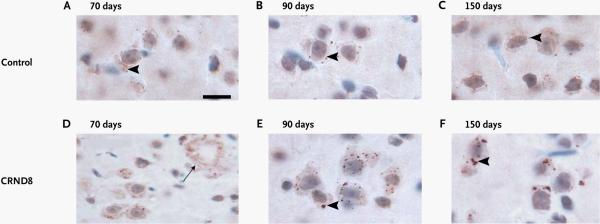
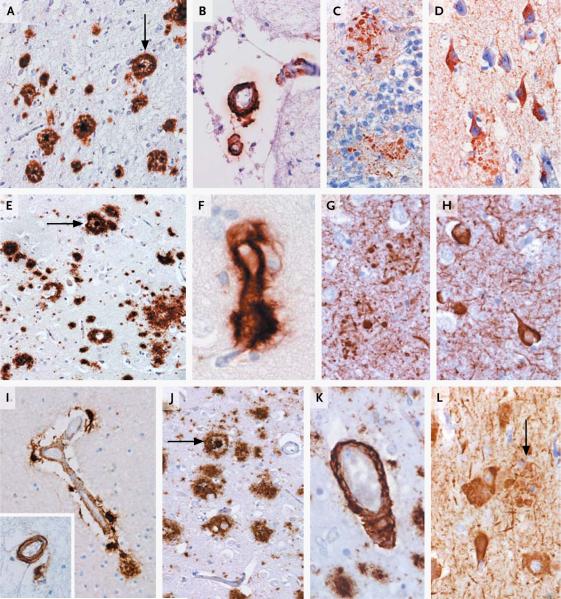
Rita Guerreiro
Rita Guerreiro, Ph.D.
1Rita Guerreiro, Ph.D., University College London (UCL) Institute of Neurology, London, and National Institute on Aging, National Institutes of Health, Bethesda, MD; Aleksandra Wojtas, M.S., Mayo Clinic, Jacksonville, FL; Jose Bras, Ph.D., UCL Institute of Neurology, London; Minerva M. Carrasquillo, Ph.D., Mayo Clinic, Jacksonville, FL; Ekaterina Rogaeva, Ph.D., Tanz Centre, University of Toronto, Toronto; Elisa Majounie, Ph.D., Mayo Clinic, Jacksonville, FL; Carlos Cruchaga, Ph.D., Washington University School of Medicine, St. Louis; Celeste Sassi, M.D., Institute of Neurology, London, and National Institute on Aging, National Institutes of Health, Bethesda, MD; John S.K. Kauwe, Ph.D., Brigham Young University, Provo, Utah; Michelle K. Lupton, Ph.D., King's College London Institute of Psychiatry, London; Mina Ryten, M.D., Ph.D., UCL Institute of Neurology, London; Kristelle Brown, Ph.D., University of Nottingham, Nottingham, United Kingdom; the EADI Consortium, the GERAD Consortium, the UKBE Consortium, James Lowe, D.M., University of Nottingham, Nottingham, United Kingdom; Perry G. Ridge, M.S., Brigham Young University and the ARUP Institute, Salt Lake City; Monia B. Hammer, M.S., National Institute on Aging, National Institutes of Health, Bethesda, MD; Yosuke Wakutani, M.D., Tanz Centre, University of Toronto, Toronto; Lilinaz Hazrati, M.D., Ph.D., Tanz Centre, University of Toronto, Toronto; Petroula Proitsi, Ph.D., King's College London Institute of Psychiatry, London; Stephen Newhouse, Ph.D., King's College London Institute of Psychiatry, London; Ebba Lohmann, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Nihan Erginel-Unaltuna, Ph.D., Institute for Experimental Medical Research, Istanbul University, Istanbul, Turkey; Christopher Medway, Ph.D., University of Nottingham, Nottingham, United Kingdom; Hasmet Hanagasi, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Claire Troakes, Ph.D., King's College London Institute of Psychiatry, London; Hakan Gurvit, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Basar Bilgic, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Safa Al-Sarraj, M.D., King's College London Institute of Psychiatry and King's College Hospital, London; Bruno Benitez, M.D., Washington University School of Medicine, St. Louis; Breanna Cooper, B.S., Washington University School of Medicine, St. Louis; David Carrell, M.S., Washington University School of Medicine, St. Louis; Murat Emre, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Fanggeng Zou, Ph.D., Mayo Clinic, Jacksonville, FL; Li Ma, M.S., Mayo Clinic, Jacksonville, FL; Melissa E. Murray, Ph.D., Mayo Clinic, Jacksonville, FL; Dennis W. Dickson, M.D., Mayo Clinic, Jacksonville, FL; Steven Younkin, M.D., Ph.D., Mayo Clinic, Jacksonville, FL; Ronald C. Petersen, Ph.D., M.D., Mayo Clinic, Rochester, MN; Christopher D. Corcoran, Sc.D., Utah State University, Logan; Yefei Cai, Ph.D., Washington University School of Medicine, St. Louis; Catarina Oliveira, M.D., Ph.D., University of Coimbra, Coimbra, Portugal; Maria Helena Ribeiro, M.S., University of Coimbra, Coimbra, Portugal; Isabel Santana, M.D., Ph.D., University of Coimbra, Coimbra, Portugal; JoAnn T. Tschanz, Ph.D., Utah State University, Logan; J. Raphael Gibbs, B.Sc., UCL Institute of Neurology, London, and National Institute on Aging, National Institutes of Health, Bethesda, MD; Maria C. Norton, Ph.D., Utah State University, Logan; Iwona Kloszewska, Ph.D., Medical University of Lodz, Lodz, Poland; Patrizia Mecocci, M.D., Ph.D., University of Perugia, Perugia, Italy; Hilkka Soininen, M.D., University of Eastern Finland and Kuopio University Hospital, Kuopio, Finland; Magda Tsolaki, M.D., Aristotle University of Thessaloniki, Thessaloniki, Greece; Bruno Vellas, M.D., Ph.D., UMR INSERM 1027, Hôpitaux de Toulouse, Toulouse, France; Ronald G. Munger, Ph.D., M.P.H., Utah State University, Logan; David M.A. Mann, Ph.D., University of Manchester, Manchester, United Kingdom; Stuart Pickering-Brown, Ph.D., University of Manchester, Manchester, United Kingdom; Simon Lovestone, Ph.D., King's College London Institute of Psychiatry, London; Jonathan Beck, B.Sc., MRC Prion Unit and UCL Institute of Neurology, London; Simon Mead, M.D., MRC Prion Unit and UCL Institute of Neurology, London; John Collinge, M.D., MRC Prion Unit and UCL Institute of Neurology, London; Linda Parsons, M.Phil., UCL Institute of Neurology, London; Jennifer Pocock, Ph.D., UCL Institute of Neurology, London; John C. Morris, M.D., Washington University School of Medicine, St. Louis; Tamas Revesz, M.D., UCL Institute of Neurology, London; Tammaryn Lashley, Ph.D., UCL Institute of Neurology, London; Nick C. Fox, M.D., UCL Institute of Neurology, London; Martin N. Rossor, M.D., UCL Institute of Neurology, London; Benjamin Grenier-Boley, M.Sc., INSERM, U744, Institut Pasteur de Lille and Université Lille-Nord de France, Lille, France; Céline Bellenguez, Ph.D., INSERM, U744, Institut Pasteur de Lille and Université Lille-Nord de France, Lille, France; Valentina Moskvina, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, School of Medicine, Cardiff University, Cardiff, United Kingdom; Rebecca Sims, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, United Kingdom; Denise Harold, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, United Kingdom; Julie Williams, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, United Kingdom; Jean-Charles Lambert, Ph.D., INSERM, U744, Institut Pasteur de Lille and Université Lille-Nord de France, Lille, France; Philippe Amouyel, M.D., Ph.D., INSERM, U744, Institut Pasteur de Lille, CHRU de Lille, and Université Lille-Nord de France, Lille, France; Neill Graff-Radford, M.B., B.Ch., Mayo Clinic, Jacksonville, FL; Alison Goate, Ph.D., Washington University School of Medicine, St. Louis; Rosa Rademakers, Ph.D., Mayo Clinic, Jacksonville, FL; Kevin Morgan, Ph.D., School of Molecular Medical Sciences, University of Nottingham, Nottingham, United Kingdom; John Powell, Ph.D., King's College London Institute of Psychiatry, London; Peter St. George-Hyslop, M.D., Tanz Centre, University of Toronto, Toronto, and Cambridge Institute for Medical Research, University of Cambridge, Cambridge, United Kingdom; Andrew Singleton, Ph.D., National Institute on Aging, National Institutes of Health, Bethesda, MD; and John Hardy, Ph.D., UCL Institute of Neurology, London
1,
Aleksandra Wojtas
Aleksandra Wojtas, M.S.
