Abstract
Animal viruses are broadly categorized structurally by the presence or absence of an envelope composed of a lipid-bilayer membrane1, attributes that profoundly affect stability, transmission, and immune recognition. Among those lacking an envelope, the Picornaviridae are a large and diverse family of positive-strand RNA viruses that includes hepatitis A virus (HAV), an ancient human pathogen that remains a common cause of enterically-transmitted hepatitis2–4. HAV infects in a stealth-like manner and replicates efficiently in the liver5. Virus-specific antibodies appear only after 3–4 weeks of infection, and typically herald its resolution3,4. Although unexplained mechanistically, both anti-HAV antibody and inactivated whole-virus vaccines prevent disease when administered as late as 2 weeks after exposure6, when virus replication is well established in the liver5. Here, we show that HAV released from cells is cloaked in host-derived membranes, thereby protecting the virion from antibody-mediated neutralization. These enveloped viruses (“eHAV”) resemble exosomes7, small vesicles that are increasingly recognized to play important roles in intercellular communications. They are fully infectious, sensitive to chloroform extraction, and circulate in the blood of infected humans. Their biogenesis is dependent upon host proteins associated with endosomal-sorting complexes required for transport (ESCRT)8, VPS4B and ALIX. While the hijacking of membranes by HAV facilitates escape from neutralizing antibodies and likely promotes virus spread within the liver, anti-capsid antibodies restrict replication following infection with eHAV, suggesting a possible explanation for post-exposure prophylaxis. Membrane hijacking by HAV blurs the classic distinction between “enveloped” and “nonenveloped” viruses, and has broad implications for mechanisms of viral egress from infected cells as well as host immune responses.
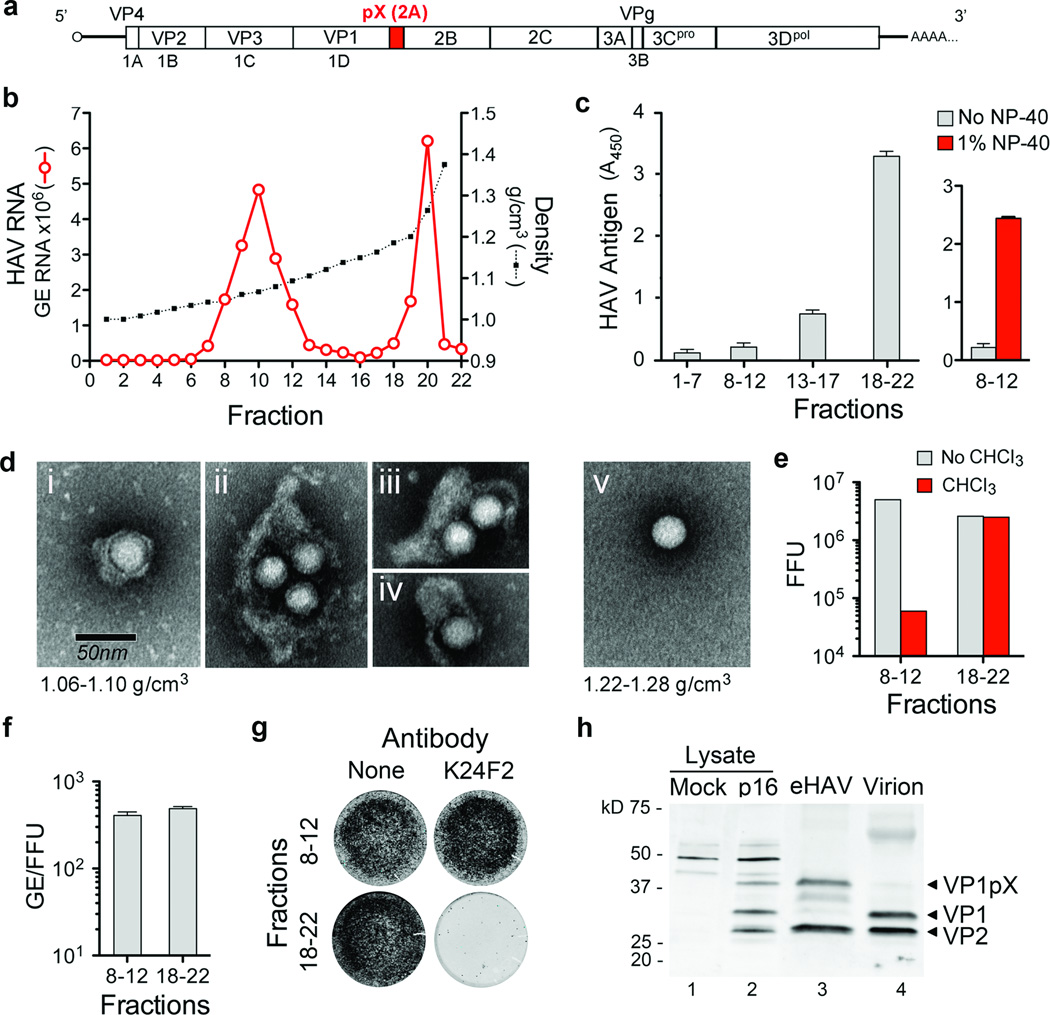
Supernatant fluids of hepatoma cell cultures infected with low passage, noncytopathic HAV9 (Fig. 1a) contain two populations of virus particles that are resolved in isopycnic iodixanol gradients (Fig. 1b). One bands at a low density consistent with membrane association (1.06–1.10 g/cm3, fractions 8–12) and is not detected in a capsid antigen ELISA (Fig. 1c, left), while the other bands at the density expected for picornaviruses (1.22–1.28 g/cm3, fractions 18–22), and is readily detected by ELISA. Electron microscopy of the light fractions revealed numerous virus-like particles enclosed in membranes (Fig. 1d, left, and Supplementary Fig. 1a) with morphology indistinguishable from ~27 nm HAV particles in dense fractions (Fig. 1d, right). These membranous structures ranged from 50–110 nm in diameter, similar to exosomes7, and contained 1–4 virus-like particles (Supplementary Fig. 1b). Consistent with this, viral RNA banded in gradients with capsid protein (VP2) and the exosome-associated protein, flotillin-1 (Supplementary Fig. 1c).
