Abstract
Estrogen and progesterone hormones are key regulators of a wide variety of biological processes. In addition to their influence on reproduction, cell differentiation and apoptosis, they affect inflammatory response, cell metabolism and most importantly, they regulate physiological breast tissue proliferation and differentiation as well as the development and progression of breast cancer. In order to assess whether genetic variants in the steroid hormone receptor gene ESR1 (estrogen receptor alpha) had an effect on sporadic breast cancer susceptibility, we assessed 7 ESR1 single nucleotide polymorphisms (SNPs) for associations with breast cancer susceptibility and clinical parameters in 221 breast cancer patients and 221 controls, respectively. We identified ESR1 intron SNP +2464 C/T (rs3020314) and ESR1 intron SNP -4576 A/C (rs1514348) to correlate with breast cancer susceptibility and progesterone receptor expression status. Patients genotyped CT for ESR1 intron SNP +2464 (rs3020314) (p ≤ 0.045) or genotyped AC for ESR1 intron SNP -4576 (rs1514348) (p ≤ 0.000026) were identified to carry a significant risk as to the development of breast cancer in the Central European Caucasian population (both together: p ≤ 0.000488). Our study could confirm previous associations and revealed new associations of SNP rs1514348 with susceptibility to breast cancer and clinical outcome, which might be used as new additional SNP markers.
Keywords: Estrogen receptor, ESR1, SNP, breast cancer
Introduction
In the Western World breast cancer affects one in eight women during their lifetime and is the most common type of invasive cancer in women worldwide. Breast cancer is the type of cancer with the highest incidence in women worldwide with the second highest mortality rate among all tumors in Europe and North America [1]. Breast cancer is an enormous public health issue, with approximately 60,000 women being diagnosed annually in Germany, leading to about 18,000 deaths [2]. Estrogen exposure has been identified as one central factor in the origination and growth of this cancer [3] and its effects on the breast epithelium is mainly mediated by estrogen receptor alpha (ESR1) [4]. ESR1 is a steroid hormone receptor with a molecular weight of about 140 kb. It is localized on chromosome 6q25.1 and comprises 8 exons [5].
ESR1 single nucleotide genetic polymorphisms have been studied in the context of numerous clinical studies with quite heterogeneous results. Several studies showed statistically significant associations between ESR1 polymorphisms and breast cancer [6-8], whereas other studies did not find any associations [9-11]. However, recent studies revealed some new ESR1 SNPs to have an influence on breast cancer susceptibility. Rs2046210, located 180 kb upstream of the transcription starting site of exon 1 of the ESR1 gene, showed positive association for breast cancer risk in Chinese women in two separate studies [12-14]. A strong association of rs741581, located within PPARGC1B, an ER co-activator, and breast cancer risk among northern Europe patients was identified by Li et al., 2011 [15]. Another strong association was detected for the ESR1 intron SNP +7807 G/T (rs7766585) in a SNP approach study on northern European patients [16]. Fletcher et al., 2011 found also a breast cancer susceptibility locus for ESR1 (ESR1 promoter SNP +1517 A/C (rs3734805) and ESR1 intron SNP +9436 G/T (rs9383938)) in the northern European population [17]. Based on these findings ESR1 and its genetic variations appear to be essential as to the pathogenesis of breast cancer and need therefore to be further investigated. We studied 7 ESR1 single nucleotide polymorphisms (SNPs) and correlated the frequency of these SNPs in a population of 221 breast cancer patients with 221 unrelated, healthy donors (controls) of the German population, in order to determine whether and to what extent specific ESR1 SNPs had an effect as to breast cancer susceptibility. In addition, we correlated SNP genotype data with clinical parameters such as the cancer histology, receptor status or the outcome of chemotherapy.
Material and methods
Patient samples
221 female breast cancer patients who were recruited from 2003 till 2010 and 221 healthy female as controls were analyzed. Breast cancer patients were 31 to 90 years old and were diagnosed at the Saarland University Hospital - Department of Gynecology, Obstetrics and Reproductive Medicine. The controls consisted of healthy women aged 20 to 30 years.
