Abstract
Droplet microfluidics, which involves micrometer-sized emulsion droplets on a microfabricated platform, is an active research endeavor that evolved out of the larger field of microfluidics. Recently, this subfield of microfluidics has started to attract greater interest because researchers have been able to demonstrate applications of droplets as miniaturized laboratories for biological measurements. This perspective explores the recent developments and the potential future biological applications of droplet microfluidics.
Keywords: Droplets, droplet microfluidics, droplet polymerase chain reaction
1. INTRODUCTION
Today’s chemical research is moving at a fast pace. Chemical analyses are expected to be rapid as well as accurate, cost-effective, reliable, and sensitive. One way of meeting these expectations has been to exploit microfluidics, which can be defined as fluidic applications that are on the scale of micrometers or smaller and are based on modern microfabrication methods. 1, 2 For over 20 years, microfluidics has been a thriving research field. 3 The field is driven by great expectations because it manipulates reaction volumes of nanoliters or less inside channels that are often narrower than the width of a human hair. By doing so, microfluidics reduces costs and consumption of reagents and samples, cuts the time spent on experiments, and increases sample throughput. It also increases accuracy, reliability and sensitivity of experiments. 4, 5
Droplet microfluidics emerged from microfluidics. It uses immiscible phases to create emulsions of monodisperse droplets that act as reaction and transport vehicles 6. Emulsions and droplets on the macroscale have long been a part of scientific studies, most famously by Lord Rayleigh in the 19th century who investigated the formation of droplets from liquid jets. 7 Droplet generation on the microscroscale had to wait for the emergence of microfluidics; the first studies of droplet breakup in microfluidic devices were reported in the early 2000’s 8, 9. The advantage of generating microscale droplets over the macroscale ones is the larger surface-to-volume ratio. The change in surface-to-volume ratio and the small length scale has a strong impact on mass and heat transfer, often allowing a significant increase in reaction kinetics in comparison to reactions on larger scales due to faster mixing, an advantage that is also generally applicable to microfluidics as a whole. However, microdroplets are dispersed in an immiscible phase, which isolates the aqueous droplet from its environment. This effectively creates self-contained microreactors. These individual microreactors allow researchers to conduct massively parallel studies of minute changes in reaction conditions by gradually changing the composition of the droplets during their formation. Furthermore, the small size of the droplets minimizes reagent volumes, thus dropping costs of the experiments. Compared to other emulsification techniques in bulk (e.g., shear emulsion in a beaker), generating droplets in microfluidic systems has the advantage of creating monodisperse sample volumes at the expense of lower volumetric throughput, because in most cases droplets are made in a serial fashion. This expense, however, is justified in droplet microfluidics, as the control over droplet size and size distribution improves control over reaction conditions and improves accuracy and tunability of conditions. As a result, droplet microfluidics is becoming increasingly appealing for chemical and biological studies which require great sensitivity.
Like its parent field, droplet microfluidics has shown similar hype cycles of expectations and challenges, although at an even faster pace. Rapid advances in the fundamental studies of droplet microfluidics showed potential applications in biological fields, such as cell biology. The expectations of droplet microfluidic applications to cutting-edge research areas, such as single-cell studies, often created new questions and required the development of new technologies (e.g., miniaturizing conventional bulk assays to nanoliter-sized sample volumes, while maintaining efficiency and sensitivity). In this sense, the field of droplet microfluidics is often at the forefront of the current technology limits. The success of droplet microfluidics as a versatile and powerful tool in biology is reflected in the successes of commercialized droplet-based assays. Companies like Bio-Rad Laboratories and Raindance Technologies are at the forefront of providing biological and clinical diagnostics tools based on droplet microfluidics.
Here, we will take a look at the field of droplet microfluidics and provide a perspective of where the field currently is and how it may develop. Other sub-fields have emerged from microfluidics over the years, including digital microfluidics, optofluidics, and paper microfluidics, but these are beyond the scope of this perspective article. The interested reader is referred to the numerous well-written reviews that cover these topics10–16.
2. SYSTEMS FOR DROPLET GENERATION
The central tenet of droplet microfluidics is the formation of droplets of precise volume and composition. These droplets can span three orders of magnitude in diameter (i.e., from hundreds of nanometers to hundreds of micrometers) and require an immiscible phase for their creation. The immiscibility of the droplets with the surrounding phase makes them self-contained microreactors, with limited loss of sample and limited potential for cross-contamination with reagents from the surrounding phases. The size of these droplets is ideal for applications requiring heating or cooling because the large surface area of the droplets with respect to the volume allow for fast heat transfer. In addition, microdroplets are ideal reaction vessels for biological studies because they are similar in size to cells and can be readily assessed by optical microscopy. Moreover, the small size of the droplets allows researchers to perform thousands of individual reactions with a few microliters of sample.
