Abstract
We previously reported that paternally-inherited human leukocyte antigen (HLA) alleles are a template for women's preference for male odors (P = 0.0007). However, it has been suggested that sequence variation in a nearby olfactory receptor (OR) cluster on chromosome 6p influences smell preference. To determine if the HLA-linked OR genes contribute to previously observed HLA-mediated behaviors, we use the odor preference data from our earlier study in combination with a new resequencing study of four functional HLA-linked OR genes in the same subjects. Our results indicate that OR alleles in the genes surveyed are not in linkage disequilibrium (LD) with HLA variation and do not explain the previous findings of HLA-associated odor preference.
Introduction
The genes of the major histocompatibility complex (MHC) play critical roles in immune response and disease resistance, as well as in mediating social and reproductive behaviors [1, 2]. In rodents, MHC loci are involved in mating preferences and kin recognition 3-6, and odor-guided kin recognition has been reported in a number of other vertebrates, including fish, birds, hamsters, beavers, and humans [3-8]. In some vertebrates MHC-mediated behaviors are olfactory mediated [1, 6, 9-12], but not in others [3, 13].
The human MHC, encoding the human leukocyte antigens (HLA), includes the most polymorphic loci in the genome and has been implicated in human mate choice [14, 15] and odor preferences [8, 16, 17]. Previous work in our laboratory demonstrated that paternally inherited HLA alleles influence a woman's preferences for male odors [16]. Specifically, women preferred the odors of men with more HLA matches to her paternally-inherited alleles (1.39±0.15 matches to most preferred versus 0.55±0.10 matches to least preferred, out of 5 possible matches; P < 0.0001 by paired t-test). Matches to alleles inherited from her mother or to the noninherited alleles from either parent did not influence her choice (P > 0.50).
The region of chromosome 6p21 containing the HLA complex has among the highest levels of linkage disequilibrium (LD) observed in the human genome [18, 19], with large stretches of LD encompassing both HLA and non-HLA loci. Therefore, the identification of specific variants, or even genes, that contribute to HLA-associated diseases or behaviors is not straightforward due to the complicated relationships among loci in the region. For example, a cluster of olfactory receptor (OR) genes is located telomeric to the HLA complex, within a region of high LD spanning over 500 kb [19]. Proteins encoded by OR loci serve as the basis for odor detection, and it has therefore been suggested that the close linkage of the OR cluster with the HLA complex could account for some of the behavioral observations attributed to HLA [16, 20-23]. This could occur if alleles in OR genes and HLA haplotypes are in sufficient LD so that the HLA alleles or haplotypes merely serve as markers for the relevant OR alleles. In addition, alleles at the HLA-linked OR genes (in the smellers) would have to specifically bind odorants produced by HLA molecules (in the donors).
To further examine this question, we resequenced four functional HLA-linked OR genes in the same subjects that participated in our previous study of HLA-associated odor preferences [16] and examined patterns of LD between the OR and HLA loci in these subjects. We then directly tested the hypothesis that variation in these HLA-linked OR genes influence women's preferences for male body odors or account for all or some of the previously observed HLA-associated preferences.
Materials and Methods
Study Sample
The 49 women participants are of European descent and were unmarried at the time of our earlier study. The average age was 25 years (range=13 to 56), and none were using hormonal contraception or had ever been pregnant. The women were unaware of the study hypothesis and the identity of the odors in question. The six male T-shirt donors ranged in age from 23 to 47 years of age (average=31.3 years). More detailed information on sample composition, odor collection, olfactory sessions, and HLA typing can be found in Jacob et al. [16].
