Abstract
Quantitation of DNA adducts could provide critical information on the relationship between exposure to tobacco smoke and cancer risk in smokers. In this study, we developed a robust and sensitive liquid chromatography-tandem mass spectrometry method for the analysis of 4-hydroxy-1-(3-pyridyl)-1-butanone (HPB1)-releasing DNA adducts in human oral cells, a non-invasive source of DNA for biomarker studies. Isolated DNA undergoes acid hydrolysis, after which samples are purified by solid-phase extraction and analyzed by LC-ESI-MS/MS. The developed method was applied for analysis of samples obtained via collection with a commercial mouthwash from 30 smokers and 15 nonsmokers. In smokers, the levels of HPB-releasing DNA adducts averaged 12.0 pmol HPB/mg DNA (detected in 20 out of 28 samples with quantifiable DNA yield) and in nonsmokers, the levels of adducts averaged 0.23 pmol/mg DNA (detected in 3 out of 15 samples). For the 30 smoking subjects, matching buccal brushings were also analyzed and HPB-releasing DNA adducts were detected in 24 out of 27 samples with quantifiable DNA yield, averaging 44.7 pmol HPB/mg DNA. The levels of adducts in buccal brushings correlated with those in mouthwash samples of smokers (R = 0.73, p < 0.0001). Potentially the method can be applied in studies of individual susceptibility to tobacco-induced cancers in humans.
Keywords: HPB, DNA adducts, oral cells
INTRODUCTION
Individual susceptibility to the effects of chemical carcinogens depends on a variety of factors, including the levels of exposure and uptake, relative efficiency of metabolic activation and detoxification pathways, and response to the damage caused by the metabolically activated forms of carcinogens to target cell macromolecules. Quantitation of DNA adducts can provide a direct measure of DNA damage caused by the exposure to a specific chemical carcinogen, thus providing important information for the evaluation of the relationship between exposure and cancer risk. Tobacco smoking causes 19 types of cancers1 and 21% of all cancer mortality worldwide.2 In addition to public education and tobacco control efforts, the ability to identify those smokers who are at higher risk of developing cancer could promote the development of preventive measures, potentially reducing the death toll of tobacco use.
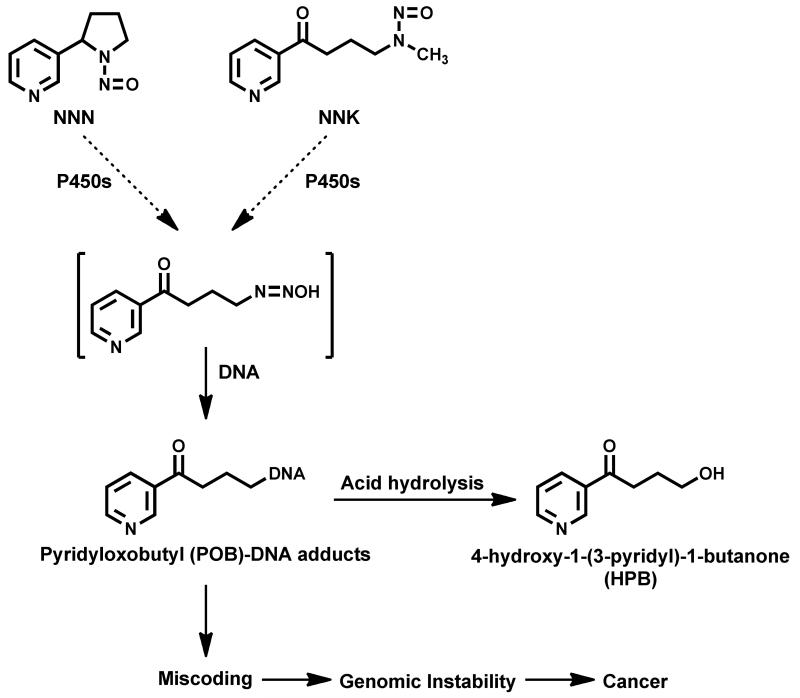
The tobacco-specific N-nitrosamines N-nitrosonornicotine (NNN) and 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) are among the most important carcinogens in unburned tobacco and cigarette smoke.3 Results of numerous animal, mechanistic, and epidemiological studies indicate that NNN and NNK play an important role in the induction by tobacco products of cancers of the lung, esophagus, oral cavity, and pancreas.3-6 A common pathway in NNK and NNN metabolism leads to the formation of a reactive pyridyloxobutyl (POB) metabolite that reacts with the 7-, O6- and N2-positions of guanine and the O2-position of cytosine and thymidine to produce well characterized DNA adducts, some of which have known miscoding properties7,8 (Scheme 1). To date there are no reports on individual POB-DNA adducts in human tissues, most likely because these adducts are present at levels that are not detectable with the available methodologies. However, it is known that acid hydrolysis of pyridyloxobutylated DNA releases 4-hydroxy-1-(3-pyridyl)-1-butanone (HPB)9,10 (Scheme 1). Numerous studies in cellular and rodent systems demonstrated the importance of HPB-releasing adducts in carcinogenesis by NNK and NNN.3,11 Since several adducts of various structures contribute to the release of HPB, analysis of this compound in hydrolyzed DNA represents an advantage over the measurement of individual adducts in human tissues. Indeed, in 1991 our group demonstrated the presence of HPB-releasing adducts in lung DNA from smokers.12 Levels of these adducts were higher than in non-smokers, as expected based on the tobacco-specificity of NNK and NNN. A much larger study examined levels of HPB-releasing adducts in human lung and clearly demonstrated significantly higher levels of HPB-releasing adducts in pulmonary DNA from lung cancer patients compared to controls.13
Scheme 1.
Pyridyloxobutyl-DNA adduct formation from NNN and NNKa
aFor more detailed information on NNN and NNK metabolism and DNA adduct formation, see ref. 3 and 7
Even though DNA adducts are central to the carcinogenic process, and their measurement potentially can provide the most direct link between cellular exposure and cancer, measurement of these biomarkers in the lung of healthy smokers is not practical. Exfoliated oral mucosa cells are emerging as a promising source of DNA for biomarker studies in humans,14-17 non-invasiveness of their collection being an important advantage. Significantly higher levels of various DNA adducts in the buccal cells of smokers, as compared to nonsmokers, have been reported,17,18 and a recent study has demonstrated that oral epithelium can serve as a surrogate tissue for assessing tobacco-induced molecular alterations in the lungs.19 Formation of POB-DNA adducts in oral mucosa of rats chronically treated with NNK and NNN has been demonstrated in our laboratory.20,21 In another study, Heling et al.22 measured HPB-releasing DNA adducts in buccal cells from nonsmokers, smokers, and smokeless tobacco users. They demonstrated that the levels of HPB-releasing DNA adducts in the oral mucosa of smokers and smokeless tobacco users are four-fold and nine-fold, respectively, higher than in non-users of tobacco.
