Abstract
ADARs (adenosine deaminases acting on RNA) are RNA editing enzymes that bind double helical RNAs and deaminate select adenosines (A). The product inosine (I) is read during translation as guanosine (G) so such changes can alter codon meaning. ADAR-catalyzed A to I changes occur in coding sequences for several proteins of importance to the nervous system. However, these sites constitute only a very small fraction of known A to I sites in the human transcriptome and the significance of editing at the vast majority sites is unknown at this time. Site-selective inhibitors of RNA editing are needed to advance our understanding of the function of editing at specific sites. Here we show that 2’-O-methyl/locked nucleic acid (LNA) mixmer antisense oligonucleotides are potent and selective inhibitors of RNA editing on two different target RNAs. These reagents are capable of binding with high affinity to RNA editing substrates and remodeling the secondary structure by a strand-invasion mechanism. The potency observed here for 2’-O-methyl/LNA mixmers suggests this backbone structure is superior to the morpholino backbone structure for inhibition of RNA editing. Finally, we demonstrate antisense inhibition of editing of the mRNA for the DNA repair glycosylase NEIL1 in cultured human cells providing a new approach to exploring the link between RNA editing and the cellular response to oxidative DNA damage.
RNA editing reactions modify, insert or delete nucleotides and can change the coding properties of an RNA molecule (1). Deamination at C6 of adenosine (A) in RNA generates inosine (I) at the corresponding nucleotide position. A to I editing is catalyzed by the ADAR family of enzymes (adenosine deaminases acting on RNA). ADARcatalyzed A to I changes occur in coding sequences for several proteins of importance to the nervous system (e.g. glutamate receptors, serotonin receptors, voltage-gated ion channels, etc.) and A to I editing is essential to proper nervous system function (reviewed in (2)). However, editing sites that cause codon changes in neurotransmitter receptors and ion channels constitute only a very small fraction of known A to I sites in the human transcriptome (reviewed in (3)). For instance, thousands of adenosine deamination sites have been found in repeating sequence elements in untranslated regions of human transcripts (4–6). In addition, several A to I sites have been identified that lead to codon changes in proteins with functions outside the nervous system, such as the K/R site in the human DNA repair enzyme NEIL1 (7–9). The biological significance of editing at the vast majority of known A to I sites is unknown at this time. Up to this point the study of the biological function of editing at specific sites has relied heavily on genetically engineered organisms (10–12). However, these experiments are expensive, laborious, time consuming and limited to genetically tractable systems. There is also limited temporal control over editing using these approaches. Inhibitors of RNA editing capable of blocking deamination at specific adenosines are needed. Such molecules will be valuable research tools to study the consequences of editing at specific sites. This is particularly significant now given the recent explosion in the number of known editing sites from high throughput sequencing efforts (7, 8). In addition, site-selective editing inhibitors could have therapeutic potential since hyper-editing at specific sites is correlated with certain disease states (13–15).
Site-specific RNA editing inhibitors need high affinity and selectivity for their target RNAs and must be able to block the activity of tight binding ADAR proteins. They must also be nontoxic, able to permeate the cell nucleus and allow translation of the mature mRNA. The lack of approaches to control editing in a site-specific manner stimulated us to address this problem. In an earlier study, we showed that a synthetic helix-threading peptide that binds near the serotonin 2c receptor editing sites was able to selectively inhibit ADAR2 editing on this RNA in vitro (16). However, the affinity, specificity and cell permeability of molecules of this type must be improved before they can be useful tools for controlling editing (17).
Different antisense strategies have been shown to be effective at controlling RNA processing events (reviewed in (18)). These include strategies that do not require RNase H activity, such as controlling splicing by masking splice sites on pre-mRNAs (19, reviewed in 20). There are several examples in the literature of successful use of this approach to control splicing, including for pre-mRNA targets in the brains of mice (21, 22). However, the ADAR reaction requires the editing site be in, or very near, stable double helical structure in the RNA and these sites are typically avoided when choosing a binding site for an antisense oligonucleotide (AON) (23, 24). Nevertheless, invasion of stable secondary structure has been reported for certain antisense reagents (25–27). Furthermore, if properly designed, one could envision an AON binding to an editing site complementary sequence and localizing the target adenosine to a single stranded region, thus inhibiting the ADAR reaction (Figure 1). In addition, a very recent report indicated that AONs with the morpholino backbone structure could inhibit A to I editing at a glutamate receptor site, albeit with low potency (28). Thus, AONs have promise for the control of RNA editing at specific sites. In this report, we show that stable RNA structures found at two different ADAR sites can be targeted by AONs, but potency can only be achieved by careful choice of backbone structure to allow for tight binding between the AON and the RNA target. Surprisingly, morpholino AONs, similar to one recently shown to inhibit editing in a cellular assay (28), were not effective in inhibiting editing in our study. Instead, a 2’-O-methyl/locked nucleic acid (LNA) mixmer worked most effectively. Use of the 2’-O-methyl/LNA backbone structure led to inhibition of editing of the human NEIL1 mRNA in the nanomolar concentration range both in vitro and in HeLa cells. Inhibition was shown to be sequence-specific for both the target substrate and the AON. Interestingly, the dynamics of binding for different target RNA structures appears to be a critical factor for defining efficacy of inhibition. Therefore, this work presents a variety of considerations when both choosing a target and designing an AON for inhibition of RNA editing.
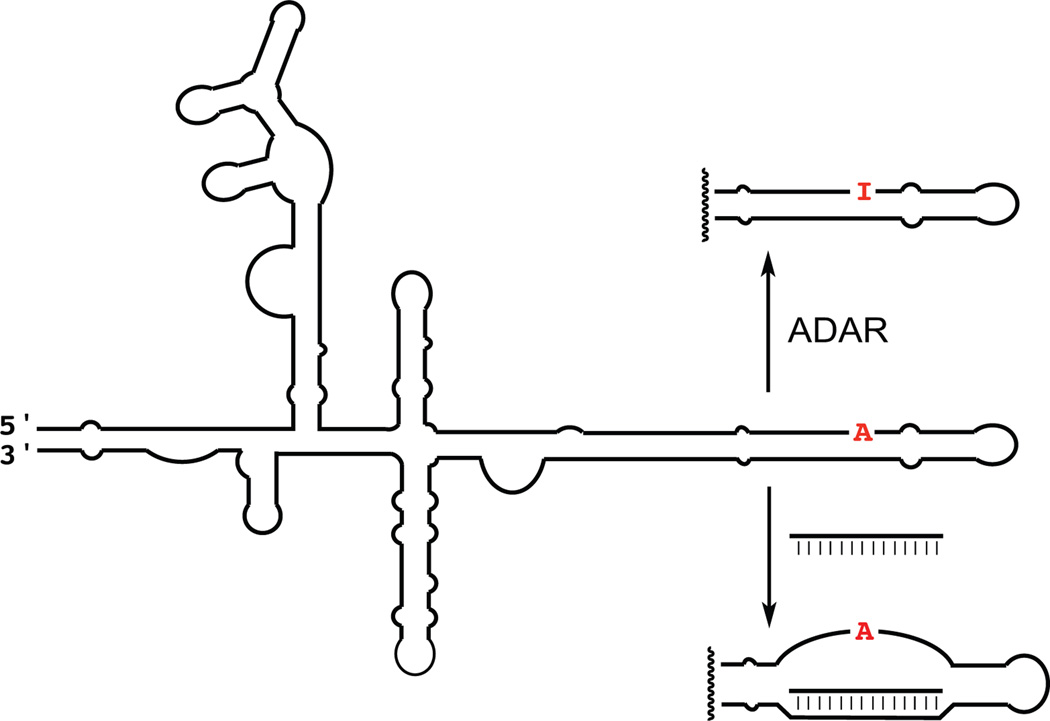
Figure 1.
