Abstract
A definitive diagnostic test for multiple sclerosis (MS) does not exist; instead physicians use a combination of medical history, magnetic resonance imaging, and cerebrospinal fluid analysis (CSF). Significant effort has been employed to identify biomarkers from CSF to facilitate MS diagnosis; however none of the proposed biomarkers have been successful to date. Urine is a proven source of metabolite biomarkers and has the potential to be a rapid, non-invasive, inexpensive, and efficient diagnostic tool for various human diseases. Nevertheless, urinary metabolites have not been extensively explored as a source of biomarkers for MS. Instead, we demonstrate that urinary metabolites have significant promise for monitoring disease-progression, and response to treatment in MS patients. NMR analysis of urine permitted the identification of metabolites that differentiate experimental autoimmune encephalomyelitis (EAE)-mice (prototypic disease model for MS) from healthy and MS drug-treated EAE mice.
Keywords: Multiple sclerosis, NMR metabolomics, biomarkers, disease diagnosis
Multiple sclerosis (MS) is a demyelinating disease of the central nervous system (CNS) characterized by selective loss of myelin sheath encapsulating the neuronal axons.1 Treatments of MS are more effective during the early course of the disease when symptoms are mild.2 Thus, the early diagnosis of MS is critical in order to quickly initiate treatments that slow the progression of the disease and improve the quality of a patient’s life.3 Unfortunately, MS is a very challenging disease to properly diagnose,4 where a misdiagnosis is a common occurrence that inevitably leads to a delay in the correct treatment.5 Significant effort has been employed to identify biomarkers from cerebrospinal fluid (CSF) to facilitate MS diagnosis, but this endeavor has proven to be challenging and has not been successful (see supplementary discussion).6 The analysis of urine samples for MS biomarkers has only been minimally investigated, but holds significant promise.7 The pathology of MS is associated with widespread demyelination, glial scarring and an inflammatory response that will inevitably result in a systematic change in the excreted metabolome. Thus, urinary metabolites may be a valuable approach for diagnosing MS and evaluating the in vivo efficacy of MS drug candidates.8
MS is believed to be an immune-mediated (autoimmune) disease requiring the mediation of T cells and/or B cells. Thus, experimental autoimmune encephalomyelitis (EAE) can be induced by immunizing mice with myelin antigens or their peptide fragments emulsified in complete Freund’s adjuvant (CFA).9 Because of similarities with respect to genetic susceptibility, environmental triggers, disease pathology, and clinical signs and disease course, the EAE model has also been used extensively in MS drug discovery research and as a disease model for MS.10 In fact, the majority of drugs being tested now in the phase II and phase III clinical trials were first examined in EAE.11 This includes fingolimod (Gilenya™, Novartis), a first-in-class orally administered drug approved for MS therapy.12, 13 Fingolimod is more efficacious than other MS treatments.14, 15 Importantly, the mechanism of action for fingolimod has been shown to be similar in both humans and the EAE-mouse model.16 Specifically, fingolimod suppresses the disease-inducing abilities of myelin-reactive T cells by multiple mechanisms.17, 18 Thus, the analysis of changes in urinary metabolites in mice resulting from fingolimod treatment is likely to translate to similar human studies.
Our studies involve the use of 6-to 8-week-old female C57Bl/6 mice which develop a chronic progressive form of paralysis when immunized with the myelin oligodendrocyte glycoprotein (MOG) 35–55 (EAE mice).19 To monitor a response to drug treatment, a group of EAE mice were treated with fingolimod. Establishment of EAE and the efficacy of fingolimod treatment were confirmed by clinical scoring for paralysis and histological evaluation of brains and spinal cords (Fig. 1a and Supplementary Table 1). The experimental design consisted of seven treatment groups (n=13) that includes healthy, saline, CFA, EAE, saline plus fingolimod (saline-treated), CFA plus fingolimod (CFA-treated) and EAE plus fingolimod (EAE-treated). Fingolimod (1 mg/kg body weight) completely prevented clinical EAE, except for one mouse with EAE score of 2 (mild EAE). Expectedly, MOG-specific T cells in EAE mice treated with fingolimod expanded comparably with those of untreated mice, but they did not induce the disease (Supplementary Fig. 1 and Fig. 1a).