Our goal in this study was to develop a novel sensitive and robust liquid chromatography (LC)-electrospray ionization (ESI)-tandem mass spectrometry (MS/MS) method for the measurement of HPB-releasing DNA adducts in human oral cells and to apply this method for the analysis of oral samples from smokers and nonsmokers. The ultimate goal is to understand whether HPB-releasing DNA adducts in oral cells of smokers could serve as a useful biomarker of NNK and NNN metabolic activation, and the risk of developing tobacco-induced cancers.
EXPERIMENTAL PROCEDURES
Caution
[13C6]NNN and [13C6]NNAL are carcinogenic and mutagenic and should be handled with extreme care, using appropriate protective clothing and ventilation at all times.
Chemicals and Enzymes
HPB and [3,3,4,4-D4]HPB ([D4]HPB) were purchased from Toronto Research Chemicals (North York, Ontario, Canada). [13C6]NNN and [13C6]NNAL were purchased from Cambridge Isotope Laboratories (Cambridge, MA). Reagents and enzymes for DNA isolation were obtained from QIAGEN Sciences (Germantown, MD). All other chemicals and solvents were purchased from Sigma-Aldrich Chemical Co. (Milwaukee, WI) or Fisher Scientific (Fairlawn, NJ).
Subjects
Mouthwash samples, buccal brushings, and urine samples from smokers were obtained from the TobPRAC Biorepository which was supported by an NCI contract with the Lombardi Comprehensive Cancer Center (Washington, DC). The entrance criteria for the TobPRAC Biorepository required subjects to smoke at least 10 cigarettes per day for at least 1 year. Nonsmoking volunteers were recruited independent of TobPRAC, at the Masonic Cancer Center, University of Minnesota. Collection of these samples was approved by the University of Minnesota Research Subjects’ Protection Programs Institutional Review Board: Human Subjects Committee.
Collection of oral and urine samples
According to the TobPRAC protocol, buccal cell brushings and mouthwash samples were collected from the same subjects at the same clinic visit. Buccal cells were collected at the beginning of the visit by brushing the oral mucosa inside one cheek with a clean toothbrush and swirling the brush in a sterile polypropylene centrifuge tube with the commercial mouthwash Listerine (containing, among other ingredients, water, 21.6% alcohol, menthol, sucralose, flavors, and colorants), to transfer the collected buccal cells from the brush into the liquid. The mouthwash samples were collected at the end of the same visit, by rinsing the mouth with 30 ml of Listerine, and expectorating the rinse into sterile polypropylene centrifuge tubes. The mouthwash samples from nonsmokers were collected by the same procedure. After the collection, both buccal brushings and mouthwash samples were centrifuged at 2,700 rpm to pellet cells; the pellets were washed with Tris-EDTA buffer (pH 7.4), and stored at −80 °C. Urine samples were collected over a 24-h period, total urine volume was recorded, and aliquots were stored at −80 °C. The requested mouthwash, buccal cell, and urine samples were shipped to our laboratory on dry ice and stored frozen until analysis.
DNA isolation
We used a modified Qiagen DNA isolation protocol to isolate DNA from mouthwash and buccal cell samples. Briefly, the samples were thawed at room temperature and centrifuged at 1500 × g for 15 min. The supernatant was discarded, the cell pellet was resuspended in 3 ml of cell lysis solution from Qiagen, homogenized, and after treating with proteinase K, RNase A, and precipitating proteins, DNA was isolated and purified as previously described.23,24 Content of guanine in isolated DNA was determined by HPLC,25 and the amount of DNA was calculated as described.26
Analysis of HPB releasing DNA adducts
To analyze HPB-releasing adducts, the isolated DNA samples were dissolved in 0.5 mL deionized water and mixed with 20 pg of [D4]HPB internal standard. A water blank (0.5 mL) and a sample of calf thymus DNA (~ 100 Sg dissolved in 0.5 mL deionized water) were added to each sample set as negative controls. To each sample, 3.6 N HCl was added to bring the final concentration of the acid to 0.8 N, and the samples were hydrolyzed at 80 °C for 3 h to release HPB.9,12,27 Additional experiments have demonstrated that no D-H exchange takes place in the internal standard under these conditions (data not shown). The hydrolysates were cooled down to room temperature, adjusted to neutral pH with 1 N NaOH, and 40 μL of each sample was transferred to clean microinsert vials for DNA quantitation by HPLC. The remaining hydrolysates were loaded on 30-mg 1cc Strata X cartridges (Phenomenex) activated with 1 mL CH3OH and pre-equilibrated with 2 mL deionized H2O. After loading samples, the cartridges were washed with 2 mL H2O and 1 mL of 10% CH3OH in H2O, the eluates going to waste. HPB was eluted with 2 mL of 100% CH3OH; the eluates were collected into clean vials, concentrated to dryness under a stream of N2, redissolved in 20 μL deionized H2O and transferred into autosampler microinsert vials. Eight μL of the prepared sample were injected for analysis.
LC-ESI-MS/MS was carried out on a Finnigan TSQ Quantum Discovery Max instrument (Thermo Electron Corp., Waltham, MA) interfaced with an Agilent Model 1100 capillary high-performance liquid chromatography system and a Model 1100 micro autosampler (Agilent, Palo Alto, CA). The high-performance liquid chromatograph was fitted with a 250 × 0.5 mm Luna C18(2) 5 μm column (Phenomenex) eluted with a gradient from 5 to 65% methanol in 15 mM ammonium acetate over a period of 30 min, then returning to 5% methanol in 3 min and holding for 15 min at this composition, at a flow rate of 10 μL/min. The column was operated at 30 °C. The first 10 min of eluent was directed to waste and the 10 to 40 min fraction was diverted to the ESI source. The retention times for HPB and [D4]HPB under these conditions were around 24 min, with [D4]HPB eluting 0.16 min earlier than HPB. The ESI source was operated in the positive ion mode, monitoring m/z 166 [M + H]+ → 106 [C5H4N–C≡O]+, m/z 166 → 134 [C5H4N–CO–CH2–CD2]+, and m/z 166 → 79 [C5H5N]+ for HPB, and m/z 170 [M + H]+ → 106, m/z 170 → 136 [C5H4N–CO–CH2–CD2]+, and m/z 170 → 79 for [D4]HPB, at 0.4 amu scan width. The collision gas was Ar at a pressure of 1 mTorr, with collision energy of 8 eV. The quadrupoles were operated at a resolution of 0.2 (Q1) and 0.7 (Q3) amu. The quantitation of HPB was based on the peak area ratio of HPB (m/z 166 → 106) to [D4]HPB (m/z 170 → 106), the constructed calibration curves, and the amount of internal standard added. Calibration curves were constructed before each analysis by using a series of standard solutions containing a constant amount of [D4]HPB (5 pg/μL) and differing amounts of HPB (0.1 – 10 pg/μL).