Allelic discrimination
Genomic DNA was purified from peripheral blood mononuclear cells (PBMCs), with the Nucleospin® Blood genomic DNA purification kit in accordance with the manufacturer’s protocol (Macherey-Nagel, Düren, Germany). The following ESR1 SNPs were analyzed: ESR1 intron SNP +8406 C/T (rs985694), ESR1 intron SNP +8916 A/T (rs7757956), ESR1 intron SNP +2464 C/T (rs3020314), ESR1 intron SNP -4576 A/C (rs1514348), ESR1 intron SNP +1619 A/G (rs2347867), ESR1 intron SNP +6362 C/T (rs6557171) and ESR1 intron SNP +8935 A/G (rs9397456). Analyses were focused on coding, nonsynonymous SNPs, which potentially influence protein structure, activity, stability or localization because of changes in amino acid sequence and on promoter SNPs, which can lead to changes in ESR1 expression levels. These SNPs were chosen because of disease associations that were previously reported in the literature, as rs985694 is associated with higher incidence of type 2 diabetes and higher fasting plasma glucose [18], rs7757956 is linked to obesity in 11 year old children [19], rs3020314 is related with susceptibility to lymphoma [20], rs1514348, rs2347867, rs6557171 and rs9397456 are associated with an increased risk for Alzheimer’s disease [21]. ESR1 SNPs with allelic frequencies of more than 15 % in Caucasians were also analyzed. Primer sequences are listed in Table 1.
Table 1.
Gene, SNPs, and Sequences
| Gene | SNP Name | Intron Number | Sequence |
|---|---|---|---|
| ESR1 | rs985694 | 4 | GCTTTATATAATACA[C/T]CCCTGAAGTTTAGAT |
| rs7757956 | 4 | CACCCAAGGCCCTGC[A/T]GGGTTCAGGCCACAC | |
| rs3020314 | 4 | GAGAGATGACAGAAG[C/T]CTCTGTTTAAGGACA | |
| rs1514348 | 2 | AGCAGGCTGAATGGA[A/C]AATGCAGACTTACCC | |
| rs2347867 | 3 | GGTAACTGAGACAGC[A/G]CCAAATAAGGAGCAG | |
| rs6557171 | 3 | TATTTTTAACAAGTT[C/T]GTCCTGTTAAATGTC | |
| rs9397456 | 4 | TTCTTTTTCCACATC[A/G]ATGTCCAGTTGCTTC |
SNP validation was carried out through allele specific SNP-Assays by TaqMan-PCR on an ABI Prism 7700 Sequence Detection System (ABI, Darmstadt, Germany) in accordance with the instructions provided by the manufacturer. The polymerase chain reaction was run in a total reaction volume of 5 μl containing an average of 10 ng/μl of genomic DNA, a final concentration of 2.5 μl TaqMan Mastermix, 0.25 μl SNP Assay [20 fold] and 2.25 μl DNA in H2O. The following PCR protocol was used: Incubation at 50°C for 5 seconds, denaturation at 95°C for 10 minutes, followed by 40 cycles of denaturation at 92°C for 15 seconds and annealing and extension at 60°C for 1 minute. After running the PCR, the genotype of each sample was automatically determined. For each PCR run two negative controls and three positive controls (homozygous wild type or variant and heterozygous) were used.
Statistical analyses
Statistical analysis of as to genotype frequencies in association with clinical parameters were performed with the Pearson’s X2 tests, on the basis of deviations of genotype frequencies in controls that were calculated with the Hardy-Weinberg formula. Genotype risks were estimated as odds ratios with 95 % confidence intervals. Unless otherwise stated, the p-values presented are for the overall differences in genotype distribution between cases and controls or clinical aspects.