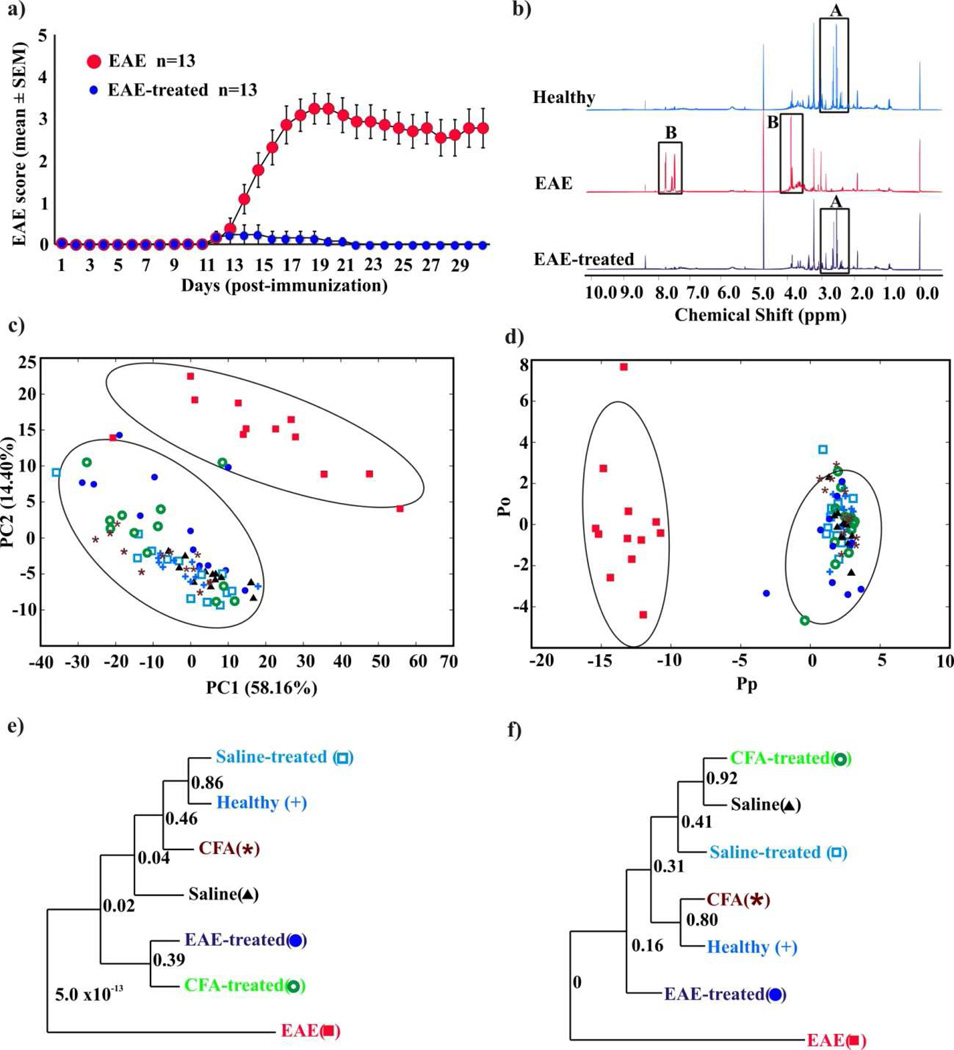
Figure 1. NMR metabolomics of EAE mice.
(a) Clinical scores of EAE and EAE-treated mice. Groups of mice were immunized with MOG 35–55 in CFA, and the animals were treated with or without fingolimod daily (1mg/kg body weight) from day 7 postimmunization through day 30. The animals were monitored for EAE signs and the disease-severity was scored. (b) Examples of 1D 1H NMR spectra of urine samples collected from EAE, healthy, and EAE-treated mice. Major spectral differences are highlighted. (c) 2D PCA and (d) 2D OPLS-DA scores plot generated from the 1D 1H NMR spectra acquired for the urine samples from the seven treatment groups namely: healthy ( ), saline (
), saline ( ), CFA (
), CFA ( ), EAE (
), EAE ( ), saline-treated (
), saline-treated ( ), CFA-treated (
), CFA-treated ( ), and EAE-treated mice (
), and EAE-treated mice ( ). The OPLS-DA used one predictive component and one orthogonal component to yield a R2X of 0.690, R2Y of 0.938 and Q2 of 0.841. The CV-ANOVA validation of the OPLS-DA class distinctions yielded a p value of 1.55e−31. The ellipses correspond to the 95% confidence limits from a normal distribution for each cluster. (e) and (f) Metabolomics tree diagrams determined from the PCA and OPLS-DA scores plots, respectively. The coloring scheme for each group in the tree diagram correlates with the data point colors in the scores plot. The p values for each node are indicated on the tree diagram.
). The OPLS-DA used one predictive component and one orthogonal component to yield a R2X of 0.690, R2Y of 0.938 and Q2 of 0.841. The CV-ANOVA validation of the OPLS-DA class distinctions yielded a p value of 1.55e−31. The ellipses correspond to the 95% confidence limits from a normal distribution for each cluster. (e) and (f) Metabolomics tree diagrams determined from the PCA and OPLS-DA scores plots, respectively. The coloring scheme for each group in the tree diagram correlates with the data point colors in the scores plot. The p values for each node are indicated on the tree diagram.
Individual urine samples were collected daily from all the mice belonging to the seven treatment groups, where sample collection began on day 7 and continued until day 30 postimmunization. Ninety one-dimensional (1D) 1H NMR spectra were acquired for the urine samples collected on day 17, when the EAE severity reached a peak (Fig. 1a). Representative NMR spectra are shown in figure 1b, where a visual comparison shows a clear difference between EAE and healthy mice; and importantly, a similarity in the pattern between healthy and EAE-treated mice. A set of NMR peaks (labeled A) were significantly decreased in the spectra for EAE mice relative to healthy mice. Conversely, two sets of NMR peaks (labeled B) were increased in the EAE mice relative to healthy mice. These spectral differences are potential biomarkers for MS. The NMR spectra for the other control groups were essentially identical to the NMR spectra for the healthy mice and EAE-treated mice (Supplementary Fig. 2) suggesting that evidence of inflammatory changes in the CNS can be captured by analyzing urinary metabolites.
Principal component analysis (PCA) of the 1D 1H NMR spectra further defines a difference in the urine profiles between healthy and EAE-mice (Fig. 1c-f). The two-dimensional (2D) PCA scores plot generated from 86 of the 1D 1H NMR spectra (4 spectra were rejected during the analysis) exhibited two distinct clusters. The rejected spectra fell significantly outside the 95% confidence limit for the EAE-mice cluster and for the PCA model (not shown), and were randomly distributed throughout the scores plot. The ellipses that correspond to the 95% confidence interval from a normal distribution for each cluster clearly define two statistically distinct classes (Fig. 1c). Correspondingly, this result indicates that the urinary metabolites for EAE mice are distinct from the healthy mice. Importantly, the EAE-treated mice cluster together with the healthy mice. This shift in the urinary metabolite profile provides a proof-of concept for two key events: (i) fingolimod was effective in suppressing the disease severity in EAE mice and (ii) the therapeutic efficacy of fingolimod was expected to change the metabolite pattern of EAE group towards the healthy group. This is consistent with the clinical scoring and histological evaluation observed for the EAE-treated mice (Fig. 1a and Supplementary Table 1).