Figure 1. Enveloped particles (eHAV) are the dominant form of virus released from infected cell cultures.
a, HAV genome organization. The polyprotein is depicted as a box with pX highlighted. b, Buoyant density of HAV particles released by Huh-7.5 cells in iodixanol gradients. eHAV bands at 1.06–1.10 g/cm3, whereas nonenveloped HAV bands at 1.22–1.28 g/cm3. GE, genome equivalents. c, (left) HAV capsid antigen was detected by ELISA only in pools of denser fractions from the gradient in b. (right) Capsid antigen was detected in light fractions after treatment with 1% NP-40. Data shown are mean OD450 ± range in duplicate assays. d, Electron microscopic images of negative-stained eHAV (i-iv from fraction 10 in b) and a nonenveloped virion (v, fraction 20 in b). e, Infectious titer of pooled fractions containing eHAV or nonenveloped virions before and after chloroform extraction. FFU, focus-forming units. f, Specific infectivity of pooled fractions containing eHAV or nonenveloped virions, calculated by dividing the HAV RNA copy number (GE, qRT-PCR) by infectious titer (FFU, IR-FIFA). Values shown are means ± range from duplicate RT-PCR reactions. g, eHAV is resistant to neutralization by anti-capsid monoclonal antibody K24F211. Antibody-virus mixtures were incubated for 1 hr at 37 °C, inoculated onto cells for 1 hr, followed by removal of the inoculum, washing ×3 with PBS, and addition of an agarose overlay. Viral antigen was visualized by infra-red immunofluorescence (IR-FIFA)10. h, Immunoblots of HAV capsid proteins (VP1 and VP2) in lysates of mock or HAV-infected cells (lanes 1 and 2), gradient-purified eHAV (lane 3), and chloroform-extracted nonenveloped virions (lane 4).
A modified plaque assay (IR-FIFA)10 revealed the membrane-wrapped particles to be infectious (Fig. 1e) with specific infectivity equivalent to virions (Fig. 1f). Chloroform extraction, a classic method for distinguishing enveloped from nonenveloped viruses, had no effect on standard virions but resulted in a 2 log10 decrease in infectious virus in the light fraction (Fig. 1e and Supplementary Fig. 2a). We call these membrane-wrapped HAV particles “enveloped HAV” (eHAV). Capsid antigen could be detected in the eHAV fraction following treatment with 1% NP-40 (Fig. 1c, right). This shifted the viral particles to an intermediate density in iodixanol gradients (1.15–1.17 g/cm3), but did not destroy infectivity (Supplementary Fig. 2b and 2c). A potent, neutralizing, monoclonal antibody (mAb), K24F211, failed to neutralize eHAV (Fig. 1g), providing further evidence for complete envelopment of the capsid.
In 12 experiments, eHAV represented 79% ±13 s.d. of virus in medium from infected cell cultures. eHAV was released from multiple cell types, and also observed with high passage, cytopathic virus (Supplementary Fig. 3). Gradient-purified eHAV contained mostly mature VP2 (Fig. 1h, lane 3, and Supplementary Fig. 2d), indicating that enveloped virions have undergone maturation cleavage of VP0 to VP4+VP2. However, while nonenveloped virions contained fully processed VP1, eHAV contained primarily unprocessed VP1pX (Fig. 1h, compare lanes 3 and 4). pX is an unusual 8 kD C-terminal extension on VP1 that is unique to HAV among picornaviruses (Fig. 1a). It functions in virion assembly and is processed from VP1 by an unidentified host protease late in the viral lifecycle12,13. pX was protected from proteinase K in eHAV particles, but rendered susceptible to digestion following treatment with NP-40 (Supplementary Fig. 2e). Thus, pX is fully enclosed in membranes. We conclude that most HAV is released enveloped in host membranes, a process we term “membrane hijacking”.