Studies of OR Genes
Four functional (single exon) OR genes were selected from each of the three LD bins that we previously identified in this region (Nicolae et al. 2005). To represent the variation in this region, we studied 1) OR2H2 (called FAT11 [20] and OR2H3 [24] in our previous studies), the only OR gene in the most proximal bin (closest to the HLA gene cluster), 2) OR5V1 from the most distal bin, and 3) OR12D2 and OR10C1 from the middle bin. PCR and sequencing primers were designed according to GenBank sequences (OR12D2, NM_013936; OR10C1, NT_113893; OR2H2, NM_007160; and OR5V1, NM_030876). DNA from one chimpanzee was amplified and sequenced using the same primers and used to determine the ancestral allele at each single nucleotide polymorphism (SNP). All genes were amplified using TagGold DNA polymerase, primers, and amplification conditions as described in Supplemental Table 1. PCR products were cleaned with exonuclease I and shrimp alkaline phosphatase (United States Biochemicals). Dye-terminator sequencing was performed with ABI Big Dye Terminator v. 3.1 Cycle Sequencing kit and products were analyzed on an ABI 3700 or 3730 automated sequencer (Applied Biosystems). The Phred-Phrap-Consed package was used to assemble and analyze all sequences [25]. All putative polymorphisms were visually inspected and individual genotypes were confirmed using Consed. All SNPs in this study were in Hardy Weinberg equilibrium and we observed no Mendelian errors.
A low frequency copy number variant in the region encompassing the OR2H2 gene has been reported [26, 27] (2 events in 270 HapMap individuals [27]). In order to investigate the possibility of copy number variation more directly, we performed quantitative PCR of genomic DNA with a known 2-copy control gene (OR12D2) in the 49 study participants. Based on the ratio of the two genes, three women appear to have more than two copies of OR2H2, however, removing these women from the analysis did not affect the results.
Haplotype Construction and Parent of Origin
Each of the women and their parents were previously genotyped for two SNPs in OR2H2 and one SNP each in OR10C1 and OR12D2 [24]. Because no family data were available for SNPs in OR5V1, the parents of each woman were genotyped by direct sequencing for the two SNPs in this gene. Haplotype construction and parent of origin were determined by direct observation of alleles in the women participants and their parents.
Data analysis
LD between OR SNPs and multi-allelic HLA genes was assessed by r2, with the average value calculated as the mean of all pairwise r2 between OR SNPs and each allele at the five HLA loci.
To test the hypothesis that diplotypes or alleles at OR loci were associated with female's preferences for male odors, we constructed test statistics that quantify the difference in preference for subjects grouped by genotype/diplotype. For diplotype comparisons, we counted the number of times each donor was ranked as the most or least preferred according to women's diplotype (for all diplotypes with at least 5 occurances). The results were analyzed by chi-squared tests. To confirm that the previously reported association between paternally-inherited HLA alleles and preferences for male odor donors can not be explained by sequence variation at the OR genes, we repeated the same analysis as in our earlier paper [16], stratified by paternally inherited alleles at the SNPs in the OR genes. For every smeller we calculated the difference between the number of paternal HLA matches with the most and least preferred donor. These differences were shown previously to be significantly larger than zero using a sign test, and we contrasted, for every SNP, the distribution of the signs between the two groups formed based on the paternally inherited allele. The comparison of the two allele groups was performed using Fisher's exact test.
Results
Twenty-two SNPs in four OR genes are present in this sample (Table 1). Eighteen result in non-synonymous/replacement changes, three in synonymous/silent changes, and one in a nonsense mutation. The OR10C1 gene may be a segregating pseudogene, as we observed a nonsense mutation on a unique haplotype background (rs17184009, Q55X).
Table 1.
Summary of twenty two SNPs identified in the four OR genes surveyed.
