Abstract
Bacterial colonization occurs in all wounds, chronic or acute and the break in epithelium integrity that defines a wound impairs the forces that shape and constrain the microbiome at that site. This review highlights the interactions between bacterial communities in the wound and the ultimate resolution of the wound or development of fibrotic lesions. Chronic wounds support complex microbial communities comprised of a wide variety of bacterial phyla, genera and species, including some fastidious anaerobic bacteria not identified using culture-based methods. Thus, the complexity of bacterial communities in wounds has historically been underestimated. There are a number of intriguing possibilities to explain these results that may also provide novel insights into changes and adaptation of bacterial metabolic networks in inflamed and wounded mucosa, including the critical role of biofilm formation. It is well accepted that the heightened state of activation of host cells in a wound that is driven by the microbiota can certainly lead to detrimental effects on wound regeneration, but the microbiota of the wound may also have beneficial effects on wound healing. Studies in experimental systems have clearly demonstrated a beneficial effect for members of the gut microbiota on regulation of systemic inflammation, which could also impact wound healing at sites outside the gastrointestinal tract. The utilization of culture-independent microbiology to characterize the microbiome of wounds and surrounding mucosa has raised many intriguing questions regarding previously held notions about the cause and effect relationships between bacterial colonization and wound repair and mechanisms involved in this symbiotic relationship.
Keywords: Bacteria, Inflammation, Skin, Gastrointestinal Tract
Introduction
The host microbiome is the entire collection of microorganisms that inhabit a host. This includes bacteria, archaea, fungi, parasites, viruses and phages. In this review, we will focus almost extensively on the bacterial microbiome. This subset of the microbiome is almost exclusively restricted to mucosal sites during health, i.e. the surfaces of the body that are exposed to the outside world. This includes the skin, gastrointestinal tract, upper airways, oral cavity and reproductive tract. However, despite being exposed to the outside world, the mucosal surfaces exert selective pressures on the composition of the bacterial microbiome. For example, while over 20 bacterial phyla have been reported for their growth on plant surfaces, only nine have been identified in the human gut [1,2].
In the context of this review article, a wound will be defined as a physical break in epithelium integrity and the subsequent host response to repair this break. Such a break in epithelium integrity impairs the forces that shape and constrain the microbiome at that site. Epithelium destruction will result in reduced production of mucus or lipids, alter anti-microbial peptide expression and activate inflammatory cascades. Mucosal surfaces are exposed to the environment; thus, wounds also offer an opportunity for non-indigenous microbes to colonize the site, as well as altering the forces that balance indigenous microbial colonization.
The initiation of wound repair normally begins very quickly after damage and involves well-integrated iterative steps in tissue regeneration that include the following [3–5]:
clotting and coagulation
secretion of extracellular signal molecules, e.g., cytokines, chemokines, growth factors and eicosanoids
leukocyte recruitment and formation of granulation tissue
fibroblast activation and proliferation
formation of new basement membrane and other extracellular matrix
new blood vessel growth
remodeling to restore normal tissue architecture.
The balance between extracellular matrix formation and degradation is a key step during the normal wound healing process and, combined with a balance between proliferation and apoptosis of fibroblasts, are major steps in determining whether injured tissues returned to their pre-injury state or develop fibrosis. A recent review by Meneghin and Hogaboam [3] provides an excellent overview on the role of infectious agents including bacteria, viruses, fungi and multicellular parasites in cellular activation and the promotion of fibrosis (including examples in pulmonary, cardiovascular, integumentary and alimentary systems). This review will focus on our current understanding of the role of the bacterial microbiome in mucosal wound repair pathology.
What is the bacterial microbiome and where is it found in the body?
Bacteria outnumber host cells by 10:1 and the metagenome of the bacterial microbiome is 100 times larger in size than the host genome [6]. Culture-independent methods have revealed a much greater diversity within the bacterial communities on the mucosa than previously identified by traditional culture-based methods [2,6]. The four most dominant phyla in the human body are the Firmicutes, Bacteroidetes, Proteobacteria and Actinobacteria. However, the relative distributions of these four phyla differ vastly between body sites. For example, Firmicutes and Bacteroidetes are the most abundant members in the gastrointestinal tract [2,6] while Proteobacteria and Actinobacteria dominate the skin [1] and Proteobacteria appear to be significant members of the airways [7–11]. Furthermore, the density of the indigenous microbiota differs along the mucosa, with highest concentration and diversity being found in the large intestine.
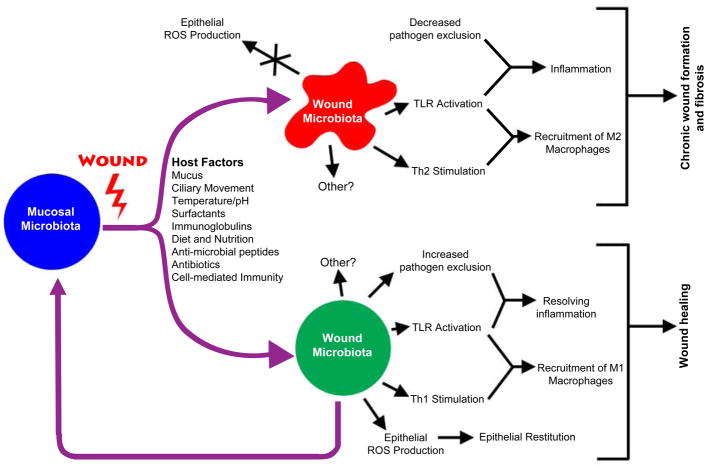
Metabolic, immunologic, secretory, and structural forces shape the bacterial communities of the mucosa [1,2,6,12] (Summarized in Fig. 1). Diet and nutrient availability are two major factors that modulate bacterial growth. Polymicrobial communities naturally form complex, three-dimensional aggregates called biofilms, which create metabolic webs of agonistic and antagonistic relationships among its membership [13,14]. Host defense/immunity also plays an important role in creating specific niches for bacterial colonization through the production of anti-microbial peptides, secretory immunoglobulins, and bactericidal effector mechanisms activated through mucosal damage [12]. Other secretory mechanisms important in modulating bacterial community structure include production of mucus, surfactants and specific lipids. All of these forces are in play on the smooth and involuted mucosal surfaces that are also subject to one or more of the following structural forces, depending on the body site: physical abrasion, peristaltic movement of solids and liquid, ciliary movement of mucus, epithelium stretch, and gradients of pH, temperature and/or oxygen concentration.