1Rita Guerreiro, Ph.D., University College London (UCL) Institute of Neurology, London, and National Institute on Aging, National Institutes of Health, Bethesda, MD; Aleksandra Wojtas, M.S., Mayo Clinic, Jacksonville, FL; Jose Bras, Ph.D., UCL Institute of Neurology, London; Minerva M. Carrasquillo, Ph.D., Mayo Clinic, Jacksonville, FL; Ekaterina Rogaeva, Ph.D., Tanz Centre, University of Toronto, Toronto; Elisa Majounie, Ph.D., Mayo Clinic, Jacksonville, FL; Carlos Cruchaga, Ph.D., Washington University School of Medicine, St. Louis; Celeste Sassi, M.D., Institute of Neurology, London, and National Institute on Aging, National Institutes of Health, Bethesda, MD; John S.K. Kauwe, Ph.D., Brigham Young University, Provo, Utah; Michelle K. Lupton, Ph.D., King's College London Institute of Psychiatry, London; Mina Ryten, M.D., Ph.D., UCL Institute of Neurology, London; Kristelle Brown, Ph.D., University of Nottingham, Nottingham, United Kingdom; the EADI Consortium, the GERAD Consortium, the UKBE Consortium, James Lowe, D.M., University of Nottingham, Nottingham, United Kingdom; Perry G. Ridge, M.S., Brigham Young University and the ARUP Institute, Salt Lake City; Monia B. Hammer, M.S., National Institute on Aging, National Institutes of Health, Bethesda, MD; Yosuke Wakutani, M.D., Tanz Centre, University of Toronto, Toronto; Lilinaz Hazrati, M.D., Ph.D., Tanz Centre, University of Toronto, Toronto; Petroula Proitsi, Ph.D., King's College London Institute of Psychiatry, London; Stephen Newhouse, Ph.D., King's College London Institute of Psychiatry, London; Ebba Lohmann, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Nihan Erginel-Unaltuna, Ph.D., Institute for Experimental Medical Research, Istanbul University, Istanbul, Turkey; Christopher Medway, Ph.D., University of Nottingham, Nottingham, United Kingdom; Hasmet Hanagasi, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Claire Troakes, Ph.D., King's College London Institute of Psychiatry, London; Hakan Gurvit, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Basar Bilgic, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Safa Al-Sarraj, M.D., King's College London Institute of Psychiatry and King's College Hospital, London; Bruno Benitez, M.D., Washington University School of Medicine, St. Louis; Breanna Cooper, B.S., Washington University School of Medicine, St. Louis; David Carrell, M.S., Washington University School of Medicine, St. Louis; Murat Emre, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Fanggeng Zou, Ph.D., Mayo Clinic, Jacksonville, FL; Li Ma, M.S., Mayo Clinic, Jacksonville, FL; Melissa E. Murray, Ph.D., Mayo Clinic, Jacksonville, FL; Dennis W. Dickson, M.D., Mayo Clinic, Jacksonville, FL; Steven Younkin, M.D., Ph.D., Mayo Clinic, Jacksonville, FL; Ronald C. Petersen, Ph.D., M.D., Mayo Clinic, Rochester, MN; Christopher D. Corcoran, Sc.D., Utah State University, Logan; Yefei Cai, Ph.D., Washington University School of Medicine, St. Louis; Catarina Oliveira, M.D., Ph.D., University of Coimbra, Coimbra, Portugal; Maria Helena Ribeiro, M.S., University of Coimbra, Coimbra, Portugal; Isabel Santana, M.D., Ph.D., University of Coimbra, Coimbra, Portugal; JoAnn T. Tschanz, Ph.D., Utah State University, Logan; J. Raphael Gibbs, B.Sc., UCL Institute of Neurology, London, and National Institute on Aging, National Institutes of Health, Bethesda, MD; Maria C. Norton, Ph.D., Utah State University, Logan; Iwona Kloszewska, Ph.D., Medical University of Lodz, Lodz, Poland; Patrizia Mecocci, M.D., Ph.D., University of Perugia, Perugia, Italy; Hilkka Soininen, M.D., University of Eastern Finland and Kuopio University Hospital, Kuopio, Finland; Magda Tsolaki, M.D., Aristotle University of Thessaloniki, Thessaloniki, Greece; Bruno Vellas, M.D., Ph.D., UMR INSERM 1027, Hôpitaux de Toulouse, Toulouse, France; Ronald G. Munger, Ph.D., M.P.H., Utah State University, Logan; David M.A. Mann, Ph.D., University of Manchester, Manchester, United Kingdom; Stuart Pickering-Brown, Ph.D., University of Manchester, Manchester, United Kingdom; Simon Lovestone, Ph.D., King's College London Institute of Psychiatry, London; Jonathan Beck, B.Sc., MRC Prion Unit and UCL Institute of Neurology, London; Simon Mead, M.D., MRC Prion Unit and UCL Institute of Neurology, London; John Collinge, M.D., MRC Prion Unit and UCL Institute of Neurology, London; Linda Parsons, M.Phil., UCL Institute of Neurology, London; Jennifer Pocock, Ph.D., UCL Institute of Neurology, London; John C. Morris, M.D., Washington University School of Medicine, St. Louis; Tamas Revesz, M.D., UCL Institute of Neurology, London; Tammaryn Lashley, Ph.D., UCL Institute of Neurology, London; Nick C. Fox, M.D., UCL Institute of Neurology, London; Martin N. Rossor, M.D., UCL Institute of Neurology, London; Benjamin Grenier-Boley, M.Sc., INSERM, U744, Institut Pasteur de Lille and Université Lille-Nord de France, Lille, France; Céline Bellenguez, Ph.D., INSERM, U744, Institut Pasteur de Lille and Université Lille-Nord de France, Lille, France; Valentina Moskvina, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, School of Medicine, Cardiff University, Cardiff, United Kingdom; Rebecca Sims, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, United Kingdom; Denise Harold, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, United Kingdom; Julie Williams, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, United Kingdom; Jean-Charles Lambert, Ph.D., INSERM, U744, Institut Pasteur de Lille and Université Lille-Nord de France, Lille, France; Philippe Amouyel, M.D., Ph.D., INSERM, U744, Institut Pasteur de Lille, CHRU de Lille, and Université Lille-Nord de France, Lille, France; Neill Graff-Radford, M.B., B.Ch., Mayo Clinic, Jacksonville, FL; Alison Goate, Ph.D., Washington University School of Medicine, St. Louis; Rosa Rademakers, Ph.D., Mayo Clinic, Jacksonville, FL; Kevin Morgan, Ph.D., School of Molecular Medical Sciences, University of Nottingham, Nottingham, United Kingdom; John Powell, Ph.D., King's College London Institute of Psychiatry, London; Peter St. George-Hyslop, M.D., Tanz Centre, University of Toronto, Toronto, and Cambridge Institute for Medical Research, University of Cambridge, Cambridge, United Kingdom; Andrew Singleton, Ph.D., National Institute on Aging, National Institutes of Health, Bethesda, MD; and John Hardy, Ph.D., UCL Institute of Neurology, London
1,
Jose Bras
Jose Bras, Ph.D.
1Rita Guerreiro, Ph.D., University College London (UCL) Institute of Neurology, London, and National Institute on Aging, National Institutes of Health, Bethesda, MD; Aleksandra Wojtas, M.S., Mayo Clinic, Jacksonville, FL; Jose Bras, Ph.D., UCL Institute of Neurology, London; Minerva M. Carrasquillo, Ph.D., Mayo Clinic, Jacksonville, FL; Ekaterina Rogaeva, Ph.D., Tanz Centre, University of Toronto, Toronto; Elisa Majounie, Ph.D., Mayo Clinic, Jacksonville, FL; Carlos Cruchaga, Ph.D., Washington University School of Medicine, St. Louis; Celeste Sassi, M.D., Institute of Neurology, London, and National Institute on Aging, National Institutes of Health, Bethesda, MD; John S.K. Kauwe, Ph.D., Brigham Young University, Provo, Utah; Michelle K. Lupton, Ph.D., King's College London Institute of Psychiatry, London; Mina Ryten, M.D., Ph.D., UCL Institute of Neurology, London; Kristelle Brown, Ph.D., University of Nottingham, Nottingham, United Kingdom; the EADI Consortium, the GERAD Consortium, the UKBE Consortium, James Lowe, D.M., University of Nottingham, Nottingham, United Kingdom; Perry G. Ridge, M.S., Brigham Young University and the ARUP Institute, Salt Lake City; Monia B. Hammer, M.S., National Institute on Aging, National Institutes of Health, Bethesda, MD; Yosuke Wakutani, M.D., Tanz Centre, University of Toronto, Toronto; Lilinaz Hazrati, M.D., Ph.D., Tanz Centre, University of Toronto, Toronto; Petroula Proitsi, Ph.D., King's College London Institute of Psychiatry, London; Stephen Newhouse, Ph.D., King's College London Institute of Psychiatry, London; Ebba Lohmann, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Nihan Erginel-Unaltuna, Ph.D., Institute for Experimental Medical Research, Istanbul University, Istanbul, Turkey; Christopher Medway, Ph.D., University of Nottingham, Nottingham, United Kingdom; Hasmet Hanagasi, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Claire Troakes, Ph.D., King's College London Institute of Psychiatry, London; Hakan Gurvit, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Basar Bilgic, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Safa Al-Sarraj, M.D., King's College London Institute of Psychiatry and King's College Hospital, London; Bruno Benitez, M.D., Washington University School of Medicine, St. Louis; Breanna Cooper, B.S., Washington University School of Medicine, St. Louis; David Carrell, M.S., Washington University School of Medicine, St. Louis; Murat Emre, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Fanggeng Zou, Ph.D., Mayo Clinic, Jacksonville, FL; Li Ma, M.S., Mayo Clinic, Jacksonville, FL; Melissa E. Murray, Ph.D., Mayo Clinic, Jacksonville, FL; Dennis W. Dickson, M.D., Mayo Clinic, Jacksonville, FL; Steven Younkin, M.D., Ph.D., Mayo Clinic, Jacksonville, FL; Ronald C. Petersen, Ph.D., M.D., Mayo Clinic, Rochester, MN; Christopher D. Corcoran, Sc.D., Utah State University, Logan; Yefei Cai, Ph.D., Washington University School of Medicine, St. Louis; Catarina Oliveira, M.D., Ph.D., University of Coimbra, Coimbra, Portugal; Maria Helena Ribeiro, M.S., University of Coimbra, Coimbra, Portugal; Isabel Santana, M.D., Ph.D., University of Coimbra, Coimbra, Portugal; JoAnn T. Tschanz, Ph.D., Utah State