| Gene | Marker | Allelesa | Effect | Frequencyb |
|---|---|---|---|---|
| OR5V1 | rs6930033 | T/G | L23F | 0.2 |
| rs9257770 | T/G | I45M | 0.04 | |
| OR12D2 | rs9257834 | G/T | V47F | 0.51 |
| rs4987411 | C/T | L56P | 0.49 | |
| rs2073154 | C/G | F113L | 0.51 | |
| rs2073153 | G/T | R120L | 0.49 | |
| rs2073152 | G/C | S121C | 0.71 | |
| rs2073151 | A/G | I159V | 0.49 | |
| rs2073150 | C/T | L255L | 0.49 | |
| OR10C1 | rs17184009 | C/T | Q55X | 0.02 |
| rs2074469 | T/C | F60L | 0.16 | |
| rs11755177 | C/T | F61F | 0.02 | |
| rs11755182 | C/A | R89S | 0.02 | |
| rs17177639 | C/T | R121C | 0.02 | |
| rs17177646 | C/T | R138W | 0.02 | |
| rs2074468 | T/C | P160S | 0.02 | |
| rs2074466 | C/A | P174Q | 0.18 | |
| rs2074464 | G/A | M246V | 0.43 | |
| rs10946991 | C/A | A254A | 0.02 | |
| rs11968123 | T/G | M310R | 0.02 | |
| OR2H2 | rs3129034 | T/C | L30F | 0.2 |
| rs1233387 | T/C | A48V | 0.55 |
Alleles shown as ancestral/derived.
Frequency of the derived allele.
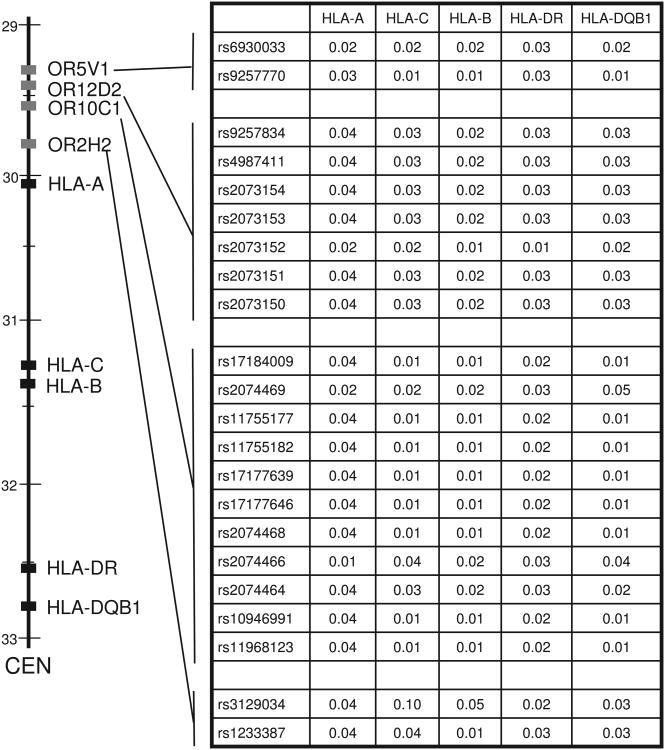
To first determine whether variation in any of the OR genes was in long range LD with HLA loci, we examined the LD pattern between OR SNPs and alleles at the five HLA genes included in our earlier study, in the same 49 women, using the linkage disequilibrium measure r2 (Figure 1). OR2H2 is the closest OR gene to the HLA complex and although it contains the SNP with the highest r2 values (rs3129034), it still shows overall very low levels of LD with alleles at HLA loci: HLA-A (mean r2 = 0.04, range = 0.00-0.17), HLA-C (mean r2 = 0.10, range = 0.02-0.23), HLA-B (mean r2 = 0.05, range = 0.00-0.21), HLA-DR (mean r2 = 0.02, range = 0.00-0.08), and HLA-DQB1 (mean r2 = 0.03, range = 0.00-0.08) (Figure 1). Overall, levels of LD between alleles at OR SNPs and five HLA genes are not sufficient to implicate OR sequence variation in the previously reported associations with HLA and smell preference (Figure 2).
Figure 1.
Linkage disequilibrium between SNPs in four 6p21-22 OR genes and five HLA loci. r2 values were averaged across all allele combinations for each pair of loci.
Figure 2.