Figure 1.
Summary of the mechanisms involved in modulating the bacterial microbiome of wounds and the effects of different wound microbiomes on the inflammatory and healing process. Specific details of the processes outlined in this figure are described in the text. ROS = reactive oxygen species; TLR = toll-like receptors.
How do you analyze the microbiome?
The adaptation of recently developed, high throughput molecular approaches to culture-independent microbiology, such as 16S rRNA gene-based pyrosequencing, is beginning to provide new insight into identifying and understanding the role of bacterial communities in the processes of wound repair, chronic wound formation and fibrosis. Of note are the recent numerous reports that chronic wounds support complex microbial communities comprised of a wide variety of bacterial phyla, genera and species, including some fastidious anaerobic bacteria not identified using culture-based methods [15–18]. The key limitation of culture-based methodologies is the inability to routinely grow out these fastidious and/or currently “unculturable” organisms. Estimates are that >60% of bacterial species in the human microbiome have not been able to be cultured and a majority of the remaining bacteria having complex and dynamic growth requirements, rendering them difficult to reproducibly isolate [19]. Most commonly, culture-independent techniques are based upon PCR amplification of the 16S rRNA gene. This gene is relatively small (approximately 1.5 Kbp), highly conserved, not subject to natural selection pressures and transmitted vertically without lateral gene transfer. It possesses nine hypervariable regions (V1-9) in which the vast majority of evolutionary changes have occurred, rendering this single gene useful for taxonomic identification [20]. Databases are available that encompass more than 500,000 16S rRNA sequences from a range of phylotypically diverse bacteria. Conserved sequence stretches within the 16S rRNA gene allow for the design of broad bacterial kingdom-specific primers, which can be used to create amplicons of individual 16S rRNA genes derived from a mixed bacterial population. High-throughput sequencing of these 16S rRNA gene amplicon libraries is possible, with the most powerful being 454-pyrosequencing, due to sequence read lengths of over 400 bp (sufficient to cover at least two adjacent 16S rRNA hypervariable regions) and output of greater than 106 high-quality sequence reads [21–23], permitting a rapid robust sampling of microbial communities [24–28].
What is the microbiome in sites of acute resolving, acute non-resolving and chronic (non-resolving) wounds?
Bacterial colonization occurs in all wounds, chronic or acute. One area of active investigation is understandind the correlation between different microbial communities in the wound and the ultimate repair of the wound (vs. the development of chronic wounds and/or fibrotic lesions). Chronic wounds often contain higher levels of culturable bacteria compared to healing wounds, and experimental colonization of wounds can delay healing [29,30]. Bacterial colonization of wounds is polymicrobial [15–17,31,32] and in the form of a biofilm [32–34]. Antibiotics typically do not sterilize wounds (even broad spectrum antibiotics) because bacterial cells in biofilms have increased resistance to biocides [35]. However, antibiotics can shift the growth balance in bacterial community composition, preventing pathogenic bacterial colonization that can lead to enhanced tissue damage and/or systemic dissemination of pathogenic bacteria. Despite this, published research does not support a positive effect of antibiotic therapy for chronic wound healing [36]. Antibiotic use for chronic wounds is often associated with selection for more resistant bacterial species. Many Pseudomonas species are extremely adept at adapting to antibiotic pressure and certain antibiotics appear to actually induce the formation of pseudomonal biofilms, perhaps accounting for the increased colonization of chronic wounds by Pseudomonadaceae [15,37].
A number of groups have used culture-independent methods to analyze bacterial wound communities and, collectively, these groups have reported the following observations about the microbiota of wounds [15–18,31,32]. First, the number and proportion of bacterial species can range greatly between individual wounds. Second, bacterial diversity as determined by culture-based methods is significantly lower than that obtained through 16S rRNA gene-based amplicon pyrosequencing. Thus, the complexity of bacterial communities in wounds has historically been underestimated. Commonly isolated organisms include Staphylococcus, Corynebacterium and Propionibacterium (although Propionibacterium are typically found at lower levels in wounds compared to healthy skin). Notable on the list of wound bacteria are the fastidious and/or anaerobic organisms Neisseriaceae, Campylobacteriaceae, and Clostridiaceae. Proteobacteria are commonly identified in wounds and largely belong to the Pseudomonadaceae, Enterobacteriaceae, Oxalobacteraceae and Neisseriaceae families. Additional work needs to be done to identify many of these non-culturable organisms at the species level. Third, the microbiota can differ between different wounds while bacterial communities at different sites within an individual wound are significantly more similar to each other than to those from different wounds [17,38]. Finally, the reliability of both culture and non-culture based analysis depends heavily on the sampling method used. For example, certain sampling techniques will not detect anaerobic bacteria, which are common in chronic wounds [39], Therefore, when studying the human microbiome, important controls need to be in place to guarantee that the chosen sampling techniques are as unbiased and comprehensive as possible.
Diabetic wounds are well documented to display defects in the steps involved in normal wound healing, resulting in chronic wounds. Colonization of diabetic vs. non-diabetic wounds is also markedly different, including an increased incidence of colonization by Streptococcus or Staphylococcus in diabetic wounds [17]. Other commonly cultured bacteria from non-healing diabetic wounds include Staphylococcus aureus, S. epidermidis, P. aeruginosa, Enterococcus spp, Peptostreptococcus spp, Bacteroides spp, and Prevotella spp [40–42]. Colonization of wounds by Staphylococcus likely impairs wound healing, as supported by both clinical associative data and experimental animal models, including reports that colonization of wounds in mice can prevent re-growth of the epithelium and the aberrant inflammatory response in the skin of diabetic db/db mice promotes colonization by Staphylococcus [40,43–47].