University, Logan; J. Raphael Gibbs, B.Sc., UCL Institute of Neurology, London, and National Institute on Aging, National Institutes of Health, Bethesda, MD; Maria C. Norton, Ph.D., Utah State University, Logan; Iwona Kloszewska, Ph.D., Medical University of Lodz, Lodz, Poland; Patrizia Mecocci, M.D., Ph.D., University of Perugia, Perugia, Italy; Hilkka Soininen, M.D., University of Eastern Finland and Kuopio University Hospital, Kuopio, Finland; Magda Tsolaki, M.D., Aristotle University of Thessaloniki, Thessaloniki, Greece; Bruno Vellas, M.D., Ph.D., UMR INSERM 1027, Hôpitaux de Toulouse, Toulouse, France; Ronald G. Munger, Ph.D., M.P.H., Utah State University, Logan; David M.A. Mann, Ph.D., University of Manchester, Manchester, United Kingdom; Stuart Pickering-Brown, Ph.D., University of Manchester, Manchester, United Kingdom; Simon Lovestone, Ph.D., King's College London Institute of Psychiatry, London; Jonathan Beck, B.Sc., MRC Prion Unit and UCL Institute of Neurology, London; Simon Mead, M.D., MRC Prion Unit and UCL Institute of Neurology, London; John Collinge, M.D., MRC Prion Unit and UCL Institute of Neurology, London; Linda Parsons, M.Phil., UCL Institute of Neurology, London; Jennifer Pocock, Ph.D., UCL Institute of Neurology, London; John C. Morris, M.D., Washington University School of Medicine, St. Louis; Tamas Revesz, M.D., UCL Institute of Neurology, London; Tammaryn Lashley, Ph.D., UCL Institute of Neurology, London; Nick C. Fox, M.D., UCL Institute of Neurology, London; Martin N. Rossor, M.D., UCL Institute of Neurology, London; Benjamin Grenier-Boley, M.Sc., INSERM, U744, Institut Pasteur de Lille and Université Lille-Nord de France, Lille, France; Céline Bellenguez, Ph.D., INSERM, U744, Institut Pasteur de Lille and Université Lille-Nord de France, Lille, France; Valentina Moskvina, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, School of Medicine, Cardiff University, Cardiff, United Kingdom; Rebecca Sims, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, United Kingdom; Denise Harold, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, United Kingdom; Julie Williams, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, United Kingdom; Jean-Charles Lambert, Ph.D., INSERM, U744, Institut Pasteur de Lille and Université Lille-Nord de France, Lille, France; Philippe Amouyel, M.D., Ph.D., INSERM, U744, Institut Pasteur de Lille, CHRU de Lille, and Université Lille-Nord de France, Lille, France; Neill Graff-Radford, M.B., B.Ch., Mayo Clinic, Jacksonville, FL; Alison Goate, Ph.D., Washington University School of Medicine, St. Louis; Rosa Rademakers, Ph.D., Mayo Clinic, Jacksonville, FL; Kevin Morgan, Ph.D., School of Molecular Medical Sciences, University of Nottingham, Nottingham, United Kingdom; John Powell, Ph.D., King's College London Institute of Psychiatry, London; Peter St. George-Hyslop, M.D., Tanz Centre, University of Toronto, Toronto, and Cambridge Institute for Medical Research, University of Cambridge, Cambridge, United Kingdom; Andrew Singleton, Ph.D., National Institute on Aging, National Institutes of Health, Bethesda, MD; and John Hardy, Ph.D., UCL Institute of Neurology, London
1,
Minerva Carrasquillo
Minerva Carrasquillo, Ph.D.
1Rita Guerreiro, Ph.D., University College London (UCL) Institute of Neurology, London, and National Institute on Aging, National Institutes of Health, Bethesda, MD; Aleksandra Wojtas, M.S., Mayo Clinic, Jacksonville, FL; Jose Bras, Ph.D., UCL Institute of Neurology, London; Minerva M. Carrasquillo, Ph.D., Mayo Clinic, Jacksonville, FL; Ekaterina Rogaeva, Ph.D., Tanz Centre, University of Toronto, Toronto; Elisa Majounie, Ph.D., Mayo Clinic, Jacksonville, FL; Carlos Cruchaga, Ph.D., Washington University School of Medicine, St. Louis; Celeste Sassi, M.D., Institute of Neurology, London, and National Institute on Aging, National Institutes of Health, Bethesda, MD; John S.K. Kauwe, Ph.D., Brigham Young University, Provo, Utah; Michelle K. Lupton, Ph.D., King's College London Institute of Psychiatry, London; Mina Ryten, M.D., Ph.D., UCL Institute of Neurology, London; Kristelle Brown, Ph.D., University of Nottingham, Nottingham, United Kingdom; the EADI Consortium, the GERAD Consortium, the UKBE Consortium, James Lowe, D.M., University of Nottingham, Nottingham, United Kingdom; Perry G. Ridge, M.S., Brigham Young University and the ARUP Institute, Salt Lake City; Monia B. Hammer, M.S., National Institute on Aging, National Institutes of Health, Bethesda, MD; Yosuke Wakutani, M.D., Tanz Centre, University of Toronto, Toronto; Lilinaz Hazrati, M.D., Ph.D., Tanz Centre, University of Toronto, Toronto; Petroula Proitsi, Ph.D., King's College London Institute of Psychiatry, London; Stephen Newhouse, Ph.D., King's College London Institute of Psychiatry, London; Ebba Lohmann, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Nihan Erginel-Unaltuna, Ph.D., Institute for Experimental Medical Research, Istanbul University, Istanbul, Turkey; Christopher Medway, Ph.D., University of Nottingham, Nottingham, United Kingdom; Hasmet Hanagasi, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Claire Troakes, Ph.D., King's College London Institute of Psychiatry, London; Hakan Gurvit, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Basar Bilgic, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Safa Al-Sarraj, M.D., King's College London Institute of Psychiatry and King's College Hospital, London; Bruno Benitez, M.D., Washington University School of Medicine, St. Louis; Breanna Cooper, B.S., Washington University School of Medicine, St. Louis; David Carrell, M.S., Washington University School of Medicine, St. Louis; Murat Emre, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Fanggeng Zou, Ph.D., Mayo Clinic, Jacksonville, FL; Li Ma, M.S., Mayo Clinic, Jacksonville, FL; Melissa E. Murray, Ph.D., Mayo Clinic, Jacksonville, FL; Dennis W. Dickson, M.D., Mayo Clinic, Jacksonville, FL; Steven Younkin, M.D., Ph.D., Mayo Clinic, Jacksonville, FL; Ronald C. Petersen, Ph.D., M.D., Mayo Clinic, Rochester, MN; Christopher D. Corcoran, Sc.D., Utah State University, Logan; Yefei Cai, Ph.D., Washington University School of Medicine, St. Louis; Catarina Oliveira, M.D., Ph.D., University of Coimbra, Coimbra, Portugal; Maria Helena Ribeiro, M.S., University of Coimbra, Coimbra, Portugal; Isabel Santana, M.D., Ph.D., University of Coimbra, Coimbra, Portugal; JoAnn T. Tschanz, Ph.D., Utah State University, Logan; J. Raphael Gibbs, B.Sc., UCL Institute of Neurology, London, and National Institute on Aging, National Institutes of Health, Bethesda, MD; Maria C. Norton, Ph.D., Utah State University, Logan; Iwona Kloszewska, Ph.D., Medical University of Lodz, Lodz, Poland; Patrizia Mecocci, M.D., Ph.D., University of Perugia, Perugia, Italy; Hilkka Soininen, M.D., University of Eastern Finland and Kuopio University Hospital, Kuopio, Finland; Magda Tsolaki, M.D., Aristotle University of Thessaloniki, Thessaloniki, Greece; Bruno Vellas, M.D., Ph.D., UMR INSERM 1027, Hôpitaux de Toulouse, Toulouse, France; Ronald G. Munger, Ph.D., M.P.H., Utah State University, Logan; David M.A. Mann, Ph.D., University of Manchester, Manchester, United Kingdom; Stuart Pickering-Brown, Ph.D., University of Manchester, Manchester, United Kingdom; Simon Lovestone, Ph.D., King's College London Institute of Psychiatry, London; Jonathan Beck, B.Sc., MRC Prion Unit and UCL Institute of Neurology, London; Simon Mead, M.D., MRC Prion Unit and UCL Institute of Neurology, London; John Collinge, M.D., MRC Prion Unit and UCL Institute of Neurology, London; Linda Parsons, M.Phil., UCL Institute of Neurology, London; Jennifer Pocock, Ph.D., UCL Institute of Neurology, London; John C. Morris, M.D., Washington University School of Medicine, St. Louis; Tamas Revesz, M.D., UCL Institute of Neurology, London; Tammaryn Lashley, Ph.D., UCL Institute of Neurology, London; Nick C. Fox, M.D., UCL Institute of Neurology, London; Martin N. Rossor, M.D., UCL Institute of Neurology, London; Benjamin Grenier-Boley, M.Sc., INSERM, U744, Institut Pasteur de Lille and Université Lille-Nord de France, Lille, France; Céline Bellenguez, Ph.D., INSERM, U744, Institut Pasteur de Lille and Université Lille-Nord de France, Lille, France; Valentina Moskvina, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, School of Medicine, Cardiff University, Cardiff, United Kingdom; Rebecca Sims, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, United Kingdom; Denise Harold, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, United Kingdom; Julie Williams, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, United Kingdom; Jean-Charles Lambert, Ph.D., INSERM, U744, Institut Pasteur de Lille and Université Lille-Nord de France, Lille, France; Philippe Amouyel, M.D., Ph.D., INSERM, U744, Institut Pasteur de Lille, CHRU de Lille, and Université Lille-Nord de France, Lille, France; Neill Graff-Radford, M.B., B.Ch., Mayo Clinic, Jacksonville, FL; Alison Goate, Ph.D., Washington University School of Medicine, St. Louis; Rosa Rademakers, Ph.D., Mayo Clinic, Jacksonville, FL; Kevin Morgan, Ph.D., School of Molecular Medical Sciences, University of Nottingham, Nottingham, United Kingdom; John Powell, Ph.D., King's College London Institute of Psychiatry, London; Peter St. George-Hyslop, M.D., Tanz Centre, University of Toronto, Toronto, and Cambridge Institute for Medical Research, University of Cambridge, Cambridge, United Kingdom; Andrew Singleton, Ph.D., National Institute on Aging, National Institutes of Health, Bethesda, MD; and John Hardy, Ph.D., UCL Institute of Neurology, London
1,
Ekaterina Rogaeva
Ekaterina Rogaeva, Ph.D.