Results
We investigated for possible significances when correlating ESR1 genotype frequencies of sporadic breast cancer patients and healthy donors with clinical parameters such as breast cancer histology, breast cancer TNM-stage, hormone receptor and Her2 receptor status, the outcome of chemotherapy and patient survival.
Our analyses identified the variant allele of the ESR1 intron SNP +2464 C/T (rs3020314) to correlate with breast cancer risk (p ≤ 0.045) as patient have higher percentages of variant allele in the homozygous (CC) and heterozygous (TC) genotype and lower percentages of the homozygous wt genotype (TT) when compared to healthy donors with odds ratio [ct/tt] = 1.53 (95 % CI 1.04-2.26), odds ratio [cc/tt] = 1.62 (95 % CI 0.68-3.85) and with significance values of p = 0.52; p = 0.045 and p = 0.022 respectively (Figure 1 and Table 2).
Figure 1.
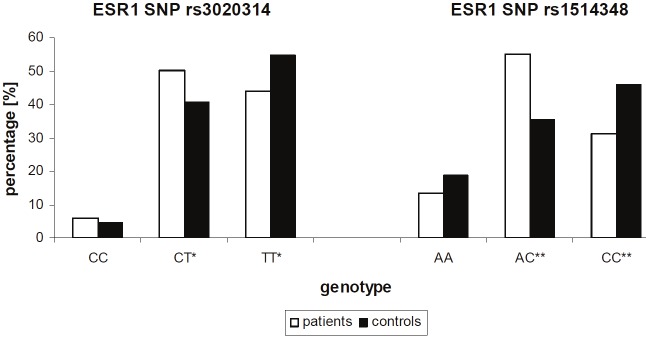
Comparison of allelic discrimination of rs3020314 and rs151434 between breast cancer patients and healthy controls. The percentage of the variant allele compared to the wt allele is higher in breast cancer patients, which is true for SNP rs302031 and rs151434. Significance of p < 0.05 *, p < 0.01 **.
Table 2.
Genotype Distributions: breast cancer patients versus controls
| Rs Number | Intron Number | Genotype | Cases | Controls | Odds ratio (95 % CI) | P-value |
|---|---|---|---|---|---|---|
| rs3020314 | 4 | CC | 13 (5.8 %) | 10 (4.5 %) | 1.62 [0.68-3.85] | 0.521 |
| CT | 111 (50.2 %) | 90 (40.7 %) | 1.53 [1.04-2.26] | 0.045 | ||
| TT | 97 (43.9 %) | 121 (54.8 %) | 1* | 0.022 | ||
| rs1514348 | 2 | AA | 30 (13.6 %) | 41 (18.6 %) | 1.07 [0.61-1.87] | 0,154 |
| AC | 122 (55.2 %) | 78 (35.5 %) | 2.28 [1.50-3.47] | 0.000026 | ||
| CC | 69 (31.2 %) | 101 (45.9 %) | 1* | 0.002 |
Reference group.
Similarly, correlations were calculated for ESR1 intron SNP -4576 A/C (rs151434), whereby breast cancer patients showed lower percentages for the wt allele (p ≤ 0.002) and higher percentages of the heterozygous genotype (p ≤ 0.000026) compared to healthy donors, with odds ratio [ac/cc] = 2.28 (95 % CI 1.50-3.47), odds ratio [aa/cc] = 1.07 (95% CI 0.61-1.87) and with even higher significances of p = 0.002 and p = 0.000026, respectively (Figure 1 and Table 2).
The other SNPs (ESR1 intron SNP +8406 C/T (rs985694), ESR1 intron SNP +8916 A/T (rs7757956), ESR1 intron SNP +1619 A/G (rs2347867), ESR1 intron SNP +6362 C/T (rs6557171) and ESR1 intron SNP +8935 A/G (rs9397456) that were analyzed failed to correlate with breast cancer risk.