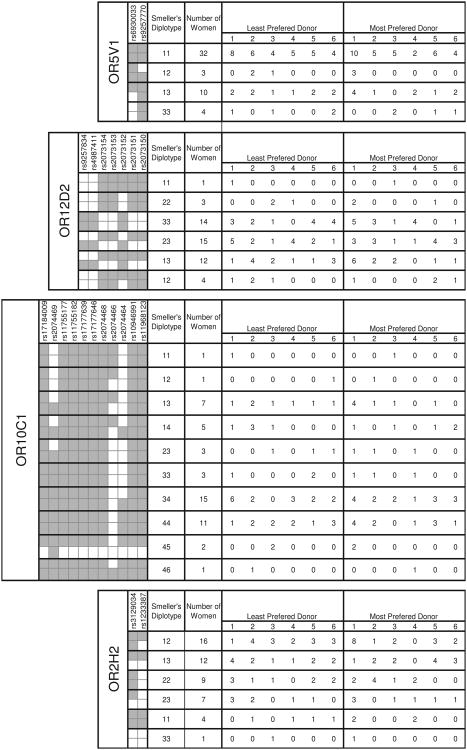
Polymorphisms and diplotypes at each of four OR genes are shown for 49 women smellers. Gray and white squares indicate ancestral and derived alleles, respectively, at each polymorphic site. Each observed haplotype (combination of alleles inherited from one parent) was assigned a number; the pair of haplotypes inherited from both parents is referred to as a diplotype. The women's ranking of each of six odor donors (labeled 1-6) as the least and most preferred are shown for women genotyped by each diplotype (from Jacob et al.) [16].
To directly rule out this possibility, we next tested the hypothesis that alleles at one or more OR genes determine preferences for male odors. Because each of the five unrelated male donors had a unique set of HLA alleles [16], we considered each male to have a unique HLA-based odor profile. No genotype at any of the OR loci was associated with women's choice of either the most or the least preferred donor (χ2 test, P > 0.05). The smellers' diplotypes (pair of haplotypes) at each OR gene and choice of the most preferred and the least preferred odor donor are shown in Figure 2. At each locus, the choice of their most and least preferred odor was random with respect to the smeller's diplotype (χ2 test, P >0.05 for all diplotypes with at least five occurrences).
Lastly, because we previously reported that women preferred the odors of men with more HLA matches to her paternally-inherited alleles (versus maternally inherited alleles or noninherited alleles from either parent), we examined whether the paternally inherited allele at each OR SNP influenced these results. Of the 22 OR SNPs, eight were not polymorphic on the paternally-inherited haplotypes. For the remaining 14 SNPs, we stratified women by their paternally-inherited OR alleles and tested if the distribution of the paternal HLA matches to least and most preferred donor was different in the two groups. Because the original analysis was done with a sign-test, we contrasted the signs (see Methods) and found no evidence that the HLA effect is dependent on the alleles inherited at the linked OR loci (P>0.05 for every SNP).
Discussion
We demonstrate here for the first time that HLA-based choices of human odors [16] are not influenced by variation in the linked OR gene cluster. We previously observed recombination between HLA-A and OR2H2 [20] and suggested that there might be insufficient LD between OR2H2 and HLA loci to account for the associations between olfactory-mediated behaviors and HLA genes per se, although this conclusion was disputed by others [23]. Here, we show not only that measures of LD between the OR and HLA loci are negligible (maximum mean r2 ≤ 0.05), but also that odor preferences are not associated directly with variation in the linked OR genes.
Our study sample provides a unique opportunity for investigating HLA- versus OR-based odor preferences, because we previously demonstrated preferences for male odors based on the number of HLA matches between the odor donors and each woman's paternally-inherited HLA alleles in this same sample [16]. The results presented here clearly show that there is insufficient LD between the HLA and OR genes to account for the observed HLA-mediated effects in humans. Although we did not survey all OR genes in the extended MHC, the haplotype structure and pattern of LD in this region [19, 24, 28, 29] make it unlikely that variation in other OR genes would provide contradictory results. Thus, we conclude that the evidence for HLA-based mate choice and odor preferences in humans is not due to standing variation at OR loci in the extended MHC on chromosome 6p.