An antisense oligonucleotide can bind and invade the secondary structure of an ADAR substrate RNA to prevent editing.
RESULTS & DISCUSSION
In vitro optimization of backbone structure
We hypothesized that an AON complementary to a segment of the duplex RNA comprising the serotonin 2C receptor (5HT2CR) pre-mRNA could bind and inhibit editing (Figure 2). This hypothesis was based upon the fact that the snoRNA HBII-52 is complementary to a section of the exonic sequence of this RNA, and is proposed to bind to this sequence and direct 2’-O-methylation (29). Therefore, this location in the RNA may be predisposed to antisense regulation.
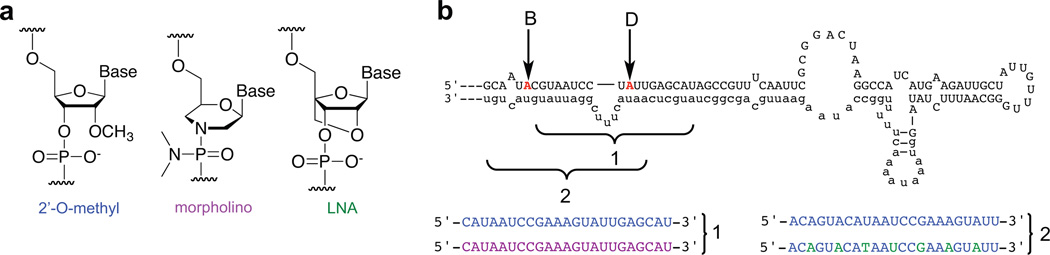
Figure 2.
5HT2CR pre-mRNA predicted secondary structure and antisense oligonucleotides used to target this editing site. a) Analog structures used in these experiments. b) Sequence and binding registers (1 or 2) of the AONs tested. Blue=2’-O-methyl, purple=morpholino, green=locked nucleic acid.
We chose to design an AON complementary to the intronic sequence instead of the exonic sequence because it may be more practical to use in future studies where inhibition of translation is undesirable. An AON bound to the intronic sequence would be expected to be removed by splicing and should not interfere with translation, whereas an AON tightly bound to exonic sequence could inhibit translation. Therefore we designed a 2’-O-methyl AON complementary to the editing complementary sequence (ECS), targeting the region of the intron directly across from the exonic sequence predicted to bind to the snoRNA HBII-52 (29). The first AON that we designed consisted entirely of 2’-O-methyl modified RNA (OMe1), because 2’-O-methyl oligonucleotides are more resistant to nuclease degradation compared to oligoribonucleotides. The intronic region corresponding to the binding register of HBII-52 contains a five-nucleotide internal loop, so the AON was 23 nt in length. We tested this AON for its ability to inhibit ADAR2 editing in a cell free assay developed previously (16). Treatment with 1 µM of the AON OMe1 inhibited editing at the D site of the 5HT2CR RNA to approximately 50% of the no inhibitor control (Figure 3a) and >90% inhibition at 5 µM (Figure 3b).
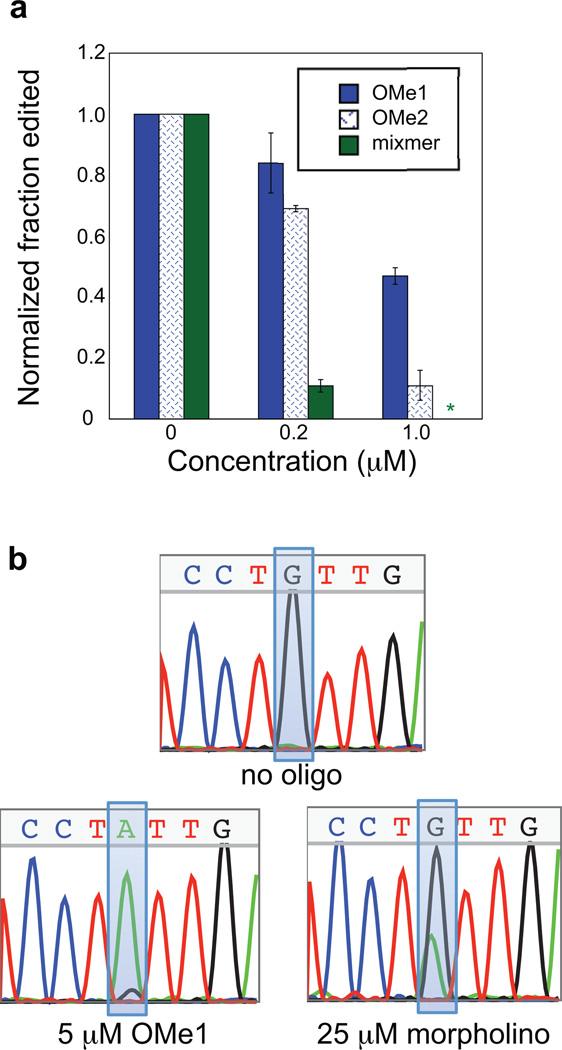
Figure 3.
In vitro inhibition of editing by AONs targeted to the 5HT2CR pre-mRNA. a) Comparison of 2 different binding registers of 2’-O-methyl AON and a locked nucleic acid/2’-O-methyl mixmer. b) Comparison of OMe1 and morpholino AONs at their respective highest concentrations tested, 5 µM OMe1 and 25 µM morpholino.
We then chose to test different backbone and sugar structures to determine the optimal one for strand-invasion. Morpholino oligonucleotides (Figure 2a) have been used to control splicing in vivo, and recent work has also shown that they can be used to inhibit editing in cell culture experiments, with cellular IC50 values around 2 µM (28). We therefore designed a morpholino oligonucleotide corresponding to the same binding register as OMe1. However, this AON displayed significantly decreased potency compared to the 2’-O-methyl oligonucleotide, with less than a factor of two change in editing even at 25 µM AON (Figure 3b). This is likely due to the morpholino’s lower affinity for complementary sequence as compared to a 2’-O-methyl oligonucleotides or oligonucleotides containing LNAs (described below) (30). We also shifted the binding register of the 2’-O-methyl AON to the edge of the predicted duplex structure to determine if an increase in potency could be realized. This AON (OMe2) at 1 µM concentration inhibited D site editing to approximately 10 % of the control lacking inhibitor (Figure 3a), indeed showing a moderate increase in potency compared to the original register. Therefore, the choice of binding register and backbone structure are critical factors when designing an AON to target an ADAR editing site.