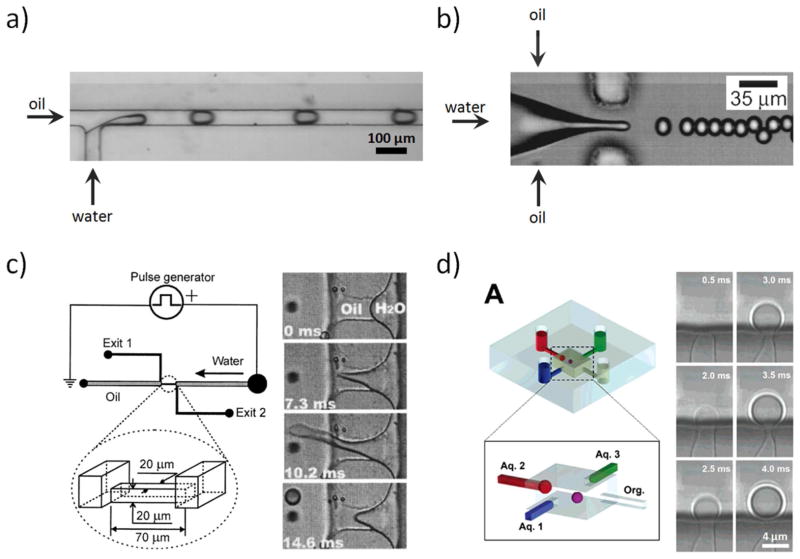
There are a variety of methods to generate droplets on microfluidic platforms, most of which can be classified as dynamic systems that rely on steady-state droplet generation. Two dynamic systems are the plug generation at T-junctions 17 and the droplet generation by hydrodynamic flow focusing 18 (Fig. 1a & 1b). The fluids used in these systems are typically driven by syringe pumps, but other approaches have been used to drive fluids, including the use of compressed air, applying a vacuum, or using centrifugal forces. 19 Sample manipulation in these systems happens in continuous flow, which is appealing for high-throughput applications where millions of monodisperse microdroplets can be generated per second.
Figure 1.
Dynamic droplet generation at (a) a T-junction and (b) by flow focusing. Reprinted with permission from reference 32. Copyright 2006, American Instiute of Physics. (c) Droplet-generation-on-demand based on electric fields. Reprinted (adapted) with permission from reference 29. Copyright (2005). (d) Other droplet-on-demand systems are based on pressure using micromanipulators. Reprinted (adapted) with permission from reference 30. Copyright (2006), American Chemical Society.
The concepts for sample handling are often developed as modular systems, where each module performs a specific task, such as droplet mixing20, splitting/fusion 21, 22, shrinking 23, freezing 24, or droplet analysis by microscopic and spectroscopic methods. Depending on the microfluidic application, these modules are serially arranged to make a lab-on-a-chip device. The droplet generation and manipulation directly coupled to droplet processing is ideal for high sample throughput, with thousands of droplets generated, processed, and analyzed per second. Because these droplets flow through the micrometer-sized channels at speeds on the order of mm/s to m/s, challenges arise when the number of generated droplets cannot be processed at the same rate in subsequent modules. In these cases, the direct coupling of different modules can negatively impact the robustness of the system and constrain the utility of the device.
There are applications that do not require large sample throughput but need greater control over sample processing, such as processes that require seconds to minutes to complete. Surgery on single cells is one example. 25, 26 Droplet-on-demand systems address the needs of these applications (Fig 1c & d). These systems give precise control over droplet size and composition at low throughput, often one droplet at a time. 27–31
3. SYSTEMS FOR DROPLET DOCKING AND STORAGE
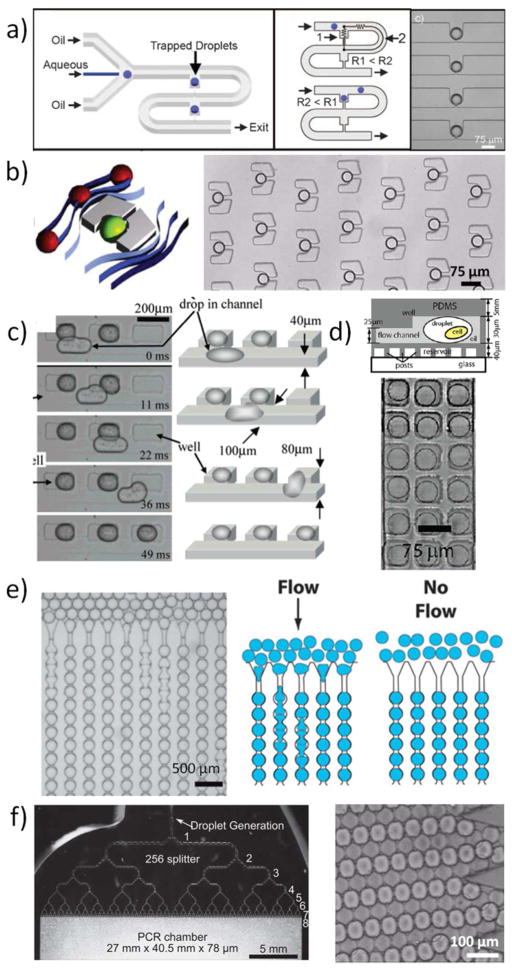
Some applications that require a time lag between droplet generation and sample reaction (e.g., kinetics, synthesis) 24, 33, 34 and data acquisition are often conducted in continuous flow where there is a long microfluidic channel directly connected to the droplet generator module. The reaction time in these cases is defined by the flow rate and the residence time of the droplets inside the channel. Other applications, however, require better control over droplets during processing. These include assays that require long term-storage of droplets, such as protein crystallization and cell culture. For these cases, the direct coupling of the droplet generation and processing modules has proven to be more challenging. Long term-storage of samples can be achieved by segmenting the sample into plugs in a microfluidic channel and subsequently stopping the flow, essentially immobilizing the plugs. However, the assays conducted in these plugs often have additional requirements, such as the mixing of samples to induce a reaction or maintaining reaction conditions. To address these requirements, various “docking” strategies to store and handle droplets have been developed. Droplet docking exploits physical means, such as vertical posts, bypass traps, or side wells, to hold a droplet in place after it is generated. Variations of docking systems have been conceived and can be classified into two groups: those that require continuous flow for the droplets to remain docked and those that do not require flow after droplet docking is completed. The former systems were initially developed for regular microfluidic perfusion-based cell culture and enzyme kinetic studies. 35, 36 These methods included bypass traps and vertical posts (Fig. 2a & b). The concept of trapping cells at vertical posts was then applied to droplets, 37–41 and combined the advantages of dynamic systems with stationary droplet analysis. The requirement of constant flow for the droplets to remain docked can be exploited to remove droplets. After the droplets are docked and the subsequent analytical steps are completed, the flow can be interrupted or reversed to clear the droplets from the traps. For the bypass traps (Fig 2a), the droplets are stored in a spatially ordered manner that facilitates droplet identification, analysis, and retrieval.