1Rita Guerreiro, Ph.D., University College London (UCL) Institute of Neurology, London, and National Institute on Aging, National Institutes of Health, Bethesda, MD; Aleksandra Wojtas, M.S., Mayo Clinic, Jacksonville, FL; Jose Bras, Ph.D., UCL Institute of Neurology, London; Minerva M. Carrasquillo, Ph.D., Mayo Clinic, Jacksonville, FL; Ekaterina Rogaeva, Ph.D., Tanz Centre, University of Toronto, Toronto; Elisa Majounie, Ph.D., Mayo Clinic, Jacksonville, FL; Carlos Cruchaga, Ph.D., Washington University School of Medicine, St. Louis; Celeste Sassi, M.D., Institute of Neurology, London, and National Institute on Aging, National Institutes of Health, Bethesda, MD; John S.K. Kauwe, Ph.D., Brigham Young University, Provo, Utah; Michelle K. Lupton, Ph.D., King's College London Institute of Psychiatry, London; Mina Ryten, M.D., Ph.D., UCL Institute of Neurology, London; Kristelle Brown, Ph.D., University of Nottingham, Nottingham, United Kingdom; the EADI Consortium, the GERAD Consortium, the UKBE Consortium, James Lowe, D.M., University of Nottingham, Nottingham, United Kingdom; Perry G. Ridge, M.S., Brigham Young University and the ARUP Institute, Salt Lake City; Monia B. Hammer, M.S., National Institute on Aging, National Institutes of Health, Bethesda, MD; Yosuke Wakutani, M.D., Tanz Centre, University of Toronto, Toronto; Lilinaz Hazrati, M.D., Ph.D., Tanz Centre, University of Toronto, Toronto; Petroula Proitsi, Ph.D., King's College London Institute of Psychiatry, London; Stephen Newhouse, Ph.D., King's College London Institute of Psychiatry, London; Ebba Lohmann, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Nihan Erginel-Unaltuna, Ph.D., Institute for Experimental Medical Research, Istanbul University, Istanbul, Turkey; Christopher Medway, Ph.D., University of Nottingham, Nottingham, United Kingdom; Hasmet Hanagasi, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Claire Troakes, Ph.D., King's College London Institute of Psychiatry, London; Hakan Gurvit, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Basar Bilgic, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Safa Al-Sarraj, M.D., King's College London Institute of Psychiatry and King's College Hospital, London; Bruno Benitez, M.D., Washington University School of Medicine, St. Louis; Breanna Cooper, B.S., Washington University School of Medicine, St. Louis; David Carrell, M.S., Washington University School of Medicine, St. Louis; Murat Emre, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Fanggeng Zou, Ph.D., Mayo Clinic, Jacksonville, FL; Li Ma, M.S., Mayo Clinic, Jacksonville, FL; Melissa E. Murray, Ph.D., Mayo Clinic, Jacksonville, FL; Dennis W. Dickson, M.D., Mayo Clinic, Jacksonville, FL; Steven Younkin, M.D., Ph.D., Mayo Clinic, Jacksonville, FL; Ronald C. Petersen, Ph.D., M.D., Mayo Clinic, Rochester, MN; Christopher D. Corcoran, Sc.D., Utah State University, Logan; Yefei Cai, Ph.D., Washington University School of Medicine, St. Louis; Catarina Oliveira, M.D., Ph.D., University of Coimbra, Coimbra, Portugal; Maria Helena Ribeiro, M.S., University of Coimbra, Coimbra, Portugal; Isabel Santana, M.D., Ph.D., University of Coimbra, Coimbra, Portugal; JoAnn T. Tschanz, Ph.D., Utah State University, Logan; J. Raphael Gibbs, B.Sc., UCL Institute of Neurology, London, and National Institute on Aging, National Institutes of Health, Bethesda, MD; Maria C. Norton, Ph.D., Utah State University, Logan; Iwona Kloszewska, Ph.D., Medical University of Lodz, Lodz, Poland; Patrizia Mecocci, M.D., Ph.D., University of Perugia, Perugia, Italy; Hilkka Soininen, M.D., University of Eastern Finland and Kuopio University Hospital, Kuopio, Finland; Magda Tsolaki, M.D., Aristotle University of Thessaloniki, Thessaloniki, Greece; Bruno Vellas, M.D., Ph.D., UMR INSERM 1027, Hôpitaux de Toulouse, Toulouse, France; Ronald G. Munger, Ph.D., M.P.H., Utah State University, Logan; David M.A. Mann, Ph.D., University of Manchester, Manchester, United Kingdom; Stuart Pickering-Brown, Ph.D., University of Manchester, Manchester, United Kingdom; Simon Lovestone, Ph.D., King's College London Institute of Psychiatry, London; Jonathan Beck, B.Sc., MRC Prion Unit and UCL Institute of Neurology, London; Simon Mead, M.D., MRC Prion Unit and UCL Institute of Neurology, London; John Collinge, M.D., MRC Prion Unit and UCL Institute of Neurology, London; Linda Parsons, M.Phil., UCL Institute of Neurology, London; Jennifer Pocock, Ph.D., UCL Institute of Neurology, London; John C. Morris, M.D., Washington University School of Medicine, St. Louis; Tamas Revesz, M.D., UCL Institute of Neurology, London; Tammaryn Lashley, Ph.D., UCL Institute of Neurology, London; Nick C. Fox, M.D., UCL Institute of Neurology, London; Martin N. Rossor, M.D., UCL Institute of Neurology, London; Benjamin Grenier-Boley, M.Sc., INSERM, U744, Institut Pasteur de Lille and Université Lille-Nord de France, Lille, France; Céline Bellenguez, Ph.D., INSERM, U744, Institut Pasteur de Lille and Université Lille-Nord de France, Lille, France; Valentina Moskvina, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, School of Medicine, Cardiff University, Cardiff, United Kingdom; Rebecca Sims, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, United Kingdom; Denise Harold, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, United Kingdom; Julie Williams, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, United Kingdom; Jean-Charles Lambert, Ph.D., INSERM, U744, Institut Pasteur de Lille and Université Lille-Nord de France, Lille, France; Philippe Amouyel, M.D., Ph.D., INSERM, U744, Institut Pasteur de Lille, CHRU de Lille, and Université Lille-Nord de France, Lille, France; Neill Graff-Radford, M.B., B.Ch., Mayo Clinic, Jacksonville, FL; Alison Goate, Ph.D., Washington University School of Medicine, St. Louis; Rosa Rademakers, Ph.D., Mayo Clinic, Jacksonville, FL; Kevin Morgan, Ph.D., School of Molecular Medical Sciences, University of Nottingham, Nottingham, United Kingdom; John Powell, Ph.D., King's College London Institute of Psychiatry, London; Peter St. George-Hyslop, M.D., Tanz Centre, University of Toronto, Toronto, and Cambridge Institute for Medical Research, University of Cambridge, Cambridge, United Kingdom; Andrew Singleton, Ph.D., National Institute on Aging, National Institutes of Health, Bethesda, MD; and John Hardy, Ph.D., UCL Institute of Neurology, London
1,
Elisa Majounie
Elisa Majounie, Ph.D.