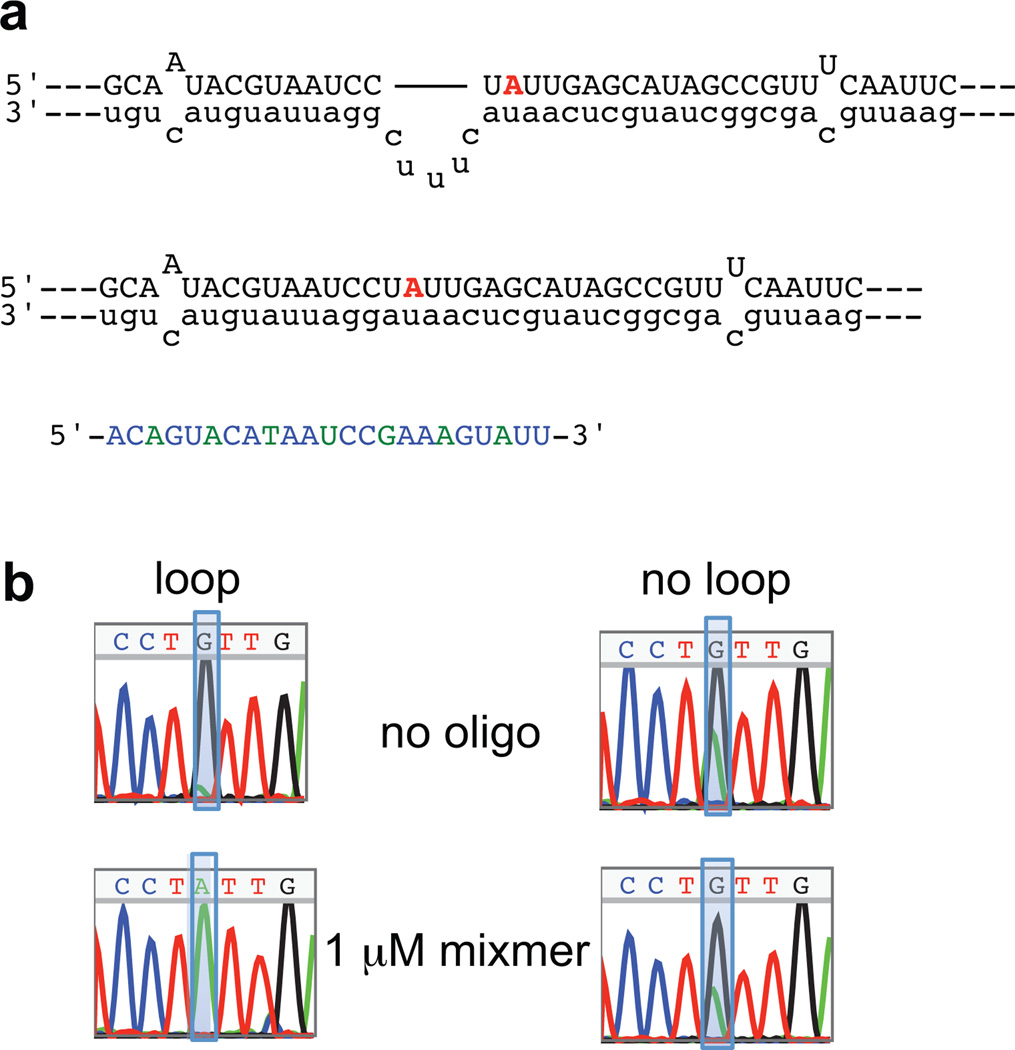
LNAs (Figure 2a) have been shown to bind to RNA with extremely high affinity (31). This is likely due to their constrained 3’-endo sugar pucker, which induces an Aform helical structure (31–33). Oligonucleotides containing LNA residues pay less of an entropic penalty upon binding to RNA due to this pre-organization (34), and also exhibit a more favorable enthalpy of binding (35). Efforts have been undertaken to define the locations within an antisense oligonucleotide that contribute the most to this enhanced binding affinity (36). In addition, mixmers consisting of LNA and 2’-O-methyl residues accumulate in the cell nucleus upon transfection (37). We therefore designed an AON consisting of approximately 30% LNAs and 70% 2’-O-methyl nucleotides, in the same binding register as OMe2. We tested inhibition of editing with this AON and found that it inhibited D site editing to 10% of the no AON control at 200 nM, and completely inhibited editing at 1 µM (Figure 3a). To determine if the observed inhibition was dependent on the target site sequence, we used an RNA target without the internal loop adjacent to the D editing site (Figure 4a). We observed only a slight inhibition of editing when the mixmer was tested with the no loop target at a concentration of 1 µM, 20 times the concentration needed to inhibit editing to 50% of control on the native target RNA (Figures 3 and 4).
Figure 4.
a) Loop and no loop control 5HT2CR pre-mRNA structures in the region of the AON binding sites. Shown with the locked nucleic acid mixmer. b) Editing of the RNAs from (a) in the presence of the mixmer.
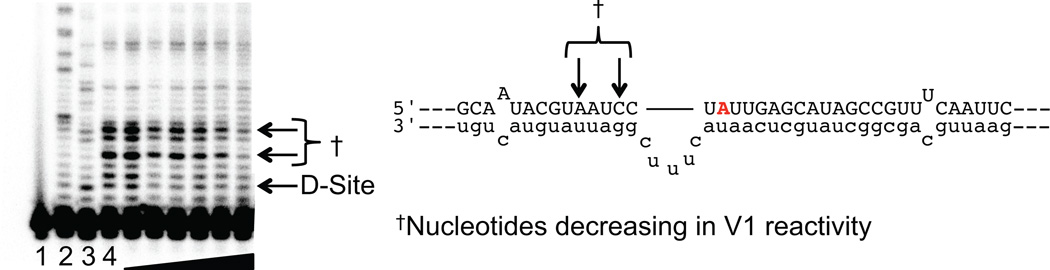
To confirm that the mixmer, our most potent AON, inhibits editing by remodeling the RNA secondary structure, we carried out a V1 nuclease footprinting experiment. V1 nuclease digests double-stranded RNA (dsRNA), so the disappearance of V1-dependent bands indicates reduced duplex structure at that location. Upon titration of the mixmer into a sample containing the RNA, we saw a change in V1-dependent bands, confirming that the secondary structure has changed (Figure 5). Therefore, the mixmer consisting of LNA and 2’-O-methyl nucleotides can inhibit editing by strand invasion of the stable structure found in an ADAR substrate RNA.
Figure 5.