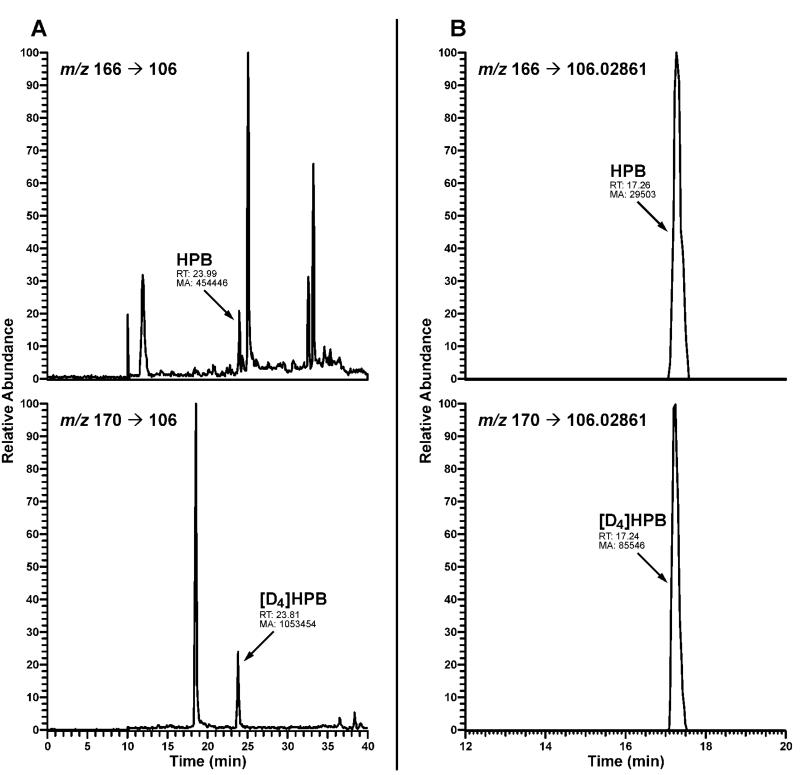
To confirm the identity of HPB in hydrolyzed oral cell DNA, several samples were also analyzed by high resolution tandem mass spectrometry with nanospray ionization using an LTQ-Orbitrap Velos instrument Orbitrap instrument. HPB was identified at m/z 166 [M + H]+ → m/z 106.02861 [C5H4N–C≡O]+ with accurate mass monitoring of the fragment ion at 5 ppm. This method is being optimized for potential routine analysis of HPB-releasing DNA adducts in human tissues and will be described in more detail in a separate publication.
Analysis of urinary biomarkers of exposure to NNN and NNK
Total NNN is a biomarker of exposure to NNN and is the sum of NNN and its N-glucuronide. Total 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol (NNAL) is a biomarker of exposure to NNK and is the sum of NNAL and its glucuronides. These biomarkers were measured in a single analysis by modifying the analytical procedures described for each biomarker.28,29 Briefly, 2 mL urine was mixed with 100 pg each [13C6]NNN and [13C6]NNAL internal standards, 15,000 units of ß-glucuronidase type IX-A from E. coli (Sigma Chemical Co., St. Louis, MO) were added, and the mixture was incubated overnight with gentle shaking at 37°C to convert any NNN- and NNAL-glucuronides to free NNN and NNAL, respectively. On the next day, the urine was purified on ChemElut cartridges (Agilent), Oasis MCX cartridges (Waters Corp., Milford, MA), and transferred to autosampler vials for analysis by LC-MS/MS as previously described.28,29 Free NNN and NNAL were also measured in a single procedure similar to that used for total NNN and total NNAL analysis, except that the treatment with ß-glucuronidase was omitted. The amounts of the corresponding glucuronides were calculated by subtracting free NNN and NNAL from total NNN and total NNAL, respectively.
Statistical Analyses
Differences between smokers and nonsmokers were assessed using the two-sample t-test (independent samples). Differences between biomarkers among smokers were assessed using the one-sample t-test (paired samples). Correlations between biomarkers among smokers are assessed using the standard Pearson’s coefficient of correlation. A p-value less than 0.05 was considered statistically significant. Calculations were performed using the SAS statistical packaged computer program
RESULTS
Development of the analytical procedure
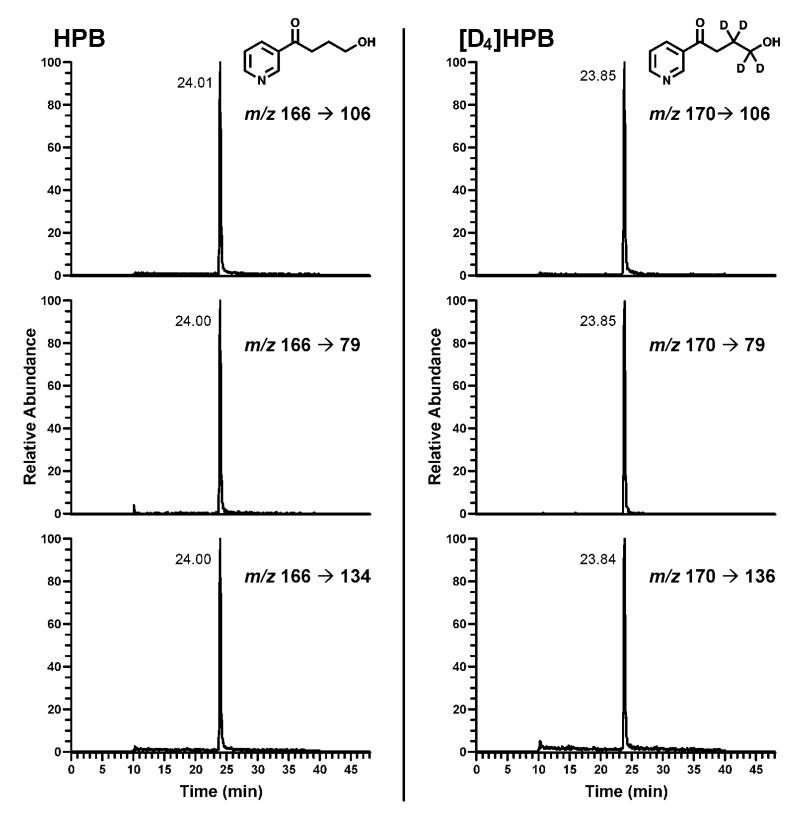
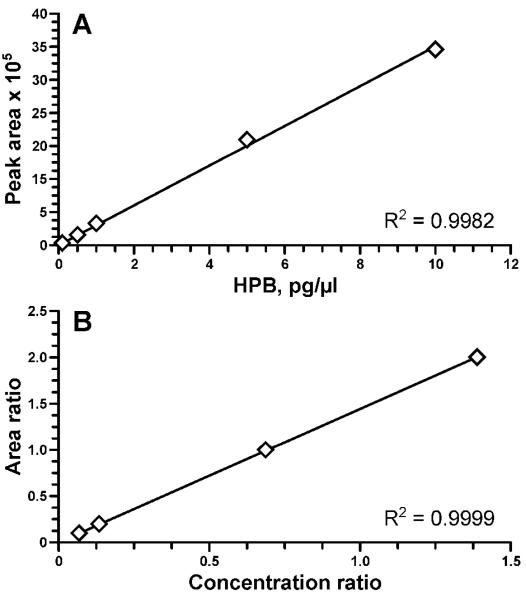
The purpose of this study was to develop a simplified analytical procedure for the direct measurement of HPB by LC-ESI-MS/MS, thus excluding the derivatization step that was traditionally used for the analysis of HPB by gas chromatography-MS.12 The LC-ESI-MS/MS method for HPB analysis was developed by using standard aqueous solutions containing HPB and [D4]HPB in the range of concentrations from 0.1 to 10 pg/μL. Based on the analysis of product scans for m/z 166 ([M+1]+ for HPB), and m/z 170 ([M+1]+ for [D4]HPB) and further optimization of the collision energy for the most abundant fragments, we selected 3 ion transitions to be monitored for each analyte. Figure 1 shows selected reaction monitoring (SRM) chromatograms obtained upon LC-ESI-MS/MS analysis of a standard mix containing 10 pg/μL each of HPB and [D4]HPB. Due to their highest signal intensities, the transitions m/z 166 → 106 and m/z 170 → 106 were selected for quantitation of HPB and [D4]HPB, respectively; the peak area ratios among the three fragments were used to confirm the identity of the analyte and the internal standard. The instrument response and the HPB/[D4]HPB ratio were linear in the 0.1-10 pg/μL range (Figure 2).