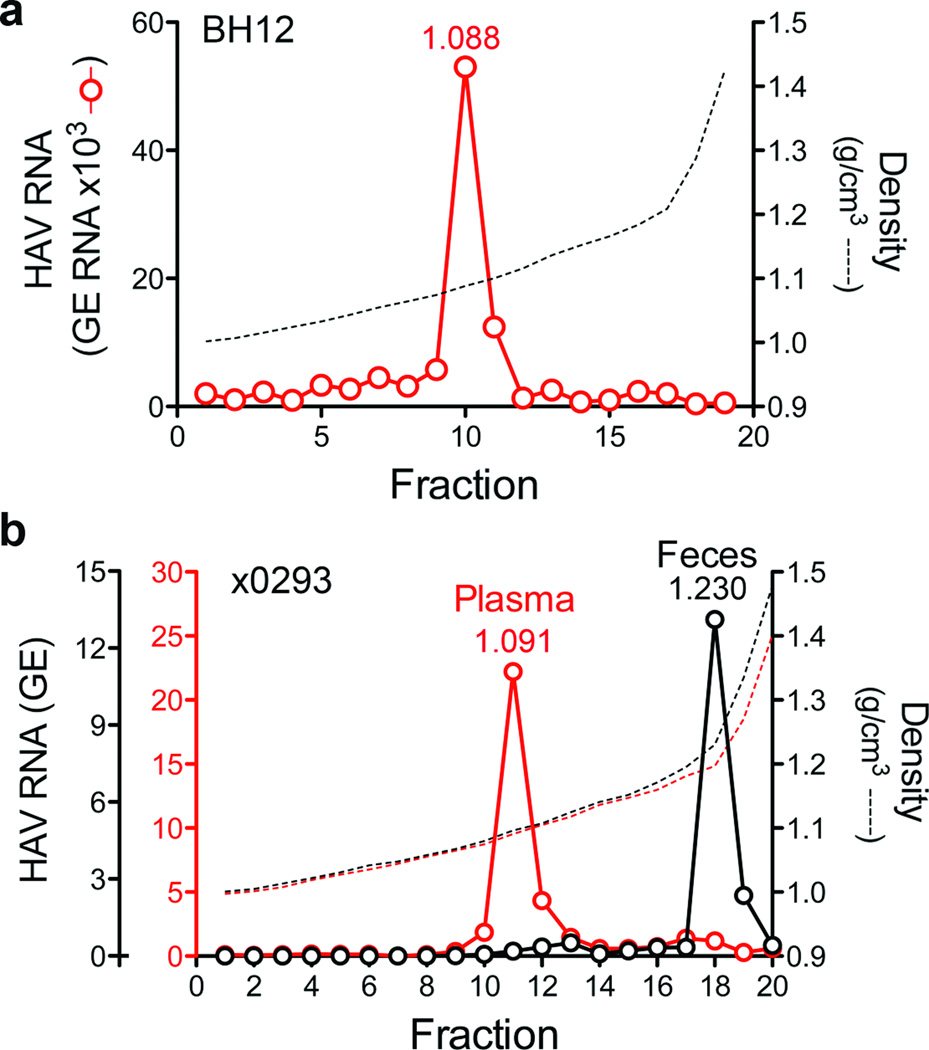
Infectious virus circulating in the blood of infected humans and chimpanzees (Pan troglodytes) possesses the buoyant density of eHAV (Fig. 2a and Supplementary Fig. 4), confirming the relevance of these observations to HAV pathogenesis. However, virus in feces is nonenveloped (Fig. 2b and Supplementary Fig. 4c). Since most fecal virus is produced in the liver14,15, the membrane may be stripped from eHAV during passage through the biliary tract to the gut. However, suspending eHAV in bile minimally altered its buoyant density (Supplementary Fig. 5a).
Figure 2. eHAV circulates in the blood of HAV-infected humans and chimpanzees.
a, Distribution of HAV RNA in an iodixanol gradient loaded with early, acute-phase serum from patient BH12. b, Buoyant density of HAV particles from plasma and feces of an experimentally-infected chimpanzee, ×02935. Fecal HAV RNA is ×105, while plasma RNA is ×102 (also see Supplementary Fig. 4). The low buoyant density of circulating virus was not due to a passive effect of serum, as the buoyant density of nonenveloped virions was not altered by suspension in 90% serum (data not shown).
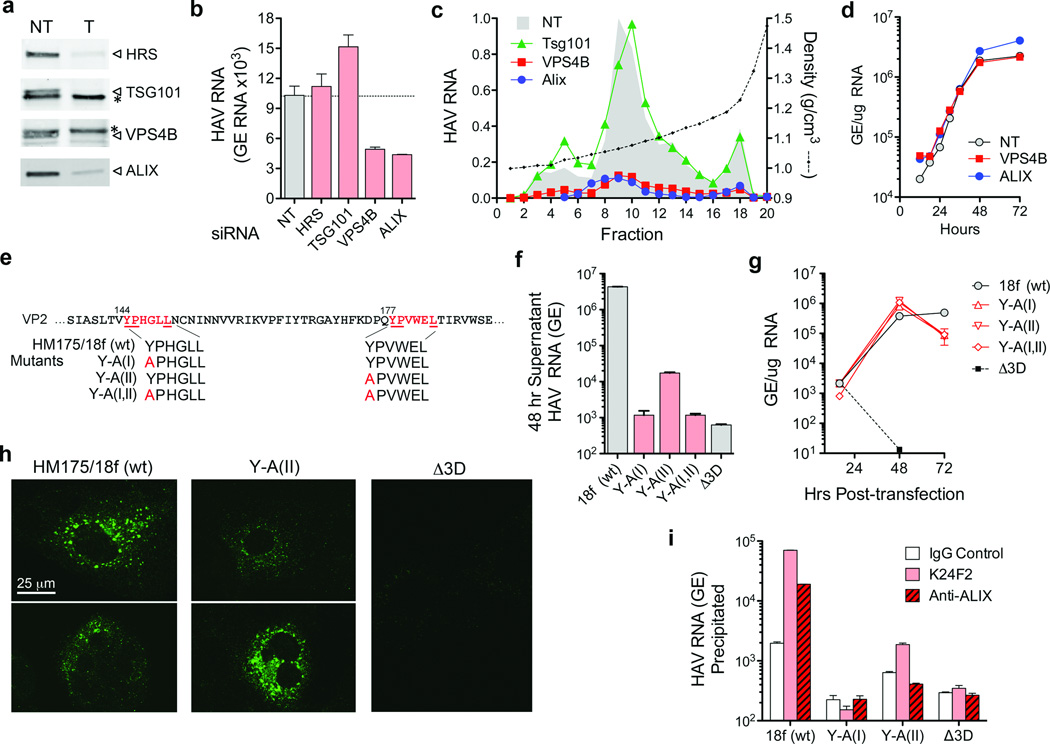
Taylor et al.16 suggested that poliovirus, a distantly related picornavirus, could be released from cells wrapped in a single membrane if entrapped in an autophagosome that fuses with the plasma membrane. However, RNAi knockdown of beclin-1, which mediates autophagosome formation, did not inhibit eHAV release (Supplementary Figs. 6a–b). In contrast, knockdown of VPS4B or ALIX, ESCRT-III associated proteins that facilitate budding of many enveloped viruses17–20, inhibited release of eHAV from cells (Figs. 3a–c, and Supplementary Figs. 6c and 7a), but not viral RNA replication (Fig. 3d), intracellular assembly of capsids, or encapsidation of viral RNA (Supplementary Figs. 7a–c). ESCRT complexes act sequentially to sort and load cargo into multivesicular bodies (MVBs)8. VPS4B is an ATPase that provides energy for the dissolution of ESCRT-III complexes at membrane fission, while ALIX contributes to exosome biogenesis and also facilitates lentivirus budding8,19,21. Knockdown of VPS4B and ALIX inhibited release of both enveloped and nonenveloped HAV (Fig. 3c), suggesting these share a common egress pathway and that nonenveloped virus in extracellular fluids might be derived from eHAV. Knockdown of HRS and TSG101, ESCRT-0 and ESCRT-I proteins involved early in cargo recruitment, did not inhibit eHAV release (Fig. 3b,c). Thus, eHAV release is dependent on ESCRT-associated proteins, but not the entire ESCRT machinery, similar to budding of some enveloped viruses8,18. Although an interaction of ALIX with the ESCRT-III protein, CHMP4B, facilitates budding of human immunodeficiency virus22, CHMP4A, -4B or -4C knockdown minimally reduced eHAV release (Supplementary Fig. 8).