The mixmer oligonucleotide causes remodeling of the 5HT2CR secondary structure as determined by V1 nuclease digest. Lane 1: no V1 nuclease; lane 2: sequencing lane with ddCTP; lane 3: sequencing lane with ddTTP; lane 4: no AON added; lanes 5–10: increasing concentrations of 5HT2CR mixmer (3 nM, 10 nM, 30 nM, 100 nM, 300 nM, 1 µM).
Inhibition of NEIL1 editing in vitro
As a test of the utility of this technique as a general tool for studying RNA editing, we wanted to measure the effect of transfected AONs on editing at different sites in the transcripts of cultured human cells. Detection of 5HT2CR editing can be challenging in immortalized cell lines due to the generally low levels of 5HT2CR expression and the alternative splicing that removes the portion of the RNA containing the editing sites (38). Instead, we focused on editing at the K/R site in the human NEIL1 pre-mRNA, a ubiquitously expressed and abundant RNA (39, 40). NEIL1 is a base excision glycosylase involved in the repair of oxidatively damaged DNA (41). An ADAR1-catalyzed editing reaction that occurs in the codon for lysine 242 of human NEIL1 converts it to an arginine codon (9). The Lys to Arg change in the protein alters NEIL1’s ability to remove various damaged base lesions from DNA, but the consequences of this editing event on the cellular response to oxidative DNA damage have yet to be fully defined (9). AON inhibitors of NEIL1 editing would be useful for exploring these effects.
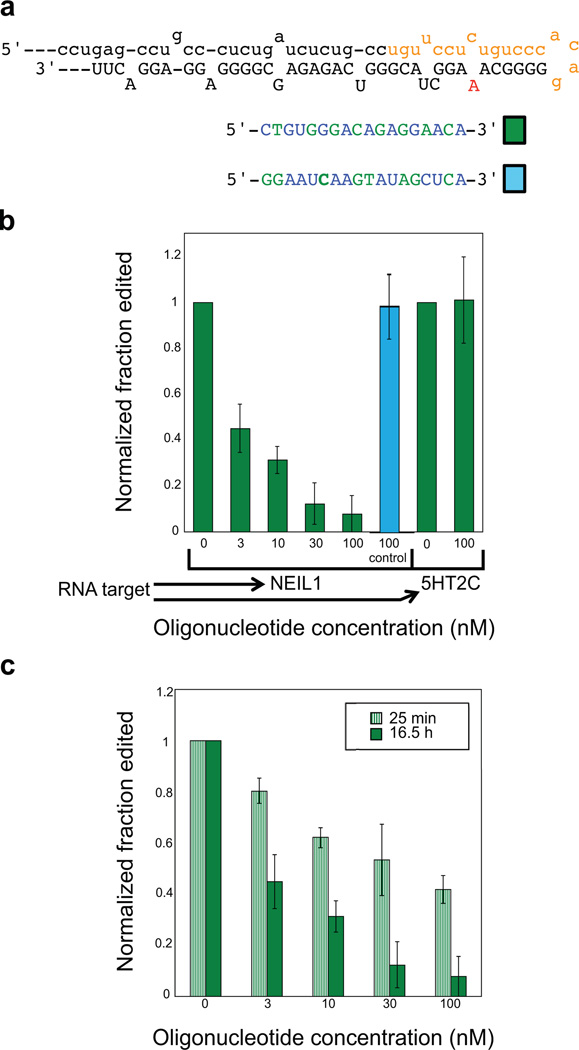
In our previous work, we showed that an RNA based on the NEIL1 pre-mRNA consisting of only 45 nucleotides was a substrate for human ADAR1 (42). Therefore, we designed a 2’-O-methyl/LNA mixmer AON (NL18) to target a site within this 45 nt including the hairpin loop and intronic sequence 5’ to the loop (Figure 6a). In in vitro ADAR1 assays (16, 42), this oligonucleotide inhibited editing of the NEIL1 recoding site but not the B site of the 5HT2CR RNA, also a known ADAR1 site (Figure 6b). In ddition, a control oligo of the same length and number of LNA residues had no effect on NEIL1 editing (Figure 6a,b). The effect of different concentrations of NL18 on ADAR1-catalyzed editing of the NEIL1 RNA in vitro indicated an IC50 < 3 nM (Figure 6b).
Figure 6.
AONs targeting the NEIL1 pre-mRNA. a) NEIL1 pre-mRNA predicted secondary structure, target and control AON sequences. Target AON binding site is shown in orange, editing site is in red. b) Inhibition of NEIL1 editing by the target AON. The control AON has no effect on NEIL1 editing, and the NEIL1 target AON has no effect on editing the 5HT2CR B site, a known ADAR1 site (43). c) Comparison of NEIL1 editing inhibition when incubated with AON for different periods of time. Data reported are the average ± standard deviation for three experiments.
Interestingly, the rate at which full inhibition is realized is lower for the NEIL1-targeted AON compared to 5HT2CR-targeted AONs. Under conditions in which full inhibition of D site editing is observed with the 5HT2CR mixmer, inhibition of NEIL1 editing by NL18 is incomplete. Instead, editing is reduced to approximately 40% of the no inhibitor control at 100 nM AON with little additional inhibition seen at 300 nM AON (Figure 6c). However, when the incubation time was increased from 25 min to 16 h to allow for AON and substrate RNA binding prior to addition of ADAR1, full inhibition was achieved at 30 nM AON (Figure 6c). Thus, while AON affinity for target is clearly important for efficient editing inhibition, the on-rate for AON binding to target also appears to be important.
A critical contributing factor to on-rate is the nucleation site available on the target RNA. The AON targeting the 5HT2CR RNA is likely to nucleate from the five nt bulge loop, while the AON targeting the NEIL1 RNA is likely to nucleate from the four nt hairpin loop. Bulge loops have been shown to induce a kink or bend in the duplex (44). AONs are better able to invade duplex structure when they are able to first bind to single stranded regions and stack onto the end of the adjacent stem, and then are able to incorporate the entire strand of a stem into the heteroduplex (45). By inducing a kink in the structure, the bulge loop may allow the 5’ end of the stem to function independently, such that the AON is able to stack against the end and then invade the entirety of that stem. These differences may help explain the difference in sensitivity to AON inhibition, as it was possible to completely inhibit editing of the D site when AON was incubated with target RNA for only 25 min. Future experiments will be necessary to dissect the thermodynamic (affinity) vs kinetic (on-rate) contributions leading to efficient editing inhibition.
