Abstract
Recent data from multiple organisms indicate that γ-tubulin has essential, but incompletely defined, functions in addition to nucleating microtubule assembly. To investigate these functions, we examined the phenotype of mipAD159, a cold-sensitive allele of the γ-tubulin gene of Aspergillus nidulans. Immunofluorescence microscopy of synchronized material revealed that at a restrictive temperature mipAD159 does not inhibit mitotic spindle formation. Anaphase A was inhibited in many nuclei, however, and after a slight delay in mitosis (∼6% of the cell cycle period), most nuclei reentered interphase without dividing. In vivo observations of chromosomes at a restrictive temperature revealed that mipAD159 caused a failure of the coordination of late mitotic events (anaphase A, anaphase B, and chromosomal disjunction) and nuclei reentered interphase quickly even though mitosis was not completed successfully. Time-lapse microscopy also revealed that transient mitotic spindle abnormalities, in particular bent spindles, were more prevalent in mipAD159 strains than in controls. In experiments in which microtubules were depolymerized with benomyl, mipAD159 nuclei exited mitosis significantly more quickly (as judged by chromosomal condensation) than nuclei in a control strain. These data reveal that γ-tubulin has an essential role in the coordination of late mitotic events, and a microtubule-independent function in mitotic checkpoint control.
INTRODUCTION
γ-Tubulin has a well-established role in the nucleation of mitotic spindle microtubules (reviewed by Wiese and Zheng, 1999; Oakley, 2000), but recent data indicate that it, and its associated proteins, may have other essential, but as yet incompletely defined, functions (Paluh et al., 2000; Vogel and Snyder, 2000; Hendrickson et al., 2001; Jung et al., 2001; Prigozhina et al., 2001; Sampaio et al., 2001; Vogel et al., 2001; Yao et al., 2001; Fujita et al., 2002; Vardy et al., 2002). In general, these data are from analyses of γ-tubulin mutations or mutations in genes that encode proteins that interact with γ-tubulin. The mutations arrest growth or have significant phenotypic effects under some conditions but do not cause an apparent inhibition of microtubule assembly. The phenotypes of the mutations are often striking. Microtubule dynamics, lengths, or organization is altered in some instances (Paluh et al., 2000; Vogel and Snyder, 2000; Jung et al., 2001; Vogel et al., 2001; Fujita et al., 2002). Organization of microtubules into a functional spindle (and, in particular, the establishment of spindle bipolarity) is compromised in others (Prigozhina et al., 2001; Sampaio et al., 2001). In still others, checkpoint control and, in particular, the relationship between mitosis and cytokinesis, seem to be altered (Hendrickson et al., 2001; Sampaio et al., 2001; Vardy et al., 2002). Given the well-established role of γ-tubulin in microtubule nucleation, however, there has been some question as to whether these phenotypes are independent of the role of γ-tubulin in microtubule nucleation or just unobvious manifestations of alterations of microtubule nucleation. For example, at equilibrium a reduction in the number of microtubule nucleation sites would lead to fewer but longer microtubules. In addition, reduction in the nucleation of mitotic spindle microtubules would clearly have checkpoint effects. One would expect, however, that reduced microtubule nucleation would activate the spindle checkpoint and cause a mitotic block, whereas, in at least some γ-tubulin mutants, nuclei proceed through an abnormal mitosis (Hendrickson et al., 2001).
Conditionally lethal mutations in the mipA, γ-tubulin, gene of Aspergillus nidulans created by charged-to-alanine scanning mutagenesis (Jung et al., 2001) are potentially powerful tools for studying γ-tubulin functions. Among these mutant alleles, some have robust but highly abnormal mitotic spindles under restrictive conditions and normal localization of γ-tubulin to the spindle pole bodies (SPBs). These data have raised the possibility that these alleles might inhibit an essential mitotic function of γ-tubulin other than microtubule nucleation. We were, consequently, interested in determining the precise effects of the mutant alleles on the progression through mitosis.
In this study, we focus on mipAD159, a cold-sensitive allele created by the replacement of aspartic acid at amino acid 159 and arginine at amino acid 160 with alanines (Jung et al., 2001). Under restrictive conditions, mipAD159 causes spindle abnormalities that include split, multipolar, and bent spindles (Jung et al., 2001), and large, obviously polyploid, nuclei are common. Although observations of material grown under restrictive conditions are valuable, it is difficult to know whether the defects seen are primary effects of mipAD159 or secondary effects caused by one or more failed mitoses. For example, multipolar spindles could be caused directly by mipAD159 or they could be caused by multiple SPBs resulting from progression through the cell cycle after a failed mitosis.
We have used reciprocal block experiments and live imaging to determine the primary defects caused by mipAD159. We have found that mipAD159 does not inhibit mitotic spindle formation, but it does disrupt the coordination of late mitotic events, resulting in severe mitotic defects. Nuclei exit mitosis quickly even if mitosis has not been completed successfully. In addition, mipAD159 accelerates mitotic exit even if microtubules are eliminated by benomyl. These results indicate that γ-tubulin has an important function in the coordination of late mitotic events and that it has a microtubule-independent role in establishing or maintaining a mitotic checkpoint block. To account for these and related data, we propose a model for how γ-tubulin and polar microtubule organizing centers, such as the centrosome and spindle pole body, function in the regulation of mitosis and the cell cycle.
MATERIALS AND METHODS
Strains and Media
UVts911 (also called NIM911) (chaA1, nimT23, adE20, biA1, wA2, cnxE16, sC12, methG1, nicA2, lacA1, choA1) (Morris, 1976) was used as a mipA+/nimT23 strain. LO715 was used as a mipAD159/nimT23 strain. It is a pyrG+ segregant of a cross between UVts911 and LO699 (mipAD159, pyrG89, fwA1, pabaA1, uaY9) (Jung et al., 2001). Strains were grown on YAG medium (5 g/l yeast extract, 20 g/l d-glucose, 15 g/l agar) supplemented with 50 μg/ml adenine HCl, 0.02 μg/ml biotin, 300 μg/ml ammonium sulfate, 50 μg/ml methionine, 2 μg/ml niacin, and 20 μg/ml choline or FYG (5 g/l yeast extract, 20 g/l d-glucose, 25 g/l pretested Burtonite 44c; TIC Gums, Belcamp, MD) with the same supplements. Experiments were carried out in YG medium (5 g/l yeast extract, 20 g/l d-glucose) with the same supplements. Comparisons of growth rates of LO715, UVts911, and LO635 (mipAD159 pabaA1, fwA1) on solid media at various temperatures indicated that there were no synthetic interactions between nimT23 and mipAD159.
Three strains were used for live imaging of chromosomes. All are progeny of a cross between LO715 and GΔ5 (yA2, riboB2, pabaA1, hhoA-gfp, hhoA::pyr4; Ramon et al., 2000), and all carry the histone H1 green fluorescent protein (GFP) fusion. LO883 carries nimT23 and is mipA+, LO905 and LO906 carry nimT23 and mipAD159. LO905 was used in some experiments and LO906 in others, and results with the two strains were indistinguishable.
Three strains were used for live imaging of mitotic spindles. All carry a fusion of the GFP to the N terminus of the tubA, α-tubulin gene. GFP-tub7 (Han et al., 2001) served as a mipA+ control. LO669 and LO672 carry mipAD159. They were constructed by crossing the original evictant carrying mipAD159 (Jung et al., 2001) to GFP-tub7. Both carry pyrG89, which is complemented by the Neurospora crassa pyr4 gene used in the GFP-tubA construction. In addition, LO672 carries pyroA4.
Microscopy
Growth of cells for microscopy, temperature shift procedures, fixation, and preparation for immunofluorescence microscopy were as described previously (Prigozhina et al., 2001). A mouse monoclonal anti-β-tubulin antibody (tu27b, originally obtained from Dr. Lester Binder, Northwestern University School of Medicine) and an affinity-purified rabbit polyclonal anti-γ-tubulin antibody (Oakley et al., 1990) were used as primary antibodies. We used fluorescein isothiocyanate-conjugated or Alexa Fluor 488-conjugated goat anti-mouse secondary antibodies (Jackson ImmunoResearch Laboratories, West Grove, PA and Molecular Probes, Eugene, OR) and CY3-conjugated goat anti-rabbit secondary antibodies (Jackson ImmunoResearch Laboratories).
A Nikon Eclipse E800 microscope was used for most fluorescence observations and image capture. Images were captured with a Princeton Instruments MicroMax charge-coupled device camera driven by IPLab Spectrum software on a MacIntosh computer. An Olympus IX71 microscope was also used for fluorescence microscopy in which case images were captured with a Hamamatsu Orca ER camera driven by Slidebook software (Intelligent Imaging Innovations, Denver, CO). Images were processed with NIH Image, Adobe Photoshop, or Slidebook, and composite figures were prepared with CorelDraw 8 (Mac). Graphing was done in CricketGraph III and graphs were prepared for publication by using CorelDraw 8 or 11.