In addition, we also observed a significant association between negative progesterone receptor status and the homozygous and heterozygous genotype of the variant allele of ESR1 intron SNP +2464 C/T (rs3020314) with odds ratio [cc/ct] = 3.22 (95 % CI 0.68-15.25), odds ratio [tt/ct] = 2.57 (95 % CI 1.35-4.87) and with p values of p = 0.008 and 0.002 respectively (Figure 2, Table 3). 67.2 % of patients who tested negatively for the expression of the progesterone receptor showed a heterozygous genotype CT and 29.5 % showed to be homozygous for the wt allele TT. On the other hand, in patients who tested positively for the expression of the progesterone receptor the heterogenous genotype CT was significantly lower (43.8 %) while the percentage of patients with the homozygous wt genotype TT was significantly higher (49.4 %) when compared to the PR negative controls (Figure 2 and Table 3).
Figure 2.
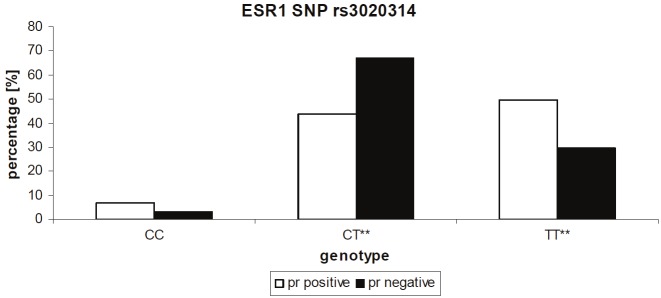
Association between rs3020314 and progesterone receptor status. The percentage of progesterone receptor positive patients decreases from wt to heterozygous and homozygous variant genotype, whereas the percentage of progesterone receptor negative patients increases from homozygous wt to the heterozygous genotype. While the percentage of the of progesterone receptor negative patients is significantly lower in wt patients (p = 0.008) it is significantly higher in patients with heterozygous genotype (p = 0.002). Significance of p < 0.05 (*), p < 0.01 (**).
Table 3.
Genotype Distributions: breast cancer patients and progesterone receptor status
| Rs Number | Intron Number | Genotype | Cases | Controls | Odds ratio (95 % CI) | P-value |
|---|---|---|---|---|---|---|
| rs3020314 | 4 | CC | 11 (6.8 %) | 2 (3.3 %) | 3.22 [0.68-15.25] | 0.310 |
| CT | 70 (43.8 %) | 41 (67.2 %) | 1* | 0.002 | ||
| TT | 79 (49.4 %) | 18 (29.5 %) | 2.57 [1.35-4.87] | 0.008 | ||
| rs1514348 | 2 | AA | 28 (17.5 %) | 2 (3.3 %) | 6.33 [1.43-27.95] | 0.006 |
| AC | 84 (52.2 %) | 38 (62.3 %) | 1* | 0.191 | ||
| CC | 48 (30 %) | 21 (34.4 %) | 1.03 [0.54-1.96] | 526 |
Reference group.
A correlation of ESR1 intron SNP -4576 A/C (1514348) with the progesterone receptor status identified a significant correlation for the homozygous variant genotype, which was higher for PR positive patients (odds ratio [aa/ac] = 6.33 (95 % CI 1.43-27.95), odds ratio [cc/ac] = 1.03 (95 % CI 0.54-1.96) and p = 0.006), while the proportion of the wt and heterozygous genotypes was nearly similar between PR positive and PR negative patients (Table 3). The other SNPs (ESR1 intron SNP +8406 C/T (rs985694), ESR1 intron SNP +8916 A/T (rs7757956), ESR1 intron SNP +1619 A/G (rs2347867), ESR1 intron SNP +6362 C/T (rs6557171) and ESR1 intron SNP +8935 A/G (rs9397456)) that were analyzed showed no correlations with the progesterone receptor status.