Orthogonal partial least-squares discriminant analysis (OPLS-DA) of the 1D 1H NMR metabolomics data was performed to further substantiate the observed difference in urinary metabolites between EAE, healthy and EAE-treated mice (Fig. 1d). Strikingly, the OPLS-DA 2D scores plot also contains two statistically distinct clusters. Importantly, a CV-ANOVA test validated the OPLS-DA model with a resulting p value of 1.6x10−31.20 This provides statistical verification that the urinary metabolites from EAE mice are distinct from both healthy and EAE-treated mice; and the corresponding similarity between healthy and EAE-treated mice. Further validation of the statistical significance of the clustering pattern in the PCA and OPLS-DA 2D scores plot was determined by generating metabolomics tree diagrams (Figs. 1e and 1f) using our PCAtoTree program, where we recently replaced bootstrap values with a Mahalanobis metric (p values).21, 22 Again, the high p values (most > 0.1) indicate that only the nodes that separate the EAE mice from the healthy and the EAE-treated mice (p value <5.0x10−13) are statistically significant. PCA and OPLS-DA scores plot generated from the NMR analysis of urine samples collected on days 23 and 30 are presented in the supplementary material (Supplementary Figs. 3–4) and yielded similar results. It is also important to note that the healthy and EAE-treated mice also clustered together and were indistinguishable from the four other negative controls, further establishing that cluster separation is a result of EAE.
The group distinction based on the observed changes in urinary metabolites is disease related as opposed to other environmental factors. The C57Bl/6 mice are inbred animals, were obtained simultaneously, were randomly distributed between each group, and were maintained under identical conditions besides the described group-specific treatments. The only other variable between the groups was the hydration and nutrient supplement (DietGel) provided to EAE and EAE-treated mice with a clinical score of 3 or higher. This is based on accepted protocols for the ethical treatment of animals. Importantly, both EAE and EAE –treated mice received the DietGel supplement, but separation was still observed in the PCA and OPLS-DA scores plot. Critically, the EAE mice treated with fingolimod still clustered with the healthy mice and other control mice, which did not receive the DietGel supplement (Fig. 1c and d).
A diet control study comparing healthy mice with or without access to a DietGel supplement resulted in urinary metabolomes distinct from EAE mice (Supplementary Fig. 5). Mice received the DietGel supplements for 5 to 7 days, similar to the EAE-mice in the EAE induction and treatment study shown in figures 1 and 2 (days 12 to 17). Clearly, the DietGel supplement is not the source of the observed changes in the EAE mice urine. A corresponding 2D PCA scores plot comparing urine samples collected over three days from healthy mice with and without the DietGel supplement resulted in a larger within group variation for mice receiving the DietGel supplement (Supplementary Fig. 5b). In fact, the distribution appears bimodal, potentially distinguishing between individual animals and their preference for the DietGel supplement over food-pellets. Correspondingly, the mice that only received the Teklad food pellets fall within the ellipse defining the 95% confidence interval for the mice receiving the DietGel supplement. Presumably, the pellet-only fed mice overlap with the mice that prefer the Teklad food pellets over the DietGel supplement. Variations were also observed between the healthy mice samples collected from day 5 to day 7 and from those of day 17 (Supplementary Fig. 5a). Nevertheless, the 2D PCA scores plot (Supplementary Fig. 5c) once again yields a clear separation between healthy and EAE mice. Furthermore, the scores plot indicates that the healthy mice with or without access to the DietGel supplement are more similar to each other than to the EAE mice. This clearly indicates that metabolite changes due to EAE are more pronounced than perturbations from dietary variations. This bodes well for being able to detect similar urinary biomarkers for MS despite an expected wider diversity in the diets of human patients.