Surprisingly, many of these studies are finding readily-culturable bacteria by culture-independent methods that are otherwise not being identified by traditional culture-based methods [15–17,31,32]. Cultivation relies heavily on selective media and can mask the presence of less numerous organisms. However, beyond the obvious Devil’s Advocate answer that these bacteria are actually dead (which evidence from a variety of sources is rendering unlikely), there are a number of intriguing possibilities to explain these results that may also provide novel insights into changes and adaptation of bacterial metabolic networks in inflamed and wounded mucosa. Within biofilms, the precipitous oxygen gradients that form microaerophilic and anaerobic regions, combined with increased gene flow among biofilm members, leads to metabolic alteration of bacterial cells [48,49]. Species known to readily grow on standard laboratory media will no longer exhibit typical phenotypes, sometimes leading to non-culturability. This phenomenon has been heavily researched in Pseudomonadaceae biofilms. For example, Pseudomonas species isolated from clinical chronic biofilms will commonly exhibit a loss of motility [50], amino acid auxotrophy [51], lack of type-III secretion and siderophore production [52], and a long list of other loss-of-function mutations [53]. There is also a robust line of investigation in understanding the mechanisms that regulate the viable-but-not-culturable (VBNC) state, where bacteria cease to multiply but remain metabolically active, [54] and this may be relevant to the lifestyle of wound bacteria.
In microbial communities, both bacterial competition and cooperation will have an impact on the pathophysiology of the host. One example of cooperation that leads to increased virulence involves siderophore-producing bacteria, such as P. aeruginosa, that are able to ‘share’ the production of iron-scavenging siderphores with non-siderophore producers [55]. A heterogenuous population of siderophore-producers and non-siderophore producing bacteria can lead to higher levels of bacterial growth. The evolution of such iron ‘cheats’ occurs more readily in low-iron conditions, revealing how the host environment shapes such microbial cooperation [56]. It should be noted that sequestration of free iron is a major mechanism of innate immunity [57,58]. However, inter-species bacterial competition also plays a role in mixed bacterial populations. For example, P. aeruginosa can lyse other bacterial species, such as S. aureus, and utilize the bacterial lysate as a source of free iron, therby increasing its ability to colonize under iron-limited conditions of the host [59,60].
An additional illustration of potential bacteria-bacteria antagonism in a wound, it has been demonstrated that Lactobacillus reuteri strain RC-14 can inhibit Staphylococcus aureus infection in a rat surgical-implant model [61]. In this study, a small sterile piece of silicone was inserted subcutaneously and inoculated with L. reuteri and/or S. aureus. The surgical incisions were then sutured and abscess formation followed. Implantation of lactobacillus alone did not induce any abscess formation and the addition of the lactobacilli prevented abscess formation induced by S. aureus. This was not true for all the lactobacilli strains tested, suggesting a strain-specific mechanism, which may involve a secreted cell-signaling molecule [62]. Many other examples exist in the literature of competing bacteria interfering with quorum-sensing pathways, bacteriocin production and virulence factor production. It remains to be determined whether such mechanisms shape the bacterial microbiome of wounds.
What is the effect of changing the localized microbiome on wound repair?
Microbial colonization or infection is hypothesized to play an important role in driving chronic inflammation, chronic wounds and the development of fibrosis [3,5,63] (summarized in Fig. 1). Pulmonary tuberculosis is one example of a chronic disease that is characterized by bacterially-induced chronic fibrosis. In this disease, there are significant numbers of activated M2 macrophages and fibroblasts in Mycobacterium tuberculosis-containing granulomas [64]. These macrophages express TLRs that when activated with the appropriate bacterial PAMPs stimulate T cells to produce cytokines and chemokines that promote a fibrotic response. Activated M2 macrophages are also found at sites of wound repair in chronic non-resolving wounds; thus, it seems likely that the microbial populations that colonize these wounds can significantly influence the activation and signal production by these macrophages (reviewed in [3,5]). In addition to leukocytes, myofibroblasts also express many TLRs, including TLRs 2-7. Stimulation of these TLRs can lead to the production of chemokines and cytokines, including CXC chemokine ligand 8 (CXCL8/IL-8), which has both neutrophil chemotactic activity as well as being a pro-angiogenesis molecule. Bacterial CpG stimulation of TLR9 can also drive interstitial fibrotic pathways. Primary fibroblasts from sites of inflammation have also been demonstrated to release large amounts of other CXC and CC chemokines including CCL5, CCL8, and CXCL6. Thus, stimulation of TLRs by PAMPs released by the microbiota of wounds may play a role in maintaining leukocytes and myofibroblasts in a heightened state of activation.
The heightened state of activation of host cells in a wound that is driven by the microbiota can certainly lead to detrimental effects on wound regeneration, but the microbiota of the wounds may also have beneficial effects on wound healing. For example, wound healing following surgical skin incision and suture was followed in germ-free and conventionalized mice [65]. The conventional mice showed greater tensile strength of the wound initially, including higher hydroxyproline concentration in the surrounding tissue than the germ-free mice. In another study, of dermal wounding in rats, inoculation of wound sites with P. aeroginosa PA01 accelerated re-epithelialization, epidermal cell proliferation, and neo-vascularization, as well as the local infiltration of neutrophils and TNF production [66]. Treatment of these rats with antibodies against neutrophils or TNF caused a significant reduction in the wound healing response. Other experimental studies have demonstrated a similar positive effect of low level wound colonization, even by potentially pathogenic microbes, depending on the level of colonization and type of wound [67–69]. While these examples suggest local benefits of the microbiota on wound healing, other studies in experimental systems have clearly demonstrated a beneficial effect for members of the gut microbiota on regulation of systemic inflammation [70,71], which could impact wound healing at sites outside the gut.
Could changes in the gastrointestinal microbiome affect local epithelium repair?
The GI epithelium is constantly undergoing wounding and repair. The process of repairing small gaps, i. e. a few individual epithelial cells, within the epithelial layer of the GI tract is known as epithelial restitution [72]. This process involves a highly localized response in the adjacent epithelium, while losses of larger contiguous epithelium regions (>10 cells) stimulates a more robust repair response, analogous to skin wound repair, and involves cell types present in the lamina propria -including leukocytes, fibroblasts and subepithelial myofibroblasts.