Figure 1.
SRM chromatogram obtained upon analysis of a standard mix containing 10 pg/μL each HPB and [D4]HPB (internal standard). Three ion transitions are being monitored for both HPB (parent ion m/z 166) and [D4]HPB (parent ion m/z 170). Signal intensities for each transition are expressed as % of the intensity of quantitation fragments (m/z 166 → 106 for HPB and m/z 170 → 106 for [D4]HPB).
Figure 2.
Linearity of the method: (A) MS/MS signal in the range 0.1-10 pg/μL of HPB; (B) linearity of HPB:[D4]HPB at constant [D4]HPB concentration (5 pg/μL) and HPB ranging from 0.1 to 10 pg/μL, at 8 μL injections.
Method characteristics
At a 3:1 signal-to-noise ratio, the on-column limit of detection was 0.5 fmol HPB for a standard solution, and the limit of quantitation at a 5:1 signal-to-noise ratio was 2.4 fmol/sample for calf thymus DNA spiked with HPB prior to acid hydrolysis and solid phase purification. Precision of the assay (coefficient of variation) was 7.1%, as determined by analyzing six aliquots of calf thymus DNA, each spiked with 50 pg HPB and further subjected to acid hydrolysis, purification, and LC-ESI-MS/MS analysis. The average accuracy of the assay was determined by spiking aliquots of calf thymus DNA and a nonsmoker’s oral cell DNA with various amounts of HPB in the range from 1 to 50 pg/sample: the measured levels of HPB (expressed as % of added HPB) averaged 102% for calf thymus DNA (R2 = 0.996) and 111% for nonsmoker’s DNA (R2 = 0.999). The recovery of the method was determined by comparing oral cell DNA samples to which [D4]HPB was added prior to their hydrolysis and processing, with those mixed with the [D4]HPB after processing, just before the analysis by LC-ESI-MS/MS. The recovery of [D4]HPB averaged 41.6% (N = 20).
Quantitation of HPB-releasing DNA adducts in human oral cells
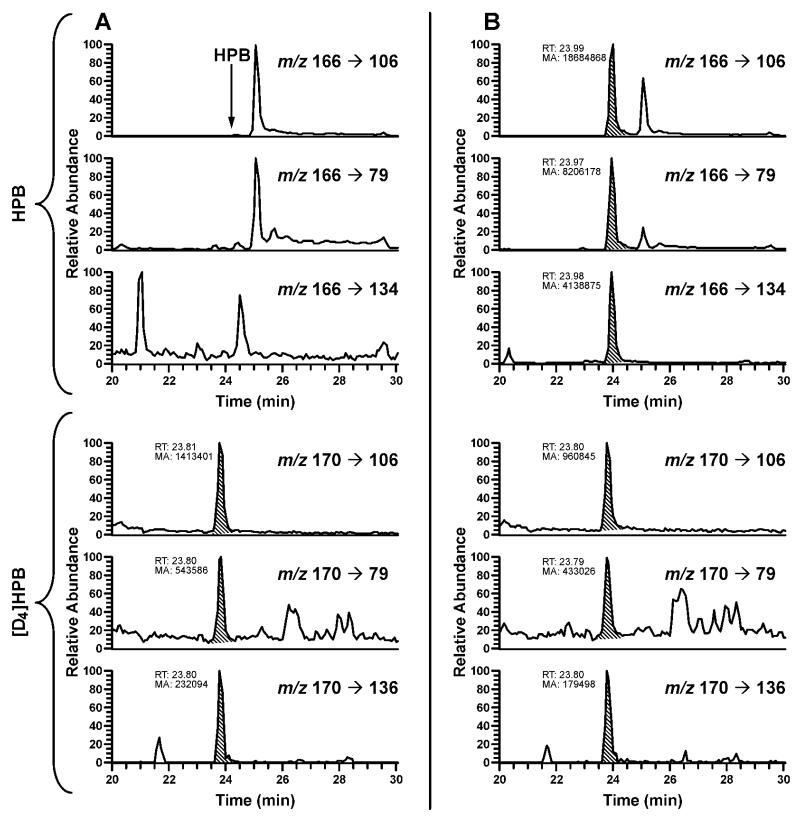
The method was first applied for analysis of oral cell samples from 30 smokers and 15 nonsmokers obtained via the mouthwash collection protocol. Typical LC-ESI-MS/MS traces obtained upon analysis of HPB-releasing DNA adducts in human oral cell DNA are shown in Figure 3. The results are summarized in Table 1. DNA yield in these samples averaged 22 μg, ranging from non-quantifiable (guanine levels below the limit of quantitation upon HPLC analysis of DNA hydrolysate) to 188 μg per sample. The average amount of DNA isolated from the freshly collected nonsmokers’ samples was significantly higher than that obtained from the frozen smokers’ cell pellets received from the NCI Biorepository: 55 vs. 5.3 μg/sample, respectively (P = 0.0001). Additional experiments revealed that freezing cell pellets prior to DNA isolation significantly reduces DNA yields (data not shown). In smokers, HPB-releasing DNA adducts were detected in 20 out of 28 samples with quantifiable DNA yield, averaging 12.0 pmol HPB/mg DNA. Identity of HPB in hydrolyzed oral cell DNA from several smokers was also confirmed with an Orbitrap detector with accurate mass monitoring of the fragment ion [C5H4N–C≡O]+ at m/z 106.02861 (Figure 4). In nonsmokers, the adducts were detected in 3 out of 15 samples, averaging 0.23 pmol/mg DNA. HPB was not detected in negative controls.
Figure 3.
Typical SRM chromatograms obtained upon analysis of HPB-releasing DNA adducts in oral cells from (A) nonsmoker and (B) smoker. For each transition, the peak corresponding to the adduct is shaded. The oral cells were obtained using the mouthwash collection protocol.
