Abstract
MicroRNAs (miRNAs) have recently been shown to play fundamental roles in diverse cellular processes and linked to variety of cancers. Dicer and Drosha are two major enzymes in the miRNA maturation process. DGCR8 is the assistant of Drosha in the microprocessor complex. In this study, we evaluated the mRNA expression profiles of major miRNA processing machinery Drosha, Dicer, and DGCR8 in human gastrointestinal (AGS, KYSE30 and HepG2) cancer cell lines. Materials and Methods: The cells were cultured and harvested, and total cellular RNA was isolated from cells. Then, first-strand cDNA was synthesized from the RNA of cells. Afterward, Quantitative analysis was performed by real-time RT-PCR using the PowerSYBR Green PCR Master Mix. Results: Expression levels of Drosha in AGS and HepG2 cells were higher than the controls, whereas, Drosha’s expression level in KYSE-30 cell line was lower. The Dicer expression levels in AGS and HepG2 cells were higher, while, its expression level in KYSE-30 cell was lower. The DGCR8 expression levels in all three cell lines were significantly higher than the control samples. Conclusion: Expression levels of the two most important enzymes of the miRNA machinery, Drosha and Dicer, and microprocessor complex component, DGCR8 were noticeably dysregulated when compared to healthy controls.
Keywords: Drosha, Dicer, DGCR8, microRNA machinery, cancer cell line
Introduction
MicroRNAs (miRNA) are 17 to 24 nucleotide (nt) RNA molecules that are considered as a group of non-protein-coding RNAs; miRNAs are capable of post-transcriptional gene regulation by binding to target mRNAs [1]. The miRNA base-pairs with target mRNA to direct gene silencing through mRNA cleavage or translation repression based on the level of complientarily between the miRNA and the mRNA target [2]. miRNAs have recently been shown to play fundamental roles in diverse developmental and cellular processes and linked to variety of cancers [3].
miRNA maturation begins in the nucleus, where the primary miRNA (pri-miRNA) transcript is transcribed by RNA polymerase II [4]. The RNA III endonuclease Drosha inside the nucleus, beside with DiGeorge syndrome critical region gene 8 (DGCR8 or Pasha) construct the microprocessor complex. This complex cuts the pri-miRNA transcript within the nucleus into several precursor miRNAs (pre-miRNAs) [5]. Drosha is the catalytic subunit of the pri-miRNA processing microprocessor complex, while DGCR8 stabilizes Drosha and recognizes the RNA substrate [6,7]. The 70-90 nt long pre-miRNAs are then transported to the cytoplasm for further processing, where Dicer, another RNase III enzyme cleaves it to produce mature miRNA [8]
Although several studies have revealed dysregulation of microRNA (miRNA) expression profiles in various cancers, there has been little research on the miRNA machinery itself. In this study, we evaluated the mRNA expression profiles of major miRNA processing machinery Drosha and Dicer, and DGCR8 in human gastrointestinal (AGS, KYSE30 and HepG2) cancer cell lines.
Materials and methods
Cell lines
The human cancer cell lines including, human gastric carcinoma (AGS), human esophageal squamous cell carcinoma (KYSE-30), and human hepatocellular carcinoma (HepG2) were purchased from Pasteur Institute of Iran Cell Bank (Tehran, Iran). All reagents and medium were prepared just before use. Normal cells were obtained from healthy people.
Cell culture
The cells were cultured in RPMI-1640 (Cat. No: 51800–035, GIBCO, UK) medium suppliented with 10% fetal bovine serum (FBS; Cat. No: 10270–106, GIBCO, UK), penicillin 100 unit/ml and streptomycin 100 μg/ml. The cells were incubated at 37°C in a water saturated atmosphere of 5% CO2 and 95% air until confluence. One week later, the cells were rioved with a solution containing 0.25 (w/v) trypsin and 0.02 (w/v) ethylenediaminetetraacetic acid.