Inhibition of NEIL1 editing in HeLa cells
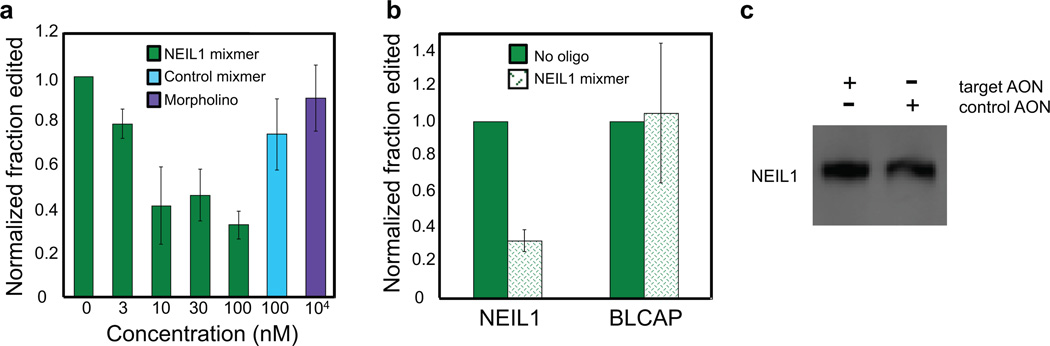
We transfected the NL18 AON into HeLa cells and monitored the resultant level of editing at different sites. We used editing of the BLCAP Y/C and Q/R sites (both primarily substrates for ADAR1) as a control to confirm the substrate-specificity of the target AON (46). Upon NL18 transfection, we observed decreased editing of the NEIL1 RNA, but no change in editing of the BLCAP Y/C site (Figure 7a,b). Substantial inhibition of NEIL1 editing was achieved at 10 nM NL18 corresponding to a reduction in editing to ~40% of that observed with transfection agent alone. Treatment with higher concentrations of AON resulted in only modest improvement (Figure 7a). Effects observed with control AON also established the AON sequence specificity (Figure 7a). In addition, we saw no inhibition of editing upon treatment with 10 µM AON of the same sequence but with morpholino backbone chemistry (Figure 7a). Our results strongly support the 2’-O-methyl/LNA mixmer backbone type as the superior choice for inhibition of editing.
Figure 7.
Inhibition of NEIL1 editing in HeLa cells using the NL18 mixmer AON or analogous morpholino AON. a) NL18 mixmer inhibits editing in HeLa cells. b) The NL18 AON does not inhibit editing of the BLCAP pre-mRNA. Data reported in (a) and (b) are the average ± standard deviation for three experiments. c) Transfection of the NL18 AON does not reduce NEIL1 protein levels compared to the control AON.
Importantly, transfection of the target AON did not significantly alter expression of the NEIL1 protein under these conditions as indicated by Western blotting with a NEIL1-specific antibody (Figure 7c). Thus, these studies have identified an AON (NL18) that, when transfected at 100 nM concentration, reduces NEIL1 editing levels in HeLa cells by approximately three fold without reducing NEIL1 protein levels. Experiments designed to evaluate the impact of this change in editing on the ratio of the K242/R242 forms of the NEIL1 protein and on the cellular response to oxidative DNA damage are currently underway in our laboratory. In addition, further optimization of NL18 may be desirable. While the NL18 AON is able to fully inhibit the ADAR1-catalzyed NEIL1 editing reaction in vitro, editing at the NEIL1 site was not fully inhibited in HeLa cells by NL18. This effect may be related to the slow onset of inhibition observed for this AON in vitro and an inability to reach equilibrium prior to target RNA turnover (Figure 6c). It will be important in future studies to optimize independently AON on-rate and affinity for the NEIL1 target RNA to determine which, if either, of these properties controls the extent of editing inhibition in transfected cells. Additional modifications to the AON beyond the 2’-Omethyl/LNA backbone, such as stabilizing base modifications or conjugation with organic cations and/or intercalators, will be useful in this regard (47–49). Focusing those additional modifications on the part of NL18 likely to be involved in nucleation (i.e. the end that binds the hairpin loop) should prove informative.
In summary, these experiments have defined parameters useful in the design of antisense oligonucleotides for inhibition of editing by the ADAR family of enzymes. While other work has provided guidelines for design of antisense oligonucleotides for splicing inhibition (reviewed in (50)), the differences in secondary structure seem to dictate distinctly different requirements. While AONs inhibiting splicing should be targeted at sequence that is not in a stable duplex structure (51), AONs inhibiting editing must be targeted to such a site. We have shown that 2’-O-methyl/LNA mixmer AONs are effective at inhibition of RNA editing on two different target RNAs. These reagents are capable of binding with high affinity to RNA editing substrates and remodeling the secondary structure by a strand-invasion mechanism. The potency observed here for 2’-O-methyl/LNA mixmers suggests this backbone structure is superior to the morpholino backbone structure for inhibition of RNA editing. Finally, antisense inhibition of editing of the NEIL1 message in cultured human cells provides a means of selectively controlling editing levels of this target and, thus, an approach to exploring the link between RNA editing and the cellular response to oxidative DNA damage.
METHODS
All primer and AON sequences may be found in Supplementary Table 1 in the Supporting Information.
Protein overexpression and purification
Human ADAR2 in yeast expression plasmid (YEpTOP2PGAL1) was overexpressed in Saccharomyces cerevisiae and purified as previously described (52–54). Human ADAR1 in yeast expression plasmid (YEpTOP2PGAL1) was overexpressed and purified as previously described (42).
Purification of oligonucleotides
2’-O-Methyl oligonucleotides were synthesized as described previously (55). LNA/2’-O-methyl mixmers (Exiqon) were purified as described previously (42). Morpholino oligonucleotides (Gene Tools) were used as received without further purification. DNA for primer extension (Bioneer) was re-suspended in nanopure water and diluted to 12 µM for 5’-end labeling.
In vitro editing of the 5HT2CR pre-mRNA
Editing of the 5HT2CR substrate RNAs with and without the internal loop was carried out as previously described (16), except that 20 nM ADAR2 was mixed with 10 nM RNA and assay buffer containing 15 mM Tris-HCl, pH 7.5, 3% (v/v) glycerol, 0.5 mM DTT, 150 mM KCl, 3 mM MgCl2, 1.5 mM EDTA, 0.003% Nonidet P-40, 160 units mL−1 RNasin (Promega), and 1.0 µg mL−1 yeast tRNAPhe. AONs were added to the reaction components and the sample was held at 30 °C for 25 min before addition of ADAR2. The reaction was carried out at 30 °C for 30 min. Editing was determined by RT-PCR and sequencing and quantified as described previously (16). Editing was normalized against the no AON control, which showed 100% editing.