The mechanisms by which the resident microbiota influence restitution and the wound repair response of the gastrointestinal epithelium are areas of active investigation [73]. Gastrointestinal restitution is modulated by numerous host factors; however, the indigenous microbiota also plays a role (positive and/or negative) in modulating the GI restitution response, such as through the production of short chain fatty acids and modification of bile acids. Epithelial cells of the GI tract express multiple TLRs on both their basolateral and apical surfaces, as well as intracellular TLRs that recognize bacterial PAMPs. The requirement for TLR-mediated signaling in the GI repair response is illustrated in a study of DSS-induced colitis [74]. In this study, acute GI epithelial damage and inflammation was aggravated in TLR2−/−, TLR4−/− and MyD88−/− mice, confirming the key role of microorganisms and microbial products in GI repair.
Epithelial restitution requires active cell migration, a process dependent on a constant turnover of focal cell-matrix adhesions (FAs). In a recent study, it was demonstrated that enteric resident bacteria can potentiate epithelial restitution via the generation of reactive oxygen species (ROS) in epithelial cells, which in turn, mediates inactivation of focal adhesion kinase phosphatases [75]. ROS generation induced oxidation of target cysteines in the redox-sensitive tyrosine phosphatases, LMW-PTP and SHP-2, which in turn resulted in increased phosphorylation of focal adhesion kinase (FAK), a key protein regulating the turnover of FAs. Phosphorylation of FAK substrate proteins, focal adhesion formation, and cell migration were all significantly enhanced by bacterial contact in both in vitro and in vivo models of wound closure.
Microbiota-host epithelial cell interactions can also modulate β-catenin signaling, a key component in regulating epithelial cell proliferation [76]. Enteric bacteria can inhibit the NF-κB pathway through the blockade of IκB-α ubiquitination, a process catalyzed by the E3-SCF(β-TrCP) ubiquitin ligase. The activity of this ubiquitin ligase is regulated via covalent modification of the Cullin-1 subunit by the ubiquitin-like protein NEDD8 and it is reported that the interaction of viable indigenous bacteria with mammalian intestinal epithelial cells can result in a rapid and reversible generation of ROS in epithelial cells that modulate neddylation of Cullin-1 and suppression of the NF-κB pathway [77]. The short chain fatty acid and bacterial fermentation product butyrate has also been shown to influence epithelial signaling via ROS-mediated changes in cullin-1 neddylation [78]. Treatment of human intestinal epithelia in vitro and human tissue ex vivo with butyrate can cause a loss of neddylated Cul-1 and modulate the ubiquitination and degradation of a target of the E3-SCF(β-TrCP) ubiquitin ligase, the NF-κB inhibitor IκB-α.
A number of indigenous bacterial species have also been demonstrated to induce ERK phosphorylation without stimulating pro-inflammatory phospho-IκB or pro-apoptotic phospho-c-Jun NH2-terminal kinase [79]. Of interest was the observation that Lactobacillus species have very potent activity. Whole bacterial cell signaling has also been recapitulated in experimental studies of epithelial cells using the bacterial peptide N-formyl-Met-Leu-Phe (N-fMLP) through the formyl peptide receptors (FPRs). This induction of extracellular signal-regulated kinase pathway signaling occurred via FPR-dependent redox modulation of dual specific phosphatase 3 [80]. The indigenous microbiota could initiate ERK signaling through rapid FPR-dependent ROS generation and subsequent modulation of MAP kinase phosphatase redox status. Epithelial ROS generation induced by Lactobacillus rhamnosus GG and N-fMLP could be abolished in the presence of selective inhibitors for G protein-coupled signaling and FPR ligand interaction. Inhibitors of ROS generation could attenuate microbiota-induced ERK signaling, implicating ROS generation in ERK pathway activation. Altogether, the studies described in the last few paragraphs raise the possibility that normal members of the bacterial microbiome stimulate the generation of ROS in intestinal epithelial cells through FPR-mediated signaling that modulates the ERK pathway, which in turn, regulates epithelial cell migration and epithelium restitution.
The indigenous microbiota can also promote wound pathology. For example, indomethacin-induced gastric ulceration in rats is dependent on the presence of a microbiota [81]. Treatment of rats with indomethacin also changes their microbiome [82], implicating a relationship between mucosal inflammation and microbiome community structure. This concept is supported by other studies of gastrointestinal inflammation in which mucosal inflammation and epithelium damage drive changes in the indigenous microbiota, resulting in the outgrowth of γ– proteobacteria, such as E. coli and Klebsiella pneumoniae [83,84]. In a model of transmissible spontaneous colitis in mice, the transmissible bacterial community required the presence of K. pneumoniae and P. mirabilis in that community for mucosal inflammation [85] Outgrowth of gamma proteobacteria in the GI microbiota appears to be a general ecological phenomena of a disturbed microbiome [86]. Thus, wounded epithelium appears to provide a selective niche for chronic colonization by γ–proteobacteria, as well as other bacteria in a polymicrobial biofilm [32–34], which in turn, may drive inflammatory and fibrogenic processes that are detrimental to controlled wound repair and tissue regeneration.
Conclusion
The utilization of culture-independent microbiology to characterize the microbiome of wounds and surrounding mucosa has raised many intriguing questions regarding previously held notions about the cause and effect relationships and mechanisms involved. Microbial colonization or infection is hypothesized to play an important role in driving chronic inflammation and the development of fibrosis, while inflammation itself can alter microbial colonization. Percival at el. have recently discussed two possible hypotheses to explain the role of bacteria in non-healing chronic wounds [87]. In both of these hypotheses, wounds naturally have bacterial biofilms that form and only when the environmental balance within the wound’s microbiota changes (due to alterations in host immune response, local pH, temperature, wound dressings, antimicrobial treatment, etc.) do these biofilms lead to infection. The first of these hypotheses is the “specific bacterial hypothesis,” which states that only a few select species of bacteria break away from the polymicrobial biofilms, cause infection and lead to delayed wound healing. The second, “non-specific bacterial” or “community” hypothesis states that it is the overall composition of the biofilms that creates a pathogenic bacterial community and leads to infection and delays in wound healing [87].