1Rita Guerreiro, Ph.D., University College London (UCL) Institute of Neurology, London, and National Institute on Aging, National Institutes of Health, Bethesda, MD; Aleksandra Wojtas, M.S., Mayo Clinic, Jacksonville, FL; Jose Bras, Ph.D., UCL Institute of Neurology, London; Minerva M. Carrasquillo, Ph.D., Mayo Clinic, Jacksonville, FL; Ekaterina Rogaeva, Ph.D., Tanz Centre, University of Toronto, Toronto; Elisa Majounie, Ph.D., Mayo Clinic, Jacksonville, FL; Carlos Cruchaga, Ph.D., Washington University School of Medicine, St. Louis; Celeste Sassi, M.D., Institute of Neurology, London, and National Institute on Aging, National Institutes of Health, Bethesda, MD; John S.K. Kauwe, Ph.D., Brigham Young University, Provo, Utah; Michelle K. Lupton, Ph.D., King's College London Institute of Psychiatry, London; Mina Ryten, M.D., Ph.D., UCL Institute of Neurology, London; Kristelle Brown, Ph.D., University of Nottingham, Nottingham, United Kingdom; the EADI Consortium, the GERAD Consortium, the UKBE Consortium, James Lowe, D.M., University of Nottingham, Nottingham, United Kingdom; Perry G. Ridge, M.S., Brigham Young University and the ARUP Institute, Salt Lake City; Monia B. Hammer, M.S., National Institute on Aging, National Institutes of Health, Bethesda, MD; Yosuke Wakutani, M.D., Tanz Centre, University of Toronto, Toronto; Lilinaz Hazrati, M.D., Ph.D., Tanz Centre, University of Toronto, Toronto; Petroula Proitsi, Ph.D., King's College London Institute of Psychiatry, London; Stephen Newhouse, Ph.D., King's College London Institute of Psychiatry, London; Ebba Lohmann, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Nihan Erginel-Unaltuna, Ph.D., Institute for Experimental Medical Research, Istanbul University, Istanbul, Turkey; Christopher Medway, Ph.D., University of Nottingham, Nottingham, United Kingdom; Hasmet Hanagasi, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Claire Troakes, Ph.D., King's College London Institute of Psychiatry, London; Hakan Gurvit, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Basar Bilgic, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Safa Al-Sarraj, M.D., King's College London Institute of Psychiatry and King's College Hospital, London; Bruno Benitez, M.D., Washington University School of Medicine, St. Louis; Breanna Cooper, B.S., Washington University School of Medicine, St. Louis; David Carrell, M.S., Washington University School of Medicine, St. Louis; Murat Emre, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Fanggeng Zou, Ph.D., Mayo Clinic, Jacksonville, FL; Li Ma, M.S., Mayo Clinic, Jacksonville, FL; Melissa E. Murray, Ph.D., Mayo Clinic, Jacksonville, FL; Dennis W. Dickson, M.D., Mayo Clinic, Jacksonville, FL; Steven Younkin, M.D., Ph.D., Mayo Clinic, Jacksonville, FL; Ronald C. Petersen, Ph.D., M.D., Mayo Clinic, Rochester, MN; Christopher D. Corcoran, Sc.D., Utah State University, Logan; Yefei Cai, Ph.D., Washington University School of Medicine, St. Louis; Catarina Oliveira, M.D., Ph.D., University of Coimbra, Coimbra, Portugal; Maria Helena Ribeiro, M.S., University of Coimbra, Coimbra, Portugal; Isabel Santana, M.D., Ph.D., University of Coimbra, Coimbra, Portugal; JoAnn T. Tschanz, Ph.D., Utah State University, Logan; J. Raphael Gibbs, B.Sc., UCL Institute of Neurology, London, and National Institute on Aging, National Institutes of Health, Bethesda, MD; Maria C. Norton, Ph.D., Utah State University, Logan; Iwona Kloszewska, Ph.D., Medical University of Lodz, Lodz, Poland; Patrizia Mecocci, M.D., Ph.D., University of Perugia, Perugia, Italy; Hilkka Soininen, M.D., University of Eastern Finland and Kuopio University Hospital, Kuopio, Finland; Magda Tsolaki, M.D., Aristotle University of Thessaloniki, Thessaloniki, Greece; Bruno Vellas, M.D., Ph.D., UMR INSERM 1027, Hôpitaux de Toulouse, Toulouse, France; Ronald G. Munger, Ph.D., M.P.H., Utah State University, Logan; David M.A. Mann, Ph.D., University of Manchester, Manchester, United Kingdom; Stuart Pickering-Brown, Ph.D., University of Manchester, Manchester, United Kingdom; Simon Lovestone, Ph.D., King's College London Institute of Psychiatry, London; Jonathan Beck, B.Sc., MRC Prion Unit and UCL Institute of Neurology, London; Simon Mead, M.D., MRC Prion Unit and UCL Institute of Neurology, London; John Collinge, M.D., MRC Prion Unit and UCL Institute of Neurology, London; Linda Parsons, M.Phil., UCL Institute of Neurology, London; Jennifer Pocock, Ph.D., UCL Institute of Neurology, London; John C. Morris, M.D., Washington University School of Medicine, St. Louis; Tamas Revesz, M.D., UCL Institute of Neurology, London; Tammaryn Lashley, Ph.D., UCL Institute of Neurology, London; Nick C. Fox, M.D., UCL Institute of Neurology, London; Martin N. Rossor, M.D., UCL Institute of Neurology, London; Benjamin Grenier-Boley, M.Sc., INSERM, U744, Institut Pasteur de Lille and Université Lille-Nord de France, Lille, France; Céline Bellenguez, Ph.D., INSERM, U744, Institut Pasteur de Lille and Université Lille-Nord de France, Lille, France; Valentina Moskvina, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, School of Medicine, Cardiff University, Cardiff, United Kingdom; Rebecca Sims, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, United Kingdom; Denise Harold, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, United Kingdom; Julie Williams, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, United Kingdom; Jean-Charles Lambert, Ph.D., INSERM, U744, Institut Pasteur de Lille and Université Lille-Nord de France, Lille, France; Philippe Amouyel, M.D., Ph.D., INSERM, U744, Institut Pasteur de Lille, CHRU de Lille, and Université Lille-Nord de France, Lille, France; Neill Graff-Radford, M.B., B.Ch., Mayo Clinic, Jacksonville, FL; Alison Goate, Ph.D., Washington University School of Medicine, St. Louis; Rosa Rademakers, Ph.D., Mayo Clinic, Jacksonville, FL; Kevin Morgan, Ph.D., School of Molecular Medical Sciences, University of Nottingham, Nottingham, United Kingdom; John Powell, Ph.D., King's College London Institute of Psychiatry, London; Peter St. George-Hyslop, M.D., Tanz Centre, University of Toronto, Toronto, and Cambridge Institute for Medical Research, University of Cambridge, Cambridge, United Kingdom; Andrew Singleton, Ph.D., National Institute on Aging, National Institutes of Health, Bethesda, MD; and John Hardy, Ph.D., UCL Institute of Neurology, London
1,
Carlos Cruchaga
Carlos Cruchaga, Ph.D.
1Rita Guerreiro, Ph.D., University College London (UCL) Institute of Neurology, London, and National Institute on Aging, National Institutes of Health, Bethesda, MD; Aleksandra Wojtas, M.S., Mayo Clinic, Jacksonville, FL; Jose Bras, Ph.D., UCL Institute of Neurology, London; Minerva M. Carrasquillo, Ph.D., Mayo Clinic, Jacksonville, FL; Ekaterina Rogaeva, Ph.D., Tanz Centre, University of Toronto, Toronto; Elisa Majounie, Ph.D., Mayo Clinic, Jacksonville, FL; Carlos Cruchaga, Ph.D., Washington University School of Medicine, St. Louis; Celeste Sassi, M.D., Institute of Neurology, London, and National Institute on Aging, National Institutes of Health, Bethesda, MD; John S.K. Kauwe, Ph.D., Brigham Young University, Provo, Utah; Michelle K. Lupton, Ph.D., King's College London Institute of Psychiatry, London; Mina Ryten, M.D., Ph.D., UCL Institute of Neurology, London; Kristelle Brown, Ph.D., University of Nottingham, Nottingham, United Kingdom; the EADI Consortium, the GERAD Consortium, the UKBE Consortium, James Lowe, D.M., University of Nottingham, Nottingham, United Kingdom; Perry G. Ridge, M.S., Brigham Young University and the ARUP Institute, Salt Lake City; Monia B. Hammer, M.S., National Institute on Aging, National Institutes of Health, Bethesda, MD; Yosuke Wakutani, M.D., Tanz Centre, University of Toronto, Toronto; Lilinaz Hazrati, M.D., Ph.D., Tanz Centre, University of Toronto, Toronto; Petroula Proitsi, Ph.D., King's College London Institute of Psychiatry, London; Stephen Newhouse, Ph.D., King's College London Institute of Psychiatry, London; Ebba Lohmann, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Nihan Erginel-Unaltuna, Ph.D., Institute for Experimental Medical Research, Istanbul University, Istanbul, Turkey; Christopher Medway, Ph.D., University of Nottingham, Nottingham, United Kingdom; Hasmet Hanagasi, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Claire Troakes, Ph.D., King's College London Institute of Psychiatry, London; Hakan Gurvit, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Basar Bilgic, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Safa Al-Sarraj, M.D., King's College London Institute of Psychiatry and King's College Hospital, London; Bruno Benitez, M.D., Washington University School of Medicine, St. Louis; Breanna Cooper, B.S., Washington University School of Medicine, St. Louis; David Carrell, M.S., Washington University School of Medicine, St. Louis; Murat Emre, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Fanggeng Zou, Ph.D., Mayo Clinic, Jacksonville, FL; Li Ma, M.S., Mayo Clinic, Jacksonville, FL; Melissa E. Murray, Ph.D., Mayo Clinic, Jacksonville, FL; Dennis W. Dickson, M.D., Mayo Clinic, Jacksonville, FL; Steven Younkin, M.D., Ph.D., Mayo Clinic, Jacksonville, FL; Ronald C. Petersen, Ph.D., M.D., Mayo Clinic, Rochester, MN; Christopher D. Corcoran, Sc.D., Utah State University, Logan; Yefei Cai, Ph.D., Washington University School of Medicine, St. Louis; Catarina Oliveira, M.D., Ph.D., University of Coimbra, Coimbra, Portugal; Maria Helena Ribeiro, M.S., University of Coimbra, Coimbra, Portugal; Isabel Santana, M.D., Ph.D., University of Coimbra, Coimbra, Portugal; JoAnn T. Tschanz, Ph.D., Utah