RNase V1 footprinting assays
In vitro transcribed RNA in 10 µL assay buffer (15 mM Tris-HCl, pH 7.4, 3% (v/v) glycerol, 0.5 mM DTT, 150 mM KCl, 3 mM MgCl2, 1.5 mM EDTA, 0.003% Nonidet P-40 and 10 µg mL−1 yeast tRNAPhe) was treated with AON. The samples were held at room temperature for 20 min then digested with 3 X 10−3 U RNase V1 (Life Technologies) for 30 min at room temperature. The reaction was quenched with 190 µL hot water then phenol-chloroform extracted and ethanol precipitated. DNA primer was 5’-end labeled using T4 Polynucleotide Kinase (New England Biolabs). Primer extension was carried out using dNTP stocks (Promega), ddNTP (TriLink BioTechnologies), and AMV Reverse Transcriptase (Promega). Sequencing lanes were generated using a 1:5 ratio of ddNTP:dNTP, using stocks with 25 µM ddNTP, 125 µM corresponding dNTP, and 500 µM every other dNTP. A 5X dNTP stock was made for primer extension of RNase V1 digested RNA such that it contained 500 µM each dNTP. RNA pellets were dissolved in 5 µL water and 5’-end labeled primer (~5000cpm) was added. For sequencing lanes, 67 nM in vitro transcribed RNA was mixed with 5’-end labeled primer (~5000 cpm) in 5 µL of water. All samples were annealed at 62 °C for 15 min then cooled on ice. Reverse transcription was initiated by the addition of 5 µL of enzyme stock (1 µL of either 5X ddNTP stock or dNTP stock, 1 µL 5X AMV RT buffer, and 5 U AMV RT) to a final reaction volume of 10 µL. The reactions were incubated at 42 °C for 45 min and quenched with 7 µL denaturing loading buffer. Primer extension products were resolved by a 12% denaturing polyacrylamide gel and visualized by storage phosphor autoradiography and a Typhoon Trio Variable Mode Imager (GE Healthcare).
In vitro editing of the NEIL1 pre-mRNA
Editing of the NEIL1 pre-mRNA was analyzed as described previously (42) with some changes (see Supplementary Figure 1 for representative TLC image). AON was mixed with RNA for either 25 min or 16.5 h prior to addition of ADAR1 to a final concentration of 50 nM. Reactions were incubated at 30 °C for 45 min before quenching with hot 1% (w/v) SDS. Editing level was normalized against the no AON control within the same assay. Data reported are the average ± standard deviation for three experiments. Editing in the absence of AON was 43 ± 4% with 25 min pre-incubation and 46 ± 8% with 16.5 h pre-incubation. ADAR1 editing of the 5HT2CR was carried out under the same conditions as used for the NEIL1 pre-mRNA except that the reaction was stopped after 25 min. Editing in the absence of AON was 48 ± 4% with 25 min pre-incubation and 56 ± 5% with 16.5 h pre-incubation.
Analysis of editing in HeLa cells
HeLa cell suspension (2 mL) was plated at a density of 1.9 X 104 cells mL−1 in a 6-well plate. The following day, cells were transfected either with NL18 using Lipofectamine 2000 (Life Technologies) or with the analogous morpholino AON using Endoporter (Gene Tools). After 24 h RNA was isolated using the RNAqueous 4PCR kit (Life Technologies). Nested RT-PCR was carried out using the Access RT-PCR kit (Promega) for the first PCR, 15 cycles after the reverse transcription, and Phusion Hot Start DNA Polymerase (ThermoScientific) for the second PCR, 20 cycles. The PCR product was purified using the Qiagen Gel Extraction Kit and sequenced. 4Peaks (v1.7) and ImageJ software (v10.2) (56) were used for quantification of editing (see above). Editing was normalized against the control with transfection agent but no AON, which for NEIL1 showed 38 ± 4% with Lipofectamine 2000 and 36 ± 3% with Endoporter and for BLCAP showed 13 ± 3% (see Supplementary Figure 2 for representative sequencing traces). Data reported are the average ± standard deviation for three experiments.
Detection of NEIL1 protein in transfected cells
Transfection for Western blotting was carried as described above. Cells were lysed with 375 µL lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% (v/v) Triton X-100, supplemented with Halt protease inhibitor cocktail (ThermoFisher)) by shaking on ice for 30 min. Protein was immunoprecipitated using NEIL1 antibody S-17 (Santa Cruz Biotechnology) and Protein A/G Plus Agarose (Santa Cruz Biotechnology). Western blotting was carried out using the same NEIL1 primary antibody at 1:200 dilution and alkaline phosphatase-conjugated secondary antibody (Santa Cruz Biotechnology) at 1:2000 dilution. The proteins were detected using ECF substrate (GE Healthcare) on a Typhoon Trio Variable Mode Imager (GE Healthcare).
Supplementary Material
ACKNOWLEDGMENTS
P. Beal acknowledges the National Institutes of Health for financial support in the form of grant R01-GM061115. R. Mizrahi was supported by training grant T32-GM08799 from NIGMS-NIH and a Graduate Research Fellowship from the National Science Foundation (1148897). The contents of this report are solely the responsibility of the authors and do not necessarily represent the official views of the NIGMS, NIH or NSF.
Footnotes
SUPPORTING INFORMATION AVAILABLE: This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Grosjean H, editor. Fine-tuning of RNA functions by modification and editing. Springer; 2005. [Google Scholar]
- 2.Hood JL, Emeson RB. Editing of neurotransmitter receptor and ion channel RNAs in the nervous system. Curr Top Microbiol Immunol. 2012;353:61–90. doi: 10.1007/82_2011_157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pullirsch D, Jantsch MF. Proteome diversification by adenosine to inosine RNA-editing. RNA Biol. 2010;7:205–212. doi: 10.4161/rna.7.2.11286. [DOI] [PubMed] [Google Scholar]
- 4.Athanasiadis A, Rich A, Maas S. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol. 2004;2:2144–2158. doi: 10.1371/journal.pbio.0020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levanon EY, Eisenberg E, Yelin R, Nemzer S, Hallegger M, Shemesh R, Fligelman ZY, Shoshan A, Pollock SR, Sztybel D, Olshansky M, Rechavi G, Jantsch MF. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat Biotechnol. 2004;22:1001–1005. doi: 10.1038/nbt996. [DOI] [PubMed] [Google Scholar]
- 6.Kim DDY, Kim TTY, Walsh T, Kobayashi Y, Matise TC, Buyske S, Gabriel A. Widespread RNA editing of embedded alu elements in the human transcriptome. Genome Res. 2004;14:1719–1725. doi: 10.1101/gr.2855504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li JB, Levanon EY, Yoon J-K, Aach J, Xie B, Leproust E, Zhang K, Gao Y, Church GM. Genome-wide identification of human RNA editing sites by parallel DNA capturing and sequencing. Science. 2009;324:1210–1213. doi: 10.1126/science.1170995. [DOI] [PubMed] [Google Scholar]