Table 1. Levels of HPB-releasing DNA adducts in smokers' and nonsmokers' oral cells collected by mouthwash.
| Subject # | DNA yield, μg |
HPB, pmol/mg DNA |
|---|---|---|
|
Smokers
| ||
| 1 | 0.27 | 21.7 |
| 2 | 0.35 | 20.4 |
| 3 | 0.32 | LOQa |
| 4 | 1.1 | 6.51 |
| 5 | 0.34 | LOD |
| 6 | 0.25 | 17.6 |
| 7 | 3.1 | 4.39 |
| 8 | 22 | 0.29 |
| 9 | 0.03 | 187 |
| 10 | 1.3 | 5.65 |
| 11 | 1.7 | 11.3 |
| 12 | 5.5 | 1.01 |
| 13 | 5.5 | LOD |
| 14 | 0.70 | LOD |
| 15 | 0.60 | 16.1 |
| 16 | 9.0 | LOQ |
| 17 | LOQ | NAb |
| 18 | 30 | 0.09 |
| 19 | 1.9 | 17.6 |
| 20 | 9.1 | 4.93 |
| 21 | LOQ | NA |
| 22 | 24 | LOQ |
| 23 | 8.6 | 0.67 |
| 24 | 2.3 | LOQ |
| 25 | 1.7 | 2.02 |
| 26 | 4.1 | 0.78 |
| 27 | 2.3 | 4.68 |
| 28 | 1.4 | LOQ |
| 29 | 7.4 | 0.88 |
| 30 | 16 | 0.34 |
|
| ||
| Average c | 5.3 | 12.0 |
| SD | 7.7 | 35.1 |
|
Nonsmokers
| ||
| 1 | 46 | LOQ |
| 2 | 188 | LOQ |
| 3 | 32 | LOQ |
| 4 | 19 | LOQ |
| 5 | 77 | LOQ |
| 6 | 72 | 0.78 |
| 7 | 36 | LOQ |
| 8 | 66 | LOQ |
| 9 | 60 | LOQ |
| 10 | 67 | 1.1 |
| 11 | 39 | LOQ |
| 12 | 29 | LOQ |
| 13 | 18 | LOQ |
| 14 | 49 | LOQ |
| 15 | 33 | 1.3 |
|
| ||
| Average | 55 | 0.23 |
| SD | 41 | 0.43 |
LOQ, below the limit of quantitation: 0.02 μg for DNA and 2.4 fmol/sample for HPB.
NA, not analyzed because DNA was not quantifiable.
To calculate average values, % of corresponding LOQ was used for samples in which DNA or HPB were not quantifiable. DNA: 0.01 μg. HPB: for each sample, LOQ (2.4 fmol/sample) was divided by DNA yield, and half of that value was used.
Figure 4.
SRM chromatograms obtained upon analysis of HPB-releasing DNA adducts in the same oral cell DNA sample from a smoker: (A) analyzed by MS/MS and (B) analyzed by an Orbitrap detector with accurate mass monitoring.
For the 30 smoking subjects, matching buccal brushings were also analyzed in order to provide insight into whether buccal mucosa cells are primarily responsible for the remarkably high levels of HPB-releasing DNA adducts measured in some smokers. The amount of DNA isolated from the buccal brushings averaged 0.9 μg, ranging from non-quantifiable to 5.1 μg/sample (Table 2). HPB-releasing DNA adducts were detected in 24 out of 27 samples with quantifiable DNA yield, averaging 44.7 pmol HPB/mg DNA. The levels of adducts in buccal brushings correlated with those in mouthwash samples (R = 0.73, p < 0.0001).
Table 2. Biomarker levels measured in buccal cells and urine of smokers.
| Buccal cells |
Urinary NNAL |
Urinary NNN |
||||
|---|---|---|---|---|---|---|
| Subject # | DNA yield, μg |
HPB, pmol/mg DNA |
Total, pmol/mg creatinine |
% Gluc | Total, pmol/mg creatinine |
% Gluc |
| 1 | LOQ | NA | 0.19 | 61 | 0.027 | 58 |
| 2 | 0.05 | 100 | 1.4 | 67 | 0.085 | 48 |
| 3 | 0.65 | 11.3 | 1.5 | 88 | 0.072 | 85 |
| 4 | 0.11 | 81.3 | 5.0 | 83 | 0.34 | 72 |
| 5 | 0.14 | 25.3 | 1.0 | 72 | 0.046 | 60 |
| 6 | LOQ | NA | 0.44 | 70 | 0.060 | 55 |
| 7 | 0.08 | 61.4 | 0.15 | 60 | 0.018 | 46 |
| 8 | 2.5 | 13.7 | 0.99 | 80 | 0.081 | 87 |
| 9 | 0.15 | 246 | 0.73 | 75 | 0.043 | 59 |
| 10 | 0.05 | 107 | 3.8 | 81 | 0.24 | 81 |
| 11 | 1.7 | 1.97 | 2.7 | 79 | 0.22 | 92 |
| 12 | 0.83 | 5.86 | 0.19 | 63 | 0.035 | 14 |
| 13 | 1.3 | 6.38 | 2.0 | 67 | 0.068 | 73 |
| 14 | 0.04 | 103 | 3.3 | 65 | 0.14 | 51 |
| 15 | 0.55 | 7.15 | 1.3 | 71 | 0.076 | 65 |
| 16 | 2.5 | 2.12 | 1.5 | 78 | 0.069 | 57 |
| 17 | 1.2 | 4.16 | 2.9 | 73 | 4.4 | 33 |
| 18 | 4.9 | 108 | 0.37 | 84 | 0.037 | 57 |
| 19 | 5.1 | 109 | 2.2 | 64 | 0.36 | 57 |
| 20 | 1.1 | LOQ | 0.39 | 56 | 0.034 | 53 |
| 21 | 0.54 | 7.72 | 1.2 | 66 | 0.058 | 48 |
| 22 | LOQ | NA | 1.2 | 64 | 0.066 | 58 |
| 23 | 0.17 | 46.3 | 4.5 | 69 | 0.25 | 70 |
| 24 | 0.25 | 26.1 | 3.6 | 82 | 0.20 | 78 |
| 25 | 1.3 | 2.98 | 2.3 | 69 | 0.12 | 60 |
| 26 | 0.32 | 19.1 | 2.2 | 81 | 0.084 | 70 |
| 27 | 0.22 | 18.4 | 0.55 | 54 | 0.050 | 42 |
| 28 | 0.05 | 84.6 | 2.7 | 80 | 0.12 | 88 |
| 29 | 0.34 | LOQ | 1.4 | 65 | 0.11 | 70 |
| 30 | 0.49 | LOQ | 0.46 | 67 | 0.054 | 71 |
|
| ||||||
| Average c | 0.89 | 45 | 1.7 | 71 | 0.25 | 62 |
| SD | 1.3 | 57 | 1.3 | 8.8 | 0.78 | 17 |
LOQ, below the limit of quantitation: 0.02 μg for DNA and 2.4 fmol/sample for HPB.
NA, not analyzed because DNA was not quantifiable.
To calculate average values, % of corresponding LOQ was used for samples in which DNA or HPB were not quantifiable. DNA: 0.01 μg. HPB: for each sample, LOQ (2.4 fmol/sample) was divided by DNA yield, and half of that value was used.