Figure 3. eHAV biogenesis requires ESCRT-associated proteins.
a, siRNA knockdown of ESCRT-associated proteins. Immunoblots of HRS, TSG101, VPS4B, and ALIX in HAV-infected cells 72 hrs after transfection with the indicated gene-specific targeting (T) or non-targeting (NT) control siRNAs. (*), nonspecific protein band. qRT-PCR assays confirmed >80% knockdown of TSG101 and VPS4B (Supplementary Fig. 6c). b, Total viral RNA detected in culture fluids 48–72 hrs after siRNA transfection. c, Relative yield and buoyant density of virus released from siRNA transfected cells (as in panel a) determined in iodixanol gradients. RNA associated with eHAV particles (fractions 8–12) was reduced 85% and 87% by depletion of VPS4B and ALIX, respectively. d, One-step growth of eHAV in cells transfected with NT and gene-specific siRNAs. Huh7.5 cells were transfected with indicated siRNA for 3 days, then infected with gradient-purified eHAV at a multiplicity of infection (m.o.i.) of 20. Cell-associated HAV RNA was assayed by qRT-PCR at the times indicated. Knockdown efficiency was confirmed at the end of the experiment (not shown). e, Tandem YPX3L ALIX-interacting motifs (red typeface) in VP2. Below are the motif sequences in wild-type (wt, HM175/18f) and mutant viral RNAs. f, Total viral RNA released into culture fluids 24–48 hrs after electroporation of Huh-7.5 cells with wild-type and mutant viral RNAs. Δ3D is a replication-incompetent subgenomic replicon RNA with a lethal frame-shift mutation in 3Dpol. g, Intracellular HAV RNA following electroporation of cells with wild-type and VP2 mutant RNAs. h, Confocal microscopy showing K24F2 detection of capsid antigen in cells electroporated 48 hrs previously with wild-type or mutant RNAs. K24F2 recognizes a conformation-dependent assembled neutralization epitope in the viral capsid11. i, Anti-capsid (K24F2) and anti-ALIX (Bethyl) antibody-mediated immunoprecipitation of encapsidated, RNase-resistant viral RNA in detergent-treated lysates of cells electroporated with wild-type or mutant HAV RNAs. RT-PCR data represent mean ± s.e.m. from 2–3 replicate assays; all results are representative of 2–3 independent experiments.
Structural proteins of enveloped viruses interact with ESCRT-related proteins via Pro-rich late (“L”) domain motifs: PPXY, P(S/T)AP, or (L)YPX1/3L8,17. Only two domains in the 2227 amino acid-long HAV polyprotein conform to these motifs: conserved, tandem YPX3L motifs separated by 28 residues in the VP2 capsid protein (Fig. 3e). YPX1/3L motifs mediate interactions with ALIX19, and their presence in VP2 is consistent with a requirement for ALIX. Disrupting either YPX3L motif by substituting Ala for Tyr (Fig. 3e) severely reduced virus release, but did not impair viral RNA replication (Figs. 3f and g). Intracellular capsid antigen was similarly reduced, suggesting the mutations impede capsid assembly, but was detected in cells transfected with Y-A(II) RNA in which the second YPX3L motif is ablated (Fig. 3h). Limited capsid assembly was confirmed by K24F2 precipitation of RNase-protected viral RNA from detergent-treated cell lysates (Fig. 3i). Consistent with a role for ALIX in eHAV biogenesis, anti-ALIX antibody precipitated encapsidated (RNase-protected) viral RNA from lysates of cells transfected with wild-type, but not Y-A(II) RNA (Fig. 3i). Furthermore, ALIX banded at the same density as eHAV in iodixanol gradients (Supplementary Fig. 7d). Although details remain to be elucidated, these data suggest HAV co-opts ALIX to facilitate its envelopment in host membranes. Where this occurs is uncertain, but HAV particles have been observed within cytoplasmic vesicles in infected liver14,23.