Examples of areas of active investigation into the role of the microbiome in wound repair in the lungs include diseases such as interstitial pulmonary fibrosis, chronic obstructive pulmonary disease (COPD), cystic fibrosis, and asthma [9–11,88]. In the gastrointestinal tract, normal members of the microbiota, especially lactobacilli and butyrate-producing bacteria, promote epithelial repair. Are there also locally resident bacteria that promote wound repair in the skin and lungs? What about sterile sites, such as the heart or liver? Could there be effects of the mucosal microbiome on wound repair of these non-mucosal sites? The study of the microbiome as a metabolically active “organ” [89–92], which can have local and systemic effects on wound repair and fibrosis, has only just begun.
Table 1.
Wound Microbiome Bacterial Species with Known Associations in Wound Repair
| Summary of Studies of Bacteria Associated with Wounds | Association |
|---|---|
| Streptococcus spp. | Higher incidence in chronic diabetic wounds |
| Staphylococcus aerueus, Staphylococcus epidermidis | Higher incidence in chronic diabetic wounds Colonization can impair the wound healing process Readibly cultured from wounds |
| Enterococcus spp. | Higher incidence in chronic diabetic wounds |
| Peptostreptococcus spp. | Higher incidence in chronic diabetic wounds |
| Bacteroides spp | Higher incidence in chronic diabetic wounds |
| Prevotella spp. | Higher incidence in chronic diabetic wounds |
| Pseudomonas aeruginosa | Higher incidence in chronic diabetic wounds Inoculation into germ-free mice wounds leads to acccelerated wound repair (re-epithelialization, epidermial cell proliferation and neo-vascularization) Readibly cultured from wounds |
| Lactobacillus spp | Induces ERK phosphorylation in the GI tract, which could regulate cell migration and epithelium restitution during wound repair |
| Lactobacillus reuteri strain RC-14 | Can inhibit colonization of other species (i.e. Staphylococcus aureus) in a strain-specific manner |
| Escherichia coli | Outgrowth in the GI tract can occur in environments of increased inflammation due to epithelial damage Readibly cultured from wounds |
| Klebsiella pneumoniae | Outgrowth in the GI tract can occur in environments of increased inflammation due to epithelial damage Necessary to induce transmissible spotaneous colitis in mice |
| Proteus mirabilis | Necessary to induce transmissible spotaneous colitis in mice |
| Corynebacteria spp. | Readibly cultured from wounds |
| Propionbacteria spp. | Readibly cultured from wounds, but in lesser levels than that found in healthy skin |
| Neisseria spp. | Fastidious and anaerobic bacteria commonly found in wounds |
| Campylobacteria spp. | Fastidious and anaerobic bacteria commonly found in wounds |
| Clostridiaceae | Fastidious and anaerobic bacteria commonly found in wounds |
This table highlights microbes with known associations to wound healing that are discussed in this review.
ERK = extracellular signal-regulated kinases; GI = gastrointestinal; TLRs = toll-like receptors
Acknowledgments
This work was in part supported by NIH grants T32AI007528 (BSS), U19AI090871 (GBH), P30DK034933 (GBH), R21AI087869 (GBH) and R01HL114447 (GBH).
Abbreviations
- ROS
Reactive oxygen species
- FAK
Focal adhesion kinase
- FAs
Focal cell-matrix adhesions
- FPR
Formyl peptide receptor
Footnotes
Conflict of Interest Statement: None of the authors have any conflicts of interest with the material presented in this review article
Bibliography
- 1.Grice EA, Segre JA. The skin microbiome. Nat Rev Microbiol. 2011;9:244–253. doi: 10.1038/nrmicro2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 3.Meneghin A, Hogaboam CM. Infectious disease, the innate immune response, and fibrosis. J Clin Invest. 2007;117:530–538. doi: 10.1172/JCI30595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reinke JM, Sorg H. Wound Repair and Regeneration. Eur Surg Res. 2012;49:35–43. doi: 10.1159/000339613. [DOI] [PubMed] [Google Scholar]
- 5.Wynn TA. Cellular and molecular mechanisms of fibrosis. J Pathol. 2008;214:199–210. doi: 10.1002/path.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peterson DA, Frank DN, Pace NR, et al. Metagenomic approaches for defining the pathogenesis of inflammatory bowel diseases. Cell Host & Microbe. 2008;3:417–427. doi: 10.1016/j.chom.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bogaert D, Keijser B, Huse S, et al. Variability and diversity of nasopharyngeal microbiota in children: a metagenomic analysis. PLoS One. 2011;6:e17035. doi: 10.1371/journal.pone.0017035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Charlson ES, Chen J, Custers-Allen R, et al. Disordered microbial communities in the upper respiratory tract of cigarette smokers. PLoS One. 2010;5:e15216. doi: 10.1371/journal.pone.0015216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Erb-Downward J, Thompson D, Han M, et al. Analysis of the lung microbiome in the “healthy” smoker and in COPD. PLoS One. 2011;6:e16384. doi: 10.1371/journal.pone.0016384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang YJ, Kim E, Cox MJ, et al. A persistent and diverse airway microbiota present during chronic obstructive pulmonary disease exacerbations. OMICS. 2010;14:9–59. doi: 10.1089/omi.2009.0100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hilty M, Burke C, Pedro H, et al. Disordered microbial communities in asthmatic airways. PLoS One. 2010;5:e8578. doi: 10.1371/journal.pone.0008578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duerkop BA, Vaishnava S, Hooper LV. Immune responses to the microbiota at the intestinal mucosal surface. Immunity. 2009;31:368–376. doi: 10.1016/j.immuni.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 13.Peters BM, Jabra-Rizk MA, O’May GA, et al. Polymicrobial interactions: impact on pathogenesis and human disease. Clin Microbiol Rev. 2012;25:193–213. doi: 10.1128/CMR.00013-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elias S, Banin E. Multi-species biofilms: living with friendly neighbors. FEMS Microbiol Rev. 2012 doi: 10.1111/j.1574-6976.2012.00325.x. [DOI] [PubMed] [Google Scholar]