For determination of chromosome mitotic indices in experiments involving benomyl treatments, LO715 and LO732 were grown at 2.5 × 106 conidia/ml in 25 ml of liquid medium in a 125-ml Erlenmeyer flask. The medium was YG plus 0.1% agar (to minimize clumping of germlings) supplemented with 50 μg/ml adenine HCl, 0.02 μg/ml biotin, 300 μg/ml ammonium sulfate, 50 μg/ml methionine, 2 μg/ml niacin, and 20 μg/ml choline. Cultures were incubated shaking in a water bath for 6.5 h at 43°C. Benomyl (Sigma-Aldrich, St. Louis, MO) was then added to a final concentration of 2.4 μg/ml, and cultures were incubated for an additional 30 min at 43°C. Flasks were then transferred to a water bath equilibrated to 20°C. Samples (900 μl) were added to 100 μl of 10% glutaraldehyde (Electron Microscopy Sciences, Fort Washington, PA) equilibrated to the culture temperature. They were fixed for 10 min at the culture temperature and then washed 2 × 10 min in double distilled water at room temperature. Samples were resuspended in 200 μl of double distilled water and sonicated for 5 min to break up clumps. Then 20 μl of 10% Nonidet P-40 (Sigma-Aldrich) was added followed by 20 μl of 200 μg/ml mithramycin in 300 mM MgCl2. Samples were scored on an Olympus IX71 microscope equipped with a 1.3 numerical aperture Planfluor objective.
Time-lapse GFP images were taken with a 100× Plan Apochromatic objective (1.40 numerical aperture) on a Nikon Eclipse TE300 inverted microscope equipped with a PerkinElmer Ultraview spinning-disk confocal system controlled by Ultraview software. Images were acquired with a Hamamatsu Orca ER camera. Images were imported into Slidebook where they were cropped and exported as Quicktime movies or Tiff series. Tiff series were made into composite figures by using Coreldraw 11.
Spindle measurements were made from saved Tiff series projections of Z-series stacks. These were imported into the Mac OSX version of ImageJ (http://rsb.info.nih.gov/ij/), the Java successor to NIH Image. The coordinates of spindle ends were saved as text files and imported into Microsoft Excel (Mac OSX) where spindle lengths were calculated. Rates of spindle elongation were determined by linear regression analysis using SPSS software (OSX).
Creation of a Deletion of the A. nidulans mad2 Homologue
The A. nidulans mad2 homolog was identified by blast searching the A. nidulans genome database (http://www-genome.wi.mit.edu/annotation/fungi/aspergillus/) with the mad2 sequence of Saccharomyces cerevisiae. The search revealed an A. nidulans gene with strong similarities to mad2 from S. cerevisiae (E value 2e-35) and mad2 homologues from Schizosaccharomyces pombe, Homo sapiens, and Mus musculus. We designate this gene md2A. This gene was deleted using a polymerase chain reaction (PCR) gene replacement procedure (Kuwayama et al., 2002). Primers were constructed based on the genomic sequence that allowed the amplification of a 2004-base pair fragment immediately 5′ to the coding region and a 1877-base pair fragment immediately 3′ to the coding region. The two primers nearest the coding sequence were designed with “tails” that allowed them to anneal to appropriate sequences flanking the A. nidulans pyrG gene. A fragment containing the pyrG gene was amplified from plasmid pPL6. The three fragments were mixed and amplified using the outside primers to form a single fragment consisting of the 5′ and 3′ flanking sequences of md2A surrounding the pyrG gene. This fragment was used to transform the A. nidulans strain G191 (pyrG89, pabaA1, fwA1, uaY9). PyrG+ transformants were selected and tested for supersensitivity to the antimicrotubule agent benomyl (the anticipated phenotype of a md2A deletant). A benomyl supersensitive transformant was chosen and tested by PCR with multiple sets of appropriate primers from within md2A to verify that md2A had been deleted. The A. nidulans klpA gene was amplified as a positive PCR control. The putative md2A deletion was further verified by transformation with wild-type md2A, which was amplified from the wild-type strain FGSC4. As expected, wild-type md2A complemented the benomyl super sensitivity and the resultant strain was indistinguishable in growth from the parental strain G191 except that it was pyrG+.
RESULTS
MipAD159 Does Not Inhibit Mitotic Spindle Formation under Restrictive Conditions, but Nuclei Reenter Interphase without Completing Mitosis Successfully
We used a mipAD159/nimT23 double mutant to observe the effects of mipAD159 on the progression through mitosis. NimT encodes a phosphatase that is essential for the activation of the A. nidulans homolog of P34cdc2 and thus for the G2-to-M transition (O'Connell et al., 1992). NimT23 is a heat-sensitive allele that blocks the cell cycle in G2 at high temperatures (42-43°C). The block is rapidly released upon a shift to permissive temperatures (<37°C), and nuclei enter mitosis rapidly and with excellent synchrony (Martin et al., 1997). MipAD159 inhibits growth almost completely at 20-25°C but allows near normal growth at high temperatures (Jung et al., 2001). Our experiments were carried out with germinating conidia (uninucleate asexual spores), which are arrested in G1 before germination. Incubation of conidia of the mipAD159/nimT23 strain at 43°C allowed them to germinate and proceed through the cell cycle until they were arrested in late G2 by nimT23. They were then shifted rapidly from 43 to 20°C. This shift released the nimT23 block and imposed the mipAD159 block. Samples were collected immediately before the shift from 43 to 20°C and at intervals afterward and were prepared for immunofluorescence microscopy. As a control, we used a strain carrying nimT23 and the wild-type mipA allele (mipA+). Germlings of this strain are blocked in G2 at 43°C, but they pass through mitosis normally upon the shift to 20°C.
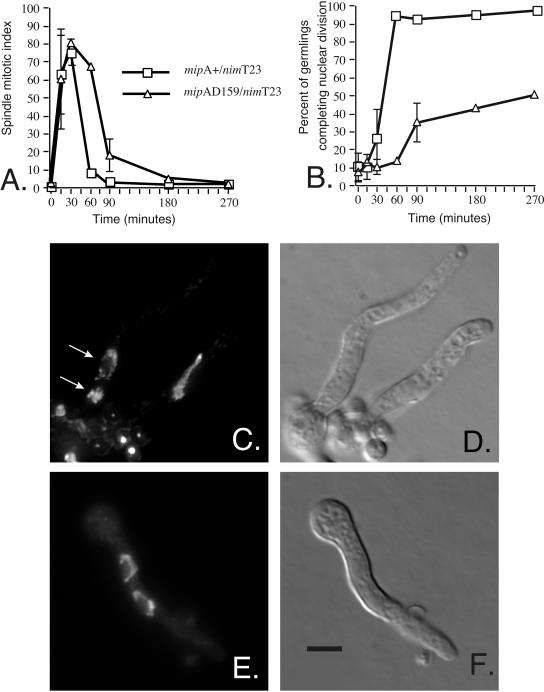
We first examined the effects of mipAD159 on mitotic spindle formation. Figure 1A shows the spindle mitotic index (percentage of germlings with mitotic spindles, abbreviated SMI) for mipAD159/nimT23 and for the mipA+/nimT23 control. The rate of assembly of mitotic spindles was essentially identical in the two strains. At 30 min after the shift, 75-80% of the germlings in each strain had mitotic spindles. At this time point, the majority of spindles in the mipAD159/nimT23 strain were normal in size and shape, and the chromatin was normal in appearance and distribution.
Figure 1.
Effects of mipAD159 on mitotic spindle formation and nuclear division. Germlings were shifted from 43 to 20°C at time 0. (A) The spindle mitotic index (percentage of germlings with mitotic spindles). Spindle formation is essentially identical in the mipA+ and mipAD159 strains, but mitotic exit (spindle disassembly) is slightly delayed (∼30 min) in the mipAD159 strain. (B) Percentage of germlings that have completed nuclear division (i.e., there are two or more nuclei in the germling). At the 60-min time point >90% of the mipA+ germlings have completed nuclear division and, as expected, this coincides with mitotic exit (A). Nuclear division does not occur in most mipAD159 germlings, however, even though nuclei exit mitosis. Values in both panels are means ± standard deviations for three experiments. Sample size for each time point was ≥400. Where bars are not shown, the standard deviations were sufficiently small that they were within the symbol. (C-F) Nuclear morphology in mipAD159 and mipA+ strains. (C) 4,6-Diamidino-2-phenylindole staining of a mipAD159 strain at the 270-min time point. Two nuclei unequal in size are present in one germling (arrows). The top nucleus is in interphase and the bottom nucleus is still in mitosis. In the germling at the right there is a nucleus in which the chromatin is probably decondensing, although mitosis has not been completed. Several ungerminated conidia are present at the lower left, and a differential interference contrast image of the same field is shown in D. The image in C is a Z-series that has been deconvolved and projected to allow all nuclei to be seen in a single image. (E) 4,6-Diamidino-2-phenylindole staining of normal nuclei in a mipA+ strain at the 270-min time point. The nuclei are similar in size and are in interphase. (F) A differential interference contrast image of the same field. C-F are the same magnification. Bar (F), 5 μm.
Although spindle formation was nearly normal in the mipAD159/nimT23 strain, completion of mitosis was not. In the mipA+/nimT23 control, the SMI dropped to <10% at 60 min after the shift to permissive temperature, and this indicates that most nuclei had returned to interphase. We verified that nuclear division had occurred by scoring the percentage of germlings with two or more nuclei. Because conidia are uninucleate, and because nimT23 causes a G2 block at 43°C, conidia that are germinated at 43°C are blocked with a single G2 nucleus before the shift to 20°C. The presence of two nuclei in a germling indicates that nuclear division has occurred. Figure 1B shows that at the 60-min time point, nuclear division had occurred in >90% of the mipA+/nimT23 germlings. Completion of mitosis was delayed to some extent in the mipAD159/nimT23 strain. The SMI lagged behind that of the mipA+/nimT23 control by ∼30 min (Figure 1A) (i.e., spindles were present longer in the germlings carrying mipAD159 than in the control). By 90 min after the shift, however, spindles were gone from most of the germlings carrying mipAD159. The disappearance of mitotic spindles in the mipAD159/nimT23 strain did not result from successful completion of mitosis, however. Less than 40% of the mipAD159/nimT23 germlings contained two nuclei at 90 min after the shift and <50% of them contained two nuclei even at 270 min after the shift (Figure 1B). In many of the germlings with two nuclei, the nuclei were unequal in size (Figure 1C) and this suggests that even when nuclear division occurred, the chromosomes were not partitioned correctly. These data demonstrate that mipAD159 causes a transient mitotic delay and that spindles disassemble and nuclei reenter interphase without successfully completing mitosis.