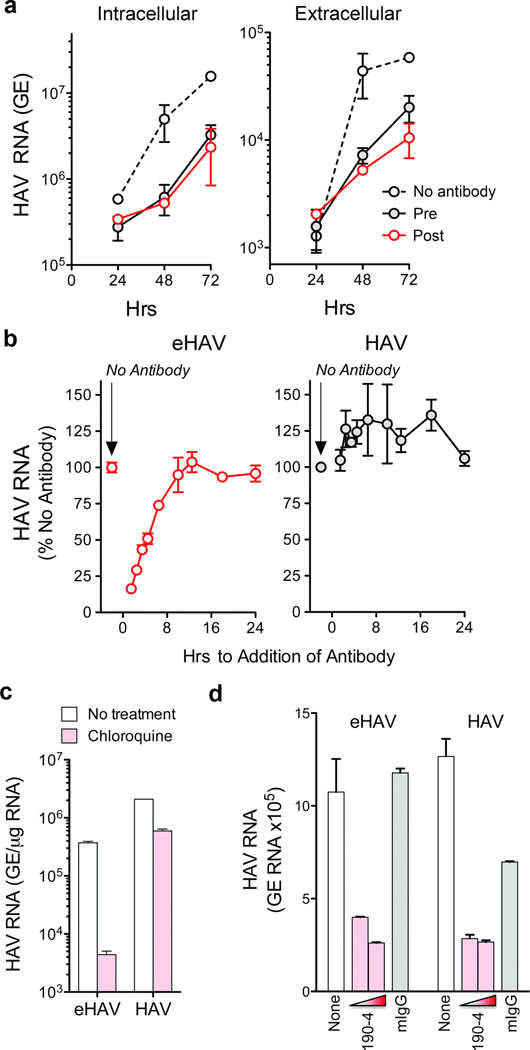
The cloaking of its capsid by membranes allows eHAV to evade neutralizing antibodies (Fig. 1g and Supplementary Fig. 9a) and explains how infectious virus co-exists with anti-HAV in serum15. However, antibodies protect against hepatitis A when passively transferred after HAV replication is well established in the liver5,6. We thus reasoned that eHAV might be neutralized after binding to or entering hepatocytes. Consistent with this, eHAV was neutralized when cells were exposed to antibody either immediately before, or up to 6 hrs after inoculation with virus (Figs. 4a and b). IgG and IgA anti-capsid mAbs neutralized eHAV when added to cultures after removal of the inoculum, but an IgM mAb, H14C4211, failed to do so despite potent neutralizing activity against nonenveloped HAV (Supplementary Fig. 9b). Collectively, these results show capsid is the target for neutralization and that eHAV membranes must rupture prior to neutralization, but not before eHAV is internalized and inaccessible to H14C42 in the medium. This suggests that eHAV may be neutralized following endocytosis. Chloroquine efficiently blocked eHAV (but not nonenveloped HAV) infection (Fig. 4c), indicating that endosomal acidification is important for eHAV entry, while antibody to the HAV receptor, TIM-1 (HAVCR-1)24, inhibited both (Fig. 4d). We found eHAV membranes to be stable at pH 5.0 (Supplementary Fig. 5b), but they could nonetheless be degraded in late endosomes and lysosomes25. If so, this would render the capsid accessible to TIM-1, which undergoes constitutive endocytosis and trafficking to late endosomes and lysosomes26, as well as anti-HAV. Many details of eHAV entry and neutralization remain to be elucidated. However, tripartite motif-containing 21 (TRIM-21), implicated in intracellular neutralization of other viruses27, appears to play no role (Supplementary Fig. 9c).
Figure 4. Extracellular eHAV is resistant to antibody-mediated neutralization, but is neutralized by antibody following infection.
a, Neutralization of eHAV in cells treated with anti-HAV before (pre) or after (post) adsorption of virus. Pre-treated cells were incubated with JC plasma (1:50–1:100) for 1 hr, washed extensively with PBS, then incubated with gradient-purified eHAV (m.o.i. ~6) for 1 hr in the absence of antibody. For post-treatment, JC was added to the medium upon refeeding after removal of the inoculum. Cells were subsequently refed at 24 hr intervals with fresh media containing no antibody. Data shown represent means ± s.e.m. from duplicate cultures, and are representative of 6 independent experiments. b, Adding anti-HAV to medium as late as 6 hrs after removal of the inoculum restricts the replication of eHAV (left panel), but not nonenveloped HAV (right panel). JC antibody was added at intervals after removal of the inoculum, and intracellular viral RNA quantified at 48 hrs. Results shown are mean ± s.e.m. of replicate RT-PCR assays and representative of 3 independent experiments. c, Chloroquine (50 µM) blocks eHAV but not HAV entry. Cells were treated for 30 min prior to inoculating virus, and harvested at 48 hr to assay intracellular HAV RNA. d, Anti-TIM-1 mAb 190-4 blocks entry of both eHAV and HAV. GL37 cells were incubated with 190-4 (10–100 µg/ml) or control IgG (100 µg/ml) for 1 hr at 37 °C, then inoculated with virus for 1 hr. Intracellular HAV RNA was assayed at 72 hrs. RT-PCR data represent mean ± s.e.m. from 2–3 replicate assays; all results are representative of 2–3 independent experiments.
In summary, our findings show how a “nonenveloped” picornavirus promotes its egress in the absence of cell lysis, and reveal a previously unrecognized strategy by which a virus cloaks itself in host membranes to evade neutralizing antibodies. The host membrane enveloping eHAV is likely to facilitate its spread within the liver. Antibodies restrict the replication of eHAV when added to cultures several hours after infection, but further studies will be needed to determine how this happens and whether this can explain why immune globulin and vaccines protect against hepatitis when administered long after exposure, when virus is already circulating in blood5. Most importantly, our results suggest that categorizing viruses into those that are enveloped and those that are not, long a tradition in virology1, is overly simplistic, and that efforts should be made to identify other examples of membrane hijacking by “nonenveloped” viruses.
Methods Summary
Noncytopathic, cell culture-adapted HM175/p16 HAV9 was propagated in Huh-7.5 cells. Reverse molecular genetics studies were carried out with pHM175/18f, a molecular clone of a related cytopathic variant28. Buoyant density was assessed in 8–40% iodixanol (Opti-Prep) gradients centrifuged at 141,000 × g for 48 hrs at 4 °C. Viral RNA was measured by qRT-PCR with primers targeting the 5’-untranslated region. Infectivity was quantified by infrared fluorescence immunofocus assay (IR-FIFA)10. For RNAi studies, cells were transfected with SmartPool siRNAs (Dharmacon) and samples collected 48–72 hrs later for viral RNA quantification. To analyze VP2-ALIX interactions, cell lysates were prepared 48 hrs after electroporation of mutant and wild-type viral RNAs, treated with RNase, and immunoprecipitated. RNA extracted from immunoprecipitates was assayed by HAV-specific qRT-PCR. For intracellular neutralization, cells were incubated with eHAV for 1 hr at 37 °C, then washed extensively. Antibodies were added at intervals, and intra- and extracellular HAV RNA quantified at 48–72 hrs. For standard neutralization assays, virus was incubated with antibodies for 1 hr at 37 °C, then inoculated onto cells.