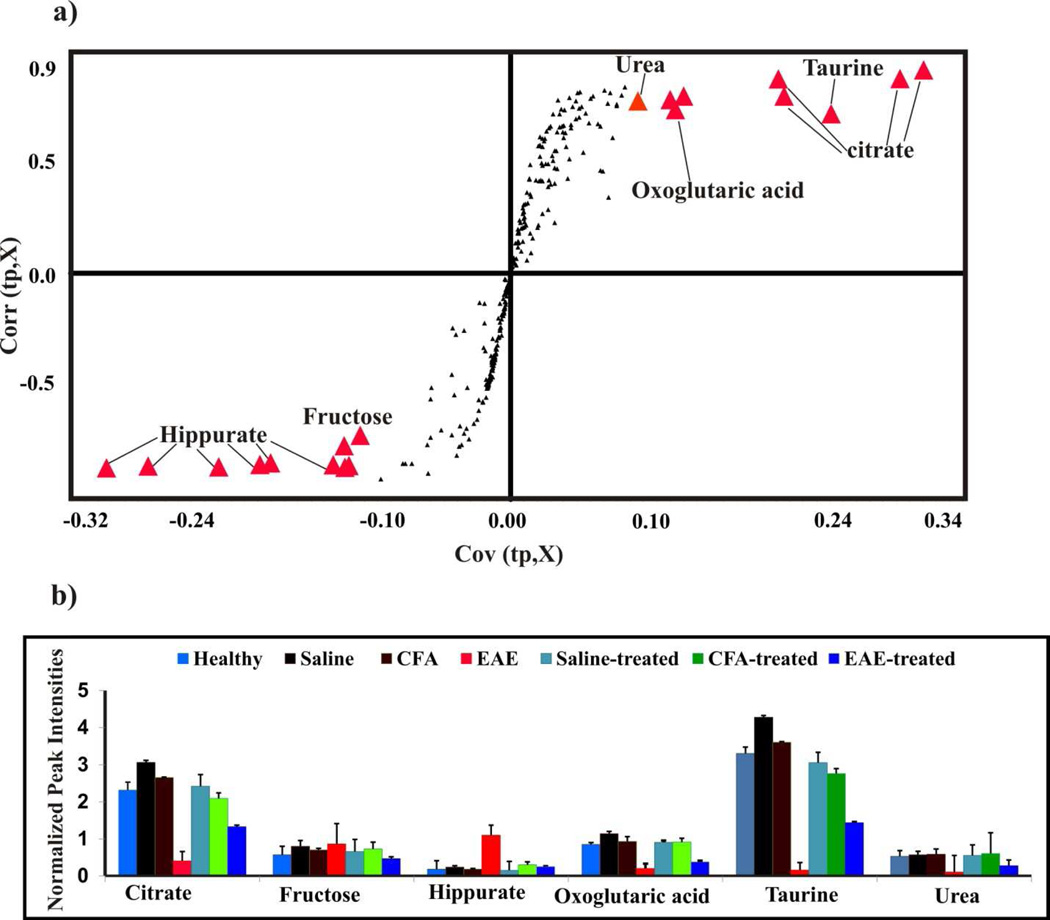
Figure 2. Potential metabolite biomarkers for EAE.
(a) S-plot generated from the OPLS-DA model presented in Figure 1d. Each NMR bin with a covariance of greater than 0.10 or less than −0.10 were identified as major contributors to class separation. These bins are highlighted as red triangles and are labeled with the assigned metabolite. (b) The normalized average peak intensities or bin integrals identified in the S-plot from (a) as major contributors to class separation are plotted as a function of treatment class and metabolite assignment. The standard deviation in the normalized average peak intensities is indicated on the bar graphs.
The class distinction observed in the PCA and OPLS-DA scores plot suggest the presence of a set of metabolites that could be used as potential biomarkers to differentiate EAE mice from healthy mice. The S-plot generated from the OPLS-DA model identifies the major spectral features that contribute to the class separation observed in the scores plot, where the corresponding metabolite assignments are labeled in figure 2a. To further validate the statistical relevance of these metabolites to differentiate between healthy and EAE mice, normalized average peak intensities were calculated for each bin and then compared between the seven treatment groups using a standard Student’s t-test (Fig. 2b). These normalized average peak intensities are proportional to metabolite concentrations. Statistically significant changes were observed in the relative metabolite concentrations between healthy and EAE mice. Specifically, hippurate (p value 1.3x10−5) and fructose (p value 2.1x10−3) were up-regulated in EAE mice compared to healthy mice. Conversely, citrate (p value 1.3x10−6), oxoglutaric acid (p value 7.0x10−7), taurine (p value 4.9x10−7) and urea (p value 5.8x10−8), were down-regulated in EAE mice compared to healthy mice. In addition, the effect of fingolimod treatment was also assessed by a similar comparison between the EAE mice with and without fingolimod treatment. Hippurate (p value 2.7x10−5) and fructose (p value 2.2x10−4) were up-regulated in EAE-mice compared to EAE-treated mice. Conversely, citrate (p value 1.2x10−5), oxoglutaric acid (p value 2.4x10−3), taurine (p value 2.5x10−6) and urea (p value 4.2x10−4) were down-regulated in EAE mice compared to EAE-treated mice (Fig. 2b).
2D 1H-13C HSQC NMR spectra were also acquired to provide a further, in-depth analysis of EAE-induced metabolite changes in urine samples (Fig. 3a). The improved resolution and correlated 1H and 13C chemical shifts increases the accuracy and the number of metabolite assignments. But, the analysis of the low naturally abundant 13C-labeled metabolites (only 1.1%) is a significant challenge that required large urine volumes (~ 500 µl) to obtain acceptable spectral signal-to-noise. Unfortunately, obtaining this much urine from a completely paralyzed EAE mice is not practically feasible. Thus, it was necessary to pool equal volume of urine samples from a group of 4 to 5 mice of identical clinical scores within a cage. Pooling samples does have an advantage since it will minimize within-group variations and maximize between-group differences by effectively averaging NMR peak intensities (metabolite concentrations) within a group.
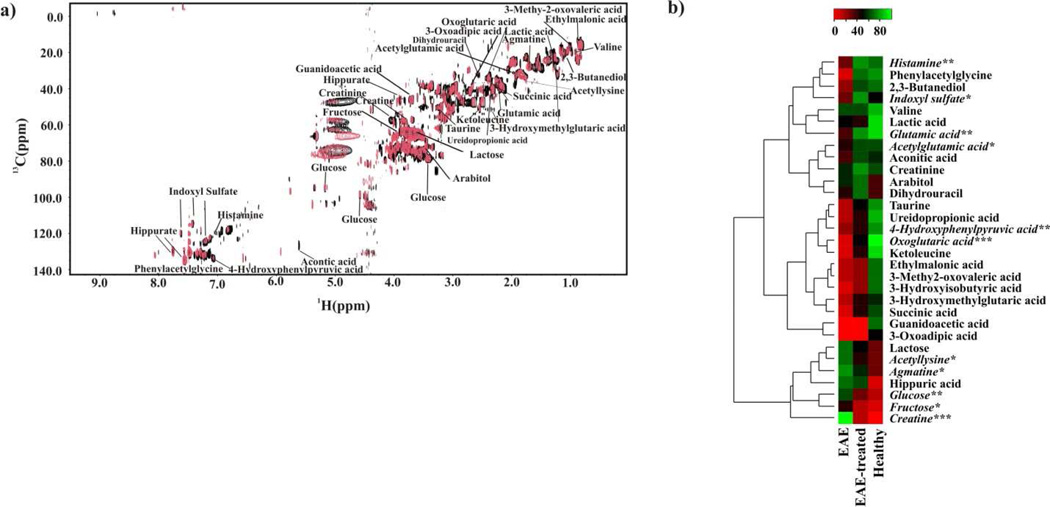
Figure 3. Detailed analysis of EAE-dependent urine metabolites.