MipAD159 Disrupts the Coordination of Late Mitotic Events
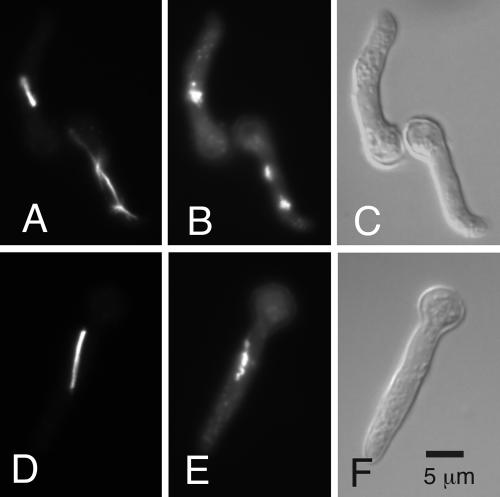
We examined the morphology of chromosomes and spindles in mipAD159/nimT23 and mipA+/nimT23 mitotic nuclei from the synchronization experiments described above. The morphology of mitosis in wild-type A. nidulans has been described in detail previously (Jung et al., 1998, and references therein). Briefly, the nuclear envelope remains intact throughout mitosis, and there are no microtubules in the nucleus in interphase. At the onset of mitosis, cytoplasmic microtubules disassemble, whereas, within the nucleus, microtubules assemble from the two adjacent SPBs and the chromatin condenses. The spindle quickly becomes bipolar and elongates slowly. In prophase, chromosomes condense into a clump around the center of the spindle (upper nucleus in Figure 2, A and B). When anaphase begins, the chromosomes move quickly to the poles (anaphase A). At this stage, chromosomes can sometimes be seen arranged along the spindle, but because this stage occurs rapidly, such configurations are rare in unsynchronized material. Spindle elongation (anaphase B) (lower nucleus in Figure 2, A and B) continues after chromosomes reach the poles and the spindle eventually disassembles in telophase as chromosomes decondense.
Figure 2.
Immunofluorescence of mitosis in mipA+ and mipAD159 strains. (A and D) Anti-β-tubulin staining. (B and E) Chromatin (4,6-diamidino-2-phenylindole). (C and F) Differential interference contrast images. A-C show the mipA+ strain. The upper germling contains a medial nuclear division (metaphase) spindle. Chromatin is in a clump surrounding the spindle. The lower germling contains an anaphase B spindle. The chromosomes have moved to the pole, astral microtubules extend from the poles and from live imaging we know that such spindles are elongating rapidly. D-F show the mipAD159 strain. The spindle is long and robust, but chromosomes have not moved to the poles but are strung out along the spindles. All images are the same magnification. Scale in F = 5 μm.
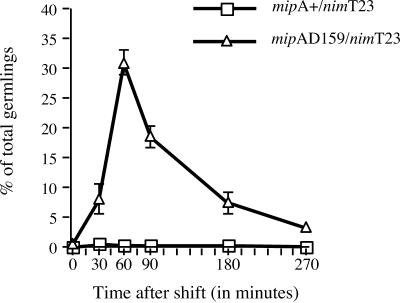
At the 15- and 30-min time points, the morphologies of most of the mipAD159/nimT23 spindles were similar to those of the mipA+/nimT23 control. At the 60- and 90-min time points, however, the percentage of cells with chromosomes arranged along the spindle (Figure 2, D and E) was dramatically elevated relative to the mipA+/nimT23 control (Figure 3). Cells in which chromosomes had reached the poles (anaphase B or telophase) were significantly less frequent in the mipAD159/nimT23 strain than in the mipA+/nimT23 control. At the 60-min time point, 29.6 ± 8.4% of mitotic cells in the mipA+/nimT23 control were in anaphase B or telophase (mean ± SD for three experiments, 200 cells scored per experiment), whereas only 0.2 ± 0.3% of the mitotic mipAD159/nimT23 cells were in anaphase B or telophase (mean ± SD for three experiments, 200 cells scored per experiment). Comparisons at later time points were not possible because the SMI was very low in the mipA+/nimT23 control. These data, in combination, demonstrate that anaphase A, movement of chromosomes to the poles, is significantly inhibited by mipAD159.
Figure 3.
Inhibition of chromosome-to-pole movement by mipAD159. Germlings were shifted from 43 to 20°C at time 0. The values shown are the percentages of total germlings with chromosomes arranged along the spindle (see Figure 2, D and E). Values are means ± standard deviations for three experiments. Sample size for each time point was ≥400. Where bars are not shown, the standard deviations were sufficiently small that they were within the symbol.
Interestingly, although movement of chromosomes to the poles was inhibited by mipAD159, spindle elongation did occur. Figure 2, D and E, shows a typical example. The chromosomes in the mipAD159/nimT23 strain have not moved to the poles, but the spindle is as long as the control spindle that is in anaphase B (Figure 2, A and B). Note that no astral microtubules are visible in the mipAD159 spindle (Figure 2D). To determine whether the lack of astral microtubules is characteristic of mipAD159 spindles, we scored 100 spindles of the mipAD159 strain at the 30-min time point and an equal number of spindles in the control. The percentages of spindles with astral microtubules were similar in the two strains (78% in the mipAD159 strain, 75% in the control). There was a visible difference, however, in the abundance of astral microtubules. In the mipAD159 strain, there were generally only one or two short astral microtubules, whereas they were longer and more abundant in the control. This might suggest that mipAD159 SPBs are defective in the nucleation of astral microtubules. At later time points, however, robust asters were present in the mipAD159 strain. At the 60-min time point, for example, 32% of the mipAD159 spindles had robust asters. It is also important to remember that astral microtubule proliferation occurs at anaphase, and it is possible that the failure of anaphase A causes astral microtubule proliferation to fail or be delayed (or that a common signal that triggers both anaphase A and astral microtubule proliferation is defective). It is also worth noting that the spindles in the mipAD159/nimT23 strain were as bright and large as those in the control strain. There was, thus, no gross reduction of microtubule nucleation by mi- pAD159. The accuracy of quantitation allowed by immunofluorescence is not sufficiently precise, however, to allow us to rule out the possibility that there is a slight reduction in the number of microtubules in the spindles of the mipAD159/nimT23 strain relative to the control strain.
Mitotic exit was abnormal in the mipAD159 strain. At the 60-min time point, 28.5% (57/200) of germlings had cytoplasmic microtubules and lacked spindles but had condensed chromosomes (some partially condensed and, thus, probably in the process of decondensing). Normally, spindle disassembly and chromosome decondensation are tightly coupled.
Although immunofluorescence of synchronized material is informative, live imaging often reveals information that cannot be obtained from fixed material. A GFP histone H1 fusion is available in A. nidulans (Ramon et al., 2000), and it showed no evidence of genetic interactions with mipAD159 (i.e., the growth rates of strains carrying mipAD159 and the fusion were indistinguishable from strains carrying mipAD159 without the fusion; our unpublished data). We were, thus, able to use this fusion to observe the effects of mipAD159 on chromosomal condensation, decondensation, and movement.
These were not synchronization experiments, but, rather, simple temperature-shift experiments. Strains were grown at a permissive temperature of 37°C and then placed at a room temperature of 24°C (a restrictive temperature for mipAD159; Jung et al., 2001) for 15 min before observation. Observations were made up to 2 h after the shift, a time that is well within the first cell cycle after the shift. Progression through mitosis was recorded at 24°C by time-lapse spinning-disk confocal microscopy. When comparing the data from these experiments to those from our synchronization experiments, it should be noted that mitosis proceeds more rapidly at 24°C than at 20°C. Because it is easy to observe chromosomal condensation in A. nidulans, it was relatively easy to identify early prophase nuclei and follow them as they progressed through mitosis. In addition, A. nidulans is coenocytic and nuclei within a single cell enter mitosis at almost but not exactly the same time. There is usually a wave of mitosis with the nucleus at one end of the cell entering mitosis first and the nucleus adjacent to it entering second, etc. If nuclei at one end of the cell are in prophase, nuclei at the other end are often in late G2, and this makes it possible to film the entire mitotic process.