- 8.Shah SP, Morin RD, Khattra J, Prentice L, Pugh T, Burleigh A, Delaney A, Gelmon K, Guliany R, Senz J, Steidl C, Holt RA, Jones S, Sun M, Leung G, Moore R, Severson T, Taylor GA, Teschendorff AE, Tse K, Turashvili G, Varhol R, Warren RL, Watson P, Zhao Y, Caldas C, Huntsman D, Hirst M, Marra MA, Aparicio S. Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature. 2009;461:809–813. doi: 10.1038/nature08489. [DOI] [PubMed] [Google Scholar]
- 9.Yeo J, Goodman RA, Schirle NT, David SS, Beal PA. RNA editing changes the lesion specificity for the DNA repair enzyme NEIL1. Proc Natl Acad Sci USA. 2010;107:20715–20719. doi: 10.1073/pnas.1009231107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Higuchi M, Maas S, Single FN, Hartner J, Rozov A, Burnashev N, Feldmeyer D, Sprengel R, Seeburg PH. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature. 2000;406:78–81. doi: 10.1038/35017558. [DOI] [PubMed] [Google Scholar]
- 11.Kawahara Y, Grimberg A, Teegarden S, Mombereau C, Liu S, Bale TL, Blendy JA, Nishikura K. Dysregulated editing of serotonin 2C receptor mRNAs results in energy dissipation and loss of fat mass. J Neurosci. 2008;28:12834–12844. doi: 10.1523/JNEUROSCI.3896-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morabito MV, Abbas AI, Hood JL, Kesterson RA, Jacobs MM, Kump DS, Hachey DL, Roth BL, Emeson RB. Mice with altered serotonin 2C receptor RNA editing display characteristics of Prader–Willi syndrome. Neurobiol Dis. 2010;39:169–180. doi: 10.1016/j.nbd.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gurevich I, Tamir H, Arango V, Dwork AJ, Mann JJ, Schmauss C. Altered editing of serotonin 2C receptor pre-mRNA in the prefrontal cortex of depressed suicide victims. Neuron. 2002;34:349–356. doi: 10.1016/s0896-6273(02)00660-8. [DOI] [PubMed] [Google Scholar]
- 14.Iwamoto K, Kato T. RNA editing of serotonin 2C receptor in human postmortem brains of major mental disorders. Neurosci Lett. 2003;346:169–172. doi: 10.1016/s0304-3940(03)00608-6. [DOI] [PubMed] [Google Scholar]
- 15.Beghini A, Ripamonti CB, Peterlongo P, Roversi G, Cairoli R, Morra E, Larizza L. RNA hyperediting and alternative splicing of hematopoietic cell phosphatase (PTPN6) gene in acute myeloid leukemia. Hum Mol Genet. 2000;9:2297–2304. doi: 10.1093/oxfordjournals.hmg.a018921. [DOI] [PubMed] [Google Scholar]
- 16.Schirle NT, Goodman RA, Krishnamurthy M, Beal PA. Selective inhibition of ADAR2-catalyzed editing of the serotonin 2c receptor premRNA by a helix-threading peptide. Org Biomol Chem. 2010;8:4898–4904. doi: 10.1039/c0ob00309c. [DOI] [PubMed] [Google Scholar]
- 17.Krishnamurthy M, Simon K, Orendt AM, Beal PA. Macrocyclic helix-threading peptides for targeting RNA. Angew Chem Int Ed. 2007;46:7044–7047. doi: 10.1002/anie.200702247. [DOI] [PubMed] [Google Scholar]
- 18.Kole R, Krainer AR, Altman S. RNA therapeutics: beyond RNA interference and antisense oligonucleotides. Nat Rev Drug Discov. 2012;11:125–140. doi: 10.1038/nrd3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dominski Z, Kole R. Restoration of correct splicing in thalassemic pre-mRNA by antisense oligonucleotides. Proc Natl Acad Sci USA. 1993;90:8673–8677. doi: 10.1073/pnas.90.18.8673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bennett CF, Swayze EE. RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu Rev Pharmacol Toxicol. 2010;50:259–293. doi: 10.1146/annurev.pharmtox.010909.105654. [DOI] [PubMed] [Google Scholar]
- 21.Williams JH, Schray RC, Patterson CA, Ayitey SO, Tallent MK, Lutz GJ. Oligonucleotide-mediated survival of motor neuron protein expression in CNS improves phenotype in a mouse model of spinal muscular atrophy. J Neurosci. 2009;29:7633–7638. doi: 10.1523/JNEUROSCI.0950-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sekiguchi M, Zushida K, Yoshida M, Maekawa M, Kamichi S, Yoshida M, Sahara Y, Yuasa S, Takeda S, Wada K. A deficit of brain dystrophin impairs specific amygdala GABAergic transmission and enhances defensive behaviour in mice. Brain. 2009;132:124–135. doi: 10.1093/brain/awn253. [DOI] [PubMed] [Google Scholar]
- 23.Walton SP, Stephanopoulos GN, Yarmush ML, Roth CM. Prediction of antisense oligonucleotide binding affinity to a structured RNA target. Biotechnol. Bioeng. 1999;65:1–9. [PubMed] [Google Scholar]
- 24.Johnson E, Srivastava R. Volatility in mRNA secondary structure as a design principle for antisense. Nucleic Acids Res. 2013;41:e43. doi: 10.1093/nar/gks902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holmes SC, Arzumanov AA, Gait MJ. Steric inhibition of human immunodeficiency virus type-1 Tat-dependent trans-activation in vitro and in cells by oligonucleotides containing 2'-O-methyl G-clamp ribonucleoside analogues. Nucleic Acids Res. 2003;31:2759–2768. doi: 10.1093/nar/gkg384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu J, Matsui M, Gagnon KT, Schwartz JC, Gabillet S, Arar K, Wu J, Bezprozvanny I, Corey DR. Allele-specific silencing of mutant huntingtin and ataxin-3 genes by targeting expanded CAG repeats in mRNAs. Nat Biotechnol. 2009;27:478–484. doi: 10.1038/nbt.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vickers T, Baker BF, Cook PD, Zounes M, Buckheit RW, Germany J, Ecker DJ. Inhibition of HIV-LTR gene expression by oligonucleotides targeted to the TAR element. Nucleic Acids Res. 1991;19:3359–3368. doi: 10.1093/nar/19.12.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Penn AC, Balik A, Greger IH. Steric antisense inhibition of AMPA receptor Q/R editing reveals tight coupling to intronic editing sites and splicing. Nucleic Acids Res. 2013;41:1113–1123. doi: 10.1093/nar/gks1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cavaillé J, Buiting K, Kiefmann M, Lalande M, Brannan CI, Horsthemke B, Bachellerie JP, Brosius J, Hüttenhofer A. Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proc Natl Acad Sci USA. 2000;97:14311–14316. doi: 10.1073/pnas.250426397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aartsma-Rus A, Kaman WE, Bremmer-Bout M, Janson AAM, den Dunnen JT, van Ommen G-JB, van Deutekom JCT. Comparative analysis of antisense oligonucleotide analogs for targeted DMD exon 46 skipping in muscle cells. Gene Ther. 2004;11:1391–1398. doi: 10.1038/sj.gt.3302313. [DOI] [PubMed] [Google Scholar]