Effect of DNA yield on HPB quantitation
Because of the large variation in DNA yields across the samples analyzed here, we tested for potential effect of the starting amount of DNA on the quantitation of HPB after its hydrolysis and purification. Six samples containing various amounts of DNA ranging from 0.025 to 100 μg were spiked with 4 pg HPB and subjected to acid hydrolysis and solid phase extraction. The measured concentrations of HPB in these samples produced a coefficient of variation of 5.5%. This experiment was repeated by spiking the same range of DNA amounts with 40 pg HPB; the coefficient of variation for measured HPB in this experiment was 3.1%.
Urinary biomarkers
Urine samples obtained from subjects who were smokers at the time of oral sample collection were analyzed for total and free NNN and NNAL. Total NNN averaged 0.25 pmol/mg creatinine and total NNAL averaged 1.7 pmol/mg creatinine, with 62% of total NNN and 71% of total NNAL being comprised by the corresponding glucuronides (Table 2). There was no correlation between urinary biomarkers and HPB-releasing DNA adducts in oral cells.
DISCUSSION
Formation of HPB-releasing DNA adducts is an important mechanism of NNN- and NNK-induced carcinogenesis in laboratory animals and is believed to play an important role in tobacco carcinogenesis in humans. The measurement of HPB-releasing DNA adducts in tobacco users potentially can provide the most direct link between the cellular exposure to these carcinogens and development of cancer. In this study, we developed a robust and sensitive LC-ESI-MS/MS method for the analysis of HPB-releasing adducts in human oral cells. The method can be applied in studies of individual susceptibility to tobacco-induced cancers in humans.
The original method for HPB-releasing DNA adduct analysis that was developed in our laboratory and used over the years by various research groups included DNA hydrolysis with acid followed by derivatization of the released HPB with pentafluorobenzoyl chloride and analysis of the derivative by gas chromatography-mass spectrometry (GC-MS).12,13,30 In this study, we developed a new method that allows direct measurement of HPB by LC-ESI-MS/MS. Monitoring the formation of three different fragments from the parent HPB ion can be particularly useful for its proper identification in the presence of interfering peaks. The method is accurate, precise, and its sensitivity is in the range of that reported for the GC-MS method.13,30
The levels of HPB-releasing DNA adducts in oral cells from smokers in this study are in agreement with those reported by Heling et al.,22 and are much higher than those found in lung or other tissues of smokers.12,31 The reasons for the differences in the levels of HPB-releasing DNA adducts between oral cells and other tissues are not known. POB-DNA adducts are formed in oral tissues of rats treated with NNN and NNK,21,32 and the ability of human cultured oral tissues to metabolically activate these carcinogens has been demonstrated in vitro.33 It is possible that HPB-releasing DNA adducts in the oral cavity of smokers are formed both as a result of the direct exposure of oral mucosa to freshly generated cigarette smoke and via the circulation of activated NNN and NNK metabolites. The ability of activated metabolites to circulate in the human body is supported by the finding that HPB-releasing hemoglobin adducts are formed from a pyridyloxobutylating NNK metabolite which travelled out of hepatocytes and into red blood cells where it reacted with hemoglobin.34 Even higher levels of the adducts were measured in buccal brushings of smokers (Table 2). Since buccal brushings contain predominantly epithelial buccal cells, while saliva contains both buccal cells and leukocytes,35,36 our results suggest that buccal cells are the main contributor to the measured HPB-releasing DNA adducts in oral samples collected by mouthwash. Further studies are needed to understand the mechanism of HPB-releasing DNA adduct formation in the oral cavity of smokers. Moreover, even though the measurement of individual POB adducts in oral cells was outside the scope of this study, it needs to be conducted in the future. Not all of the POB-DNA adducts with characterized structures are capable of releasing HPB; and structures of additional adducts that contribute to its release are yet to be established.11,37 However, it was established that 7-pyridyloxobutyl-deoxyguanosine (7-POB-dGuo) accounts for 30-35% of the HPB-releasing adducts in DNA pyridyloxobutylated by 4-(acetoxymethylnitrosamino)-1-(3-pyridyl)-1-butanone.11,37 Therefore, given the high levels of HPB-releasing DNA adducts detected in some samples in this study, it is possible that 7-POB-dGuo can be detected in oral cells of some smokers.
Our finding that the levels of HPB-releasing adducts in oral cells of smokers did not correlate with urinary total NNN or total NNAL is in agreement with the previously reported lack of a direct relationship between the levels of HPB-releasing hemoglobin adducts and the extent of exposure to NNN and NNK.38 It is possible that smokers with higher levels of adducts metabolically activate NNN and NNK to reactive electrophiles more efficiently, or repair NNN- and NNK-induced DNA damage less efficiently, than smokers with lower levels of adducts. A combination of these factors with smoking patterns and history could also be responsible for the observed large inter-individual variation of oral HPB-releasing adduct levels among smokers. This large variation may be indicative of the differences in the individual risk of developing cancer in smokers.
In a study by Hözle et al., HPB-releasing DNA adducts were detected in lung tissues of 8 out of 11 nonsmokers, and the levels were significantly lower than those measured in smokers.13 In a later study by the same group, HPB-releasing DNA adduct levels in lung, esophagus, and cardia from tumor-free smokers were not significantly different from values in nonsmokers.39 In contrast to these reports, only a few mouthwash samples collected from nonsmoking volunteers in this study had detectable levels of HPB. This could be due to differences in sample preparation and analysis procedures and in sampling approaches.
While small sample size is a limitation of this study, our primary goal was to develop and characterize a new analytical procedure for the measurement of HPB-releasing DNA adducts and to test its applicability to the analysis of human oral cell DNA. Studies are underway to apply this methodology to larger sets of samples collected from smokers and smokeless tobacco users. Investigation of the HPB-releasing DNA adducts in oral cells after smoking or smokeless tobacco use cessation could also provide important insights into the role of these adducts in tobacco carcinogenesis. Even though we did not observe any effect of the amount of DNA on HPB quantitation, variation in DNA yields across oral samples analyzed in this study is another limitation. It should be noted that, due to differences in oral health status and other individual characteristics, inter-individual variations in oral cell DNA yields most likely cannot be avoided. However, a standardized approach to sample collection and processing should be used in the future for comparisons of HPB-releasing DNA adduct levels among various population groups. Furthermore, analysis of individual POB-DNA adducts should be performed on samples that contain high levels of HPB-releasing DNA adducts.
In summary, we have developed a sensitive and robust LC-ESI-MS/MS method for the analysis of HPB-releasing DNA adducts in human oral cells, and demonstrated applicability of the method to the analysis of mouthwash samples and buccal cell brushings from smokers and nonsmokers. The demonstrated differences in the levels of oral HPB-releasing DNA adducts between smokers and nonsmokers, as well as the large inter-individual variation among smokers, suggest that this biomarker can be used as a measure of tobacco-specific DNA damage. Further studies are needed to examine applicability of this biomarker as an indicator of individual cancer risk in tobacco users.
Acknowledgements
We thank Ryan Baumgartner and Catalin Marian for technical support, and Bob Carlson for editorial assistance.