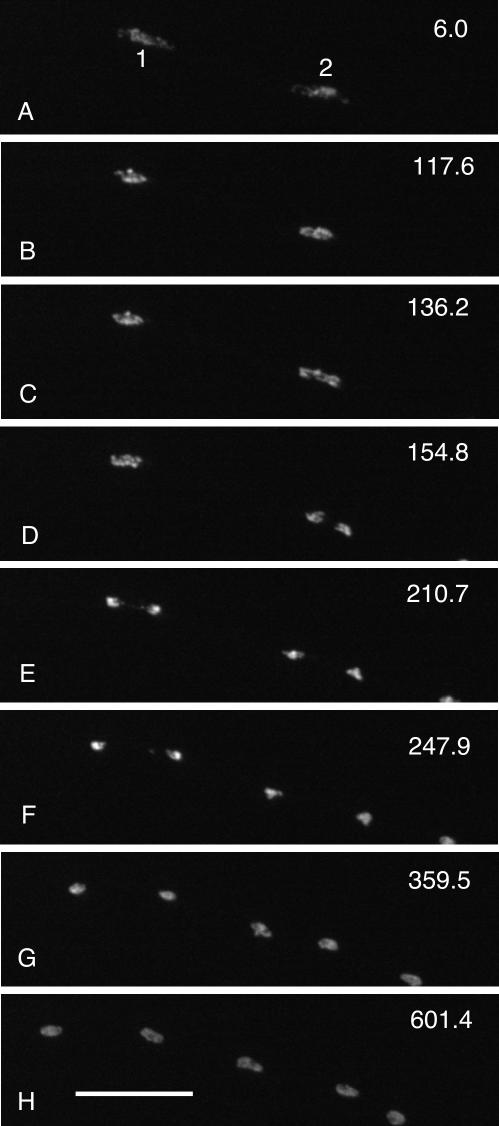
We first recorded four-dimensional videos of mitosis in 20 hyphae of the control (mipA+) strain LO883 by spinning-disk confocal microscopy. Some hyphae grew along the coverslip on which they rested, whereas others grew away from the coverslip. Thus, in some hyphae, it was possible to observe two or more nuclei, whereas in other hyphae, nuclei were in such different focal planes that only one could be observed. Z-series image stacks were collected at ∼15-s intervals, and each stack required 2-3 s to collect. To follow the behavior of all chromosomes, we made a maximum intensity projection of each stack and each projection became a single frame in a time-lapse video. We found that mitosis was remarkably consistent in all nuclei observed, and our observations were completely consistent with previous observations of fixed material (Jung et al., 1998). Figure 4 and Fig4video1.mov (online supplemental material) show a typical example. The chromosomes initially condensed into a single mass (Figure 4, A and B). They then moved rapidly to the poles (anaphase A) (Figure 4, B-E) forming two masses (Figure 4, D-F). The two masses moved further apart (anaphase B) and each decondensed forming interphase nuclei with clear regions that correspond to nucleoli (Figure 4, G and H). The nuclei then moved to establish an even spacing among them. Because of the consistency of this pattern, we were confident that we could determine whether chromosomal condensation, decondensation, or movement was abnormal in the mipAD159 strain.
Figure 4.
Images from Fig4video1 (online supplemental material) of mitosis in a mipA+ strain. Chromosomes are labeled with a histone H1 GFP fusion and each panel is a maximum intensity projection of a Z-series stack. The time (in seconds) for the completion of each Z-series stack is in the upper right of each panel. In A, two nuclei (designated 1 and 2) in the same hypha are entering mitosis. In B, the chromatin in each nucleus has condensed into a mass, and nucleus 2 is just beginning anaphase. In C, nucleus 2 is in anaphase. In D, nucleus 2 is in anaphase B and nucleus 1 is just beginning anaphase. In E, nucleus 1 is in mid-anaphase, whereas nucleus 2 is late anaphase. In F, nucleus 2 begins telophase, whereas nucleus 1 is in late anaphase. In G and H, chromatin in both nuclei decondense as the nuclei reenter interphase. All figures are the same magnification. Bar (H), 10 μm.
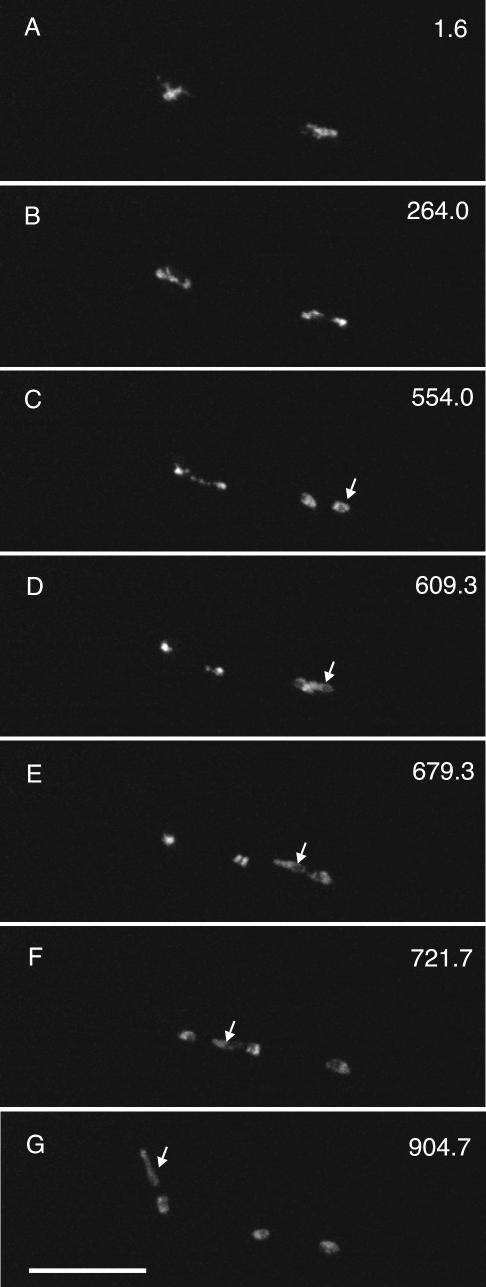
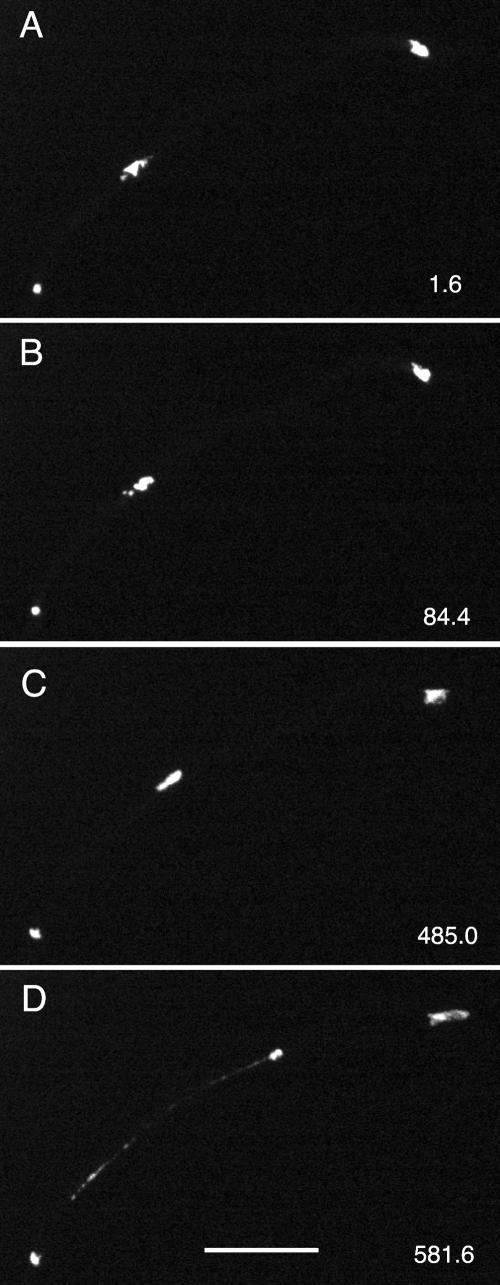
We recorded mitosis in 52 hyphae of strains LO905 and LO906, which carry mipAD159. Early mitotic events seemed normal. Chromosomes condensed and formed a single mass. There were obvious abnormalities, however, in late mitotic events in most (43/52 or 83%) cases. Anaphase A was inhibited at least transiently in 17/52 (33%) cases (Figures 5 and 6 and Fig5video2.mov and Fig6video3.mov). Chromosomal segregation was visibly abnormal in 36/52 (69%) cases. Among these, obvious nondisjunction (often with one or more lagging chromosomes that were stretched as the chromatin masses moved apart) occurred in 20/52 (38%) cases (Figure 6 and Fig6video3.mov). In some cases (11/52 or 21%), nuclei and chromatin were stretched after nuclei reentered interphase (Figure 7 and Fig7video4.mov). We believe that this is due to the fact that these nuclei have two SPBs, and the dynein-dependent forces that normally act on the microtubules attached to the SPBs of the daughter nuclei after nuclear division now act on the two SPBs in the single nucleus, pulling them apart and causing the nucleus to stretch. In 24/52 hyphae (46%), nuclei reentered interphase without completing mitosis. Of these, nuclei in 7/52 hyphae simply reentered interphase without entering anaphase, whereas in 18/52 hyphae nuclei entered anaphase but did not complete mitosis successfully before reentering interphase. (The numerical discrepancy is due to the fact that, in one hypha, one nucleus went into interphase without entering anaphase and another nucleus entered anaphase before reentering interphase.) In 26/52 hyphae, nuclei displayed abnormal movement after mitosis (Figure 5 and Fig5video2.mov).
Figure 5.
Inhibition of anaphase A and abnormal nuclear movement in a mipAD159 strain. Each panel is taken from Fig5video2 (online supplemental material) and is a maximum intensity projection of a Z-series stack. The time (in seconds) for the completion of each Z-series stack is in the upper right of each panel. Two mitotic nuclei in the same hypha are shown. The nucleus at the right completes anaphase A nearly normally (B) but the nucleus at the left is delayed in the completion of anaphase A (B and C). One of the two daughter nuclei (arrows in C-G) from the nucleus that completes mitosis first exhibits unusual postmitotic movement. It bumps into and apparently moves past its sister nucleus and then past the two daughter nuclei from the other nucleus. All figures are the same magnification. Bar (G), 10 μm.
Figure 6.