Additional Methods
Reagents and antibodies
Chemical reagents were purchased from Sigma unless otherwise noted. A human convalescent plasma (JC plasma) sample with high-titer anti-HAV antibody, collected 90 days after the onset of acute hepatitis A, was described previously11,29. Other antibodies were obtained from: anti-TSG101 (Abcam), anti-HRS (Bethyl) anti-ALIX (Pierce and Bethyl), anti-VPS4B (Sigma), anti-beclin-1 (Cell Signaling), anti-flotillin-1 (BD Biosciences), and anti-HAV mAbs K2-4F2 and K3-2F2 (Commonwealth Serum Laboratories, Victoria, Australia). The sources of other anti-HAV mAbs have been described previously11. Anti-TIM-1 mAb 190-4 was a gift from Gerardo Kaplan (Center for Biologics Evaluation and Research, U.S. Food and Drug Administration). Guinea pig anti-peptide antibodies to HAV VP1 (residues 211–228) and VP2 (residues 240–259) were a gift from David Sangar (formerly Wellcome Biotech).
Cells and viruses
Huh-7.5 and HepG2 (both human hepatoma cells), MRC-5 (human lung fibroblasts), FRhK-4 (fetal rhesus monkey kidney cells), BS-C-1, and GL3730 (both African green monkey kidney) cells were cultured in DMEM supplemented with 10% FBS. Cell-culture adapted variants of the HM175 strain of HAV, HM175/p169,31 (noncytopathic) and HM175/18f28,32 (rapidly replicating, cytopathic), have been described previously9,32. Infectious virus titers were determined by a modification of the IR-FIFA described by Counihan et al.10 carried out in FRhK-4 cells.
Human samples and clinical information
All human samples and related clinical information were obtained with the approval of the Institutional Review Boards of the Seoul National University Bundang Hospital and the University of North Carolina at Chapel Hill.
Chimpanzee samples
All chimpanzee materials were archived samples collected during previous studies5 prior to January, 2011.
HAV capsid antigen ELISA assay
HAV capsid antigen was detected using an ELISA modification of a previously described radioimmunoassay33. Immobilized antigen captured by JC antibody was detected by sequential incubation of ELISA plates with K24F2 (1:1000 dilution) and HRP-conjugated goat-anti-mouse antibody (1:10,000) at room temperature for 1 hr each. Following washing with PBS-T and substrate (TMB) addition, the OD450 was determined with a Synergy 2 (BioTek) microplate reader.
qRT-PCR assay for HAV RNA
Total cellular RNA was extracted from cell lysates with the RNeasy Kit (Qiagen); RNA from culture supernatants and gradient fractions was extracted with the QiaAmp viral RNA Isolation Kit (Qiagen). HAV RNA was quantified by real-time qRT-PCR against a synthetic RNA standard as previously described5. Intracellular viral RNA was normalized to total RNA abundance or GAPDH mRNA level determined in a SYBR green one-step qRT-PCR assay (Bio-Rad). For experiments involving electroporation of synthetic viral RNAs, a two-step qRT-PCR procedure was carried out in which cDNA was synthesized using oligo(dT)20 as primer, followed by RNaseH digestion and qPCR assay with primers targeting the HAV 5’ untranslated RNA segment.
Immunoblotting
Cells were lysed with CelLytic Lysis Buffer (Invitrogen) in the presence of protease inhibitor cocktail (Roche). Proteins were extracted from gradient fractions by TCA precipitation. Immunoblots were carried out using standard procedures and the indicated antibodies. Protein bands were visualized with an Odyssey Infrared Imaging System (LI-COR Biosciences).
Immunofluorescence microscopy
Huh-7.5 cells seeded on 4-well chamber slides were fixed with 4% paraformaldehyde, permeabilized with 0.2% Triton X-100 in PBS, and blocked with 10% goat serum. Cells were then incubated with mAb K24F2 (1:300 dilution in 1% BSA) for 1 hr at room temperature, washed, and incubated with FITC-conjugated goat-anti-mouse antibody (1:300, Southern Biotech) for 1 hr. Images were collected using a Zeiss 510 Meta laser-scanning confocal microscope.
Isopycnic gradient centrifugation of virus
Cell culture supernatant fluids were centrifuged at 1,000 × g for 10 min at 4 °C to remove cells and debris, further clarified by 2× centrifugation at 10,000 × g for 30 min, and concentrated by ultracentrifugation at 100,000 × g for 1 hr at 4 °C. The resulting pellet was resuspended in PBS, loaded onto a 8–40% iodixanol (Opti-Prep) step gradient, and centrifuged at 141,000 × g in a Superspin 630 rotor for 48 hrs at 4 °C in a Sorvall Ultra-80 ultracentrifuge. Approximately 20 fractions were collected from the top of the gradient; density was determined using a refractometer.
Rate-zonal centrifugation of HAV in sucrose gradients
Lysates were prepared from HAV (HM175/p16)-infected Huh-7.5 cells, loaded onto a 7.5–45% (wt/vol) sucrose gradient in Hank’s balanced buffer, and centrifuged at 4°C in a Beckman SW55i rotor for 3 hrs at 42,000 rpm (167,000 × g). Fractions were assayed by HAV antigen ELISA and HAV RNA-specific qRT-PCR to identify pentamers, procapsids and mature HAV virions.