RNA isolation
Total cellular RNA was isolated from cells using Trizol reagent (Cat. No: 15596-026, Invitrogen, CA, USA), following this procedure; cultured cells were harvested and centrifuged (1000 rpm, 10 min) for rioving supernatant media; afterward 0.8ml of Trizol reagent were added per 5x106 cell, after pipetting, 0.2ml of chloroform were added and tubes were shaken vigorously, after that samples were centrifuged at 12000g for 15min at 4°C, then upper aqueous phase was transferred to a new tube by angling the tube at 45° and pipetting the solution out, subsequently 0.5ml of isopropanol were added and incubated at room tiperature for 10min, next samples were centrifuged at 12000g for 10min at 4°C, after that supernatants were rioved and pellet of the tubes were washed and vortexed with 1ml of 75% ethanol and were centrifuged at 7500g for 5min at 4°C, finally supernatants were discarded and RNA pellets were air dried for 10min; and were re-suspended in 40μl of RNAase free water. Prepared RNAs were stored at -80°C refrigerator until downstream application.
Reverse transcription and cDNA synthesis
First-strand cDNA was synthesized from the RNA of cells by RevertAid First Strand cDNA Synthesis Kit, Fermentas (Cat No: #K1621, Maryland, USA) according to the manufacturer’s protocol. Briefly, 4μl of isolated RNA from cells were mixed with 1μl of random hexamer primer and 7μl RNAase-free water and incubated at 65°C for 5min. then micro-tubes were chilled on ice, afterward, mixture of (reaction buffer 4μl + RNAase inhibitor 1μl + dNTP mix 2μl + reverse transcriptase 1μl) was added on each sample. Immediately samples were incubated at 25°C for 5min followed by 42°C for 60min; the reaction was terminated by heating at 70°C for 5min. Reverse transcription was performed in the final volume of 20μL.
Real-time quantitative RT-PCR
Primers were used from previous studies [9,10] that Sand, et al, evaluated Drosha, Dicer, DGCR8 and RISC components in their studies. Primers (Drosha, Dicer, DGCR8 and the RPL38 as a housekeeping gene) were designed using Primer Express 3.0 (PE Applied Biosystis, Foster City, CA, USA). See Supplientary Table 1 for the details of primers used in quantitative real-time PCR. For accuracy and specificity all primers were blasted in NCBI website: http://www.ncbi.nlm.nih.gov/tools/primer-blast/. Primers were produced by the custom oligonucleotide synthesis service, Metabion (Martinsried, Germany).
Table 1.
Primers used in quantitative PCR, the amplicon sizes and the melting tiperature (Tm) of each reaction
| Target | Sequence | Amplicon Size (bp) | Tm (°C) |
|---|---|---|---|
| Dicer F | 5′-TTAACCTTTTGGTGTTTGATGAGTGT-3′ | 94 | 58.5 |
| Dicer R | 5′-GGACATGATGGACAATTTTCACA-3′ | 57.1 | |
| Drosha F | 5′-CATGTCACAGAATGTCGTTCCA-3′ | 115 | 58.4 |
| Drosha R | 5′- GGGTGAAGCAGCCTCAGATTT-3′ | 59.8 | |
| DGCR8 F | 5′-GCAAGATGCACCCACAAAGA-3′ | 93 | 57.3 |
| DGCR8 R | 5′-TTGAGGACACGCTGCATGTAC-3′ | 59.8 | |
| RPL38 F | 5′-TCACTGACAAAGAGAAGGCAGAGA-3′ | 88 | 61 |
| RPL38 R | 5′-TCAGTGTGTCTGGTTCATTTCAGTT-3′ | 59.7 |
F: forward primer, R: reverse primer, bp: base pairs, RPL38: ribosomal protein (housekeeping gene).
Quantitative analysis was performed by real-time RT-PCR using the PowerSYBR Green PCR Master Mix (Cat. No: 4309155, Applied Biosystis, Foster City, CA, USA) and StepOne Real-Time PCR Systi (Applied Biosystis 7500, Foster City, CA, USA). Each reaction mixture contained a total volume of 25μl (master mix 12.5μl, cDNA 3μl, primer 3μl, and H2O 6.5μl).
The quantitative RT-PCR conditions were: 50°C for 2 minutes, 95°C for 10 minutes, then 60 cycles of 95°C for 30 seconds, and 60°C 30 seconds and 72°C for 30 seconds.