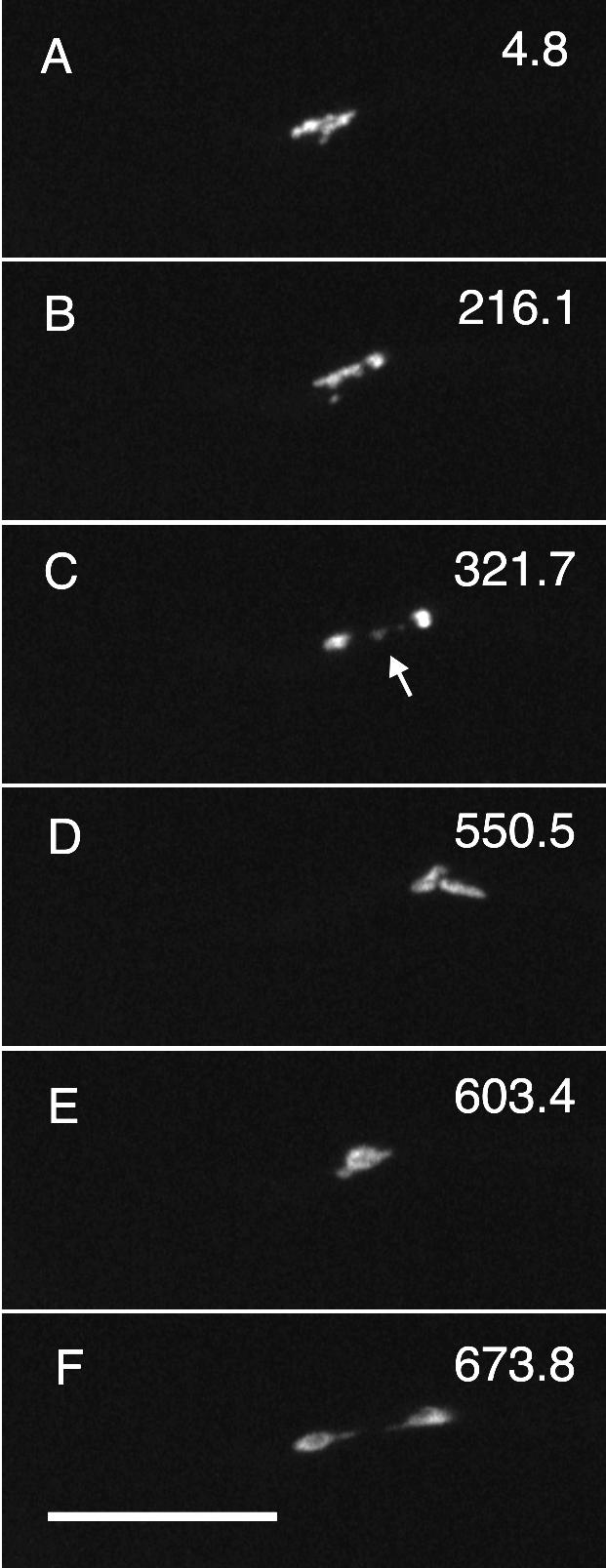
Nondisjunction in a mipAD159 strain. Each panel is taken from Fig6video3 (online supplemental material) and is a maximum intensity projection of a Z-series stack. The time (in seconds) for the completion of each Z-series stack is in the upper right of each panel. The nucleus is in anaphase A in A and remains in anaphase A for an abnormally long time (B). At least one chromosome fails to disjoin (arrow in C). The two separated chromatin masses move back into a single clump (D and E). As the chromatin decondenses the chromatin is pulled into two masses with a thin chromatin thread between them (F). All figures are the same magnification. Bar (F), 10 μm.
Figure 7.
Premature mitotic exit and chromatin stretching in a mipAD159 strain. Images are from Fig7video4. Chromosomes are labeled with a histone H1 GFP fusion and each panel is a maximum intensity projection of a Z-series stack. The time (in seconds) for the completion of each Z-series stack is in the lower right of each panel. The arrows in A indicate two mitotic nuclei. The nucleus in the upper right simply reenters interphase and undergoes some strange almost amoeboid movement as seen in Fig7video4.mov. The other nucleus is delayed in anaphase A (B). The chromosomes then begin to decondense (C) and in D the chromatin is stretched into a long, thin thread-like structure. All images are the same magnification. Bar (D), 10 μm.
It is important to note that failed mitosis and interphase reentry in mipAD159 nuclei did not take a great deal longer than normal mitosis in the mipA+ control. This can be observed in Fig4video1.mov, Fig5video2.mov, Fig6video3.mov, and Fig7video4.mov. Recording and playback conditions for all videos were the same, so the wild type and mutant can be compared directly. It is difficulty to give precise figures for mitotic duration because it varied to some extent among the mipAD159 mitoses recorded, and it was sometimes difficult to define the end of mitosis in the mipAD159 strain (because the morphology was highly abnormal). Mitosis in the mipA+ strain took ∼6-8 min under our conditions, however, and in the mipAD159 strain it took <15 min, with 10-12 min being typical. There was, thus, only a slight delay in mitosis.
It is also worth noting that chromosomal movement was evident in all mipAD159 nuclei. In many cases, chromosomes were severely stretched. We can assume, therefore, that most chromosomes attached to mitotic spindle microtubules and that force was exerted on them. In the cases where chromosomal stretching was observed, the forces were apparently very significant.
We observed mitotic spindle behavior by four-dimensional imaging using a GFP-α-tubulin fusion. These observations were more difficult than the GFP-Histone H1 imaging because the GFP-α-tubulin was less bright and faded more quickly. We were able, however, to obtain informative time-lapse sequences of mipA+ and mipAD159 strains at 23-25°C. Spindle behavior in the mipA+ control was very consistent (Figure 8, A-D, and Fig8video5.mov). Cytoplasmic microtubules disassembled as nuclei entered mitosis. Although a few cytoplasmic microtubules were sometimes present in early mitosis, they quickly disappeared. After spindles became bipolar, there was a period of slow spindle elongation during which astral microtubules were few and short lived. In anaphase, astral microtubules became more abundant and the spindle elongated rapidly. In favorable image series, astral microtubules from adjacent spindles could be seen to interact in late anaphase. It is possible that this interaction may play a role in the establishment of nuclear spacing after mitosis.
Figure 8.
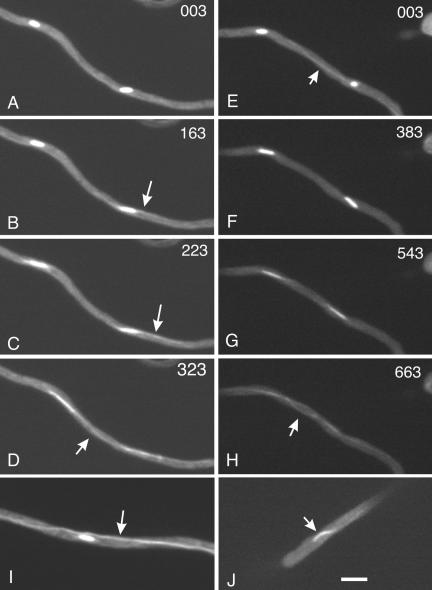
Spindle behavior in wild-type and mipAD159 strains. A-D are from Fig8video5, and E-H are from Fig8video6. The time in seconds for the completion of each Z-series stack is in the upper right of each panel. Two spindles are shown in A. They elongate slowly and astral microtubules extending from one spindle pole start to be visible in B (arrow). Astral microtubules (arrow) elongate as the spindles begin to elongate rapidly in anaphase (C). In D, the spindles have lengthened and astral microtubules are faintly visible between two adjacent spindles (arrow). E-H show normal mitotic spindles in a mipAD159 strain. Recording of this set of images was started at an earlier point in mitosis than for A-D and the spindles faded to greater extent by the end of mitosis. Two spindles are shown. A remnant cytoplasmic microtubule is visible in E, but it subsequently disassembles. Spindles elongate slowly initially and then rapidly in anaphase (G). Interacting astral microtubules are visible (although faint) in H. I, a mipAD159 spindle in which a cytoplasmic microtubule (or bundle of microtubules) has not broken down. Unlike the hypha shown in E-H, these cytoplasmic microtubules did not disassemble during mitosis. J shows a bent anaphase spindle in a mipAD159 strain. All panels are the same magnification. Bar (J), 5 μm.
We were able to make useful time-lapse series of 84 mipA+ nuclei. In 83, mitosis was normal as described above. In one case, cytoplasmic microtubules failed to disassemble completely in mitosis, but mitosis was still apparently completed successfully. We made useful time-lapse images of 63 mipAD159 nuclei. Spindle behavior in 40 of these was apparently normal (Figure 8, E-H, and Fig8video6.mov). In 13 cases the spindles became bent (Figure 8J). The bending was generally transient and occurred rapidly (in <1 min). In one case, the spindle bowed out on both sides. In eight cases, cytoplasmic microtubules failed to break down during mitosis (Figure 8I). In nine cases, the spindles failed to enter anaphase and complete mitosis.
We were also interested in whether spindle elongation rates were similar in normal spindles in the mipAD159 strains and in the mipA+ control. We were able to measure spindle elongation rates in 11 favorable mipA+ nuclei and 10 favorable mipAD159 nuclei. The rate for the slower (preanaphase) movement in the mipA+ control was 0.19 ± 0.07 μm/min (mean ± SD) and 0.15 ± 0.08 μm/min in the mipAD159 mutants. The rates of anaphase elongation were 1.33 ± 0.57 μm/min in the mipA+ control and 1.55 ± 0.81 μm/min in the mutant strains. The values for the mutant strains were not significantly different from the mipA+ control (Student's two-sample t test, p = 0.19 for preanaphase movement, p = 0.49 for anaphase movement).