Immunoprecipitation of HAV
Cells (2 × 106) were lysed in the presence of detergent with CelLytic lysis buffer (Invitrogen), treated with RNase A 50 µg/ml (Qiagen) and RNase T1 25 U/ml (Ambion) at 37 °C for 15 min, and incubated with antigen-specific and isotype-matched antibodies overnight at 4 °C. Protein G beads were added and incubated for additional 2 hrs. Beads were extensively washed with lysis buffer and captured immunoprecipitates subjected to RNA extraction for qRT-PCR, or immunoblotting, as above.
Virus isolation from iodixanol gradient fractions
Huh-7.5 cells in 12-well culture plates were inoculated with 10–100 µl peak iodixanol gradient fractions containing eHAV recovered from human or chimpanzee serum/plasma. Cells were refed weekly, and split 1:3 at 2 weeks. RNA was extracted from supernatant culture fluids 8, 23, and 30 days after inoculation and assayed for HAV RNA by qRT-PCR as described above. To confirm viral isolates, RNA was extracted from supernatant culture fluids using the QIAamp UltraSens Virus Kit (Qiagen) and cDNA transcribed using random primers (Superscript III First-Strand Synthesis System, Invitrogen). HAV cDNA was amplified by nested RT-PCR with primer sets bracketing the VP1pX-2B junction (nts 2908–3306 in wild-type HM-175) (PrimeSTAR, TaKaRa). Second-round PCR products were isolated from agarose gels (Mini-Elute Gel Extraction kit, Qiagen) and directly sequenced.
Generation and characterization of HAV VP2 mutants
Mutagenesis of the infectious cDNA clone, pT7-HM175/18f, containing the complete sequence of HM175/18f virus28, was accomplished using the Quick Change II Site-directed Mutagenesis Kit (Agilent). PCR-derived fragments were sequenced to ensure sequence fidelity. Plasmids were linearized and viral RNA synthesized with the T7 Megascript Kit (Ambion). Synthetic viral RNA (10 µg) was mixed with 5 × 106 Huh-7.5 cells in a 4-mm cuvette and pulsed once at 250 V, 950 µF, and 100 Ω in a Gene Pulser Xcell Total electroporation system (Bio-Rad). Cells and supernatants were harvested at subsequent intervals for RNA extraction and qRT-PCR assay.
Electron Microscopy
Gradient fractions (2.5 µl each) were spread onto the surface of a glow-discharged 400 mesh Formvar/carbon coated copper grid and allow to adsorb for 5 minutes. The grid was inverted onto a droplet of 1% glutaraldehyde in 0.15 M sodium phosphate, pH 7.4, for 1 min, followed by rinsing with deionized water and staining with 3% ammonium molybdate, pH 7.0. The grid was air-dried before examination in a LEO EM910 transmission electron microscope (Carl Zeiss SMT, Inc., Peabody, MA) operating at 80kV. Digital images were acquired using a Gatan Orius SC1000 CCD Digital Camera and Digital Micrograph 3.11.0 software (Gatan, Inc., Pleasanton, CA).
siRNA knockdown of protein expression
Huh-7.5 cells were transfected with gene-specific SmartPool siRNAs (Dharmacon, see Supplementary Table 1) using SilentFect (Bio-Rad) transfection reagent. Knockdown efficiency was determined at 48–72 hrs by qRT-PCR (SYBR Green One-step qRT-PCR Kit, BioRad) measurement of mRNAs, or by immunoblotting as described.
Supplementary Material
Acknowledgements
We thank Mara Duncan for helpful discussions and staff of the Michael Hooker Microscopy Facility for assistance with confocal microscopy. We are grateful to Gerardo Kaplan for the gift of 190-4 antibody and GL-37 cells, Michael Fried for anti-HAV-negative human sera, Anthony Blikslager for porcine bile, and Charles Rice for Huh-7.5 cells. This work was supported in part by a grant from the National Institute of Health (R01-AI103083) and the University Cancer Research Fund.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions. Z.F., C.H., K.L.M., F.H., V.M. and LF-P. designed experiments, collected and analyzed data; S-H.J., C.W., and R.L. provided unique human and primate samples for analysis, Z.F. and S.M.L. conceived the study, analyzed data, and wrote the paper. All authors discussed the results and commented on the manuscript.
References
- 1.Harrison SC. In: Fields Virology. Knipe DM, et al., editors. Lippincott Williams & Wilkins; 2007. pp. 59–98. Ch. 3. [Google Scholar]
- 2.Feng Z, Lemon SM. In: The Picornaviruses. Ehrenfeld E, Domingo E, Roos RP, editors. Washington, DC: ASM Press; 2010. pp. 383–396. Ch. 25. [Google Scholar]
- 3.Lemon SM. Type A viral hepatitis: new developments in an old disease. New Engl. J. Med. 1985;313:1059–1067. doi: 10.1056/NEJM198510243131706. [DOI] [PubMed] [Google Scholar]
- 4.Martin A, Lemon SM. In: Hepatitis Viruses. Ou J, editor. Kluwer Academic Publishers; 2002. pp. 23–50. [Google Scholar]
- 5.Lanford RE, et al. Acute hepatitis A virus infection is associated with a limited type I interferon response and persistence of intrahepatic viral RNA. Proc Natl Acad Sci U S A. 2011;108:11223–11228. doi: 10.1073/pnas.1101939108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Victor JC, et al. Hepatitis A vaccine versus immune globulin for postexposure prophylaxis. N Engl J Med. 2007;357:1685–1694. doi: 10.1056/NEJMoa070546. [DOI] [PubMed] [Google Scholar]
- 7.Bobrie A, Colombo M, Raposo G, Thery C. Exosome secretion: molecular mechanisms and roles in immune responses. Traffic. 2011;12:1659–1668. doi: 10.1111/j.1600-0854.2011.01225.x. [DOI] [PubMed] [Google Scholar]
- 8.Hurley JH. The ESCRT complexes. Crit Rev Biochem Mol Biol. 2010;45:463–487. doi: 10.3109/10409238.2010.502516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jansen RW, Newbold JE, Lemon SM. Complete nucleotide sequence of a cell culture-adapted variant of hepatitis A virus: comparison with wild-type virus with restricted capacity for in vitro replication. Virology. 1988;163:299–307. doi: 10.1016/0042-6822(88)90270-x. [DOI] [PubMed] [Google Scholar]