- 15.Price LB, Liu CM, Melendez JH, et al. Community analysis of chronic wound bacteria using 16S rRNA gene-based pyrosequencing: impact of diabetes and antibiotics on chronic wound microbiota. PLoS One. 2009;4:e6462. doi: 10.1371/journal.pone.0006462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dowd SE, Sun Y, Secor PR, et al. Survey of bacterial diversity in chronic wounds using pyrosequencing, DGGE, and full ribosome shotgun sequencing. BMC Microbiol. 2008;8:43. doi: 10.1186/1471-2180-8-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Price LB, Liu CM, Frankel YM, et al. Macroscale spatial variation in chronic wound microbiota: a cross-sectional study. Wound Repair Regen. 2011;19:80–88. doi: 10.1111/j.1524-475X.2010.00628.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grice EA, Kong HH, Conlan S, et al. Topographical and temporal diversity of the human skin microbiome. Science. 2009;324:1190–1192. doi: 10.1126/science.1171700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Young VB, Schmidt TM. Gi Microbiota and Regulation of the Immune System. Springer-Verlag Berlin; Berlin: 2008. Overview of the gastrointestinal microbiota; pp. 29–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ashelford KE, Chuzhanova NA, Fry JC, et al. At least 1 in 20 16S rRNA sequence records currently held in public repositories is estimated to contain substantial anomalies. Appl Environ Microbiol. 2005;71:7724–7736. doi: 10.1128/AEM.71.12.7724-7736.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Margulies M, Egholm M, Altman WE, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ronaghi M. Pyrosequencing sheds light on DNA sequencing. Genome Res. 2001;11:3–11. doi: 10.1101/gr.11.1.3. [DOI] [PubMed] [Google Scholar]
- 23.Hamady M, Knight R. Microbial community profiling for human microbiome projects: Tools, techniques, and challenges. Genome Research. 2009;19:1141–1152. doi: 10.1101/gr.085464.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hall N. Advanced sequencing technologies and their wider impact in microbiology. J Exp Biol. 2007;210:1518–1525. doi: 10.1242/jeb.001370. [DOI] [PubMed] [Google Scholar]
- 25.Liu Z, Lozupone C, Hamady M, et al. Short pyrosequencing reads suffice for accurate microbial community analysis. Nucleic Acids Res. 2007;35:e120. doi: 10.1093/nar/gkm541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lozupone CA, Knight R. Global patterns in bacterial diversity. Proc Natl Acad Sci U S A. 2007;104:11436–11440. doi: 10.1073/pnas.0611525104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sogin ML, Morrison HG, Huber JA, et al. Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc Natl Acad Sci U S A. 2006;103:12115–12120. doi: 10.1073/pnas.0605127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Antonopoulos DA, Huse SM, Morrison HG, et al. Reproducible Community Dynamics of the Gastrointestinal Microbiota following Antibiotic Perturbation. Infection and Immunity. 2009;77:2367–2375. doi: 10.1128/IAI.01520-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Siddiqui AR, Bernstein JM. Chronic wound infection: facts and controversies. Clin Dermatol. 2010;28:519–526. doi: 10.1016/j.clindermatol.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 30.Bucknall TE. The effect of local infection upon wound healing: an experimental study. Br J Surg. 1980;67:851–855. doi: 10.1002/bjs.1800671205. [DOI] [PubMed] [Google Scholar]
- 31.Frank DN, Wysocki A, Specht-Glick DD, et al. Microbial diversity in chronic open wounds. Wound Repair Regen. 2009;17:163–172. doi: 10.1111/j.1524-475X.2009.00472.x. [DOI] [PubMed] [Google Scholar]
- 32.Han A, Zenilman JM, Melendez JH, et al. The importance of a multifaceted approach to characterizing the microbial flora of chronic wounds. Wound Repair Regen. 2011;19:532–541. doi: 10.1111/j.1524-475X.2011.00720.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bjarnsholt T, Kirketerp-Moller K, Jensen PO, et al. Why chronic wounds will not heal: a novel hypothesis. Wound Repair Regen. 2008;16:2–10. doi: 10.1111/j.1524-475X.2007.00283.x. [DOI] [PubMed] [Google Scholar]
- 34.Rickard AH, Colacino KR, Manton KM, et al. Production of cell-cell signalling molecules by bacteria isolated from human chronic wounds. J Appl Microbiol. 2010;108:1509–1522. doi: 10.1111/j.1365-2672.2009.04554.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Drenkard E. Antimicrobial resistance of Pseudomonas aeruginosa biofilms. Microbes Infect. 2003;5:1213–1219. doi: 10.1016/j.micinf.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 36.O’Meara S, Al-Kurdi D, Ologun Y, et al. Antibiotics and antiseptics for venous leg ulcers. Cochrane Database Syst Rev. 2010:CD003557. doi: 10.1002/14651858.CD003557.pub3. [DOI] [PubMed] [Google Scholar]
- 37.Hoffman LR, D’Argenio DA, MacCoss MJ, et al. Aminoglycoside antibiotics induce bacterial biofilm formation. Nature. 2005;436:1171–1175. doi: 10.1038/nature03912. [DOI] [PubMed] [Google Scholar]
- 38.Davies CE, Wilson MJ, Hill KE, et al. Use of molecular techniques to study microbial diversity in the skin: chronic wounds reevaluated. Wound Repair Regen. 2001;9:332–340. doi: 10.1046/j.1524-475x.2001.00332.x. [DOI] [PubMed] [Google Scholar]