Figure 2.
Systems for droplet docking and storage: a) in-flow docking systems with by-pass traps (Reprinted (adapted) with permission from reference 40. Copyright (2009) Wiley-VCH Verlag GmbH & Co.) and b) docking by vertical posts (Reprinted (adapted) with permission from reference 39. Copyright (2006) American Chemical Society), c) droplet storage in side wells (Reprinted (adapted) with permission from reference 42. Copyright (2007) American Chemical Society) and d) top wells (Reprinted (adapted) with permission from reference 43. Copyright (2009) American Chemical Society), e) droplet immobilization (Adapted with permission from reference 44. Copyright 2009), and f) storage in large open reservoirs (Adapted with permission from references 45, 46. Copyright 2011).
Other droplet-docking systems developed are based on wells placed downstream from the droplet generator module and at the side or at the top of a main channel carrying sample plugs. The plugs dock when they pass by the wells by changing to their least energy-intensive shape in the form of spherical droplets (Fig. 2c & d). 42, 43 Another method for docking droplets is to use constricted channels (Fig. 2e). 44 The droplets flow through these constrictions but when the flow is stopped, the droplets become immobilized. All of these docking methods were developed for specific applications, with the intention of keeping the droplets docked and separated from each other. Therefore, these methods required complex channel geometries. A simplified approach for droplet storage that is less dependent on channel geometry was later developed: A large reservoir or tube takes advantage of high droplet density and the spherical shape of unconfined droplets for dense packing and storage (Fig. 2f). 45, 46 This method of droplet storage mainly relies on the use of surfactants that prevent droplets from merging. The approach is well-suited for high-throughput applications.
Overall, the different droplet generation, docking and storage methods fulfill specific needs and have their own advantages and disadvantages. For example, in-flow docking systems are ideal for applications requiring sample perfusion, but demand constant flow for the droplets to remain docked. Docking droplets in side and top wells requires no constant flow once the docking is completed, but droplet removal from the wells can be challenging. Droplet immobilization addresses the need for no-flow docking and potential droplet removal, but requires precise control over droplet size to match the immobilizer module’s geometry. Storage in large reservoirs by dense packing improves throughput, but can be challenging for identification of individual droplets in the dense package of droplets.
However, despite the compelling methods, designs, and procedures that have been developed so far, droplet microfluidics has had limited success in industrial applications. The inherent challenges of combining individual modules of microfluidic droplet manipulations are reflected in the most successful use of droplet microfluidics in commercial applications to date: the polymerase chain reaction (PCR). PCR is a well-established biochemical method that is used to determine the presence or absence of a specific DNA sequence in a sample by enzymatically amplifying the target DNA sequence. The volumes required in traditional PCR methods are on the order of microliters and the samples need to be repeatedly heated and cooled (a process known as thermocycling) to allow the amplification of a target sequence above a detection limit. PCR is an ideal assay for droplet microfluidics for several reasons. The reaction volumes are reduced by several orders of magnitude, which significantly lowers costs of reagents. The number of data sets can be significantly increased by analyzing tens of thousands of micro-PCR results, thus improving the statistics of the analysis. In addition, the reaction kinetics are faster in droplets than in bulk because the mass and heat transfer is better due to the improved surface-to-volume ratio. The transfer improvement results in better template conversion and improves reaction efficiency.