Although we were not able to observe the behavior of chromosomes and the mitotic spindle at the same time, we believe we can correlate spindle and chromosome behavior in the mipAD159 strains with reasonable confidence. First, our chromosome and spindle imaging both reveal that ∼14% of nuclei never enter anaphase (13.4% from chromosome imaging, 14.2% from spindle imaging). Second, observations of chromosomes are probably a more sensitive indicator of anaphase delays or nondisjunction than are spindle observations. The fraction of spindles that were morphologically normal and elongated normally was higher than the fraction of nuclei in which chromosomes behaved normally. We believe that spindles in nuclei where there is only a transient anaphase delay, or in which only one chromosome fails to disjoin seem morphologically normal and have rates of elongation that are within the normal range. Third, more severe nondisjunction (several chromosomes failing to disjoin) may result in bent, or bowed spindles as the spindles attempt to elongate but cannot do so because kinetochore microtubules and nondisjoined chromosomes form a continuous connection between the two poles. If only one chromosome fails to disjoin, it stretches, but if several fail to disjoin the spindle bends. In cases of improper chromosomal segregation, nuclei may exit mitosis before nuclear division is complete.
MipAD159 Causes a Premature, Microtubule-independent Inactivation of the Spindle and/or Mitotic Exit Checkpoints
Because γ-tubulin is important for nucleation of spindle microtubules, a γ-tubulin mutation might, in theory, be expected to cause reduced nucleation of spindle microtubules, resulting in a nonfunctional or poorly functional spindle. Such spindles might activate the spindle checkpoint and cause a mitotic delay. Although our synchronization and live imaging experiments did reveal a slight delay in mitotic exit, the delay was surprisingly brief [slightly >30 min in a cell cycle that takes approximately 8 h at 20°C (i.e., 1/16 of a cell cycle) and even less in the live imaging experiments at 24°C]. In addition, most nuclei exited mitosis without dividing successfully. This suggested that mipAD159 might cause premature mitotic exit under restrictive conditions. This could happen in two general ways. One possibility is that the effects on mitotic exit are related to the microtubule nucleation or attachment function of γ-tubulin. For example, mipAD159 might cause the attachment of the kinetochore microtubules to the SPB to be weak under restrictive conditions. Spindle assembly and attachment of spindle microtubules to kinetochores would be normal and the spindle checkpoint would be inactivated, allowing mitosis to proceed. Mitosis would be unsuccessful, however, because kinetochores microtubules would break away from the SPB and chromosomes would not be pulled to the poles. A second possibility is that γ-tubulin has a function in checkpoint regulation that is independent of its microtubule nucleation and/or attachment functions.
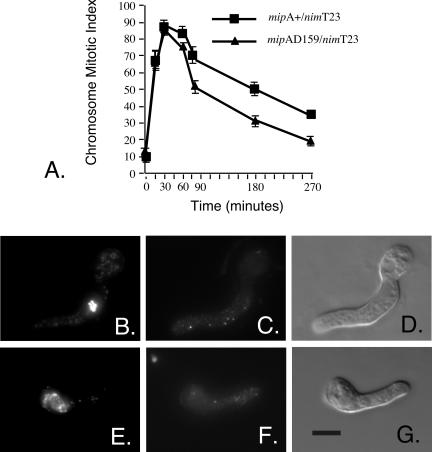
To distinguish between these possibilities, we performed synchronization experiments similar to the ones recounted previously, except that benomyl was added to a final concentration of 2.4 μg/ml 30 min before the shift from 43 to 20°C. Benomyl at this concentration rapidly eliminates microtubules (Ovechkina et al., 1998) and when cells enter mitosis the absence of spindle microtubules activates the spindle checkpoint. If mipAD159 causes a premature inactivation of the spindle and/or mitotic exit checkpoints, and this premature inactivation is microtubule independent, a strain carrying mipAD159 would be expected to exit mitosis sooner than a mipA± control strain in the presence of benomyl. If the checkpoint effects are related to microtubule nucleation or attachment, the strain carrying mipAD159 should exit mitosis at the same time as the mipA+ control. Because mitotic spindles were absent, we scored entry into and exit from mitosis by monitoring chromosomal condensation. We also verified by immunofluorescence microscopy that microtubules were, indeed, absent (our unpublished data).
The results are shown in Figure 9. The mipAD159/nimT23 strain and the mipA+/nimT23 control strain entered mitosis identically. A comparison of Figure 9 and Figure 1A shows that, as expected, both were blocked in mitosis by benomyl (i.e., both strains remained in mitosis longer than when spindles were present), but the strain carrying mipAD159 exited mitosis significantly sooner than the mipA+ control strain. The differences at the 90-, 180- and 270-min time points were highly significant. The p values from a two-sample t test were 0.009 at 90 min, 0.008 at 180 min, and 0.003 at 270 min. On average, nuclei in the strain carrying mipAD159 exited mitosis ∼90 min sooner than nuclei in the mipA+ control. As expected for benomyl-treated material, mitotic exit was not due to successful completion of mitosis in either strain. In some cases, mitotic exit resulted in large polyploid nuclei, whereas in other cases micronuclei formed around individual clumps of chromatin. There was no apparent difference in the morphology of mipA+ and mipAD159 nuclei after interphase reentry. The mipAD159 nuclei simply reentered sooner. These data indicate that mipAD159 causes a premature inactivation of the spindle and/or mitotic exit checkpoints by a mechanism that does not require spindle formation or microtubule assembly. This mechanism is extremely unlikely to be a function of the role of γ-tubulin in microtubule nucleation or attachment to the SPB.
Figure 9.
MipAD159 causes premature mitotic exit in the presence of the antimicrotubule agent benomyl. (A) Benomyl was added 30 min before the shift from 43 to 20°C at time 0. The chromosome mitotic index is the percentage of germlings with condensed chromosomes. The values are mean ± SE for four experiments. The sample size for each strain for each experiment was ≥100. Where error bars are not shown, they fell within the symbol. (B-G) Nuclear morphologies and absence of microtubules in the presence of benomyl. Images are from a mipAD159 strain at the 60-min time point. B shows 4,6-diamidino-2-phenylindole staining of a germling in which the chromosomes are condensed. C shows anti-β-tubulin staining of the same germling. Microtubules are absent. B and C are projections of Z-series stacks that had been deconvolved. D is a differential interference contrast image of the same germling. E shows 4,6-diamidino-2-phenylindole staining of another germling from the sample in which the chromosomes are almost completely decondensed. F shows the absence of microtubules in the germling. E and F are projections of Z-series stacks that had been deconvolved. G is a differential interference image of the same germling. All images are the same magnification. Bar (G), 5 μm.
MipAD159 Is Synthetically Lethal with Spindle Checkpoint Mutations
Because of the checkpoint effects of mipAD159, we decided to examine interactions of mipAD159 with spindle checkpoint mutations. We replaced the A. nidulans mad2 homologue (which we now designate md2A) with the A. nidulans pyrG gene, creating md2AΔ. Efimov and Morris (1998) have replaced the A. nidulans bub1 homologue sldA and bub3 homologue sldB with the pyr4 gene of N. crassa. These replacements were designated sldAΔ and sldBΔ. In each case the replacements are not lethal but cause increased sensitivity to antimicrotubule agents such as benomyl.
We crossed strains carrying these spindle checkpoint mutations to a strain carrying mipAD159 and looked for double mutant progeny which should be cold-sensitive and benomyl supersensitive. No such progeny were found among at least 100 segregants tested from each cross, although the other expected classes of progeny (wild-type and mipAD159 and checkpoint single mutants) were present. These results suggested that the double mutants were probably lethal, but it was also formally possible that they were phenotypically identical to another class of progeny.
To distinguish between these possibilities, we took advantage of closely linked markers to follow mipAD159 and the checkpoint mutations through crosses. The pyr4 and pyrG genes that were used to replace md2A, sldA and sldB, complement pyrG89, which prevents growth in the absence of added uracil or uridine. MipA is ∼0.3 cM from the riboB gene (Weil et al., 1986). For simplicity, we will only discuss in detail results with md2AΔ. We crossed strain LO699 (mi- pAD159, pyrG89, pabaA1) to strain LO1207 (md2AΔ, pyrG89, riboB2, pabaA1). All the progeny of this cross carry pyrG89 and only md2AΔ progeny carry pyrG+ (which was used to replace md2A). Thus, all segregants that are able to grow in the absence of uridine or uracil will carry md2AΔ. Likewise, because mipAD159 is tightly linked to riboB+, almost all riboB+ progeny will carry mipAD159. If the mipAD159, md2AΔ double mutant is inviable, pyrG+, riboB+ progeny should be very rare (occurring only if crossing over occurs between mipA and riboB, producing mipA+, riboB+ segregants). If, however, the double mutant is viable such progeny should account for ∼25% of total segregants. We found no pyrG+, riboB+ progeny among 100 segregants tested. To increase the number of ascospores tested, we inoculated 2 × 104 ascospores from this cross onto minimal medium supplemented with the common nutritional requirement paraamino benzoic acid but not with riboflavin, uridine, or uracil. Because only pyrG+, riboB+ progeny can grow on this medium, this procedure would select for rare recombinants. Only five colonies grew. This is slightly fewer than the number expected from crossovers between mipA and riboB. These colonies were very sick, did not produce viable conidia, and were not tested further. Essentially identical results were obtained with sldAΔ and sldBΔ. We conclude that mipAD159 is synthetically lethal with deletions of the A. nidulans homologues of mad2, bub1, and bub2.