- 31.Singh SK, Nielsen P, Koshkin AA, Wengel J. LNA (locked nucleic acids): synthesis and high-affinity nucleic acid recognition. Chem Commun. 1998:455–456. [Google Scholar]
- 32.Koshkin AA, Singh SK, Nielsen P, Rajwanshi VK, Kumar R, Meldgaard M, Olsen CE, Wengel J. LNA (Locked Nucleic Acids): Synthesis of the adenine, cytosine, guanine, 5-methylcytosine, thymine and uracil bicyclonucleoside monomers, oligomerisation, and unprecedented nucleic acid recognition. Tetrahedron. 1998;54:3607–3630. [Google Scholar]
- 33.Obika S, Nanbu D, Hari Y, Morio K, In Y, Ishida T, Imanishi T. Synthesis of 2′-O,4′-C-methyleneuridine and -cytidine. Novel bicyclic nucleosides having a fixed C3'-endo sugar puckering. Tetrahedron Lett. 1997;38:8735–8738. [Google Scholar]
- 34.Petersen M, Nielsen CB, Nielsen KE, Jensen GA, Bondensgaard K, Singh SK, Rajwanshi VK, Koshkin AA, Dahl BM, Wengel J, Jacobsen JP. The conformations of locked nucleic acids (LNA) J Mol Recognit. 2000;13:44–53. doi: 10.1002/(SICI)1099-1352(200001/02)13:1<44::AID-JMR486>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 35.Kaur H, Wengel J, Maiti S. Thermodynamics of DNA-RNA heteroduplex formation: effects of locked nucleic acid nucleotides incorporated into the DNA strand. Biochemistry. 2008;47:1218–1227. doi: 10.1021/bi700996z. [DOI] [PubMed] [Google Scholar]
- 36.Kierzek E, Ciesielska A, Pasternak K, Mathews DH, Turner DH, Kierzek R. The influence of locked nucleic acid residues on the thermodynamic properties of 2'-O-methyl RNA/RNA heteroduplexes. Nucleic Acids Res. 2005;33:5082–5093. doi: 10.1093/nar/gki789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arzumanov A, Stetsenko DA, Malakhov AD, Reichelt S, Sørensen MD, Babu BR, Wengel J, Gait MJ. A structure-activity study of the inhibition of HIV-1 Tat-dependent trans-activation by mixmer 2'-O-methyl oligoribonucleotides containing locked nucleic acid (LNA), alpha-L-LNA, or 2'-thio- LNA residues. Oligonucleotides. 2003;13:435–453. doi: 10.1089/154545703322860762. [DOI] [PubMed] [Google Scholar]
- 38.Maas S, Patt S, Schrey M, Rich A. Underediting of glutamate receptor GluR-B mRNA in malignant gliomas. Proc Natl Acad Sci USA. 2001;98:14687–14692. doi: 10.1073/pnas.251531398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takao M, Kanno S-I, Kobayashi K, Zhang Q-M, Yonei S, van der Horst GTJ, Yasui A. A back-up glycosylase in Nth1 knock-out mice is a functional Nei (endonuclease VIII) homologue. J Biol Chem. 2002;277:42205–42213. doi: 10.1074/jbc.M206884200. [DOI] [PubMed] [Google Scholar]
- 40.Hazra TK, Izumi T, Boldogh I, Imhoff B, Kow YW, Jaruga P, Dizdaroglu M, Mitra S. Identification and characterization of a human DNA glycosylase for repair of modified bases in oxidatively damaged DNA. Proc Natl Acad Sci USA. 2002;99:3523–3528. doi: 10.1073/pnas.062053799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.David SS, O'Shea VL, Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447:941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mizrahi RA, Phelps KJ, Ching AY, Beal PA. Nucleoside analog studies indicate mechanistic differences between RNA-editing adenosine deaminases. Nucleic Acids Res. 2012;40:9825–9835. doi: 10.1093/nar/gks752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang W, Wang Q, Kanes SJ, Murray JM, Nishikura K. Altered RNA editing of serotonin 5-HT2C receptor induced by interferon: implications for depression associated with cytokine therapy. Brain Res Mol Brain Res. 2004;124:70–78. doi: 10.1016/j.molbrainres.2004.02.010. [DOI] [PubMed] [Google Scholar]
- 44.Bhattacharyya A, Murchie AIH, Lilley DMJ. RNA bulges and the helical periodicity of double-stranded RNA. Nature. 1990;343:484–487. doi: 10.1038/343484a0. [DOI] [PubMed] [Google Scholar]
- 45.Mir KU, Southern EM. Determining the influence of structure on hybridization using oligonucleotide arrays. Nat Biotechnol. 1999;17:788–792. doi: 10.1038/11732. [DOI] [PubMed] [Google Scholar]
- 46.Riedmann EM, Schopoff S, Hartner JC, Jantsch MF. Specificity of ADAR-mediated RNA editing in newly identified targets. RNA. 2008;14:1110–1118. doi: 10.1261/rna.923308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Freier SM, Altmann KH. The ups and downs of nucleic acid duplex stability: structure-stability studies on chemically-modified DNA:RNA duplexes. Nucleic Acids Res. 1997;25:4429–4443. doi: 10.1093/nar/25.22.4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Harrison JG, Balasubramanian S. Synthesis and hybridization analysis of a small library of peptide-oligonucleotide conjugates. Nucleic Acids Res. 1998;26:3136–3145. doi: 10.1093/nar/26.13.3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Noir R, Kotera M, Pons B, Remy J-S, Behr J-P. Oligonucleotide-oligospermine conjugates (zip nucleic acids): a convenient means of finely tuning hybridization temperatures. J Am Chem Soc. 2008;130:13500–13505. doi: 10.1021/ja804727a. [DOI] [PubMed] [Google Scholar]
- 50.Aartsma-Rus A. Overview on AON Design. Methods Mol Biol. 2012;867:117–129. doi: 10.1007/978-1-61779-767-5_8. [DOI] [PubMed] [Google Scholar]
- 51.Wee KB, Pramono ZAD, Wang JL, MacDorman KF, Lai PS, Yee WC. Dynamics of co-transcriptional pre-mRNA folding influences the induction of dystrophin exon skipping by antisense oligonucleotides. PLoS One. 2008;3:e1844. doi: 10.1371/journal.pone.0001844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Haudenschild BL, Maydanovych O, Véliz EA, Macbeth MR, Bass BL, Beal PA. A transition state analogue for an RNA-editing reaction. J Am Chem Soc. 2004;126:11213–11219. doi: 10.1021/ja0472073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ley HL., III . Ph.D. Dissertation. Salt Lake City, UT: University of Utah; 2001. Editing of Hepatitis Delta Virus Antigenomic RNA by Recombinant Human Adenosine Deaminases That Act on RNA. [Google Scholar]
- 54.Macbeth MR, Lingam AT, Bass BL. Evidence for auto-inhibition by the N terminus of hADAR2 and activation by dsRNA binding. RNA. 2004;10:1563–1571. doi: 10.1261/rna.7920904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pokharel S, Jayalath P, Maydanovych O, Goodman RA, Wang SC, Tantillo DJ, Beal PA. Matching active site and substrate structures for an RNA editing reaction. J Am Chem Soc. 2009;131:11882–11891. doi: 10.1021/ja9034076. [DOI] [PubMed] [Google Scholar]
- 56.Abramoff MD, Magalhães PJ, Ram SJ. Image processing with ImageJ. Biophot Int. 2004;11:36–42. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.