Discussion
In the study presented herein, we correlated seven ESR1 SNPs with breast cancer susceptibility and clinical parameters in 221 breast cancer patients. Especially for two SNPs, ESR1 intron SNP +2464 C/T (rs3020314) and ESR1 intron SNP -4576 A/C (rs1514348), we identified highly significant associations with breast cancer susceptibility and with the progesterone receptor status. Also, we found that the heterozygous genotype (CT) of ESR1 intron SNP +2464 C/T (rs3020314) not only correlates with greater risk to come down with breast cancer but also has a higher probability of a progesterone receptor negative breast cancer. On the other hand individuals genotyped TT for ESR1 intron SNP +2464 C/T (rs3020314) are considered to have a larger probability to develop progesterone receptor positive breast cancer.
Alison M. Dunning et al. investigated genetic variation and breast cancer risk [22-25]. They showed that ESR1 intron SNP +2464 C/T (rs3020314) has an overall effect on breast cancer, including estrogen receptor status, histology, staging and grading [24] and is associated with breast cancer risk, what was explained by putative induced alterations in alternative mRNA splice variants [23].
Similar to our results they observed higher breast cancer susceptibility for the ESR1 intron SNP +2464 C/T (rs3020314) in individuals with the genotype CT in comparison to individuals with the genotype TT in the European population [23]. In our analyses we could additionally specify the declared overall effect on breast cancer. Interestingly we did not observe any association between ESR1 intron SNP +2464 C/T (rs3020314) and the estrogen receptor status, histology, staging or grading, but we saw a significant association to the progesterone receptor status, as described above.
ESR1 intron SNP -4576 A/C (rs1514348) was subject of a study which looked for the survival after the diagnosis of breast cancer [26], but no association to survival was found and neither did we. ESR1 intron SNP -4576 A/C (rs1514348) was never investigated for possible association to breast cancer susceptibility and we detected higher breast cancer susceptibility with the genotype AC in comparison to the wt genotype (CC) in the European population. Based on our results it is more common to develop positive progesterone receptor breast cancer with the AA genotype. With regard to these results the ESR1 intron SNP -4576 A/C (rs1514348) can be used as additional marker for breast cancer susceptibility.
Finally it becomes obvious that the discussion of SNPs and their relation to breast cancer comes up with many different approaches, for example whether a SNP modifies the breast cancer susceptibility, or whether a SNP modifies clinical parameters. A polymorphism may not only alter the susceptibility to acquire breast cancer, but has multifarious associations on a breast cancer patient at all.
Its to point out that we could not only confirm previous associations but that our analyses revealed new associations of ESR1 intron SNP -4576 A/C (rs1514348) with susceptibility to breast cancer and clinical outcome, which might be used as new additional SNP markers. So our results present a next step towards a comprehensive SNP marker set for breast cancer susceptibility and clinical outcome and individualized cancer therapy.