Funding Sources. This study was supported by NCI grants CA-81301, CA-138338, and N01-PC-64402 (TobPRAC).
Footnotes
Abbreviations: [D4]HPB, [3,3,4,4-D4] 4-hydroxy-1-(3-pyridyl)-1-butanone; GC-MS, gas chromatography-mass spectrometry; HPB, 4-hydroxy-1-(3-pyridyl)-1-butanone; LC-ESIMS/MS, liquid chromatography-electrospray ionization-tandem mass spectrometry; NNAL, 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol; NNN, N’-nitrosonornicotine; NNK, 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone; POB, 4-(3-pyridyl)-4-oxobut-1-yl; SRM, selected reaction monitoring.
References
- 1.Secretan B, Straif K, Baan R, Grosse Y, El Ghissassi F, Bouvard V, Benbrahim-Tallaa L, Guha N, Freeman C, Galichet L, Cogliano V. A review of human carcinogens - Part E: tobacco, areca nut, alcohol, coal smoke, and salted fish. Lancet Oncology. 2009;10:1033–1034. doi: 10.1016/s1470-2045(09)70326-2. [DOI] [PubMed] [Google Scholar]
- 2.Boyle P, Levin P. World Cancer Report, 2008. Lyon; France: 2004. [Google Scholar]
- 3.Hecht SS. Biochemistry, biology, and carcinogenicity of tobacco-specific N-nitrosamines. Chem. Res. Toxicol. 1998;11:559–603. doi: 10.1021/tx980005y. [DOI] [PubMed] [Google Scholar]
- 4.Yuan JM, Koh WP, Murphy SE, Fan Y, Wang R, Carmella SG, Han S, Wickham K, Gao YT, Yu MC, Hecht SS. Urinary levels of tobacco-specific nitrosamine metabolites in relation to lung cancer development in two prospective cohorts of cigarette smokers. Cancer Res. 2009;69:2990–2995. doi: 10.1158/0008-5472.CAN-08-4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yuan JM, Knezevich AD, Wang R, Gao YT, Hecht SS, Stepanov I. Urinary levels of the tobacco-specific carcinogen N’-nitrosonornicotine and its glucuronide are strongly associated with esophageal cancer risk in smokers. Carcinogenesis. 2011;32:1366–1371. doi: 10.1093/carcin/bgr125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Church TR, Anderson KE, Caporaso NE, Geisser MS, Le C, Zhang Y, Benoit AR, Carmella SG, Hecht SS. A prospectively measured serum biomarker for a tobacco-specific carcinogen and lung cancer in smokers. Cancer Epidemiol. Biomarkers & Prev. 2009;18:260–266. doi: 10.1158/1055-9965.EPI-08-0718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mijal RS, Loktionova NA, Vu CC, Pegg AE, Peterson LA. O6-pyridyloxobutylguanine adducts contribute to the mutagenic properties of pyridyloxobutylating agents. Chem. Res. Toxicol. 2005;18:1619–1625. doi: 10.1021/tx050139t. [DOI] [PubMed] [Google Scholar]
- 8.Gowda ASP, Krishnegowda G, Suo Z, Amin S, Spratt TE. Low fidelity bypass of O2-(3-pyridyl)-4-oxobutylthymine, the most persistent bulky adduct produced by the tobacco specific nitrosamine 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone by model DNA polymerases. Chemical Research in Toxicology. 2012;25:1195–1202. doi: 10.1021/tx200483g. [DOI] [PubMed] [Google Scholar]
- 9.Hecht SS, Spratt TE, Trushin N. Evidence for 4-(3-pyridyl)-4-oxobutylation of DNA in F344 rats treated with the tobacco specific nitrosamines 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone and N’-nitrosonornicotine. Carcinogenesis. 1988;9:161–165. doi: 10.1093/carcin/9.1.161. [DOI] [PubMed] [Google Scholar]
- 10.Peterson LA, Hecht SS. O6-Methylguanine is a critical determinant of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone tumorigenesis in A/J mouse lung. Cancer Res. 1991;51:5557–5564. [PubMed] [Google Scholar]
- 11.Wang M, Cheng G, Sturla SJ, Shi Y, McIntee EJ, Villalta PW, Upadhyaya P, Hecht SS. Identification of adducts formed by pyridyloxobutylation of deoxyguanosine and DNA by 4-(acetoxymethylnitrosamino)-1-(3-pyridyl)-1-butanone, a chemically activated form of tobacco-specific carcinogens. Chem. Res. Toxicol. 2003;16:616–626. doi: 10.1021/tx034003b. [DOI] [PubMed] [Google Scholar]
- 12.Foiles PG, Akerkar SA, Carmella SG, Kagan M, Stoner GD, Resau JH, Hecht SS. Mass spectrometric analysis of tobacco-specific nitrosamine-DNA adducts in smokers and nonsmokers. Chem. Res. Toxicol. 1991;4:364–368. doi: 10.1021/tx00021a017. [DOI] [PubMed] [Google Scholar]
- 13.Hölzle D, Schlöbe D, Tricker AR, Richter E. Mass spectrometric analysis of 4-hydroxy-1-(3-pyridyl)-1-butanone-releasing DNA adducts in human lung. Toxicology. 2007;232:277–285. doi: 10.1016/j.tox.2007.01.016. [DOI] [PubMed] [Google Scholar]
- 14.Garcia-Closas M, Egan KM, Abruzzo J, Newcomb PA, Titus-Ernstoff L, Franklin T, Bender PK, Beck JC, LeMarchand L, Lum A, Alavanja M, Hayes RB, Rutter J, Buetow K, Brinton LA, Rothman N. Collection of genomic DNA from adults in epidemiological studies by buccal cytobrush and mouthwash. Cancer Epidemiology, Biomarkers & Prevention. 2001;10:687–696. [PubMed] [Google Scholar]
- 15.Tan D, Goerlitz DS, Dumitrescu RG, Han D, Seillier-Moiseiwitsch F, Spernak SM, Orden RA, Chen J, Goldman R, Shields PG. Associations between cigarette smoking and mitochondrial DNA abnormalities in buccal cells. Carcinogenesis. 2008;29:1170–1177. doi: 10.1093/carcin/bgn034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Borthakur G, Butryee C, Stacewicz-Sapuntzakis M, Bowen PE. Exfoliated buccal mucosa cells as a source of DNA to study oxidative stress. Cancer Epidemiology, Biomarkers & Prevention. 2008;17:212–219. doi: 10.1158/1055-9965.EPI-07-0706. [DOI] [PubMed] [Google Scholar]
- 17.Proia NK, Paszkiewicz GM, Nasca MA, Franke GE, Pauly JL. Smoking and smokeless tobacco-associated human buccal cell mutations and their association with oral cancer--a review. Cancer Epidemiol Biomarkers Prev. 2006;15:1061–1077. doi: 10.1158/1055-9965.EPI-05-0983. [DOI] [PubMed] [Google Scholar]
- 18.Phillips DH. Smoking-related DNA and protein adducts in human tissues. Carcinogenesis. 2002;23:1979–2004. doi: 10.1093/carcin/23.12.1979. [DOI] [PubMed] [Google Scholar]