State University, Logan; J. Raphael Gibbs, B.Sc., UCL Institute of Neurology, London, and National Institute on Aging, National Institutes of Health, Bethesda, MD; Maria C. Norton, Ph.D., Utah State University, Logan; Iwona Kloszewska, Ph.D., Medical University of Lodz, Lodz, Poland; Patrizia Mecocci, M.D., Ph.D., University of Perugia, Perugia, Italy; Hilkka Soininen, M.D., University of Eastern Finland and Kuopio University Hospital, Kuopio, Finland; Magda Tsolaki, M.D., Aristotle University of Thessaloniki, Thessaloniki, Greece; Bruno Vellas, M.D., Ph.D., UMR INSERM 1027, Hôpitaux de Toulouse, Toulouse, France; Ronald G. Munger, Ph.D., M.P.H., Utah State University, Logan; David M.A. Mann, Ph.D., University of Manchester, Manchester, United Kingdom; Stuart Pickering-Brown, Ph.D., University of Manchester, Manchester, United Kingdom; Simon Lovestone, Ph.D., King's College London Institute of Psychiatry, London; Jonathan Beck, B.Sc., MRC Prion Unit and UCL Institute of Neurology, London; Simon Mead, M.D., MRC Prion Unit and UCL Institute of Neurology, London; John Collinge, M.D., MRC Prion Unit and UCL Institute of Neurology, London; Linda Parsons, M.Phil., UCL Institute of Neurology, London; Jennifer Pocock, Ph.D., UCL Institute of Neurology, London; John C. Morris, M.D., Washington University School of Medicine, St. Louis; Tamas Revesz, M.D., UCL Institute of Neurology, London; Tammaryn Lashley, Ph.D., UCL Institute of Neurology, London; Nick C. Fox, M.D., UCL Institute of Neurology, London; Martin N. Rossor, M.D., UCL Institute of Neurology, London; Benjamin Grenier-Boley, M.Sc., INSERM, U744, Institut Pasteur de Lille and Université Lille-Nord de France, Lille, France; Céline Bellenguez, Ph.D., INSERM, U744, Institut Pasteur de Lille and Université Lille-Nord de France, Lille, France; Valentina Moskvina, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, School of Medicine, Cardiff University, Cardiff, United Kingdom; Rebecca Sims, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, United Kingdom; Denise Harold, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, United Kingdom; Julie Williams, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, United Kingdom; Jean-Charles Lambert, Ph.D., INSERM, U744, Institut Pasteur de Lille and Université Lille-Nord de France, Lille, France; Philippe Amouyel, M.D., Ph.D., INSERM, U744, Institut Pasteur de Lille, CHRU de Lille, and Université Lille-Nord de France, Lille, France; Neill Graff-Radford, M.B., B.Ch., Mayo Clinic, Jacksonville, FL; Alison Goate, Ph.D., Washington University School of Medicine, St. Louis; Rosa Rademakers, Ph.D., Mayo Clinic, Jacksonville, FL; Kevin Morgan, Ph.D., School of Molecular Medical Sciences, University of Nottingham, Nottingham, United Kingdom; John Powell, Ph.D., King's College London Institute of Psychiatry, London; Peter St. George-Hyslop, M.D., Tanz Centre, University of Toronto, Toronto, and Cambridge Institute for Medical Research, University of Cambridge, Cambridge, United Kingdom; Andrew Singleton, Ph.D., National Institute on Aging, National Institutes of Health, Bethesda, MD; and John Hardy, Ph.D., UCL Institute of Neurology, London
1,
Celeste Sassi
Celeste Sassi, M.D.
1Rita Guerreiro, Ph.D., University College London (UCL) Institute of Neurology, London, and National Institute on Aging, National Institutes of Health, Bethesda, MD; Aleksandra Wojtas, M.S., Mayo Clinic, Jacksonville, FL; Jose Bras, Ph.D., UCL Institute of Neurology, London; Minerva M. Carrasquillo, Ph.D., Mayo Clinic, Jacksonville, FL; Ekaterina Rogaeva, Ph.D., Tanz Centre, University of Toronto, Toronto; Elisa Majounie, Ph.D., Mayo Clinic, Jacksonville, FL; Carlos Cruchaga, Ph.D., Washington University School of Medicine, St. Louis; Celeste Sassi, M.D., Institute of Neurology, London, and National Institute on Aging, National Institutes of Health, Bethesda, MD; John S.K. Kauwe, Ph.D., Brigham Young University, Provo, Utah; Michelle K. Lupton, Ph.D., King's College London Institute of Psychiatry, London; Mina Ryten, M.D., Ph.D., UCL Institute of Neurology, London; Kristelle Brown, Ph.D., University of Nottingham, Nottingham, United Kingdom; the EADI Consortium, the GERAD Consortium, the UKBE Consortium, James Lowe, D.M., University of Nottingham, Nottingham, United Kingdom; Perry G. Ridge, M.S., Brigham Young University and the ARUP Institute, Salt Lake City; Monia B. Hammer, M.S., National Institute on Aging, National Institutes of Health, Bethesda, MD; Yosuke Wakutani, M.D., Tanz Centre, University of Toronto, Toronto; Lilinaz Hazrati, M.D., Ph.D., Tanz Centre, University of Toronto, Toronto; Petroula Proitsi, Ph.D., King's College London Institute of Psychiatry, London; Stephen Newhouse, Ph.D., King's College London Institute of Psychiatry, London; Ebba Lohmann, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Nihan Erginel-Unaltuna, Ph.D., Institute for Experimental Medical Research, Istanbul University, Istanbul, Turkey; Christopher Medway, Ph.D., University of Nottingham, Nottingham, United Kingdom; Hasmet Hanagasi, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Claire Troakes, Ph.D., King's College London Institute of Psychiatry, London; Hakan Gurvit, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Basar Bilgic, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Safa Al-Sarraj, M.D., King's College London Institute of Psychiatry and King's College Hospital, London; Bruno Benitez, M.D., Washington University School of Medicine, St. Louis; Breanna Cooper, B.S., Washington University School of Medicine, St. Louis; David Carrell, M.S., Washington University School of Medicine, St. Louis; Murat Emre, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Fanggeng Zou, Ph.D., Mayo Clinic, Jacksonville, FL; Li Ma, M.S., Mayo Clinic, Jacksonville, FL; Melissa E. Murray, Ph.D., Mayo Clinic, Jacksonville, FL; Dennis W. Dickson, M.D., Mayo Clinic, Jacksonville, FL; Steven Younkin, M.D., Ph.D., Mayo Clinic, Jacksonville, FL; Ronald C. Petersen, Ph.D., M.D., Mayo Clinic, Rochester, MN; Christopher D. Corcoran, Sc.D., Utah State University, Logan; Yefei Cai, Ph.D., Washington University School of Medicine, St. Louis; Catarina Oliveira, M.D., Ph.D., University of Coimbra, Coimbra, Portugal; Maria Helena Ribeiro, M.S., University of Coimbra, Coimbra, Portugal; Isabel Santana, M.D., Ph.D., University of Coimbra, Coimbra, Portugal; JoAnn T. Tschanz, Ph.D., Utah State University, Logan; J. Raphael Gibbs, B.Sc., UCL Institute of Neurology, London, and National Institute on Aging, National Institutes of Health, Bethesda, MD; Maria C. Norton, Ph.D., Utah State University, Logan; Iwona Kloszewska, Ph.D., Medical University of Lodz, Lodz, Poland; Patrizia Mecocci, M.D., Ph.D., University of Perugia, Perugia, Italy; Hilkka Soininen, M.D., University of Eastern Finland and Kuopio University Hospital, Kuopio, Finland; Magda Tsolaki, M.D., Aristotle University of Thessaloniki, Thessaloniki, Greece; Bruno Vellas, M.D., Ph.D., UMR INSERM 1027, Hôpitaux de Toulouse, Toulouse, France; Ronald G. Munger, Ph.D., M.P.H., Utah State University, Logan; David M.A. Mann, Ph.D., University of Manchester, Manchester, United Kingdom; Stuart Pickering-Brown, Ph.D., University of Manchester, Manchester, United Kingdom; Simon Lovestone, Ph.D., King's College London Institute of Psychiatry, London; Jonathan Beck, B.Sc., MRC Prion Unit and UCL Institute of Neurology, London; Simon Mead, M.D., MRC Prion Unit and UCL Institute of Neurology, London; John Collinge, M.D., MRC Prion Unit and UCL Institute of Neurology, London; Linda Parsons, M.Phil., UCL Institute of Neurology, London; Jennifer Pocock, Ph.D., UCL Institute of Neurology, London; John C. Morris, M.D., Washington University School of Medicine, St. Louis; Tamas Revesz, M.D., UCL Institute of Neurology, London; Tammaryn Lashley, Ph.D., UCL Institute of Neurology, London; Nick C. Fox, M.D., UCL Institute of Neurology, London; Martin N. Rossor, M.D., UCL Institute of Neurology, London; Benjamin Grenier-Boley, M.Sc., INSERM, U744, Institut Pasteur de Lille and Université Lille-Nord de France, Lille, France; Céline Bellenguez, Ph.D., INSERM, U744, Institut Pasteur de Lille and Université Lille-Nord de France, Lille, France; Valentina Moskvina, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, School of Medicine, Cardiff University, Cardiff, United Kingdom; Rebecca Sims, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, United Kingdom; Denise Harold, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, United Kingdom; Julie Williams, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, United Kingdom; Jean-Charles Lambert, Ph.D., INSERM, U744, Institut Pasteur de Lille and Université Lille-Nord de France, Lille, France; Philippe Amouyel, M.D., Ph.D., INSERM, U744, Institut Pasteur de Lille, CHRU de Lille, and Université Lille-Nord de France, Lille, France; Neill Graff-Radford, M.B., B.Ch., Mayo Clinic, Jacksonville, FL; Alison Goate, Ph.D., Washington University School of Medicine, St. Louis; Rosa Rademakers, Ph.D., Mayo Clinic, Jacksonville, FL; Kevin Morgan, Ph.D., School of Molecular Medical Sciences, University of Nottingham, Nottingham, United Kingdom; John Powell, Ph.D., King's College London Institute of Psychiatry, London; Peter St. George-Hyslop, M.D., Tanz Centre, University of Toronto, Toronto, and Cambridge Institute for Medical Research, University of Cambridge, Cambridge, United Kingdom; Andrew Singleton, Ph.D., National Institute on Aging, National Institutes of Health, Bethesda, MD; and John Hardy, Ph.D., UCL Institute of Neurology, London
1,
John SK Kauwe
John SK Kauwe, Ph.D.