References
- 1.Kamangar F, Dores GM, Anderson WF. Patterns of cancer incidence, mortality, and prevalence across five continents: defining priorities to reduce cancer disparities in different geographic regions of the world. J. Clin. Oncol. 2006;24:2137–2150. doi: 10.1200/JCO.2005.05.2308. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J, Autier P, Boniol M, Heanue M, Colombet M, Boyle P. Estimates of the cancer incidence and mortality in Europe in 2006. Ann Oncol. 2007;18:581–592. doi: 10.1093/annonc/mdl498. [DOI] [PubMed] [Google Scholar]
- 3.Yager JD, Davidson NE. Estrogen carcinogenesis in breast cancer. N Engl J Med. 2006;354:270–282. doi: 10.1056/NEJMra050776. [DOI] [PubMed] [Google Scholar]
- 4.Song RX, Santen RJ. Membrane initiated estrogen signaling in breast cancer. Biol Reprod. 2006;75:9–16. doi: 10.1095/biolreprod.105.050070. [DOI] [PubMed] [Google Scholar]
- 5.Fernandez LP, Milne RL, Barroso E, Cuadros M, Arias JI, Ruibal A, Benitez J, Ribas G. Estrogen and progesterone receptor gene polymorphisms and sporadic breast cancer risk: a Spanish case-control study. Int J Cancer. 2006;119:467–471. doi: 10.1002/ijc.21847. [DOI] [PubMed] [Google Scholar]
- 6.Gold B, Kalush F, Bergeron J, Scott K, Mitra N, Wilson K, Ellis N, Huang H, Chen M, Lippert R, Halldorsson BV, Woodworth B, White T, Clark AG, Parl FF, Broder S, Dean M, Offit K. Estrogen receptor genotypes and haplotypes associated with breast cancer risk. Cancer Res. 2004;64:8891–8900. doi: 10.1158/0008-5472.CAN-04-1256. [DOI] [PubMed] [Google Scholar]
- 7.Wang J, Higuchi R, Modugno F, Li J, Umblas N, Lee J, Lui LY, Ziv E, Tice JA, Cummings SR, Rhees B. Estrogen receptor alpha haplotypes and breast cancer risk in older Caucasian women. Breast Cancer Res Treat. 2007;106:273–280. doi: 10.1007/s10549-007-9497-8. [DOI] [PubMed] [Google Scholar]
- 8.Zheng W, Long J, Gao YT, Li C, Zheng Y, Xiang YB, Wen W, Levy S, Deming SL, Haines JL, Gu K, Fair AM, Cai Q, Lu W, Shu XO. Genome-wide association study identifies a new breast cancer susceptibility locus at 6q25.1. Nat Genet. 2009;41:324–328. doi: 10.1038/ng.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Onay VU, Briollais L, Knight JA, Shi E, Wang Y, Wells S, Li H, Rajendram I, Andrulis IL, Ozcelik H. SNP-SNP interactions in breast cancer susceptibility. BMC Cancer. 2006;6:114. doi: 10.1186/1471-2407-6-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schubert EL, Lee MK, Newman B, King MC. Single nucleotide polymorphisms (SNPs) in the estrogen receptor gene and breast cancer susceptibility. J Steroid Biochem Mol Biol. 1999;71:21–27. doi: 10.1016/s0960-0760(99)00126-0. [DOI] [PubMed] [Google Scholar]
- 11.Wedren S, Lovmar L, Humphreys K, Magnusson C, Melhus H, Syvanen AC, Kindmark A, Landegren U, Fermer ML, Stiger F, Persson I, Baron J, Weiderpass E. Oestrogen receptor alpha gene haplotype and postmenopausal breast cancer risk: a case control study. Breast Cancer Res. 2004;6:R437–449. doi: 10.1186/bcr811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cai Q, Wen W, Qu S, Li G, Egan KM, Chen K, Deming SL, Shen H, Shen CY, Gammon MD, Blot WJ, Matsuo K, Haiman CA, Khoo US, Iwasaki M, Santella RM, Zhang L, Fair AM, Hu Z, Wu PE, Signorello LB, Titus-Ernstoff L, Tajima K, Henderson BE, Chan KY, Kasuga Y, Newcomb PA, Zheng H, Cui Y, Wang F, Shieh YL, Iwata