- 39.Gjodsbol K, Skindersoe ME, Christensen JJ, et al. No need for biopsies: comparison of three sample techniques for wound microbiota determination. Int Wound J. 2012;9:295–302. doi: 10.1111/j.1742-481X.2011.00883.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grice EA, Snitkin ES, Yockey LJ, et al. Longitudinal shift in diabetic wound microbiota correlates with prolonged skin defense response. Proc Natl Acad Sci U S A. 2010;107:14799–14804. doi: 10.1073/pnas.1004204107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gardner SE, Frantz RA. Wound bioburden and infection-related complications in diabetic foot ulcers. Biol Res Nurs. 2008;10:44–53. doi: 10.1177/1099800408319056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bowler PG, Duerden BI, Armstrong DG. Wound microbiology and associated approaches to wound management. Clin Microbiol Rev. 2001;14:244–269. doi: 10.1128/CMR.14.2.244-269.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bowler PG, Davies BJ. The microbiology of infected and noninfected leg ulcers. Int J Dermatol. 1999;38:573–578. doi: 10.1046/j.1365-4362.1999.00738.x. [DOI] [PubMed] [Google Scholar]
- 44.Citron DM, Goldstein EJ, Merriam CV, et al. Bacteriology of moderate-to-severe diabetic foot infections and in vitro activity of antimicrobial agents. J Clin Microbiol. 2007;45:2819–2828. doi: 10.1128/JCM.00551-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sopata M, Luczak J, Ciupinska M. Effect of bacteriological status on pressure ulcer healing in patients with advanced cancer. J Wound Care. 2002;11:107–110. doi: 10.12968/jowc.2002.11.3.26678. [DOI] [PubMed] [Google Scholar]
- 46.Schierle CF, De la Garza M, Mustoe TA, et al. Staphylococcal biofilms impair wound healing by delaying reepithelialization in a murine cutaneous wound model. Wound Repair Regen. 2009;17:354–359. doi: 10.1111/j.1524-475X.2009.00489.x. [DOI] [PubMed] [Google Scholar]
- 47.Park S, Rich J, Hanses F, et al. Defects in innate immunity predispose C57BL/6J-Leprdb/Leprdb mice to infection by Staphylococcus aureus. Infect Immun. 2009;77:1008–1014. doi: 10.1128/IAI.00976-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hassett DJ, Cuppoletti J, Trapnell B, et al. Anaerobic metabolism and quorum sensing by Pseudomonas aeruginosa biofilms in chronically infected cystic fibrosis airways: rethinking antibiotic treatment strategies and drug targets. Adv Drug Deliv Rev. 2002;54:1425–1443. doi: 10.1016/s0169-409x(02)00152-7. [DOI] [PubMed] [Google Scholar]
- 49.Hentzer M, Eberl L, Givskov M. Transcriptome analysis of Pseudomonas aeruginosa biofilm development: anaerobic respiration and iron limitation. Biofilms. 2005;2:37–61. [Google Scholar]
- 50.Mahenthiralingam E, Campbell ME, Speert DP. Nonmotility and phagocytic resistance of Pseudomonas aeruginosa isolates from chronically colonized patients with cystic fibrosis. Infect Immun. 1994;62:596–605. doi: 10.1128/iai.62.2.596-605.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barth AL, Pitt TL. Auxotrophic variants of Pseudomonas aeruginosa are selected from prototrophic wild-type strains in respiratory infections in patients with cystic fibrosis. J Clin Microbiol. 1995;33:37–40. doi: 10.1128/jcm.33.1.37-40.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Spencer DH, Kas A, Smith EE, et al. Whole-genome sequence variation among multiple isolates of Pseudomonas aeruginosa. J Bacteriol. 2003;185:1316–1325. doi: 10.1128/JB.185.4.1316-1325.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smith EE, Buckley DG, Wu Z, et al. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc Natl Acad Sci U S A. 2006;103:8487–8492. doi: 10.1073/pnas.0602138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Oliver JD. Recent findings on the viable but nonculturable state in pathogenic bacteria. FEMS Microbiol Rev. 2010;34:415–425. doi: 10.1111/j.1574-6976.2009.00200.x. [DOI] [PubMed] [Google Scholar]
- 55.Harrison F, Browning LE, Vos M, et al. Cooperation and virulence in acute Pseudomonas aeruginosa infections. BMC Biol. 2006;4:21. doi: 10.1186/1741-7007-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Harrison F, Paul J, Massey RC, et al. Interspecific competition and siderophore-mediated cooperation in Pseudomonas aeruginosa. ISME J. 2008;2:49–55. doi: 10.1038/ismej.2007.96. [DOI] [PubMed] [Google Scholar]
- 57.Radtke AL, O’Riordan MX. Intracellular innate resistance to bacterial pathogens. Cell Microbiol. 2006;8:1720–1729. doi: 10.1111/j.1462-5822.2006.00795.x. [DOI] [PubMed] [Google Scholar]
- 58.Johnson EE, Wessling-Resnick M. Iron metabolism and the innate immune response to infection. Microbes Infect. 2012;14:207–216. doi: 10.1016/j.micinf.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mashburn LM, Jett AM, Akins DR, et al. Staphylococcus aureus serves as an iron source for Pseudomonas aeruginosa during in vivo coculture. J Bacteriol. 2005;187:554–566. doi: 10.1128/JB.187.2.554-566.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Palmer KL, Mashburn LM, Singh PK, et al. Cystic fibrosis sputum supports growth and cues key aspects of Pseudomonas aeruginosa physiology. J Bacteriol. 2005;187:5267–5277. doi: 10.1128/JB.187.15.5267-5277.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gan BS, Kim J, Reid G, et al. Lactobacillus fermentum RC-14 inhibits Staphylococcus aureus infection of surgical implants in rats. J Infect Dis. 2002;185:1369–1372. doi: 10.1086/340126. [DOI] [PubMed] [Google Scholar]
- 62.Laughton JM, Devillard E, Heinrichs DE, et al. Inhibition of expression of a staphylococcal superantigen-like protein by a soluble factor from Lactobacillus reuteri. Microbiology. 2006;152:1155–1167. doi: 10.1099/mic.0.28654-0. [DOI] [PubMed] [Google Scholar]
- 63.Grice EA, Segre JA. Interaction of the microbiome with the innate immune response in chronic wounds. Adv Exp Med Biol. 2012;946:55–68. doi: 10.1007/978-1-4614-0106-3_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cosma CL, Sherman DR, Ramakrishnan L. The secret lives of the pathogenic mycobacteria. Annu Rev Microbiol. 2003;57:641–676. doi: 10.1146/annurev.micro.57.030502.091033. [DOI] [PubMed] [Google Scholar]
- 65.Okada M. The influence of intestinal flora on wound healing in mice. Surg Today. 1994;24:347–355. doi: 10.1007/BF02348566. [DOI] [PubMed] [Google Scholar]