Relative amounts of target mRNA in test sample was calculated and normalized to the corresponding RPL38 mRNA transcript level as a housekeeping gene. The comparative Ct method was used to evaluate expression as previously described by Livak and Schmittgen [11].
Statistical analysis
Data analysis was performed using a graph and data analysis software package (SigmaPlot 12.0, Systat Software, Inc.). Data were analyzed by paired t-tests to compare cell lines with healthy controls.
Results
Expression levels of Drosha in AGS and HepG2 cells were higher than the controls (P = 0.011 and P = <0.001 respectively). On the contrary, Drosha’s expression levels in KYSE-30 cell line were lower in comparison to the control (Figure 1, P = 0.038).
Figure 1.
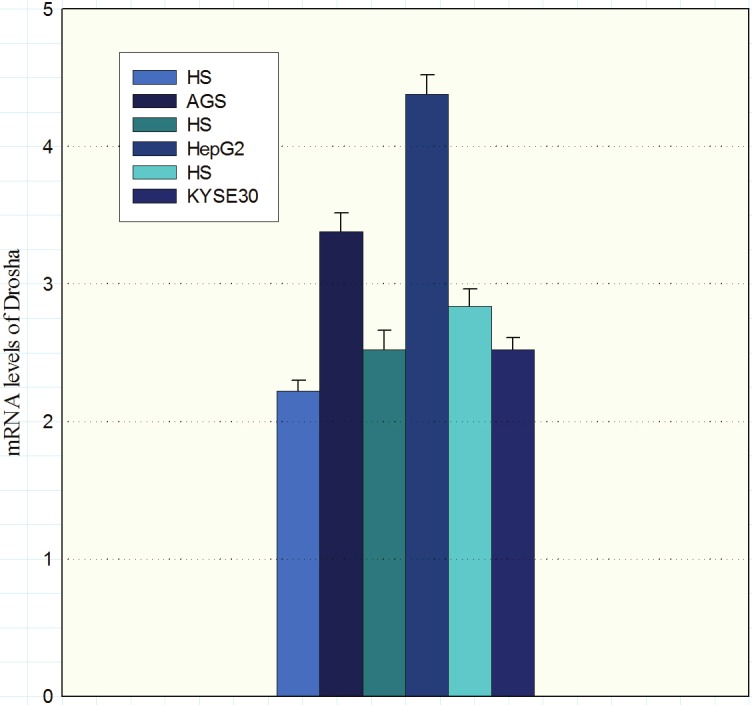
Expression of Drosha mRNA in cell lines AGS, HepG2 and KYSE30 was evaluated by quantitative RT-PCR. Values presented in the log scale. (HS: healthy subjects).
The Dicer expression levels in AGS and HepG2 cells were higher than the controls (P = 0.013 and P = 0.009 respectively). In contrast, Dicer expression levels in KYSE-30 cell line were lower in comparison to the control (Figure 2, P = <0.001).
Figure 2.
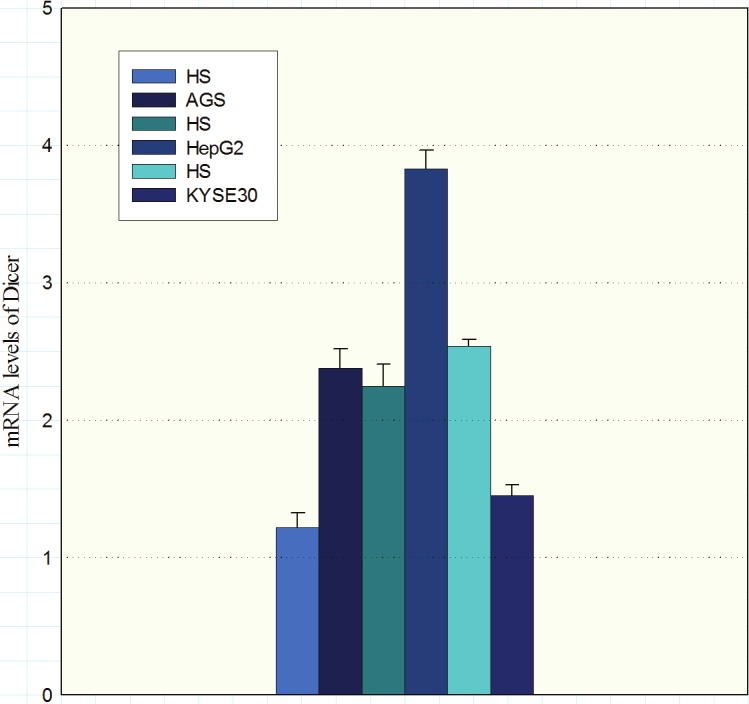
Expression of Dicer mRNA in cell lines AGS, HepG2 and KYSE30 was evaluated by quantitative RT-PCR. Values presented in the log scale. (HS: healthy subjects).