(a) An overlay of 2D 1H-13C HSQC NMR spectra acquired from healthy (black) and EAE (red) mice urine samples. Metabolite assignments are indicated on the spectrum. (b) Heat map generated from the normalized relative intensity of the corresponding metabolite peak obtained from the 2D 1H-13C HSQC NMR spectrum. The dendrogram represents a hierarchal clustering of the metabolites according to relative concentration changes between healthy, EAE and EAE-treated mice. Metabolites significantly altered in the urine from EAE, but not in EAE-treated mice are in italics (p <0.05 *, p <0.01**, p <0.001***).
Urine samples were collected from triplicate sets of EAE, EAE-treated and healthy mice, and then used to acquire the corresponding 2D 1H-13C HSQC NMR spectra. Normalized differences in average peak intensities relative to healthy mice are summarized in the heat-map depicted in figure 3b. Over 32 metabolites were identified with different concentrations between EAE and healthy mice, which also contained the metabolites identified from the OPLS-DA S-plot analysis. Interestingly, some of the identified metabolites were previously implicated in the pathogenesis of MS and EAE (histamine, glutamate)23, 24 or identified as markers of metabolic diseases with associated neurological problems (3-hydroxyisobutyric acid, 3-ureidopropionate, guanidinoacetate).25–27 Also, one of the identified metabolites, indoxyl sulfate, is likely to originate from the gut microflora.28 This is consistent with the previous observation that EAE-induction and progression is influenced by gut microorganisms as opposed to attenuation of EAE in germ-free mice.29 Other potential metabolites identified in our NMR analysis of urine samples have also been associated with EAE, MS, and other neurological diseases (see supplementary discussion).
MS is accompanied by various pathological features like inflammation, demyelination, and axonal damage. This inherent complexity of MS makes it challenging to identify a single biomarker to monitor the progression and treatment of the disease. Alternatively, our NMR analysis of urine samples demonstrates that a diverse set of metabolites can be used to differentiate between healthy and EAE mice. These metabolite changes can also be used to monitor the recovery of EAE mice upon treatment with fingolimod. Also, the 1D 1H NMR analysis of urine samples takes only about 10 minutes per sample and completely lacks any of the risks or side effects associated with the analysis of CSF. Thus, the NMR analysis of urine holds the promise of being an easy, fast, and safe diagnostic tool for MS. Additionally, the NMR analysis of urine may be a valuable approach for evaluating the in vivo efficacy of drug-leads; and for designing patient-specific treatments.
Methods
Peptide synthesis
MOG 35–55 (MEVGWYRSPFSRVVHLYRNGK) was synthesized on 9-fluorenylmethyloxy-carbonyl chemistry (Neopeptide, Cambridge, MA) to a purity of more than 90% as verified by HPLC and mass spectroscopy. The peptide was dissolved in 1x phosphate buffered saline, and stored at −20°C until used. The MOG 35–55 peptide was used to induce EAE in mice.
Mice
Ninety 6 to 8-week-old female C57Bl/6 (H-2b) mice were obtained from the Jackson Laboratory (Bar Harbor, ME). C57Bl/6 (H-2b) mice are inbred animals and individual variations in the gut flora are not expected. Furthermore, the animals are maintained in the same environment and received the same food. All the mice were located in the same room within the University of Nebraska-Lincoln (UNL) Life Sciences Annex, managed and maintained by the Institutional Animal Care Program in accordance with the animal protocol guidelines of UNL. The mice were provided with the Teklad global 16% protein rodent diet (Harlan Laboratories, Indianapolis, IN). The mice were randomly classified into twenty-one individually ventilated cages. Fifteen of the cages had four animals while the other six cages had five animals. The mice were acclimatized for three days before the start of the experiment.
During the experiment all the mice were given ad libtum access to food and water. Correspondingly, the amount of food and water consumed by an individual mouse was not regulated or quantified. Also, the mice were not fasted during urine collection or at any other time during the experiment. In this manner, the metabolomics experiment captures the natural variation in nutrient consummation per animal and provides a more realistic metabolomic background. Additionally, if a cage containing either an EAE group or EAE group under fingolimod treatment contained a mouse exhibiting a clinical score of 3 or above, the entire cage was supplemented with a nutrient fortified hydration gel (moisture, 73.4%; DietGel® 76A, ClearH2O, Portland, ME). The DietGel combines hydration and standard nutrients (76A maintenance diet formulation)30 in a palatable form for compromised rodents. Importantly, all the mice in the cage still have access to the Teklad global 16% protein rodent diet in addition to the nutrient fortified hydration gel. The EAE and EAE-treated mice received supplemental DietGel on day 12. The EAE mice received the supplemental DietGel for the remainder of the experiment; whereas, the EAE–treated mice stopped receiving the supplemental DietGel from day 15. All of the EAE mice treated with fingolimod had adequately recovered by day 15.