DISCUSSION
We have investigated the phenotype of the mutant γ-tubulin allele mipAD159 to enhance our understanding of essential functions of γ-tubulin in addition to its established role in microtubule nucleation. Our data demonstrate that, at restrictive temperatures, the γ-tubulin encoded by the mipAD159 allele supports assembly of mitotic spindles at a normal rate. The spindles are normal in appearance, and live imaging demonstrates that they are able to move chromosomes. Anaphase and telophase are abnormal in the majority of nuclei, however, and nuclei reenter interphase without completing mitosis successfully. Our data suggest that the split and bent spindles noted previously in unsynchronized material (Jung et al., 2001) are primary defects of mipAD159 and probably result from the spindles attempting to elongate with multiple chromosomes failing to disjoin. The multipolar spindles seen by Jung et al. (2001) are probably due in large part to the fact that multiple SPB are present in nuclei that have failed to complete mitosis and when these nuclei progress through the cell cycle and enter mitosis again, multipolar spindles form.
Our data indicate that mipAD159 causes defects in at least two important and related mitotic processes. One is checkpoint regulation, and the second is the coordination of late mitotic events. In our live imaging experiments, we observed that nuclei often reentered interphase relatively quickly even though mitosis had not been successfully completed. This was also evident from our synchronization experiments. Perhaps most significantly, in our experiments using benomyl to disassemble microtubules, mipAD159 nuclei reentered interphase significantly sooner than mipA+ control nuclei. This result reveals that mipAD159 causes premature inactivation of the spindle and/or mitotic exit checkpoints even when microtubules are absent. Because benomyl activates the spindle checkpoint, and we have scored mitotic exit, premature mitotic exit could be caused by inactivation of spindle or mitotic exit checkpoints or both. We have found that deletions of three spindle checkpoint genes, the A. nidulans homologues of mad2, bub1, and bub3, are synthetically lethal with mipAD159. This indicates that the spindle checkpoint is probably intact in mipAD159 strains and serves to correct some of the problems caused by mipAD159. When the spindle checkpoint is inactivated by any of these mutations the mipAD159 defects are not corrected and death results. Our data seem to be most consistent with the possibility that mipAD159 affects the mitotic exit checkpoint [see discussion of the data of Vardy et al. (2002) below]. In any case, our results demonstrate clearly that γ-tubulin has an important role in checkpoint regulation that is separate from its established role in microtubule nucleation.
Although the checkpoint defect caused by mipAD159 is certainly important, checkpoint defects do not usually result in a strong inhibition of growth (as is caused by mipAD159 at restrictive temperatures) unless mitotic spindle function is compromised (e.g., Efimov and Morris, 1998). If the mitotic apparatus functions correctly, mitosis will be completed successfully in most nuclei even in the absence of functional mitotic checkpoints. Checkpoint defects could contribute to growth inhibition, however, if mipAD159 spindles have defects, not visible by immunofluorescence, that hamper spindle function. In this case, mitosis would be partially inhibited due to these defects and the checkpoint defect would allow premature mitotic exit even though chromosomes had not segregated properly. The resulting aneuploidy would eventually lead to inviability.
Our live imaging experiments suggest, however, that mipAD159 may cause a more general defect in the coordination of late mitotic events (chromosomal disjunction, anaphase A, anaphase B, and chromosomal decondensation). For mitosis to be completed successfully, multiple mitotic events or processes must be coordinated correctly. Checkpoint regulation is one aspect of this coordination, but other types of coordination must also occur. For example, the spindle checkpoint may sense that the necessary conditions have been met for anaphase to begin, but for anaphase to be carried out correctly, chromosomal disjunction, chromosome movement to the pole, spindle elongation, and, perhaps, other processes must be coordinated. If these processes occur in an incorrect order and/or at inappropriate times, correct chromosomal segregation will be compromised. We have found that in dividing mipAD159 nuclei, anaphase B sometimes occurs before anaphase A or chromosomal disjunction. We believe that this is responsible for the high frequency of chromosomes strung out along the spindle that we observed in fixed material. We have also observed that anaphase A and B sometimes occur before chromosomal disjunction has been completed, that chromosomal decondensation sometimes occurs before anaphase is completed and that spindles sometimes disassemble and cytoplasmic microtubules reassemble before chromosomes decondense. Together, these data indicate that mipAD159 disrupts the coordination of important late mitotic events at restrictive temperatures, and it follows that γ-tubulin normally plays an important role in the coordination of these events. The growth inhibition caused by mipAD159 at restrictive temperatures is probably due to the breakdown of the coordination of late mitotic events, which causes defects in chromosomal segregation. This is exacerbated by the checkpoint defect that allows premature reentry into interphase without mitosis being completed successfully. The net result is extensive aneuploidy and loss of viability.
It is worth noting that although mipAD159 causes a checkpoint defect, strains carrying this allele are not abnormally sensitive to antimicrotubule agents (Jung et al., 2001). Perhaps some mipAD159 defects (e.g., persistence of cytoplasmic microtubules in mitosis) are corrected by antimicrotubule agents, whereas others (checkpoint defects) are exacerbated, and the net result is no great change in sensitivity.
MipAD159 resembles previously reported γ-tubulin mutations in some ways. For example, a cold-sensitive γ-tubulin allele of Schizosaccharomyces pombe, gtb1-PL301, causes chromosomal segregation to fail at restrictive temperatures even though robust spindles form (Paluh et al., 2000). Gtb1-PL301 is synthetically lethal with a null mutation in pkl1, which encodes a C-terminal motor domain kinesin-like protein (Paluh et al., 2000), whereas the cold-sensitivity of mipAD159 is enhanced by a deletion of klpA, which encodes the homologous kinesin-like protein in A. nidulans (Prigozhina et al., 2001). Another A. nidulans γ-tubulin mutation, mipAD123, also inhibits anaphase A and interacts synthetically with the klpA deletion. Perhaps most germane, Hendrickson et al. (2001) found that some human γ-tubulin mutants in S. pombe allowed anaphase and cytokinesis to proceed in spite of spindle abnormalities such that aneuploid or aploid cells were formed. These data suggested that γ-tubulin might play a role in the coordination of mitotic events and that γ-tubulin might have a role in spindle checkpoint regulation. It is important to note, however, that the spindle checkpoint is eventually inactivated in wild-type cells even when spindles are absent (Figure 9). One possibility with the data of Hendrickson et al. (2001) was, thus, that these γ-tubulin mutations were partially inhibiting microtubule nucleation such that dysfunctional spindles were formed that did not segregate chromosomes properly. If so, the spindle checkpoint would have eventually been inactivated and abnormalities in chromosomal segregation would have resulted. Our real-time imaging demonstrates that at restrictive temperatures, mipAD159 causes coordination of mitotic events to fail rapidly, without a prolonged mitotic block.
Also relevant are data from mutations in genes that encode γ-tubulin interacting proteins in S. pombe. Vardy and Toda (2000) isolated and thoroughly characterized mutations in the Alp4 and Alp6 genes of S. pombe. These genes encode two proteins that form a complex with γ-tubulin. Homologues of these genes have been identified in several phylogenetically diverse organisms and they are probably universal in eukaryotes (reviewed by Wiese and Zheng, 1999; Oakley, 2000). Mutations in Alp4 and Alp6, not surprisingly, caused a number of mitotic abnormalities. Perhaps surprisingly, however, Alp4 and Alp6 mutants did not display a typical cell cycle arrest but passed through later mitotic stages, septation, and cytokinesis. Based on these data, Vardy and Toda (2000) proposed that checkpoint pathways dependent on functional γ-tubulin complexes exist and are evolutionarily conserved even to vertebrates. These data were recently extended and clarified (Vardy et al., 2002). An Alp4 mutation (alp4-1891) causes a failure of the recruitment of the γ-tubulin complex to the SPB under restrictive conditions and this causes monopolar spindles. The alp4-1891 allele does not inactivate the Mad2 spindle checkpoint but, rather, causes untimely activation of the septation initiation network triggering septation in spite of the fact that mitosis has not occurred correctly. The phenotypes of mipAD159 and alp4-1891 differ in important ways. The mutant γ-tubulin encoded by mipAD159 localizes to the SPB, nucleates microtubule assembly and bipolar spindles form. As we discuss below, however, the phenotypes of mipAD159 and alp4-1891 could be mechanistically related.
One thing that has hindered acceptance of a role for γ-tubulin and associated proteins in cell cycle regulation has been the absence of a mechanistic model that would account for the data. The data of Vardy et al. (2002) provide an important clue as to what might be happening mechanistically. Alp4-1891 prevents recruitment of γ-tubulin complexes to the SPB. The Sid1 kinase then binds prematurely to the SPB, and this allows inappropriate activation of the septation initiation network.
It has been suggested that the SPB has an important signaling function (e.g., Pereira and Schiebel, 2001; Visintin and Amon, 2001; Lange, 2002). Many cell cycle regulatory proteins, including proteins that function in the control of mitosis, localize to the SPB or centrosome for at least part of the cell cycle. The dynamics of the association between the centrosome and two proteins with prominent roles in mitosis (MAD2 and Cdc20) have been measured (Howell et al., 2000; Kallio et al., 2002). In both cases, the association is very dynamic and the proteins have a short residence time at the centrosome. Based on these findings and our data, we propose the following speculative model. We suggest that the localization of proteins involved in the regulation of mitosis and the cell cycle to polar microtubule organizing centers (MTOCs) is important to their functioning. Binding of these proteins to polar MTOC causes the local concentration of the proteins to be much higher than it would be if they were dispersed evenly through the nucleoplasm and/or the cytoplasm. This high local concentration facilitates interactions among these proteins (e.g., phosphorylation of substrates by kinases). Cell cycle regulation would be, at least partially, a function of when regulatory proteins arrive at the polar MTOC and their residence times. The polar MTOC is a particularly good site for these putative interactions because some (perhaps many) of the regulatory proteins could be transported to the SPB by minus-end-directed motors. Some of the proteins that localize to the polar MTOC might bind to γ-tubulin, whereas others might bind to other proteins. With respect to our data, perhaps mipAD159 alters the binding, at the SPB, of proteins involved in mitotic coordination and checkpoint control. In this regard, it is worth noting that mipAD159 is recessive (Jung et al., 2001), which implies that at restrictive temperatures the γ-tubulin encoded by this allele lacks a function possessed by wild-type γ-tubulin. The fact that it allows inappropriate mitotic progression, including inactivating checkpoints, indicates that the missing function is required for the inhibition of mitotic and/or cell cycle progression when proper conditions have not been met.