The DGCR8 expression levels in all three cell lines were significantly higher than the control samples (Figure 3). There was a statistically significant difference between AGS and HepG2 and healthy subjects (P = <0.001), and also between KYSE30 and normal controls (P= 0.013).
Figure 3.
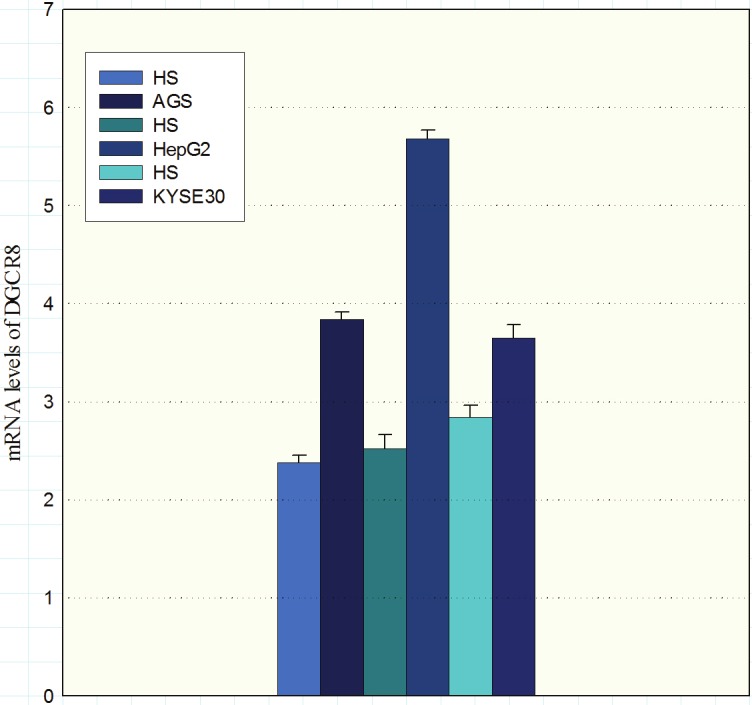
Expression of DGCR8 mRNA in cell lines AGS, HepG2 and KYSE30 was evaluated by quantitative RT-PCR. Values presented in the log scale. (HS: healthy subjects).
Discussion
Changes in the microRNA machinery components play crucial roles in the carcinogenesis of a variety of tumors [12]. These alterations are thought to elucidate abnormal miRNA profiles in various cancers. Both down and up-regulation of miRNAs have been reported, however, it is not clear whether the observed up- or downregulation of miRNA expression simply reflects malignant weakening of the tumor or directly causes tumor triggering and progression [9]. Hence, there are studies that have been proven both down and up-regulation of these machinery components in a variety of cancers [10,13-17]. Since miRNAs control both oncogenesis and tumor suppressors, it riains doubtful whether miRNAs may control carcinogenesis.
To analyze expression levels of important miRNA machinery components, we evaluated expression levels of two essential enzymes in the miRNA maturation process, Dicer and Drosha. And also DGCR8 which is a double-stranded RNA binding protein (dsRBP) and determines the cleavage sites on the pri-miRNA. Furthermore, there are also other factors that are necessary for appropriate pri-miRNA processing and are the subject of current research [18]. They include RNA-induced silencing complex (RISC) components including, argonaute-1 (AGO1), argonaute-2 (AGO2), as well as double-stranded RNA-binding proteins PACT, TARBP1, and TARBP2 [19]. Nevertheless, in this pilot study, we investigated the expression of Dicer, Drosha and DGCR8, while the other factors could be the subject of further studies.