The droplet digital PCR (ddPCR) system, originally developed by QuantaLife and now marketed by BioRad Laboratories, initially uses dynamic droplet generation by flow-focusing. However, the droplets are collected off-chip in Eppendorf tubes for thermocycling and then transferred back into a chip cytometer for data acquisition and analysis. The integration of droplet thermocycling and dynamic droplet generation was shown to be challenging. Thermocycling of samples requires precise control over temperature and time for the different stages of PCR. Continuous flow systems often required a feed-back control to account for changes in temperature and residence time, depending on the flow velocity and size of the droplets. As a result, the incorporation of heating modules in microfluidic chips was very complex, both in terms of channel design and temperature control. Generating the droplets on chip, collecting them in Eppendorf tubes, and subsequently thermocycling of the droplets in traditional thermocyclers was shown to be an easier route to commercial success.
4. NEXT-GENERATION DROPLET PLATFORMS
A new generation of droplet microfluidics has recently begun. Earlier systems pre-formed droplets on-chip and then deposited them elsewhere on the same chip for processing, but these next-generation systems form and store droplets simultaneously on the same site. Because the droplet size is determined by the storage chamber geometry, these systems provide a greater tunability over droplet volumes and this simplifies the equipment needed to generate droplets. For example, the geometries of the microfluidic chambers influence the droplet formation and digitization, which is different from the older dynamic droplet generators which are dominated by flow rates of the immiscible phases. The new generation of devices is preferentially wetted by the oil phase, thus maintaining many of the favorable properties of droplet-based systems, such as interfacial control, while utilizing some of the favorable features of geometry-induced digitization.
Two next-generation systems include the SlipChip (Fig. 3a)47, 48 and the self digitization (SD) chip (Fig. 3b). 49, 50 While they share many features in common, the actual mechanisms of droplet formation and digitization are unique to each. The SlipChip consists of two separate plates, each containing a pattern of wells and ducts. The plates are aligned under oil, which eliminates air bubbles, pre-wets the device surface, and serves as a lubricating layer to help controlled slipping of the plates relative to each other. In one alignment, there is a fluidic path for the aqueous solution to enter (under simple air pressure/actuation) and fill the main fluidic path while oil drains out between the two plates. The device is then slipped to a new alignment that isolates the wells and creates digitized droplets. One unique feature of this system is the ability to perform multistep processes.
Figure 3.
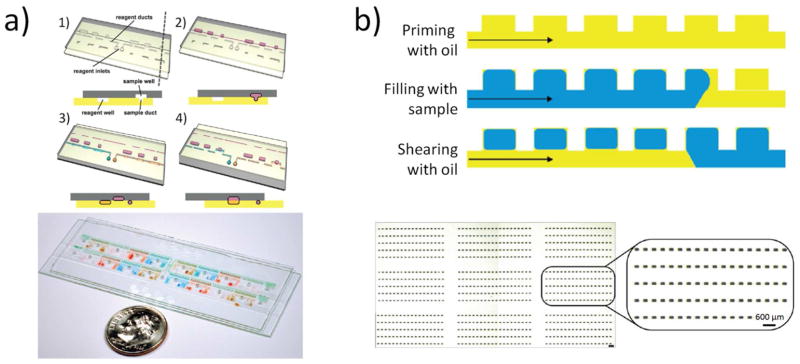
a) SlipChip (Reprinted (adapted) with permission from reference 48. Copyright (2010) American Chemical Society) and b) SD Chip (Reprinted (adapted) with permission from reference 49. Copyright (2010) American Chemical Society).
The SD chip consists of an array of chambers connected by a continuous channel network. The entire device is pre-filled with oil, and the aqueous solution is loaded using simple air pressure/actuation. The geometry of the device and interfacial properties of the aqueous solution spontaneously drives the aqueous solution into the side chambers. After the aqueous solution is loaded, more oil flows in to isolate the wells from each other, thus digitizing the aqueous droplets. This system does not require moving parts, is capable of achieving very high well density, and allows filling of the aqueous sample without any loss.
5. BIOLOGICAL APPLICATIONS
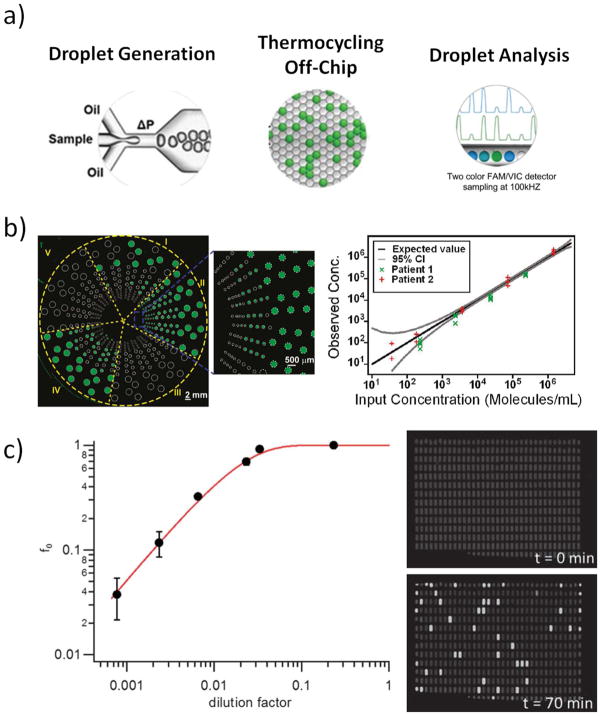
Droplet microfluidics are now overcoming the initial challenges for commercial success, most prominently in the form of digital PCR. 51, 52 Droplet digital PCR (ddPCR) is currently marketed by Bio-RAD Laboratories (Fig. 4) and Raindance Technologies. Both systems use dynamic droplet generation to create monodisperse droplets but carry out the PCR thermocycling step off-chip. The final analysis of the PCR products is conducted in a microfluidic channel. Commercial ddPCR systems are being more accepted by the research community because they are a powerful tool for studies on biomarkers. 53, 54 A similar approach, although not commercialized yet, is a microfabricated emulsion generator array (MEGA) chip 55 which uses pneumatic systems to create nanoliter droplets. Like the commercial ddPCR systems, droplets generated by the MEGA system are thermocycled off-chip and subsequently analyzed in a conventional flow cytometer.