Nuclear movement and positioning are precisely regulated phenomena in wild-type A. nidulans, and the abnormal nuclear movements after mitosis are as striking as the abnormalities in mitotic progression. It is possible that some of the abnormal nuclear movements in some hyphae are due to the failure of cytoplasmic microtubule disassembly. Normally, when astral microtubules assemble at the end of mitosis, there are few or no remnant cytoplasmic microtubules and astral microtubules from adjacent spindles interact. It is reasonable to believe that this interaction facilitates normal nuclear spacing. If cytoplasmic microtubules are already present when astral microtubules assemble, the assembling astral microtubules might interact with them. As nuclear division is completed, the interaction could power movement of daughter nuclei even moving them past other nuclei as we have observed. This could happen if cytoplasmic microtubules fail to disassemble or if nuclei in the same cytoplasm exit mitosis asynchronously such that some nuclei have nucleated extensive networks of cytoplasmic microtubules before other nuclei exit mitosis.
Note added in proof: Relevant to our explanation for the phenotypes of mipAD159, we note that Hinchcliffe et al. (Science 291, 1547-1550) have pointed out that “core centrosomal structures could bind cell cycle regulatory molecules in a way that activates their function or raises their local concentration to the point that essential reactions occur in a timely fashion.”
Supplementary Material
Acknowledgments
We thank Casey Jowdy for performing a portion of the videomicroscopy, Michael Munz for assistance with cultures, Dr. Maria Symeonidou-Sideris for help with the SPSS statistics package, Dr. Edyta Szewczyk for helping make the md2A deletion, and Dr. Claudio Scazzocchio for the GFP histone H1 strain. This work was supported by National Institutes of Health Grants GM-31837 to B.R.O. and GM-42564 to S.A.O.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03-06-0405. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-06-0405.
Online version of this article contains supplementary video material for some figures. Online version is available at www.molbiolcell.org.
References
- Efimov, V.P., and Morris, N.R. (1998). A screen for dynein synthetic lethals in Aspergillus nidulans identifies spindle assembly checkpoint genes and other genes involved in mitosis. Genetics 149, 101-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita, A., Vardy, L., Garcia, M.A., and Toda, T. (2002). A fourth component of the fission yeast γ-tubulin complex, Alp16, is required for cytoplasmic microtubule integrity and becomes indispensable when γ-tubulin function is compromised. Mol. Biol. Cell 13, 2360-2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, G., Liu, B., Zhang, J., Zuo, W., Morris, N.R., and Xiang, X. (2001). The Aspergillus cytoplasmic dynein heavy chain and NUDF localize to microtubule ends and affect microtubule dynamics. Curr. Biol. 11, 719-724. [DOI] [PubMed] [Google Scholar]
- Hendrickson, T.W., Yao, J., Bhadury, S., Corbett, A.H., and Joshi, H.C. (2001). Conditional mutations in γ-tubulin reveal its involvement in chromosome segregation and cytokinesis. Mol. Biol. Cell 12, 2469-2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell, B.J., Hoffman, D.B., Fang, G., Murray, A.W., and Salmon, E.D. (2000). Visualization of Mad2 dynamics at kinetochores, along spindle fibers, and at spindle poles in living cells. J. Cell Biol. 150, 1233-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung, M.K., May, G.S., and Oakley, B.R. (1998). Mitosis in wild-type and β-tubulin mutant strains of Aspergillus nidulans. Fungal Genet. Biol. 24, 146-160. [DOI] [PubMed] [Google Scholar]
- Jung, M.K., Prigozhina, N., Oakley, C.E., Nogales, E., and Oakley, B.R. (2001). Alanine-scanning mutagenesis of Aspergillus γ-tubulin yields diverse and novel phenotypes. Mol. Biol. Cell 12, 2119-2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallio, M.J., Beardmore, V.A., Weinstein, J., and Gorbsky, G.J. (2002). Rapid microtubule-independent dynamics of Cdc20 at kineochores and centrosomes in mammalian cells. J. Cell Biol. 158, 841-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwayama, H., Obara, S., Morio, T., Katoh, M., Urushihara, H., and Tanaka, Y. (2002). PCR-mediated generation of a gene disruption construct without the use of DNA ligase and plasmid vectors. Nucleic Acids Res. 30, e2 1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange, B.M.H. (2002). Integration of the centrosome in cell cycle control, stress response and signal transduction pathways. Curr. Opin. Cell Biol. 14, 35-43. [DOI] [PubMed] [Google Scholar]
- Martin, M.A., Osmani, S.A., and Oakley, B.R. (1997). The role of γ-tubulin in mitotic spindle formation and cell cycle progression in Aspergillus nidulans. J. Cell Sci. 110, 623-633. [DOI] [PubMed] [Google Scholar]
- Morris, N.R. (1976). Mitotic Mutants of Aspergillus nidulans. Genet. Res. 26, 237-254. [DOI] [PubMed] [Google Scholar]
- O'Connell, M.J., Osmani, A.H., Morris, N.R., and Osmani, S.A. (1992). An extra copy of nimEcyclinB elevates pre-MPF levels and partially suppresses mutation of nimTcdc25 in Aspergillus nidulans. EMBO. J. 11, 2139-2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley, B.R. (2000). γ-Tubulin. Curr. Top. Dev. Biol. 49, 27-54. [DOI] [PubMed] [Google Scholar]
- Oakley, B.R., Oakley, C.E., Yoon, Y., and Jung, M.K. (1990). γTubulin is a component of the spindle-pole-body that is essential for microtubule function in Aspergillus nidulans. Cell 61, 1289-1301. [DOI] [PubMed] [Google Scholar]
- Ovechkina, Y., Pettit, R.K., Chichacz, Z.A., Pettit, G.R., and Oakley, B.R. (1998). Spongistatin 1 has antimicrotubule activity in Aspergillus nidulans. Mol. Biol. Cell 9, 273a. [Google Scholar]
- Paluh, J.L., Nogales, E., Oakley, B.R., McDonald, K., Pidoux, A., and Cande, W.Z. (2000). A mutation in γ-tubulin alters microtubule dynamics and organization and is synthetically lethal with the kinesin-like protein Pkl1p. Mol. Biol. Cell 11, 1225-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira, G., and Schiebel, E. (2001). The role of the yeast spindle pole body and the mammalian centrosome in regulating late mitotic events. Curr. Opin. Cell Biol. 13, 762-769. [DOI] [PubMed] [Google Scholar]
- Prigozhina, N.L., Walker, R.A., Oakley, C.E., and Oakley, B.R. (2001). γ-Tubulin and the C-terminal motor domain kinesin-like protein, KLPA, function in the establishment of spindle bipolarity in Aspergillus nidulans. Mol. Biol. Cell 12, 3161-3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramon, A., Muro-Pastor, M.I., Scazzocchio, C., and Gonzalez, R. (2000). Deletion of the unique gene encoding a typical histone H1 has no apparent phenotype in Aspergillus nidulans. Mol. Microbiol. 35, 223-233. [DOI] [PubMed] [Google Scholar]
- Sampaio, P., Rebollo, E., Varmark, H., Sunkel, C.E., and Gonzalez, C. (2001). Organized microtubule arrays in γ-tubulin-depleted Drosophila spermatocytes. Curr. Biol. 11, 1788-1793. [DOI] [PubMed] [Google Scholar]
- Vardy, L., Fujita, A., and Toda, T. (2002). The γ-tubulin complex protein Alp4 provides a link between the metaphase checkpoint and cytokinesis in fission yeast. Genes Cells 7, 365-373. [DOI] [PubMed] [Google Scholar]
- Vardy, L., and Toda, T. (2000). The fission yeast γ-tubulin complex is required in G1 phase and is a component of the spindle assembly checkpoint. EMBO. J. 19, 6098-6111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visintin, R., and Amon, A. (2001). Regulation of the mitotic exit protein kinases Cdc15 and Dbf2. Mol. Biol. Cell 12, 2961-2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel, J., and Snyder, M. (2000). The carboxy terminus of Tub4p is required for γ-tubulin function in budding yeast. J. Cell Sci. 113, 3871-3882. [DOI] [PubMed] [Google Scholar]
- Vogel, J., Drapkin, B., Oomen, J., Beach, D., Bloom, K., and Snyder, M. (2001). Phosphorylation of γ-tubulin regulates microtubule organization in budding yeast. Dev. Cell 1, 621-631. [DOI] [PubMed] [Google Scholar]
- Weil, C.F., Oakley, C.E., and Oakley, B.R. (1986). Isolation of mip (microtubule-interacting protein) mutations of Aspergillus nidulans. Mol. Cell. Biol. 6, 2963-2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiese, C., and Zheng, Y. (1999). γ-Tubulin complexes and their interaction with microtubule-organizing centers. Curr. Opin. Struct. Biol. 9, 250-259. [DOI] [PubMed] [Google Scholar]
- Yao, J., Uzawa, S., Hendrickson, T.W., and Joshi, H.C. (2001). Gamma-tubulin plays a role in the spindle assembly checkpoint and mitotic exit. Mol. Biol. Cell 12, 173a. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.