmann W, Pellicena P, Miller WT, Clarkson B, Kuriyan J. Structural mechanism for STI-571 inhibition of abelson tyrosine kinase. Science. 2000;289:1938–1942. doi: 10.1126/science.289.5486.1938. [DOI] [PubMed] [Google Scholar]
- 38.DiMauro EF, Newcomb J, Nunes JJ, Bemis JE, Boucher C, Buchanan JL, Buckner WH, Cee VJ, Chai L, Deak HL, Epstein LF, Faust T, Gallant P, Geuns-Meyer SD, Gore A, Gu Y, Henkle B, Hodous BL, Hsieh F, Huang X, Kim JL, Lee JH, Martin MW, Masse CE, McGowan DC, Metz D, Mohn D, Morgenstern KA, Oliveira-dos-Santos A, Patel VF, Powers D, Rose PE, Schneider S, Tomlinson SA, Tudor YY, Turci SM, Welcher AA, White RD, Zhao H, Zhu L, Zhu X. Discovery of aminoquinazolines as potent, orally bioavailable inhibitors of Lck: synthesis, SAR, and in vivo anti-inflammatory activity. J. Med. Chem. 2006;49:5671–5686. doi: 10.1021/jm0605482. [DOI] [PubMed] [Google Scholar]
- 39.Ranjitkar P, Brock AM, Maly DJ. Affinity Reagents that Target a Specific Inactive Form of Protein Kinases. Chem. Biol. 2010;17:195–206. doi: 10.1016/j.chembiol.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ranjitkar P, Maly DJ. Affinity purification of protein kinases that adopt a specific inactive conformation. Methods Mol. Biol. 2012;928:143–151. doi: 10.1007/978-1-62703-008-3_11. [DOI] [PubMed] [Google Scholar]
- 41.Seeliger MA, Ranjitkar P, Kasap C, Shan Y, Shaw DE, Shah NP, Kuriyan J, Maly DJ. Equally Potent Inhibition of c-Src and Abl by Compounds that Recognize Inactive Kinase Conformations. Cancer Res. 2009;69:2384–2392. doi: 10.1158/0008-5472.CAN-08-3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simard JR, Kluter S, Grutter C, Getlik M, Rabiller M, Rode HB, Rauh D. A new screening assay for allosteric inhibitors of cSrc. Nat. Chem. Biol. 2009;5:394–396. doi: 10.1038/nchembio.162. [DOI] [PubMed] [Google Scholar]
- 43.Dar AC, Lopez MS, Shokat KM. Small molecule recognition of c-Src via the Imatinib-binding conformation. Chem. Biol. 2008;15:1015–1022. doi: 10.1016/j.chembiol.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Choi Y, Syeda F, Walker JR, Finerty PJ, Cuerrier D, Wojciechowski A, Liu Q, Dhe-Paganon S, Gray NS. Discovery and structural analysis of Eph receptor tyrosine kinase inhibitors. Bioorg. Med. Chem. Lett. 2009;19:4467–4470. doi: 10.1016/j.bmcl.2009.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.