1Rita Guerreiro, Ph.D., University College London (UCL) Institute of Neurology, London, and National Institute on Aging, National Institutes of Health, Bethesda, MD; Aleksandra Wojtas, M.S., Mayo Clinic, Jacksonville, FL; Jose Bras, Ph.D., UCL Institute of Neurology, London; Minerva M. Carrasquillo, Ph.D., Mayo Clinic, Jacksonville, FL; Ekaterina Rogaeva, Ph.D., Tanz Centre, University of Toronto, Toronto; Elisa Majounie, Ph.D., Mayo Clinic, Jacksonville, FL; Carlos Cruchaga, Ph.D., Washington University School of Medicine, St. Louis; Celeste Sassi, M.D., Institute of Neurology, London, and National Institute on Aging, National Institutes of Health, Bethesda, MD; John S.K. Kauwe, Ph.D., Brigham Young University, Provo, Utah; Michelle K. Lupton, Ph.D., King's College London Institute of Psychiatry, London; Mina Ryten, M.D., Ph.D., UCL Institute of Neurology, London; Kristelle Brown, Ph.D., University of Nottingham, Nottingham, United Kingdom; the EADI Consortium, the GERAD Consortium, the UKBE Consortium, James Lowe, D.M., University of Nottingham, Nottingham, United Kingdom; Perry G. Ridge, M.S., Brigham Young University and the ARUP Institute, Salt Lake City; Monia B. Hammer, M.S., National Institute on Aging, National Institutes of Health, Bethesda, MD; Yosuke Wakutani, M.D., Tanz Centre, University of Toronto, Toronto; Lilinaz Hazrati, M.D., Ph.D., Tanz Centre, University of Toronto, Toronto; Petroula Proitsi, Ph.D., King's College London Institute of Psychiatry, London; Stephen Newhouse, Ph.D., King's College London Institute of Psychiatry, London; Ebba Lohmann, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Nihan Erginel-Unaltuna, Ph.D., Institute for Experimental Medical Research, Istanbul University, Istanbul, Turkey; Christopher Medway, Ph.D., University of Nottingham, Nottingham, United Kingdom; Hasmet Hanagasi, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Claire Troakes, Ph.D., King's College London Institute of Psychiatry, London; Hakan Gurvit, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Basar Bilgic, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Safa Al-Sarraj, M.D., King's College London Institute of Psychiatry and King's College Hospital, London; Bruno Benitez, M.D., Washington University School of Medicine, St. Louis; Breanna Cooper, B.S., Washington University School of Medicine, St. Louis; David Carrell, M.S., Washington University School of Medicine, St. Louis; Murat Emre, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Fanggeng Zou, Ph.D., Mayo Clinic, Jacksonville, FL; Li Ma, M.S., Mayo Clinic, Jacksonville, FL; Melissa E. Murray, Ph.D., Mayo Clinic, Jacksonville, FL; Dennis W. Dickson, M.D., Mayo Clinic, Jacksonville, FL; Steven Younkin, M.D., Ph.D., Mayo Clinic, Jacksonville, FL; Ronald C. Petersen, Ph.D., M.D., Mayo Clinic, Rochester, MN; Christopher D. Corcoran, Sc.D., Utah State University, Logan; Yefei Cai, Ph.D., Washington University School of Medicine, St. Louis; Catarina Oliveira, M.D., Ph.D., University of Coimbra, Coimbra, Portugal; Maria Helena Ribeiro, M.S., University of Coimbra, Coimbra, Portugal; Isabel Santana, M.D., Ph.D., University of Coimbra, Coimbra, Portugal; JoAnn T. Tschanz, Ph.D., Utah State University, Logan; J. Raphael Gibbs, B.Sc., UCL Institute of Neurology, London, and National Institute on Aging, National Institutes of Health, Bethesda, MD; Maria C. Norton, Ph.D., Utah State University, Logan; Iwona Kloszewska, Ph.D., Medical University of Lodz, Lodz, Poland; Patrizia Mecocci, M.D., Ph.D., University of Perugia, Perugia, Italy; Hilkka Soininen, M.D., University of Eastern Finland and Kuopio University Hospital, Kuopio, Finland; Magda Tsolaki, M.D., Aristotle University of Thessaloniki, Thessaloniki, Greece; Bruno Vellas, M.D., Ph.D., UMR INSERM 1027, Hôpitaux de Toulouse, Toulouse, France; Ronald G. Munger, Ph.D., M.P.H., Utah State University, Logan; David M.A. Mann, Ph.D., University of Manchester, Manchester, United Kingdom; Stuart Pickering-Brown, Ph.D., University of Manchester, Manchester, United Kingdom; Simon Lovestone, Ph.D., King's College London Institute of Psychiatry, London; Jonathan Beck, B.Sc., MRC Prion Unit and UCL Institute of Neurology, London; Simon Mead, M.D., MRC Prion Unit and UCL Institute of Neurology, London; John Collinge, M.D., MRC Prion Unit and UCL Institute of Neurology, London; Linda Parsons, M.Phil., UCL Institute of Neurology, London; Jennifer Pocock, Ph.D., UCL Institute of Neurology, London; John C. Morris, M.D., Washington University School of Medicine, St. Louis; Tamas Revesz, M.D., UCL Institute of Neurology, London; Tammaryn Lashley, Ph.D., UCL Institute of Neurology, London; Nick C. Fox, M.D., UCL Institute of Neurology, London; Martin N. Rossor, M.D., UCL Institute of Neurology, London; Benjamin Grenier-Boley, M.Sc., INSERM, U744, Institut Pasteur de Lille and Université Lille-Nord de France, Lille, France; Céline Bellenguez, Ph.D., INSERM, U744, Institut Pasteur de Lille and Université Lille-Nord de France, Lille, France; Valentina Moskvina, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, School of Medicine, Cardiff University, Cardiff, United Kingdom; Rebecca Sims, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, United Kingdom; Denise Harold, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, United Kingdom; Julie Williams, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, United Kingdom; Jean-Charles Lambert, Ph.D., INSERM, U744, Institut Pasteur de Lille and Université Lille-Nord de France, Lille, France; Philippe Amouyel, M.D., Ph.D., INSERM, U744, Institut Pasteur de Lille, CHRU de Lille, and Université Lille-Nord de France, Lille, France; Neill Graff-Radford, M.B., B.Ch., Mayo Clinic, Jacksonville, FL; Alison Goate, Ph.D., Washington University School of Medicine, St. Louis; Rosa Rademakers, Ph.D., Mayo Clinic, Jacksonville, FL; Kevin Morgan, Ph.D., School of Molecular Medical Sciences, University of Nottingham, Nottingham, United Kingdom; John Powell, Ph.D., King's College London Institute of Psychiatry, London; Peter St. George-Hyslop, M.D., Tanz Centre, University of Toronto, Toronto, and Cambridge Institute for Medical Research, University of Cambridge, Cambridge, United Kingdom; Andrew Singleton, Ph.D., National Institute on Aging, National Institutes of Health, Bethesda, MD; and John Hardy, Ph.D., UCL Institute of Neurology, London
1,
Steven Younkin
Steven Younkin, M.D., Ph.D.