Supplementary Material
Acknowledgments
The authors wish to thank Yoav Gilad for primer design and helpful discussions regarding the manuscript. This work was supported by from the National Institutes of Health [HD21244 to C.O.], T32 HL007605 [E.E.T.], and a Dennis W. Jahnigen/American Geriatrics Society fellowship [J.M.P.].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Boehm T, Zufall F. MHC peptides and the sensory evaluation of genotype. Trends in neurosciences. 2006 Feb;29(2):100–7. doi: 10.1016/j.tins.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 2.Loisel D, Alberts S, Ober C. Functional significance of MHC variation in mate choice, reproductive outcome, and disease risk. Evolution in Health and Disease. (2nd) 2007 [Google Scholar]
- 3.Ekblom R, Saether SA, Grahn M, Fiske P, Kalas JA, Hoglund J. Major histocompatibility complex variation and mate choice in a lekking bird, the great snipe (Gallinago media) Mol Ecol. 2004 Dec;13(12):3821–8. doi: 10.1111/j.1365-294X.2004.02361.x. [DOI] [PubMed] [Google Scholar]
- 4.Gilbert AN, Yamazaki K, Beauchamp GK, Thomas L. Olfactory discrimination of mouse strains (Mus musculus) and major histocompatibility types by humans (Homo sapiens) J Comp Psychol. 1986 Sep;100(3):262–5. [PubMed] [Google Scholar]
- 5.Mateo JM, Johnston RE. Kin recognition and the ‘armpit effect’: evidence of self-referent phenotype matching. Proc Biol Sci. 2000 Apr 7;267(1444):695–700. doi: 10.1098/rspb.2000.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reusch TB, Haberli MA, Aeschlimann PB, Milinski M. Female sticklebacks count alleles in a strategy of sexual selection explaining MHC polymorphism. Nature. 2001 Nov 15;414(6861):300–2. doi: 10.1038/35104547. [DOI] [PubMed] [Google Scholar]
- 7.Sun L, Uuml, Ller-Schwarze D. Sibling recognition in the beaver: a field test for phenotype matching. Anim Behav. 1997 Sep;54(3):493–502. doi: 10.1006/anbe.1996.0440. [DOI] [PubMed] [Google Scholar]
- 8.Wedekind C, Seebeck T, Bettens F, Paepke AJ. MHC-dependent mate preferences in humans. Proc Biol Sci. 1995 Jun 22;260(1359):245–9. doi: 10.1098/rspb.1995.0087. [DOI] [PubMed] [Google Scholar]
- 9.Eggert F, Luszyk D, Haberkorn K, Wobst B, Vostrowsky O, Westphal E, et al. The major histocompatibility complex and the chemosensory signalling of individuality in humans. Genetica. 1998;104(3):265–73. doi: 10.1023/a:1026431303879. [DOI] [PubMed] [Google Scholar]
- 10.Hurst JL, Payne CE, Nevison CM, Marie AD, Humphries RE, Robertson DH, et al. Individual recognition in mice mediated by major urinary proteins. Nature. 2001 Dec 6;414(6864):631–4. doi: 10.1038/414631a. [DOI] [PubMed] [Google Scholar]
- 11.Olsson M, Madsen T, Nordby J, Wapstra E, Ujvari B, Wittsell H. Major histocompatibility complex and mate choice in sand lizards. Proc Biol Sci. 2003 Nov 7;270(2):S254–6. doi: 10.1098/rsbl.2003.0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamazaki K, Yamaguchi M, Baranoski L, Bard J, Boyse EA, Thomas L. Recognition among mice. Evidence from the use of a Y-maze differentially scented by congenic mice of different major histocompatibility types. J Exp Med. 1979 Oct 1;150(4):755–60. doi: 10.1084/jem.150.4.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zelano B, Edwards SV. An MHC component to kin recognition and mate choice in birds: predictions, progress, and prospects. The American Naturalist. 2002;160:S225–S37. doi: 10.1086/342897. [DOI] [PubMed] [Google Scholar]