- 19.Bhutani M, Pathak AK, Fan YH, Liu DD, Lee JJ, Tang H, Kurie JM, Morice RC, Kim ES, Hong WK, Mao L. Oral epithelium as a surrogate tissue for assessing smoking-induced molecular alterations in the lungs. Cancer Prevention Research. 2008;1:39–44. doi: 10.1158/1940-6207.CAPR-08-0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang S, Wang M, Villalta PW, Lindgren BR, Upadhyaya P, Lao Y, Hecht SS. Analysis of pyridyloxobutyl and pyridylhydroxybutyl DNA adducts in extrahepatic tissues of F344 rats treated chronically with 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone and enantiomers of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol. Chem. Res. Toxicol. 2009;22:926–936. doi: 10.1021/tx900015d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang S, Wang M, Villalta PW, Lindgren BR, Lao Y, Hecht SS. Quantitation of pyridyloxobutyl DNA adducts in nasal and oral mucosa of rats treated chronically with enantiomers of N’-nitrosonornicotine. Chem. Res. Toxicol. 2009;22:949–956. doi: 10.1021/tx900040j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heling A-K, Schutte-Borkovec K, Heppel C, Fagerstrom K, Richter E. Pyridyloxobutylated DNA adducts in oral mucosa of nonsmokers, smokers and snuff dippers in relation to other biomarkers of exposure. AACR Meeting Abstracts April 2008. 2008 Abstract 1884. [Google Scholar]
- 23.Wang M, Yu N, Chen L, Villalta PW, Hochalter JB, Hecht SS. Identification of an acetaldehyde adduct in human liver DNA and quantitation as N2-ethyldeoxyguanosine. Chem. Res. Toxicol. 2006;19:319–324. doi: 10.1021/tx0502948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang S, Villalta PW, Wang M, Hecht SS. Analysis of crotonaldehyde- and acetaldehyde-derived 1,N2-propanodeoxyguanosine adducts in DNA from human tissues using liquid chromatography-electrsopray ionization-tandem mass spectrometry. Chem. Res. Toxicol. 2006;19:1386–1392. doi: 10.1021/tx060154d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lao Y, Villalta PW, Sturla SJ, Wang M, Hecht SS. Quantitation of pyridyloxobutyl DNA adducts of tobacco-specific nitrosamines in rat tissue DNA by high performance liquid chromatography-electrospray ionization-tandem mass spectrometry. Chem. Res. Toxicol. 2006;19:674–682. doi: 10.1021/tx050351x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lao Y, Yu N, Kassie F, Villalta PW, Hecht SS. Formation and accumulation of pyridyloxobutyl DNA adducts in F344 rats chronically treated with 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone and enantiomers of its metabolite, 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol. Chem. Res. Toxicol. 2007;20:235–245. doi: 10.1021/tx060207r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang L, Spratt TE, Liu XK, Hecht SS, Pegg AE, Peterson LA. Pyridyloxobutyl adduct O6-[4-oxo-4-(3-pyridyl)butyl]guanine is present in 4-(acetoxymethylnitrosamino)-1-(3-pyridyl)-1-butanone-treated DNA and is a substrate for O6-alkylguanine-DNA alkyltransferase. Chem. Res. Toxicol. 1997;10:562–567. doi: 10.1021/tx9602067. [DOI] [PubMed] [Google Scholar]
- 28.Stepanov I, Carmella SG, Briggs A, Hertsgaard L, Lindgren B, Hatsukami DK, Hecht SS. Presence of the carcinogen N’-nitrosonornicotine in the urine of some users of oral nicotine replacement therapy products. Cancer Res. 2009;69:8236–8240. doi: 10.1158/0008-5472.CAN-09-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stepanov I, Upadhyaya P, Feuer R, Jensen J, Hatsukami DK, Hecht SS. Extensive metabolic activation of the tobacco-specific carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone in smokers. Cancer Epidemiol. Biomarkers & Prev. 2008;17:1764–1773. doi: 10.1158/1055-9965.EPI-07-2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heppel CW, Heling AK, Richter E. Ultrasensitive method for the determination of 4-hydroxy-1-(3-pyridyl)-1-butanone-releasing DNA adducts by gas chromatography-high resolution mass spectrometry in mucosal biopsies of the lower esophagus. Anal. Bioanal. Chem. 2009;393:1525–1530. doi: 10.1007/s00216-008-2566-y. [DOI] [PubMed] [Google Scholar]
- 31.Schlöbe D, Hölzle D, Hatz D, Von Meyer L, Tricker AR, Richter E. 4-Hydroxy-1-(3-pyridyl)-1-butanone-releasing DNA adducts in lung, lower esophagus and cardia of sudden death victims. Toxicology. 2008;245:154–161. doi: 10.1016/j.tox.2007.12.021. [DOI] [PubMed] [Google Scholar]
- 32.Zhang S, Wang M, Villalta PW, Lindgren BR, Upadhyaya P, Lao Y, Hecht SS. Analysis of pyridyloxobutyl and pyridylhydroxybutyl DNA adducts in extrahepatic tissues of F344 rats treated chronically with 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone and enantiomers of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol. Chem. Res. Toxicol. 2009;22:926–936. doi: 10.1021/tx900015d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Castonguay A, Stoner GD, Schut HAJ, Hecht SS. Metabolism of tobacco-specific N-nitrosamines by cultured human tissues. Proc. Natl. Acad. Sci., USA. 1983;80:6694–6697. doi: 10.1073/pnas.80.21.6694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murphy SE, Coletta KA. Two types of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone hemoglobin adducts, from metabolites which migrate into or are formed in red blood cells. Cancer Res. 1993;53:777–783. [PubMed] [Google Scholar]
- 35.Klinkhamer JM. Human oral leukocytes. Journal of the American Society of Periodontists. 1963;1:109–117. [Google Scholar]
- 36.Osswald K, Mittas A, Glei M, Pool-Zobel BL. New revival of an old biomarker: characterization of buccal cells and determination of genetic damage in the isolated fraction of viable leukocytes. Mutat. Res. 2003;544:321–329. doi: 10.1016/j.mrrev.2003.06.008. [DOI] [PubMed] [Google Scholar]
- 37.Hecht SS. DNA adduct formation from tobacco-specific N-nitrosamines. Mutat. Res. 1999;424:127–142. doi: 10.1016/s0027-5107(99)00014-7. [DOI] [PubMed] [Google Scholar]
- 38.Hecht SS, Carmella SG, Foiles PG, Murphy SE. Biomarkers for human uptake and metabolic activation of tobacco-specific nitrosamines. Cancer Res. [Suppl. ] 1994;54:1912s–1917s. [PubMed] [Google Scholar]
- 39.Schlöbe D, Hölzle D, Hatz D, Von Meyer L, Tricker AR, Richter E. 4-Hydroxy-1-(3-pyridyl)-1-butanone-releasing DNA adducts in lung, lower esophagus and cardia of sudden death victims. Toxicology. 2008;245:154–161. doi: 10.1016/j.tox.2007.12.021. [DOI] [PubMed] [Google Scholar]