Figure 4.
a) ddPCR in a commercial format available from Bio-RAD Laboratories. Reprinted (adapted) with permission from reference 54. Copyright (2011) American Chemical Society. b) Multiplex dPCR with a large dynamic range in a SlipChip (Reprinted (adapted with permission from reference 57. Copyright (2011) American Chemical Society) and c) digital loop-mediated DNA amplification (dLAMP) in a SD chip 50. (Reproduced by permission of The Royal Society of Chemistry).
The current application of the SlipChip and SD Chip is droplet-based PCR. In contrast to the methods mentioned earlier, these systems allow droplet generation, cycling, and analysis on the same chip without the need to transfer samples or use complex valving systems. If the chip is appropriately designed, the data read-out can be done by conventional plate readers. This capability, combined with the simplicity of droplet generation, may be advantageous for biomedical researchers who are not experts in microfluidics. These systems recently also have been shown to be versatile and powerful tools for multiplex PCR 56, 57 and digital loop-mediated DNA amplification (dLAMP). 50, 58
There is growing interest in using droplet microfluidics as a pre-sampling tool for molecular analyses and screening by mass spectrometry, in particular in combining droplet microfluidics with electrospray ionization to improve sample transfer into the mass spectrometer. 59–63 These studies are currently at the proof-of-concept stage and improving the sensitivity of the droplet systems for mass spectrometry is part of ongoing efforts.
The potential for droplet microfluidics to be a powerful tool for single-cell studies has been shown with demonstrations of single-cell screening assays. 64, 65 These assays can be used to produce monoclonal antibodies from individual hybridoma cell clones. 66
Other applications of droplet microfluidics include the analysis of protein-crystal growth 42, 49, 67 and enzyme kinetics 43, 68, as well as studies of directed evolution by in vitro selection. 69 There are also applications in small-molecule synthesis in microdroplets 70 and drug discovery. 71, 72
Droplet microfluidics has broad applicability in various fields of biomedical research. More applications have been presented and are beyond the scope of this perspective article. The interested reader is referred to comprehensive reviews on droplet microfluidics. 73–77
6. OUTLOOK
We presented a perspective view of the current developments in the field of droplet microfluidics, which is going through a rapid expansion. Droplet microfluidics has much to offer but also there are many challenges to overcome, especially in the development of robust, easy-to-use, and cost effective devices that will be employed routinely by the biomedical research and clinical community. At present, the steady state methods used for generating droplets in a high-throughput fashion cannot be easily adapted to making droplet streams with entirely different contents (e.g., not a gradient of a given content), but such a capability will advance this area. Additionally, while many droplet manipulation techniques have been developed thus far, the dominant approach to analyzing the contents of droplets still relies on fluorescence due to its ability to detect a few molecules in each droplet in a high-throughput fashion 78. This field will greatly benefit from other methods of droplet analysis that can offer comparable sensitivity and throughput as fluorescence, especially ones that can offer more chemical information 61–63.
Droplet digital PCR represents one platform that has now crossed the divide between proof-of-concept academic research and a commercial product. However, ddPCR utilizes only a fairly basic and rudimentary capability offered by droplet microfluidics: microchannels are used mostly to form monodispersed droplets that do not coalesce in an Eppendorf tube. Future commercial products that utilize more sophisticated droplet microfluidic manipulations will offer new capabilities for the biomedical community, but these new tools will have to address issues of robustness and crosstalk in coupling different droplet manipulation modules as well as ease of use and cost. The next generation droplet platforms appear promising, but given the many potential failure points in crossing the divide between proof-of-concept experiments and a robust device that will be widely adopted by non-technologists, it is still too early to determine their chances of success. The intense interest and effort devoted to advancing this field and the success this field has already enjoyed over a relatively short period signals the high expectation and promise droplet microfluidic holds and the broad and lasting impact it may make in shaping how future experiments are performed in biomedical research.
Acknowledgments
We thank NSF (CHE0844688) and NIH (GM103459) for support of this work.