EAE induction and treatment
The experimental design consisted of a healthy group (n=12) and six treatment groups (n=13). These include saline, complete Freund’s adjuvant (CFA), EAE, saline plus fingolimod (saline-treated), CFA plus fingolimod (CFA-treated) and EAE plus fingolimod (EAE-treated). Each group of mice was divided into three cages, respectively containing 4, 4 and 5 (or 4 for healthy) animals. The cages for each experimental group were selected randomly. To induce EAE, peptide emulsions were prepared by mixing MOG 35–55 in CFA containing Mycobacterium tuberculosis H37RA extract (Difco Laboratories, Detroit, MI) to a final concentration of 5 mg/ml. Each animal received 200 µg of peptide-emulsion subcutaneously in the inguinal and sternal regions. In addition, pertussis toxin (List Biological Laboratories, Campbell, CA) was administered (200 ng/mouse) intraperitoneally on day 0 and day 2 post-immunization.31–33 Seven days post-immunization, fingolimod dissolved in 1x sterile normal saline (working dilution of 0.2 mg/ml), was administered intraperitoneally to the animals corresponding to treated groups as indicated above at 1 mg/kg body weight daily until day 30.
Urine collection and clinical scoring
Urine samples were collected both prior to, and after disease induction. Urine collections occurred three times daily (10–11 AM; 2–3 PM and 10–11 PM) from each animal by expressing the bladder. The samples collected from each batch of animals were pooled on a daily basis and preserved at −80°C until further analysis. In addition, the samples collected from individual animals on days 17, 23 and 30 post-immunization were also preserved as separate aliquots. The immunized mice were monitored for clinical signs of EAE and scored as described previously:31, 34 0, no signs of disease; 1, limp tail or hind limb weakness; 2, limp tail and hind limb weakness; 3, partial paralysis of hind limbs; 4, complete paralysis of hind limbs and 5, moribund or dead.
NMR sample preparation
The samples for the 1D 1H NMR experiments were prepared by adding 600 µl of a 50 mM phosphate buffer in 99.8% D2O (Isotec, St. Louis, MO) at pH 7.2 (uncorrected) to 25 µL of urine collected from each mouse. The six treatment groups contained 13 mice per group and the healthy group contained 12 mice for a total of 90 NMR urine samples. The samples for the 2D 1H-13C HSQC experiments were prepared by adding 100 µL of a 50 mM phosphate buffer in 99.8% D2O at pH 7.2 (uncorrected) and 500 µL of urine pooled from a group of 4 to 5 healthy or EAE-mice. Each pooled group of mice was assigned the same clinical assessment score and the animals were placed in the same cage. The urine was collected at the same time on a daily basis. A triplicate set of NMR samples were prepared for each group for the 2D 1H-13C HSQC experiments.
NMR data collection and analysis
All NMR experiments were performed with Bruker AVANCE DRX 500 MHz spectrometer equipped with 5 mm triple-resonance cryogenic probe (1H, 13C, 15N) with a Z-axis gradient. A BACS-120 sample changer with Bruker ICON-NMR software was used to automate the NMR data collection. The 1D 1H NMR data was collected at 298K with 32K data points, a spectrum width of 5483 Hz, 128 scans and 16 dummy scans using an excitation sculpting pulse sequence to remove the solvent peak.35 The 2D 1H-13C HSQC NMR spectrum was collected at 298K with 512 scans, 32 dummy scan and a 1.5 s relaxation delay. The spectrum was collected with 2K data points and a spectrum width of 4734 Hz in the direct dimension and 64 data points and a spectrum width of 18864 Hz in the indirect dimension. ACD/1D NMR manager version 12.0 (Advanced Chemistry Development, Inc.) was used to process the 1D 1H NMR spectra. Intelligent binning was used to integrate each region with a bucket size of 0.025 ppm. The buckets were normalized by the intensity of the 3-(trimethylsilyl)propionic acid-2,2,3,3-d4(TMSP) peak. Each NMR spectrum was mean centered and auto-scaled by the standard deviation as described in Zhang et al.36
NMRPipe37 was used to process the 2D 1H-13C HSQC spectra. Peak-picking and peak-matching were accomplished using NMRViewJ Version 8.0.38 Peak-intensities were normalized for each 2D 1H-13C HSQC NMR spectrum by dividing each peak-intensity by the average peak-intensity for a given spectrum. Each NMR peak for each metabolite from the triplicate set of 2D 1H-13C HSQC spectra was further normalized to the maximum peak intensity for the metabolite. The maximum peak-intensity for each metabolite was scaled to 100. Chemical shift references from the Human Metabolomics Database were used to assign each NMR peak to a metabolite.39
Statistical analysis
Principal component analysis (PCA), orthogonal partial least square discriminant analysis (OPLS-DA), and S-plots were generated using SIMCA P+ 12 (UMETRICS). The tree diagram and p values for the dendrograms were generated using a new updated version of our PCAtoTree software previously described by Werth et al.21, 22 The new method is based on the UPGMA tree generation algorithm with multivariate normal modeling of the dataset and use of a Mahalanobis metric (p values) during calculation of tree node distances. A standard Student’s t-test calculated in Excel was used to determine if differences between groups or metabolite concentrations were statistically significant (p value > 0.05).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part from the NIH National Center for Research Resources (P20 RR-17675) and the University of Nebraska Research Council. The research was performed in facilities renovated with support from the National Institutes of Health (grant number RR015468-01).