It has been established that Drosha expression were correlated with poor prognostic factors in prostate cancer and esophageal carcinoma, besides, reduction of Drosha expression by means of siRNA has been reported to reduce cellular proliferation in esophageal cancer cell lines [20]. There are several probable explanations for the divergent expression patterns of Drosha in various tumors and how they correlate with other pathological parameters. Here, we found expression levels of Drosha in AGS and HepG2 cells were up-regulated (P = 0.011 and P = <0.001); on the other hand, it was down-regulated in KYSE-30 cell line (P = 0.038).
Regardless of growing evidence that Dicer mRNA levels differ in tumors, the regulation of this gene is indefinite. Dicer gene mutations have been found in humans, and alterations of the Dicer gene were detected in some pre-cancerous and invasive lung adenocarcinomas [21]. Additionally, there are other studies that suggest miRNA processing may be hindered in tumors with low Dicer and low Drosha expression, which could lead to a poor clinical outcome [13,22]. In our study we found that the expression of Dicer in AGS and HepG2 cells were higher than the controls (P = 0.013 and P = 0.009), on the other side, we have seen it was down-regulated in KYSE-30 cell line (P = <0.001).
DGCR8 is an RNA-binding protein that assists Drosha in the processing of miRNAs. Our experiment showed that DGCR8 expression up-regulated in AGS and HepG2 cells (P = <0.001), and also in KYSE30 cell line (P= 0.013).
To sum up all the facts, expression levels of the two most important enzymes of the miRNA machinery, Drosha and Dicer, and microprocessor complex component, DGCR8 were clearly dysregulated when compared to healthy controls. Our results are consistent with other findings and propose that miRNAs are involved in the carcinogenesis, and iphasize the importance of miRNA expression profiles to further investigate the role of miRNA dysregulation in human cancers.
However, this cannot be assessed based on our data alone. Hence, this was an in vitro study; it would be much more informative to design other experiments on animal tumor models and especially human tumors.
Acknowledgement
This study was supported by a financial grant from the research deputy for Ardabil University of Medical Sciences.
References
- 1.Sand M, Sand D, Altmeyer P, Bechara FG. MicroRNA in non-melanoma skin cancer. Cancer Biomark. 2012;11:253–257. doi: 10.3233/CBM-2012-0274. [DOI] [PubMed] [Google Scholar]
- 2.Macfarlane LA, Murphy PR. MicroRNA: Biogenesis, Function and Role in Cancer. Curr Genomics. 2010;11:537–561. doi: 10.2174/138920210793175895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sand M, Gambichler T, Sand D, Skrygan M, Altmeyer P, Bechara FG. MicroRNAs and the skin: tiny players in the body’s largest organ. J Dermatol Sci. 2009;53:169–175. doi: 10.1016/j.jdermsci.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 4.Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 2002;21:4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 6.Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Landthaler M, Yalcin A, Tuschl T. The human DiGeorge syndrome critical region gene 8 and Its D. melanogaster homolog are required for miRNA biogenesis. Curr Biol. 2004;14:2162–2167. doi: 10.1016/j.cub.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 8.Kim VN. MicroRNA precursors in motion: exportin-5 mediates their nuclear export. Trends Cell Biol. 2004;14:156–159. doi: 10.1016/j.tcb.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 9.Sand M, Gambichler T, Skrygan M, Sand D, Scola N, Altmeyer P, Bechara FG. Expression levels of the microRNA processing enzymes Drosha and dicer in epithelial skin cancer. Cancer Invest. 2010;28:649–653. doi: 10.3109/07357901003630918. [DOI] [PubMed] [Google Scholar]