References
- 1.Gravesen P, Branebjerg J, Jensen OJJ. Micromech Microeng. 1993;3:168–182. doi: 10.1088/0960-1317/3/4/002. [DOI] [Google Scholar]
- 2.Xia Y, Whitesides GM. Annu Rev Mater Sci. 1998;28:153–184. doi: 10.1002/(SICI)1521-3773(19980316)37:5<550::AID-ANIE550>3.0.CO;2-G. [DOI] [Google Scholar]
- 3.Mukhopadhyay R. Anal Chem. 2009;81:4169–73. doi: 10.1021/ac900638w. [DOI] [PubMed] [Google Scholar]
- 4.Sinclair J, Pihl J, Olofsson J, Karlsson M, Jardemark K, Chiu DT, Orwar O. Anal Chem. 2002;74:6133–8. doi: 10.1021/ac026133f. [DOI] [PubMed] [Google Scholar]
- 5.Pihl J, Karlsson M, Chiu DT. Drug Discov Today. 2005;10:1377–83. doi: 10.1016/S1359-6446(05)03571-3. [DOI] [PubMed] [Google Scholar]
- 6.Mukhopadhyay R. Anal Chem. 2006;78:1401–4. doi: 10.1021/ac069373u. [DOI] [PubMed] [Google Scholar]
- 7.Rayleigh L. Proc R Soc London. 1879;29:71–97. doi: 10.1098/rspl.1879.0015. [DOI] [Google Scholar]
- 8.Thorsen T, Roberts R, Arnold F, Quake S. Phys Rev Lett. 2001;86:4163–4166. doi: 10.1103/PhysRevLett.86.4163. [DOI] [PubMed] [Google Scholar]
- 9.Nisisako T, Torii T, Higuchi T. Lab Chip. 2002;2:24–6. doi: 10.1039/B108740C. [DOI] [PubMed] [Google Scholar]
- 10.Pamme N. Lab Chip. 2006;6:24–38. doi: 10.1039/B513005K. [DOI] [PubMed] [Google Scholar]
- 11.Di Carlo D. Lab Chip. 2009;9:3038–46. doi: 10.1039/B912547G. [DOI] [PubMed] [Google Scholar]
- 12.Cho SH, Godin JM, Chen C-H, Qiao W, Lee H, Lo Y-H. Biomicrofluidics. 2010;4:043001. doi: 10.1063/1.3511706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martinez AW, Phillips ST, Whitesides GM, Carrilho E. Anal Chem. 2010;82:3–10. doi: 10.1021/ac9013989. [DOI] [PubMed] [Google Scholar]
- 14.Kuo JS, Chiu DT. Annu Rev Anal Chem. 2011;4:275–96. doi: 10.1146/annurev-anchem-061010-113926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friend J, Yeo LY. Rev Mod Phys. 2011;83:647–704. doi: 10.1103/RevModPhys.83.647. [DOI] [Google Scholar]
- 16.Choi K, Ng AH, Fobel R, Wheeler AR. Annu Rev Anal Chem. 2012;5:413–40. doi: 10.1146/annurev-anchem-062011-143028. [DOI] [PubMed] [Google Scholar]
- 17.Garstecki P, Fuerstman MJ, Stone HA, Whitesides GM. Lab Chip. 2006;6:437–46. doi: 10.1039/B510841A. [DOI] [PubMed] [Google Scholar]
- 18.Anna SL, Bontoux N, Stone HA. Appl Phys Lett. 2003;82:364–366. doi: 10.1063/1.1537519. [DOI] [Google Scholar]
- 19.Haeberle S, Zengerle R, Ducree J. Microfluid Nanofluid. 2007;3:65–75. doi: 10.1007/s10404-006-0106-7. [DOI] [Google Scholar]
- 20.Song H, Tice JD, Ismagilov RF. Angew Chem Int Ed. 2003;42:768–72. doi: 10.1002/anie.200390203. [DOI] [PubMed] [Google Scholar]
- 21.Skelley AM, Kirak O, Suh H, Jaenisch R, Voldman J. Nat Methods. 2009;6:147–52. doi: 10.1038/nmeth.1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baroud CN, Gallaire F, Dangla R. Lab Chip. 2010;10:2032–45. doi: 10.1039/c001191f. [DOI] [PubMed] [Google Scholar]
- 23.Jeffries GD, Kuo JS, Chiu DT. Angew Chem Int Ed. 2007;46:1326–8. doi: 10.1002/anie.200603072. [DOI] [PubMed] [Google Scholar]
- 24.Sgro AE, Allen PB, Chiu DT. Anal Chem. 2007;79:4845–51. doi: 10.1021/ac062458a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jeffries GD, Edgar JS, Zhao Y, Shelby JP, Fong C, Chiu DT. Nano Lett. 2007;7:415–20. doi: 10.1021/nl0626784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zeigler MB, Chiu DT. Photochem Photobiol. 2009;85:1218–24. doi: 10.1111/j.1751-1097.2009.00581.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chiu DT, Lorenz RM. Acc Chem Res. 2009;42:649–58. doi: 10.1021/ar8002464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He M, Edgar JS, Jeffries GD, Lorenz RM, Shelby JP, Chiu DT. Anal Chem. 2005;77:1539–44. doi: 10.1021/ac0480850. [DOI] [PubMed] [Google Scholar]