Footnotes
ASSOCIATED CONTENT
Supporting information
This material is available free of charge via the Internet at http://pubs.acs.org
Author Contributions
J. R. and R. P. designed research; T. G. and M. T. W. performed NMR experiments and performed statistical analysis, C. M., A. G., T. G. and S. T. performed the animal studies; D. S. performed the histology; T. G. and R. P. analyzed NMR and statistical data; and T. G., C. M., A. G., V. K., E. D., J. R. and R. P. analyzed data and wrote the paper.
The authors declare no competing financial interest.
References
- 1.Sospedra M, Martin R. Immunology of multiple sclerosis. Annu Rev Immunol. 2005;23:683–747. doi: 10.1146/annurev.immunol.23.021704.115707. [DOI] [PubMed] [Google Scholar]
- 2.Marrie RA, Cohen JA. Interferons in secondary progressive multiple sclerosis. Informa Healthcare; 2007. pp. 393–407. [Google Scholar]
- 3.Miller JR. The importance of early diagnosis of multiple sclerosis. J Manag Care Pharm. 2004;10:S4–S11. [PubMed] [Google Scholar]
- 4.Rolak LA, Fleming JO. The differential diagnosis of multiple sclerosis. Neurologist. 2007;13:57–72. doi: 10.1097/01.nrl.0000254705.39956.34. [DOI] [PubMed] [Google Scholar]
- 5.Rudick RA, Miller AE. Multiple sclerosis or multiple possibilities: the continuing problem of misdiagnosis. Neurology. 2012;78:1904–1906. doi: 10.1212/WNL.0b013e318259e2e2. [DOI] [PubMed] [Google Scholar]
- 6.Lourenco AST, Baldeiras I, Graos M, Duarte CB. Proteomics-based technologies in the discovery of biomarkers for Multiple Sclerosis in the cerebrospinal fluid. Curr. Mol. Med. 2011;11:326–349. doi: 10.2174/156652411795677981. [DOI] [PubMed] [Google Scholar]
- 7.Dobson R. Urine: An under-studied source of biomarkers in multiple sclerosis? Multiple Sclerosis and Related Disorders. 2012;1:76–80. doi: 10.1016/j.msard.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 8.Gebregiworgis T, Powers R. Application of NMR Metabolomics to Search for Human Disease Biomarkers. Comb Chem High Throughput Screen. 2012;15:595–610. doi: 10.2174/138620712802650522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De RNK, Ben-Nun A. Experimental autoimmune encephalomyelitis induced by various antigens of the central nervous system: overview and relevance to multiple sclerosis. In: Shoenfeld Y, editor. Decade of Autoimmunity. Elsevier; 1999. pp. 169–177. [Google Scholar]
- 10.Glabinski AR, Tani M, Tuohy VK, Ransohoff RM. Murine experimental autoimmune encephalomyelitis: a model of immune-mediated inflammation and multiple sclerosis. Methods Enzymol. 1997;288:182–190. doi: 10.1016/s0076-6879(97)88015-9. [DOI] [PubMed] [Google Scholar]
- 11.Aktas O, Hartung H-P. Oral therapies for multiple sclerosis: fingolimod and cladribine. Hot Top. Neurol. Psychiatry. 2010;9:29–35. doi: 10.1016/S1474-4422(10)70088-0. [DOI] [PubMed] [Google Scholar]
- 12.Brinkmann V, Billich A, Baumruker T, Heining P, Schmouder R, Francis G, Aradhye S, Burtin P. Fingolimod (FTY720): Discovery and development of an oral drug to treat multiple sclerosis. Nat. Rev. Drug Discovery. 2010;9:883–897. doi: 10.1038/nrd3248. [DOI] [PubMed] [Google Scholar]
- 13.Perumal J, Khan O. Emerging disease-modifying therapies in multiple sclerosis. Curr Treat Options Neurol. 2012;14:256–263. doi: 10.1007/s11940-012-0173-x. [DOI] [PubMed] [Google Scholar]
- 14.Doggrell SA. Oral fingolimod for relapsing-remitting multiple sclerosis. Exper Opin. Pharmacother. 2010;11:1777–1781. doi: 10.1517/14656566.2010.481671. [DOI] [PubMed] [Google Scholar]
- 15.Chiba K, Kataoka H, Seki N, Shimano K, Koyama M, Fukunari A, Sugahara K, Sugita T. Fingolimod (FTY720), sphingosine 1-phosphate receptor modulator, shows superior efficacy as compared with interferon-β in mouse experimental autoimmune encephalomyelitis. Int. Immunopharmacol. 2011;11:366–372. doi: 10.1016/j.intimp.2010.10.005. [DOI] [PubMed] [Google Scholar]
- 16.Aktas O, Kuery P, Kieseier B, Hartung H-P. Fingolimod is a potential novel therapy for multiple sclerosis. Nat. Rev. Neurol. 2010;6:373–382. doi: 10.1038/nrneurol.2010.76. [DOI] [PubMed] [Google Scholar]
- 17.Papadopoulos D, Rundle J, Patel R, Marshall I, Stretton J, Eaton R, Richardson JC, Gonzalez MI, Philpott KL, Reynolds R. FTY720 ameliorates MOGinduced experimental autoimmune encephalomyelitis by suppressing both cellular and humoral immune responses. J. Neurosci. Res. 2010;88:346–359. doi: 10.1002/jnr.22196. [DOI] [PubMed] [Google Scholar]
- 18.Chun J, Hartung H-P. Mechanism of Action of Oral Fingolimod (FTY720) in Multiple Sclerosis. Clin. Neuropharmacol. 2010;33:91–101. doi: 10.1097/WNF.0b013e3181cbf825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mendel I, Kerlero de Rosbo N, Ben-Nun A. A myelin oligodendrocyte glycoprotein peptide induces typical chronic experimental autoimmune encephalomyelitis in H-2b mice: fine specificity and T cell receptor V beta expression of encephalitogenic T cells. Eur J Immunol. 1995;25:1951–1959. doi: 10.1002/eji.1830250723. [DOI] [PubMed] [Google Scholar]