H, Le Marchand L, Chan SY, Shrubsole MJ, Trentham-Dietz A, Tsugane S, Garcia-Closas M, Long J, Li C, Shi J, Huang B, Xiang YB, Gao YT, Lu W, Shu XO, Zheng W. Replication and functional genomic analyses of the breast cancer susceptibility locus at 6q25.1 generalize its importance in women of chinese, Japanese, and European ancestry. Cancer Res. 2011;71:1344–1355. doi: 10.1158/0008-5472.CAN-10-2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ding SL, Yu JC, Chen ST, Hsu GC, Hsu HM, Ho JY, Lin YH, Chang CC, Fann CS, Cheng CW, Wu PE, Shen CY. Diverse associations between ESR1 polymorphism and breast cancer development and progression. Clin Cancer Res. 2010;16:3473–3484. doi: 10.1158/1078-0432.CCR-09-3092. [DOI] [PubMed] [Google Scholar]
- 14.Han J, Jiang T, Bai H, Gu H, Dong J, Ma H, Hu Z, Shen H. Genetic variants of 6q25 and breast cancer susceptibility: a two-stage fine mapping study in a Chinese population. Breast Cancer Res Treat. 2011;129:901–907. doi: 10.1007/s10549-011-1527-x. [DOI] [PubMed] [Google Scholar]
- 15.Li Y, Wedren S, Li G, Charn TH, Desai KV, Bonnard C, Czene K, Humphreys K, Darabi H, Einarsdottir K, Heikkinen T, Aittomaki K, Blomqvist C, Chia KS, Nevanlinna H, Hall P, Liu ET, Liu J. Genetic variation of ESR1 and its co-activator PPARGC1B is synergistic in augmenting the risk of estrogen receptor-positive breast cancer. Breast Cancer Res. 2011;13:R10. doi: 10.1186/bcr2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harlid S, Ivarsson MI, Butt S, Hussain S, Grzybowska E, Eyfjord JE, Lenner P, Forsti A, Hemminki K, Manjer J, Dillner J, Carlson J. A candidate CpG SNP approach identifies a breast cancer associated ESR1-SNP. Int J Cancer. 2011;129:1689–1698. doi: 10.1002/ijc.25786. [DOI] [PubMed] [Google Scholar]
- 17.Fletcher O, Johnson N, Orr N, Hosking FJ, Gibson LJ, Walker K, Zelenika D, Gut I, Heath S, Palles C, Coupland B, Broderick P, Schoemaker M, Jones M, Williamson J, Chilcott-Burns S, Tomczyk K, Simpson G, Jacobs KB, Chanock SJ, Hunter DJ, Tomlinson IP, Swerdlow A, Ashworth A, Ross G, dos Santos Silva I, Lathrop M, Houlston RS, Peto J. Novel breast cancer susceptibility locus at 9q31.2: results of a genome-wide association study. J Natl Cancer Inst. 2011;103:425–435. doi: 10.1093/jnci/djq563. [DOI] [PubMed] [Google Scholar]
- 18.Dahlman I, Vaxillaire M, Nilsson M, Lecoeur C, Gu HF, Cavalcanti-Proenca C, Efendic S, Ostenson CG, Brismar K, Charpentier G, Gustafsson JA, Froguel P, Dahlman-Wright K, Steffensen KR. Estrogen receptor alpha gene variants associate with type 2 diabetes and fasting plasma glucose. Pharmacogenet Genomics. 2008;18:967–975. doi: 10.1097/FPC.0b013e32831101ef. [DOI] [PubMed] [Google Scholar]
- 19.Tobias JH, Steer CD, Vilarino-Guell C, Brown MA. Effect of an estrogen receptor-alpha intron 4 polymorphism on fat mass in 11-year-old children. J Clin Endocrinol Metab. 2007;92:2286–2291. doi: 10.1210/jc.2006-2447. [DOI] [PubMed] [Google Scholar]
- 20.Skibola CF, Bracci PM, Halperin E, Nieters A, Hubbard A, Paynter RA, Skibola DR, Agana L, Becker N, Tressler P, Forrest MS, Sankararaman S, Conde L, Holly EA, Smith MT. Polymorphisms in the estrogen receptor 1 and vitamin C and matrix metalloproteinase gene families are associated with susceptibility to lymphoma. PLoS One. 2008;3:e2816. doi: 10.1371/journal.pone.0002816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma SL, Tang NL, Tam CW, Lui VW, Lau ES, Zhang YP, Chiu HF, Lam LC. Polymorphisms of the estrogen receptor alpha (ESR1) gene and the risk of Alzheimer’s disease in a southern Chinese community. Int Psychogeriatr. 2009;21:977–986. doi: 10.1017/S1041610209990068. [DOI] [PubMed] [Google Scholar]