- 10.Sand M, Skrygan M, Georgas D, Arenz C, Gambichler T, Sand D, Altmeyer P, Bechara FG. Expression levels of the microRNA maturing microprocessor complex component DGCR8 and the RNA-induced silencing complex (RISC) components argonaute-1, argonaute-2, PACT, TARBP1, and TARBP2 in epithelial skin cancer. Mol Carcinog. 2012;51:916–922. doi: 10.1002/mc.20861. [DOI] [PubMed] [Google Scholar]
- 11.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 12.Horikawa Y, Wood CG, Yang H, Zhao H, Ye Y, Gu J, Lin J, Habuchi T, Wu X. Single nucleotide polymorphisms of microRNA machinery genes modify the risk of renal cell carcinoma. Clin Cancer Res. 2008;14:7956–7962. doi: 10.1158/1078-0432.CCR-08-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Merritt WM, Lin YG, Han LY, Kamat AA, Spannuth WA, Schmandt R, Urbauer D, Pennacchio LA, Cheng JF, Nick AM, Deavers MT, Mourad-Zeidan A, Wang H, Mueller P, Lenburg ME, Gray JW, Mok S, Birrer MJ, Lopez-Berestein G, Coleman RL, Bar-Eli M, Sood AK. Dicer, Drosha, and outcomes in patients with ovarian cancer. N Engl J Med. 2008;359:2641–2650. doi: 10.1056/NEJMoa0803785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han L, Zhang A, Zhou X, Xu P, Wang GX, Pu PY, Kang CS. Downregulation of Dicer enhances tumor cell proliferation and invasion. Int J Oncol. 2010;37:299–305. doi: 10.3892/ijo_00000678. [DOI] [PubMed] [Google Scholar]
- 15.Jakymiw A, Patel RS, Deming N, Bhattacharyya I, Shah P, Lamont RJ, Stewart CM, Cohen DM, Chan EK. Overexpression of dicer as a result of reduced let-7 MicroRNA levels contributes to increased cell proliferation of oral cancer cells. Genes Chromosomes Cancer. 2010;49:549–559. doi: 10.1002/gcc.20765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Papachristou DJ, Korpetinou A, Giannopoulou E, Antonacopoulou AG, Papadaki H, Grivas P, Scopa CD, Kalofonos HP. Expression of the ribonucleases Drosha, Dicer, and Ago2 in colorectal carcinomas. Virchows Arch. 2011;459:431–440. doi: 10.1007/s00428-011-1119-5. [DOI] [PubMed] [Google Scholar]
- 17.Valencak J, Schmid K, Trautinger F, Wallnofer W, Muellauer L, Soleiman A, Knobler R, Haitel A, Pehamberger H, Raderer M. High expression of Dicer reveals a negative prognostic influence in certain subtypes of primary cutaneous T cell lymphomas. J Dermatol Sci. 2011;64:185–190. doi: 10.1016/j.jdermsci.2011.08.011. [DOI] [PubMed] [Google Scholar]
- 18.Perron MP, Provost P. Protein components of the microRNA pathway and human diseases. Methods Mol Biol. 2009;487:369–385. doi: 10.1007/978-1-60327-547-7_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Kouwenhove M, Kedde M, Agami R. MicroRNA regulation by RNA-binding proteins and its implications for cancer. Nat Rev Cancer. 2011;11:644–656. doi: 10.1038/nrc3107. [DOI] [PubMed] [Google Scholar]
- 20.Sugito N, Ishiguro H, Kuwabara Y, Kimura M, Mitsui A, Kurehara H, Ando T, Mori R, Takashima N, Ogawa R, Fujii Y. RNASEN regulates cell proliferation and affects survival in esophageal cancer patients. Clin Cancer Res. 2006;12:7322–7328. doi: 10.1158/1078-0432.CCR-06-0515. [DOI] [PubMed] [Google Scholar]
- 21.Chiosea S, Jelezcova E, Chandran U, Luo J, Mantha G, Sobol RW, Dacic S. Overexpression of Dicer in precursor lesions of lung adenocarcinoma. Cancer Res. 2007;67:2345–2350. doi: 10.1158/0008-5472.CAN-06-3533. [DOI] [PubMed] [Google Scholar]
- 22.Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR, Marion P, Salazar F, Kay MA. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]