- 20.Eriksson L, Trygg J, Wold S. CV-ANOVA for significance testing of PLS and OPLS® models. J. Chemom. 2008;22:594–600. [Google Scholar]
- 21.Werth MT, Halouska S, Shortridge MD, Zhang B, Powers R. Analysis of metabolomic PCA data using tree diagrams. Anal. Biochem. 2010;399:58–63. doi: 10.1016/j.ab.2009.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Worley B, Halouska S, Powers R. Utilities for Quantifying Separation in PCA/PLSA-DA Scores. Anal. Biochem. accepted. 2012 doi: 10.1016/j.ab.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jadidi-Niaragh F, Mirshafiey A. Histamine and histamine receptors in pathogenesis and treatment of multiple sclerosis. Neuropharmacology. 2010;59:180–189. doi: 10.1016/j.neuropharm.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 24.Frigo M, Cogo MG, Fusco ML, Gardinetti M, Frigeni B. Glutamate and multiple sclerosis. Curr. Med. Chem. 2012;19:1295–1299. doi: 10.2174/092986712799462559. [DOI] [PubMed] [Google Scholar]
- 25.Sasaki M, Yamada N, Fukumizu M, Sugai K. Basal ganglia lesions in a patient with 3-hydroxyisobutyric aciduria. Brain Dev. 2006;28:600–603. doi: 10.1016/j.braindev.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 26.Kolker S, Okun JG, Horster F, Assmann B, Ahlemeyer B, Kohlmuller D, Exner-Camps S, Mayatepek E, Krieglstein J, Hoffmann GF. 3-Ureidopropionate contributes to the neuropathology of 3-ureidopropionase deficiency and severe propionic aciduria: a hypothesis. J. Neurosci. Res. 2001;66:666–673. doi: 10.1002/jnr.10012. [DOI] [PubMed] [Google Scholar]
- 27.Gordon N. Guanidinoacetate methyltransferase deficiency (GAMT) Brain Dev. 2010;32:79–81. doi: 10.1016/j.braindev.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 28.Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, Siuzdak G. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc. Natl. Acad. Ssci. U.S.A. 2009;106:3698–3703. doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee YK, Menezes JS, Umesaki Y, Mazmanian SK. Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc. Natl. Acad. Ssci. U.S.A. 2011;108:4615–4622. doi: 10.1073/pnas.1000082107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reeves PG. Components of the AIN-93 diets as improvements in the AIN-76A diet. J. Nutr. 1997;127:838S–841S. doi: 10.1093/jn/127.5.838S. [DOI] [PubMed] [Google Scholar]
- 31.Massilamany C, Steffen D, Reddy J. An epitope from Acanthamoeba castellanii that cross-react with proteolipid protein 139–151-reactive T cells induces autoimmune encephalomyelitis in SJL mice. J Neuroimmunol. 2010;219:17–24. doi: 10.1016/j.jneuroim.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 32.Massilamany C, Thulasingam S, Steffen D, Reddy J. Gender differences in CNS autoimmunity induced by mimicry epitope for PLP 139–151 in SJL mice. J Neuroimmunol. 2011;230:95–104. doi: 10.1016/j.jneuroim.2010.09.011. [DOI] [PubMed] [Google Scholar]
- 33.Massilamany C, Upadhyaya B, Gangaplara A, Kuszynski C, Reddy J. Detection of autoreactive CD4 T cells using major histocompatibility complex class II dextramers. BMC Immunol. 2011;12:40. doi: 10.1186/1471-2172-12-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mendel I, de RNK, Ben-Nun A. A myelin oligodendrocyte glycoprotein peptide induces typical chronic experimental autoimmune encephalomyelitis in H-2b mice: fine specificity and T cell receptor Vβ expression of encephalitogenic T cells. Eur. J. Immunol. 1995;25:1951–1959. doi: 10.1002/eji.1830250723. [DOI] [PubMed] [Google Scholar]
- 35.Nguyen BD, Meng X, Donovan KJ, Shaka AJ. SOGGY: Solventoptimized double gradient spectroscopy for water suppression. A comparison with some existing techniques. J. Magn. Reson. 2007;184:263–274. doi: 10.1016/j.jmr.2006.10.014. [DOI] [PubMed] [Google Scholar]
- 36.Zhang B, Halouska S, Schiaffo CE, Sadykov MR, Somerville GA, Powers R. NMR Analysis of a Stress Response Metabolic Signaling Network. J. Proteome Res. 2011;10:3743–3754. doi: 10.1021/pr200360w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 38.Johnson BA. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol Biol. 2004;278:313–352. doi: 10.1385/1-59259-809-9:313. [DOI] [PubMed] [Google Scholar]
- 39.Wishart DS, Knox C, Guo AC, Eisner R, Young N, Gautam B, Hau DD, Psychogios N, Dong E, Bouatra S, Mandal R, Sinelnikov I, Xia J, Jia L, Cruz JA, Lim E, Sobsey CA, Shrivastava S, Huang P, Liu P, Fang L, Peng J, Fradette R, Cheng D, Tzur D, Clements M, Lewis A, De Souza A, Zuniga A, Dawe M, Xiong Y, Clive D, Greiner R, Nazyrova A, Shaykhutdinov R, Li L, Vogel HJ, Forsythe I. HMDB: a knowledgebase for the human metabolome. Nucleic Acids Res. 2009;37:D603–D610. doi: 10.1093/nar/gkn810. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.