- 29.He M, Kuo JS, Chiu DT. Appl Phys Lett. 2005;87:031916. doi: 10.1063/1.1997280. [DOI] [Google Scholar]
- 30.Lorenz RM, Edgar JS, Jeffries GD, Chiu DT. Anal Chem. 2006;78:6433–9. doi: 10.1021/ac060748l. [DOI] [PubMed] [Google Scholar]
- 31.Lorenz RM, Fiorini GS, Jeffries GD, Lim DS, He M, Chiu DT. Anal Chim Acta. 2008;630:124–30. doi: 10.1016/j.aca.2008.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anna SL, Mayer HC. Phys Fluids. 2006;16:121512. doi: 10.1063/1.2397023. [DOI] [Google Scholar]
- 33.Tice JD, Lyon AD, Ismagilov RF. Anal Chim Acta. 2004;507:73–77. doi: 10.1016/j.aca.2003.11.024. [DOI] [Google Scholar]
- 34.Nie Z, Xu S, Seo M, Lewis PC, Kumacheva EJ. Am Chem Soc. 2005;127:8058–63. doi: 10.1021/ja042494w. [DOI] [PubMed] [Google Scholar]
- 35.Di Carlo D, Aghdam N, Lee LP. Anal Chem. 2006;78:4925–30. doi: 10.1021/ac060541s. [DOI] [PubMed] [Google Scholar]
- 36.Di Carlo D, Wu LY, Lee LP. Lab Chip. 2006;6:1445–9. doi: 10.1039/B605937F. [DOI] [PubMed] [Google Scholar]
- 37.Lau BT, Baitz CA, Dong XP, Hansen CL. J Am Chem Soc. 2007;129:454–5. doi: 10.1021/ja065855b. [DOI] [PubMed] [Google Scholar]
- 38.Shi W, Qin J, Ye N, Lin B. Lab Chip. 2008;8:1432–5. doi: 10.1039/b808753a. [DOI] [PubMed] [Google Scholar]
- 39.Huebner A, Bratton D, Whyte G, Yang M, Demello AJ, Abell C, Hollfelder F. Lab Chip. 2009;9:692–8. doi: 10.1039/B813709A. [DOI] [PubMed] [Google Scholar]
- 40.Edgar JS, Milne G, Zhao Y, Pabbati CP, Lim DS, Chiu DT. Angew Chem Int Ed. 2009;48:2719–22. doi: 10.1002/anie.200805396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bithi SS, Vanapalli SA. Biomicrofluidics. 2010;4:044110–1. doi: 10.1063/1.3523053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shim JU, Cristobal G, Link DR, Thorsen T, Jia Y, Piattelli K, Fraden S. J Am Chem Soc. 2007;129:8825–35. doi: 10.1021/ja071820f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shim JU, Olguin LF, Whyte G, Scott D, Babtie A, Abell C, Huck WT, Hollfelder F. J Am Chem Soc. 2009;131:15251–6. doi: 10.1021/ja904823z. [DOI] [PubMed] [Google Scholar]
- 44.Schmitz CHJ, Rowat AC, Köster S, Weitz DA. Lab Chip. 2009;9:44–49. doi: 10.1039/B809670H. [DOI] [PubMed] [Google Scholar]
- 45.Hatch AC, Fisher JS, Pentoney SL, Yang DL, Lee AP. Lab Chip. 2011;11:2509–17. doi: 10.1039/c0lc00553c. [DOI] [PubMed] [Google Scholar]
- 46.Hatch AC, Fisher JS, Tovar AR, Hsieh AT, Lin R, Pentoney SL, Yang DL, Lee AP. Lab Chip. 2011;11:3838–45. doi: 10.1039/c1lc20561g. [DOI] [PubMed] [Google Scholar]
- 47.Du W, Li L, Nichols KP, Ismagilov RF. Lab Chip. 2009;9:2286–92. doi: 10.1039/B908978K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li L, Du W, Ismagilov RJ. Am Chem Soc. 2010;132:106–11. doi: 10.1021/ja908555n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cohen DE, Schneider T, Wang M, Chiu DT. Anal Chem. 2010;82:5707–5717. doi: 10.1021/ac100713u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gansen A, Herrick AM, Dimov IK, Lee LP, Chiu DT. Lab Chip. 2012;12:2247–54. doi: 10.1039/c2lc21247a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vogelstein B, Kinzler KW. Proc Natl Acad Sci U S A. 1999;96:9236–41. doi: 10.1073/pnas.96.16.9236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baker M. Nature. 2010;7:351–356. doi: 10.1038/nmeth0510-351. [DOI] [Google Scholar]
- 53.Devonshire AS, Sanders R, Wilkes TM, Taylor MS, Foy CA, Huggett JF. Methods. 2013;59:89–100. doi: 10.1016/j.ymeth.2012.07.021. [DOI] [PubMed] [Google Scholar]