- 66.Kanno E, Kawakami K, Ritsu M, et al. Wound healing in skin promoted by inoculation with Pseudomonas aeruginosa PAO1: The critical role of tumor necrosis factor-alpha secreted from infiltrating neutrophils. Wound Repair Regen. 2011;19:608–621. doi: 10.1111/j.1524-475X.2011.00721.x. [DOI] [PubMed] [Google Scholar]
- 67.Laato M, Niinikoski J, Lehtonen OP, et al. Accelerated tissue repair induced by micrococcus varians. Surg Gynecol Obstet. 1987;164:340–344. [PubMed] [Google Scholar]
- 68.Laato M, Niinikoski J, Lundberg C, et al. Inflammatory reaction and blood flow in experimental wounds inoculated with Staphylococcus aureus. Eur Surg Res. 1988;20:33–38. doi: 10.1159/000128738. [DOI] [PubMed] [Google Scholar]
- 69.Levenson SM, Kan-Gruber D, Gruber C, et al. Wound healing accelerated by Staphylococcus aureus. Arch Surg. 1983;118:310–320. doi: 10.1001/archsurg.1983.01390030042007. [DOI] [PubMed] [Google Scholar]
- 70.Souza DG, Vieira AT, Soares AC, et al. The essential role of the intestinal microbiota in facilitating acute inflammatory responses. J Immunol. 2004;173:4137–4146. doi: 10.4049/jimmunol.173.6.4137. [DOI] [PubMed] [Google Scholar]
- 71.Noverr MC, Noggle RM, Toews GB, et al. Role of antibiotics and fungal microbiota in driving pulmonary allergic responses. Infect Immun. 2004;72:4996–5003. doi: 10.1128/IAI.72.9.4996-5003.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dignass AU. Mechanisms and modulation of intestinal epithelial repair. Inflamm Bowel Dis. 2001;7:68–77. doi: 10.1097/00054725-200102000-00014. [DOI] [PubMed] [Google Scholar]
- 73.Karrasch T, Jobin C. Wound healing responses at the gastrointestinal epithelium: a close look at novel regulatory factors and investigative approaches. Z Gastroenterol. 2009;47:1221–1229. doi: 10.1055/s-0028-1109766. [DOI] [PubMed] [Google Scholar]
- 74.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, et al. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 75.Swanson PA, 2nd, Kumar A, Samarin S, et al. Enteric commensal bacteria potentiate epithelial restitution via reactive oxygen species-mediated inactivation of focal adhesion kinase phosphatases. Proc Natl Acad Sci U S A. 2011;108:8803–8808. doi: 10.1073/pnas.1010042108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sun J, Hobert ME, Rao AS, et al. Bacterial activation of beta-catenin signaling in human epithelia. Am J Physiol Gastrointest Liver Physiol. 2004;287:G220–227. doi: 10.1152/ajpgi.00498.2003. [DOI] [PubMed] [Google Scholar]
- 77.Collier-Hyams LS, Sloane V, Batten BC, et al. Cutting edge: bacterial modulation of epithelial signaling via changes in neddylation of cullin-1. J Immunol. 2005;175:4194–4198. doi: 10.4049/jimmunol.175.7.4194. [DOI] [PubMed] [Google Scholar]
- 78.Kumar A, Wu H, Collier-Hyams LS, et al. The bacterial fermentation product butyrate influences epithelial signaling via reactive oxygen species-mediated changes in cullin-1 neddylation. J Immunol. 2009;182:538–546. doi: 10.4049/jimmunol.182.1.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wentworth CC, Jones RM, Kwon YM, et al. Commensal-epithelial signaling mediated via formyl peptide receptors. Am J Pathol. 2010;177:2782–2790. doi: 10.2353/ajpath.2010.100529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wentworth CC, Alam A, Jones RM, et al. Enteric commensal bacteria induce extracellular signal-regulated kinase pathway signaling via formyl peptide receptor-dependent redox modulation of dual specific phosphatase 3. J Biol Chem. 2011;286:38448–38455. doi: 10.1074/jbc.M111.268938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Satoh H, Guth PH, Grossman MI. Role of bacteria in gastric ulceration produced by indomethacin in the rat: cytoprotective action of antibiotics. Gastroenterology. 1983;84:483–489. [PubMed] [Google Scholar]
- 82.Dalby AB, Frank DN, St Amand AL, et al. Culture-independent analysis of indomethacin-induced alterations in the rat gastrointestinal microbiota. Appl Environ Microbiol. 2006;72:6707–6715. doi: 10.1128/AEM.00378-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lau HY, Huffnagle GB, Moore TA. Host and microbiota factors that control Klebsiella pneumoniae mucosal colonization in mice. Microbes Infect. 2008;10:1283–1290. doi: 10.1016/j.micinf.2008.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lupp C, Robertson ML, Wickham ME, et al. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe. 2007;2:119–129. doi: 10.1016/j.chom.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 85.Garrett WS, Gallini CA, Yatsunenko T, et al. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe. 2010;8:292–300. doi: 10.1016/j.chom.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Looft T, Allen HK. Collateral effects of antibiotics on mammalian gut microbiomes. Gut Microbes. 2012:3. doi: 10.4161/gmic.21288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Percival SL, Thomas JG, Williams DW. Biofilms and bacterial imbalances in chronic wounds: anti-Koch. Int Wound J. 2010;7:169–175. doi: 10.1111/j.1742-481X.2010.00668.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Han MK, Huang YJ, Lipuma JJ, et al. Significance of the microbiome in obstructive lung disease. Thorax. 2012;67:456–463. doi: 10.1136/thoraxjnl-2011-201183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Arck P, Handjiski B, Hagen E, et al. Is there a ‘gut-brain-skin axis’? Exp Dermatol. 2010;19:401–405. doi: 10.1111/j.1600-0625.2009.01060.x. [DOI] [PubMed] [Google Scholar]
- 90.Sekirov I, Russell SL, Antunes LC, et al. Gut microbiota in health and disease. Physiol Rev. 2010;90:859–904. doi: 10.1152/physrev.00045.2009. [DOI] [PubMed] [Google Scholar]
- 91.Noverr MC, Huffnagle GB. Does the microbiota regulate immune responses outside the gut? Trends Microbiol. 2004;12:562–568. doi: 10.1016/j.tim.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 92.O’Hara AM, Shanahan F. The gut flora as a forgotten organ. EMBO Rep. 2006;7:688–693. doi: 10.1038/sj.embor.7400731. [DOI] [PMC free article] [PubMed] [Google Scholar]