- 22.Cox A, Dunning AM, Garcia-Closas M, Balasubramanian S, Reed MW, Pooley KA, Scollen S, Baynes C, Ponder BA, Chanock S, Lissowska J, Brinton L, Peplonska B, Southey MC, Hopper JL, McCredie MR, Giles GG, Fletcher O, Johnson N, dos Santos Silva I, Gibson L, Bojesen SE, Nordestgaard BG, Axelsson CK, Torres D, Hamann U, Justenhoven C, Brauch H, Chang-Claude J, Kropp S, Risch A, Wang-Gohrke S, Schurmann P, Bogdanova N, Dork T, Fagerholm R, Aaltonen K, Blomqvist C, Nevanlinna H, Seal S, Renwick A, Stratton MR, Rahman N, Sangrajrang S, Hughes D, Odefrey F, Brennan P, Spurdle AB, Chenevix-Trench G, Beesley J, Mannermaa A, Hartikainen J, Kataja V, Kosma VM, Couch FJ, Olson JE, Goode EL, Broeks A, Schmidt MK, Hogervorst FB, Van’t Veer LJ, Kang D, Yoo KY, Noh DY, Ahn SH, Wedren S, Hall P, Low YL, Liu J, Milne RL, Ribas G, Gonzalez-Neira A, Benitez J, Sigurdson AJ, Stredrick DL, Alexander BH, Struewing JP, Pharoah PD, Easton DF. A common coding variant in CASP8 is associated with breast cancer risk. Nat Genet. 2007;39:352–358. doi: 10.1038/ng1981. [DOI] [PubMed] [Google Scholar]
- 23.Dunning AM, Healey CS, Baynes C, Maia AT, Scollen S, Vega A, Rodriguez R, Barbosa-Morais NL, Ponder BA, Low YL, Bingham S, Haiman CA, Le Marchand L, Broeks A, Schmidt MK, Hopper J, Southey M, Beckmann MW, Fasching PA, Peto J, Johnson N, Bojesen SE, Nordestgaard B, Milne RL, Benitez J, Hamann U, Ko Y, Schmutzler RK, Burwinkel B, Schurmann P, Dork T, Heikkinen T, Nevanlinna H, Lindblom A, Margolin S, Mannermaa A, Kosma VM, Chen X, Spurdle A, Change-Claude J, Flesch-Janys D, Couch FJ, Olson JE, Severi G, Baglietto L, Borresen-Dale AL, Kristensen V, Hunter DJ, Hankinson SE, Devilee P, Vreeswijk M, Lissowska J, Brinton L, Liu J, Hall P, Kang D, Yoo KY, Shen CY, Yu JC, Anton-Culver H, Ziogoas A, Sigurdson A, Struewing J, Easton DF, Garcia-Closas M, Humphreys MK, Morrison J, Pharoah PD, Pooley KA, Chenevix-Trench G. Association of ESR1 gene tagging SNPs with breast cancer risk. Hum Mol Genet. 2009;18:1131–1139. doi: 10.1093/hmg/ddn429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mavaddat N, Dunning AM, Ponder BA, Easton DF, Pharoah PD. Common genetic variation in candidate genes and susceptibility to subtypes of breast cancer. Cancer Epidemiol Biomarkers Prev. 2009;18:255–259. doi: 10.1158/1055-9965.EPI-08-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pharoah PD, Tyrer J, Dunning AM, Easton DF, Ponder BA. Association between common variation in 120 candidate genes and breast cancer risk. PLoS Genet. 2007;3:e42. doi: 10.1371/journal.pgen.0030042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Udler MS, Azzato EM, Healey CS, Ahmed S, Pooley KA, Greenberg D, Shah M, Teschendorff AE, Caldas C, Dunning AM, Ostrander EA, Caporaso NE, Easton D, Pharoah PD. Common germline polymorphisms in COMT, CYP19A1, ESR1, PGR, SULT1E1 and STS and survival after a diagnosis of breast cancer. Int J Cancer. 2009;125:2687–2696. doi: 10.1002/ijc.24678. [DOI] [PubMed] [Google Scholar]