- 54.Pinheiro LB, Coleman VA, Hindson CM, Herrmann J, Hindson BJ, Bhat S, Emslie KR. Anal Chem. 2012;84:1003–11. doi: 10.1021/ac202578x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zeng Y, Novak R, Shuga J, Smith MT, Mathies RA. Anal Chem. 2010;82:3183–90. doi: 10.1021/ac902683t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shen F, Du W, Davydova EK, Karymov MA, Pandey J, Ismagilov RF. Anal Chem. 2010;82:4606–4612. doi: 10.1021/ac1007249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shen F, Sun B, Kreutz JE, Davydova EK, Du W, Reddy PL, Joseph LJ, Ismagilov RF. J Am Chem Soc. 2011;133:17705–12. doi: 10.1021/ja2060116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shen F, Davydova EK, Du W, Kreutz JE, Piepenburg O, Ismagilov RF. Anal Chem. 2011;83:3533–3540. doi: 10.1021/ac200247e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kelly RT, Page JS, Marginean I, Tang K, Smith RD. Angew Chem Int Ed. 2009;48:6832–5. doi: 10.1002/anie.200902501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kelly RT, Tang K, Irimia D, Toner M, Smith RD. Anal Chem. 2008;80:3824–31. doi: 10.1021/ac8000786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ji J, Nie L, Qiao L, Li Y, Guo L, Liu B, Yang P, Girault HH. Lab Chip. 2012;12:2625–9. doi: 10.1039/c2lc40206h. [DOI] [PubMed] [Google Scholar]
- 62.Sun S, Slaney TR, Kennedy RT. Anal Chem. 2012;84:5794–800. doi: 10.1021/ac3011389. [DOI] [PubMed] [Google Scholar]
- 63.Baker CA, Roper MG. Anal Chem. 2012;84:2955–60. doi: 10.1021/ac300100b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Courtois F, Olguin LF, Whyte G, Bratton D, Huck WT, Abell C, Hollfelder F. Chembiochem. 2008;9:439–46. doi: 10.1002/cbic.200700536. [DOI] [PubMed] [Google Scholar]
- 65.Brouzes E, Medkova M, Savenelli N, Marran D, Twardowski M, Hutchison JB, Rothberg JM, Link DR, Perrimon N, Samuels ML. Proc Natl Acad Sci U S A. 2009;106:14195–200. doi: 10.1073/pnas.0903542106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.El Debs B, Utharala R, Balyasnikova IV, Griffiths AD, Merten CA. Proc Natl Acad Sci U S A. 2012;109:11570–5. doi: 10.1073/pnas.1204514109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li L, Du W, Ismagilov RF. J Am Chem Soc. 2010;132:112–9. doi: 10.1021/ja908558m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Venancio-Marques A, Liu Y-J, Diguet A, Maio Td, Gautier A, Baigl D. ACS Synth Biol. 2012;1:526–531. doi: 10.1021/sb300010a. [DOI] [PubMed] [Google Scholar]
- 69.Fallah-Araghi A, Baret JC, Ryckelynck M, Griffiths AD. Lab Chip. 2012;12:882–91. doi: 10.1039/c2lc21035e. [DOI] [PubMed] [Google Scholar]
- 70.Matosevic S, Paegel BM. J Am Chem Soc. 2011;133:2798–800. doi: 10.1021/ja109137s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gañan-Calvo AM, Martin-Banderas L, Gonzalez-Prieto R, Rodriguez-Gil A, Berdun-Alvarez T, Cebolla A, Chavez S, Flores-Mosquera M. Int J Pharm. 2006;324:19–26. doi: 10.1016/j.ijpharm.2006.05.032. [DOI] [PubMed] [Google Scholar]
- 72.Häfeli UO, Saatchi K, Elischer P, Misri R, Bokharaei M, Labiris NR, Stoeber B. Biomacromolecules. 2010;11:561–567. doi: 10.1021/bm9010722. [DOI] [PubMed] [Google Scholar]
- 73.Teh SY, Lin R, Hung LH, Lee AP. Lab Chip. 2008;8:198–220. doi: 10.1039/B715524G. [DOI] [PubMed] [Google Scholar]
- 74.Chiu DT, Lorenz RM, Jeffries GD. Anal Chem. 2009;81:5111–8. doi: 10.1021/ac900306q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chiu DT. Anal Bioanal Chem. 2010;397:1618–2642. doi: 10.1007/s00216-010-3686-8. [DOI] [PubMed] [Google Scholar]
- 76.Theberge AB, Courtois F, Schaerli Y, Fischlechner M, Abell C, Hollfelder F, Huck WT. Angew Chem Int Ed. 2010;49:5846–68. doi: 10.1002/anie.200906653. [DOI] [PubMed] [Google Scholar]
- 77.Pompano RR, Liu W, Du W, Ismagilov RF. Annu Rev Anal Chem. 2011;4:59–81. doi: 10.1146/annurev.anchem.012809.102303. [DOI] [PubMed] [Google Scholar]
- 78.Jeffries GD, Lorenz RM, Chiu DT. Anal Chem. 2010;82:9948–54. doi: 10.1021/ac102173m. [DOI] [PMC free article] [PubMed] [Google Scholar]