- 14.Ober C, Weitkamp LR, Cox N, Dytch H, Kostyu D, Elias S. HLA and mate choice in humans. Am J Hum Genet. 1997 Sep;61(3):497–504. doi: 10.1086/515511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chaix R, Cao C, Donnelly P. Is mate choice in humans MHC-dependent? 2008 doi: 10.1371/journal.pgen.1000184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jacob S, McClintock MK, Zelano B, Ober C. Paternally inherited HLA alleles are associated with women's choice of male odor. Nat Genet. 2002 Feb;30(2):175–9. doi: 10.1038/ng830. [DOI] [PubMed] [Google Scholar]
- 17.Wedekind C, Furi S. Body odour preferences in men and women: do they aim for specific MHC combinations or simply heterozygosity? Proc Biol Sci. 1997 Oct 22;264(1387):1471–9. doi: 10.1098/rspb.1997.0204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Bakker PI, McVean G, Sabeti PC, Miretti MM, Green T, Marchini J, et al. A high-resolution HLA and SNP haplotype map for disease association studies in the extended human MHC. Nat Genet. 2006 Oct;38(10):1166–72. doi: 10.1038/ng1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miretti MM, Walsh EC, Ke X, Delgado M, Griffiths M, Hunt S, et al. A high-resolution linkage-disequilibrium map of the human major histocompatibility complex and first generation of tag single-nucleotide polymorphisms. Am J Hum Genet. 2005 Apr;76(4):634–46. doi: 10.1086/429393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eklund AC, Belchak MM, Lapidos K, Raha-Chowdhury R, Ober C. Polymorphisms in the HLA-linked olfactory receptor genes in the Hutterites. Hum Immunol. 2000 Jul;61(7):711–7. doi: 10.1016/s0198-8859(00)00132-4. [DOI] [PubMed] [Google Scholar]
- 21.Fan W, Cai W, Parimoo S, Schwarz DC, Lennon GG, Weissman SM. Identification of seven new human MHC class I region genes around the HLA-F locus. Immunogenetics. 1996;44(2):97–103. doi: 10.1007/BF02660056. [DOI] [PubMed] [Google Scholar]
- 22.Yamazaki K, Boyse EA, Mike V, Thaler HT, Mathieson BJ, Abbott J, et al. Control of mating preferences in mice by genes in the major histocompatibility complex. J Exp Med. 1976 Nov 2;144(5):1324–35. doi: 10.1084/jem.144.5.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ziegler A, Ehlers A, Forbes S, Trowsdale J, Volz A, Younger R, et al. Polymorphisms in olfactory receptor genes: a cautionary note. Hum Immunol. 2000 Dec;61(12):1281–4. doi: 10.1016/s0198-8859(00)00219-6. [DOI] [PubMed] [Google Scholar]
- 24.Nicolae D, Cox NJ, Lester LA, Schneider D, Tan Z, Billstrand C, et al. Fine mapping and positional candidate studies identify HLA-G as an asthma susceptibility gene on chromosome 6p21. Am J Hum Genet. 2005 Feb;76(2):349–57. doi: 10.1086/427763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nickerson DA, Tobe VO, Taylor SL. PolyPhred: automating the detection and genotyping of single nucleotide substitutions using fluorescence-based resequencing. Nucleic Acids Res. 1997 Jul 15;25(14):2745–51. doi: 10.1093/nar/25.14.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kidd JM, Cooper GM, Donahue WF, Hayden HS, Sampas N, Graves T, et al. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008 May 1;453(7191):56–64. doi: 10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, et al. Global variation in copy number in the human genome. Nature. 2006 Nov 23;444(7118):444–54. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Younger RM, Amadou C, Bethel G, Ehlers A, Lindahl KF, Forbes S, et al. Characterization of clustered MHC-linked olfactory receptor genes in human and mouse. Genome research. 2001 Apr;11(4):519–30. doi: 10.1101/gr.160301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu HX, Chia JM, Bourque G, Wong MV, Chan SH, Ren EC. A population-based LD map of the human chromosome 6p. Immunogenetics. 2005 Sep;57(8):559–65. doi: 10.1007/s00251-005-0002-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.