- 10.Counihan NA, Daniel LM, Chojnacki J, Anderson DA. Infrared fluorescent immunofocus assay (IR-FIFA) for the quantitation of non-cytopathic and minimally cytopathic viruses. J Virol Methods. 2006;133:62–69. doi: 10.1016/j.jviromet.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 11.Ping LH, Lemon SM. Antigenic structure of human hepatitis A virus defined by analysis of escape mutants selected against murine monoclonal antibodies. J Virol. 1992;66:2208–2216. doi: 10.1128/jvi.66.4.2208-2216.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohen L, Benichou D, Martin A. Analysis of deletion mutants indicates that the 2A polypeptide of hepatitis A virus participates in virion morphogenesis. J Virol. 2002;76:7495–7505. doi: 10.1128/JVI.76.15.7495-7505.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Graff J, et al. Hepatitis A virus capsid protein VP1 has a heterogeneous C terminus. J. Virol. 1999;73:6015–6023. doi: 10.1128/jvi.73.7.6015-6023.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schulman AN, et al. Hepatitis A antigen particles in liver, bile, and stool of chimpanzees. J Infect Dis. 1976;134:80–84. doi: 10.1093/infdis/134.1.80. [DOI] [PubMed] [Google Scholar]
- 15.Asher LVS, et al. Pathogenesis of hepatitis A in orally inoculated owl monkeys (Aotus trivergatus) J Med Virol. 1995;47:260–268. doi: 10.1002/jmv.1890470312. [DOI] [PubMed] [Google Scholar]
- 16.Taylor MP, Burgon TB, Kirkegaard K, Jackson WT. Role of microtubules in extracellular release of poliovirus. J Virol. 2009;83:6599–6609. doi: 10.1128/JVI.01819-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ren X, Hurley JH. Proline-rich regions and motifs in trafficking: from ESCRT interaction to viral exploitation. Traffic. 2011;12:1282–1290. doi: 10.1111/j.1600-0854.2011.01208.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen BJ, Lamb RA. Mechanisms for enveloped virus budding: can some viruses do without an ESCRT? Virology. 2008;372:221–232. doi: 10.1016/j.virol.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sette P, et al. The Phe105 loop of Alix Bro1 domain plays a key role in HIV-1 release. Structure. 2011;19:1485–1495. doi: 10.1016/j.str.2011.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fujii K, Hurley JH, Freed EO. Beyond Tsg101: the role of Alix in 'ESCRTing' HIV-1. Nat Rev Microbiol. 2007;5:912–916. doi: 10.1038/nrmicro1790. [DOI] [PubMed] [Google Scholar]
- 21.Baietti MF, et al. Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat Cell Biol. 2012;14:677–685. doi: 10.1038/ncb2502. [DOI] [PubMed] [Google Scholar]
- 22.Morita E, et al. ESCRT-III protein requirements for HIV-1 budding. Cell Host Microbe. 2011;9:235–242. doi: 10.1016/j.chom.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shimizu YK, et al. Detection of hepatitis A antigen in human liver. Infect. Immun. 1982;36:320–324. doi: 10.1128/iai.36.1.320-324.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feigelstock D, et al. The human homolog of HAVcr-1 codes for a hepatitis A virus cellular receptor. J. Virol. 1998;72:6621–6628. doi: 10.1128/jvi.72.8.6621-6628.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kolter T, Sandhoff K. Lysosomal degradation of membrane lipids. FEBS Lett. 2010;584:1700–1712. doi: 10.1016/j.febslet.2009.10.021. [DOI] [PubMed] [Google Scholar]
- 26.Balasubramanian S, et al. TIM family proteins promote the lysosomal degradation of the nuclear receptor NUR77. Sci Signal. 2012;5:ra90. doi: 10.1126/scisignal.2003200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mallery DL, et al. Antibodies mediate intracellular immunity through tripartite motif-containing 21 (TRIM21) Proc Natl Acad Sci U S A. 2010;107:19985–19990. doi: 10.1073/pnas.1014074107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang HC, et al. An infectious cDNA clone of a cytopathic hepatitis A virus: Genomic regions associated with rapid replication and cytopathic effect. Virology. 1995;212:686–697. doi: 10.1006/viro.1995.1526. [DOI] [PubMed] [Google Scholar]
Additional References
- 29.Lemon SM, et al. Specific immunoglobulin M response to hepatitis A virus determined by solid-phase radioimmunoassay. Infect. Immun. 1980;28:927–936. doi: 10.1128/iai.28.3.927-936.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feigelstock D, Thompson P, Mattoo P, Kaplan GG. Polymorphisms of the hepatitis A virus cellular receptor 1 in African green monkey kidney cells result in antigenic variants that do not react with protective monoclonal antibody 190/4. J. Virol. 1998;72:6218–6222. doi: 10.1128/jvi.72.7.6218-6222.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taylor KL, et al. Attenuation phenotype of a cell culture-adapted variant of hepatitis A virus (HM175/p16) in susceptible new world owl monkeys. J Infect Dis. 1993;168:592–601. doi: 10.1093/infdis/168.3.592. [DOI] [PubMed] [Google Scholar]
- 32.Lemon SM, et al. Antigenic and genetic variation in cytopathic hepatitis A virus variants arising during persistent infection: evidence for genetic recombination. J. Virol. 1991;65:2056–2065. doi: 10.1128/jvi.65.4.2056-2065.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lemon SM, et al. Transmission of hepatitis A virus among recently captured Panamanian owl monkeys. J. Med. Virol. 1982;10:25–36. doi: 10.1002/jmv.1890100105. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.