1Rita Guerreiro, Ph.D., University College London (UCL) Institute of Neurology, London, and National Institute on Aging, National Institutes of Health, Bethesda, MD; Aleksandra Wojtas, M.S., Mayo Clinic, Jacksonville, FL; Jose Bras, Ph.D., UCL Institute of Neurology, London; Minerva M. Carrasquillo, Ph.D., Mayo Clinic, Jacksonville, FL; Ekaterina Rogaeva, Ph.D., Tanz Centre, University of Toronto, Toronto; Elisa Majounie, Ph.D., Mayo Clinic, Jacksonville, FL; Carlos Cruchaga, Ph.D., Washington University School of Medicine, St. Louis; Celeste Sassi, M.D., Institute of Neurology, London, and National Institute on Aging, National Institutes of Health, Bethesda, MD; John S.K. Kauwe, Ph.D., Brigham Young University, Provo, Utah; Michelle K. Lupton, Ph.D., King's College London Institute of Psychiatry, London; Mina Ryten, M.D., Ph.D., UCL Institute of Neurology, London; Kristelle Brown, Ph.D., University of Nottingham, Nottingham, United Kingdom; the EADI Consortium, the GERAD Consortium, the UKBE Consortium, James Lowe, D.M., University of Nottingham, Nottingham, United Kingdom; Perry G. Ridge, M.S., Brigham Young University and the ARUP Institute, Salt Lake City; Monia B. Hammer, M.S., National Institute on Aging, National Institutes of Health, Bethesda, MD; Yosuke Wakutani, M.D., Tanz Centre, University of Toronto, Toronto; Lilinaz Hazrati, M.D., Ph.D., Tanz Centre, University of Toronto, Toronto; Petroula Proitsi, Ph.D., King's College London Institute of Psychiatry, London; Stephen Newhouse, Ph.D., King's College London Institute of Psychiatry, London; Ebba Lohmann, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Nihan Erginel-Unaltuna, Ph.D., Institute for Experimental Medical Research, Istanbul University, Istanbul, Turkey; Christopher Medway, Ph.D., University of Nottingham, Nottingham, United Kingdom; Hasmet Hanagasi, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Claire Troakes, Ph.D., King's College London Institute of Psychiatry, London; Hakan Gurvit, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Basar Bilgic, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Safa Al-Sarraj, M.D., King's College London Institute of Psychiatry and King's College Hospital, London; Bruno Benitez, M.D., Washington University School of Medicine, St. Louis; Breanna Cooper, B.S., Washington University School of Medicine, St. Louis; David Carrell, M.S., Washington University School of Medicine, St. Louis; Murat Emre, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Fanggeng Zou, Ph.D., Mayo Clinic, Jacksonville, FL; Li Ma, M.S., Mayo Clinic, Jacksonville, FL; Melissa E. Murray, Ph.D., Mayo Clinic, Jacksonville, FL; Dennis W. Dickson, M.D., Mayo Clinic, Jacksonville, FL; Steven Younkin, M.D., Ph.D., Mayo Clinic, Jacksonville, FL; Ronald C. Petersen, Ph.D., M.D., Mayo Clinic, Rochester, MN; Christopher D. Corcoran, Sc.D., Utah State University, Logan; Yefei Cai, Ph.D., Washington University School of Medicine, St. Louis; Catarina Oliveira, M.D., Ph.D., University of Coimbra, Coimbra, Portugal; Maria Helena Ribeiro, M.S., University of Coimbra, Coimbra, Portugal; Isabel Santana, M.D., Ph.D., University of Coimbra, Coimbra, Portugal; JoAnn T. Tschanz, Ph.D., Utah State University, Logan; J. Raphael Gibbs, B.Sc., UCL Institute of Neurology, London, and National Institute on Aging, National Institutes of Health, Bethesda, MD; Maria C. Norton, Ph.D., Utah State University, Logan; Iwona Kloszewska, Ph.D., Medical University of Lodz, Lodz, Poland; Patrizia Mecocci, M.D., Ph.D., University of Perugia, Perugia, Italy; Hilkka Soininen, M.D., University of Eastern Finland and Kuopio University Hospital, Kuopio, Finland; Magda Tsolaki, M.D., Aristotle University of Thessaloniki, Thessaloniki, Greece; Bruno Vellas, M.D., Ph.D., UMR INSERM 1027, Hôpitaux de Toulouse, Toulouse, France; Ronald G. Munger, Ph.D., M.P.H., Utah State University, Logan; David M.A. Mann, Ph.D., University of Manchester, Manchester, United Kingdom; Stuart Pickering-Brown, Ph.D., University of Manchester, Manchester, United Kingdom; Simon Lovestone, Ph.D., King's College London Institute of Psychiatry, London; Jonathan Beck, B.Sc., MRC Prion Unit and UCL Institute of Neurology, London; Simon Mead, M.D., MRC Prion Unit and UCL Institute of Neurology, London; John Collinge, M.D., MRC Prion Unit and UCL Institute of Neurology, London; Linda Parsons, M.Phil., UCL Institute of Neurology, London; Jennifer Pocock, Ph.D., UCL Institute of Neurology, London; John C. Morris, M.D., Washington University School of Medicine, St. Louis; Tamas Revesz, M.D., UCL Institute of Neurology, London; Tammaryn Lashley, Ph.D., UCL Institute of Neurology, London; Nick C. Fox, M.D., UCL Institute of Neurology, London; Martin N. Rossor, M.D., UCL Institute of Neurology, London; Benjamin Grenier-Boley, M.Sc., INSERM, U744, Institut Pasteur de Lille and Université Lille-Nord de France, Lille, France; Céline Bellenguez, Ph.D., INSERM, U744, Institut Pasteur de Lille and Université Lille-Nord de France, Lille, France; Valentina Moskvina, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, School of Medicine, Cardiff University, Cardiff, United Kingdom; Rebecca Sims, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, United Kingdom; Denise Harold, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, United Kingdom; Julie Williams, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, United Kingdom; Jean-Charles Lambert, Ph.D., INSERM, U744, Institut Pasteur de Lille and Université Lille-Nord de France, Lille, France; Philippe Amouyel, M.D., Ph.D., INSERM, U744, Institut Pasteur de Lille, CHRU de Lille, and Université Lille-Nord de France, Lille, France; Neill Graff-Radford, M.B., B.Ch., Mayo Clinic, Jacksonville, FL; Alison Goate, Ph.D., Washington University School of Medicine, St. Louis; Rosa Rademakers, Ph.D., Mayo Clinic, Jacksonville, FL; Kevin Morgan, Ph.D., School of Molecular Medical Sciences, University of Nottingham, Nottingham, United Kingdom; John Powell, Ph.D., King's College London Institute of Psychiatry, London; Peter St. George-Hyslop, M.D., Tanz Centre, University of Toronto, Toronto, and Cambridge Institute for Medical Research, University of Cambridge, Cambridge, United Kingdom; Andrew Singleton, Ph.D., National Institute on Aging, National Institutes of Health, Bethesda, MD; and John Hardy, Ph.D., UCL Institute of Neurology, London
1,
Lilinaz Hazrati
Lilinaz Hazrati, M.D., Ph.D.
1Rita Guerreiro, Ph.D., University College London (UCL) Institute of Neurology, London, and National Institute on Aging, National Institutes of Health, Bethesda, MD; Aleksandra Wojtas, M.S., Mayo Clinic, Jacksonville, FL; Jose Bras, Ph.D., UCL Institute of Neurology, London; Minerva M. Carrasquillo, Ph.D., Mayo Clinic, Jacksonville, FL; Ekaterina Rogaeva, Ph.D., Tanz Centre, University of Toronto, Toronto; Elisa Majounie, Ph.D., Mayo Clinic, Jacksonville, FL; Carlos Cruchaga, Ph.D., Washington University School of Medicine, St. Louis; Celeste Sassi, M.D., Institute of Neurology, London, and National Institute on Aging, National Institutes of Health, Bethesda, MD; John S.K. Kauwe, Ph.D., Brigham Young University, Provo, Utah; Michelle K. Lupton, Ph.D., King's College London Institute of Psychiatry, London; Mina Ryten, M.D., Ph.D., UCL Institute of Neurology, London; Kristelle Brown, Ph.D., University of Nottingham, Nottingham, United Kingdom; the EADI Consortium, the GERAD Consortium, the UKBE Consortium, James Lowe, D.M., University of Nottingham, Nottingham, United Kingdom; Perry G. Ridge, M.S., Brigham Young University and the ARUP Institute, Salt Lake City; Monia B. Hammer, M.S., National Institute on Aging, National Institutes of Health, Bethesda, MD; Yosuke Wakutani, M.D., Tanz Centre, University of Toronto, Toronto; Lilinaz Hazrati, M.D., Ph.D., Tanz Centre, University of Toronto, Toronto; Petroula Proitsi, Ph.D., King's College London Institute of Psychiatry, London; Stephen Newhouse, Ph.D., King's College London Institute of Psychiatry, London; Ebba Lohmann, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Nihan Erginel-Unaltuna, Ph.D., Institute for Experimental Medical Research, Istanbul University, Istanbul, Turkey; Christopher Medway, Ph.D., University of Nottingham, Nottingham, United Kingdom; Hasmet Hanagasi, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Claire Troakes, Ph.D., King's College London Institute of Psychiatry, London; Hakan Gurvit, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Basar Bilgic, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Safa Al-Sarraj, M.D., King's College London Institute of Psychiatry and King's College Hospital, London; Bruno Benitez, M.D., Washington University School of Medicine, St. Louis; Breanna Cooper, B.S., Washington University School of Medicine, St. Louis; David Carrell, M.S., Washington University School of Medicine, St. Louis; Murat Emre, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Fanggeng Zou, Ph.D., Mayo Clinic, Jacksonville, FL; Li Ma, M.S., Mayo Clinic, Jacksonville, FL; Melissa E. Murray, Ph.D., Mayo Clinic, Jacksonville, FL; Dennis W. Dickson, M.D., Mayo Clinic, Jacksonville, FL; Steven Younkin, M.D., Ph.D., Mayo Clinic, Jacksonville, FL; Ronald C. Petersen, Ph.D., M.D., Mayo Clinic, Rochester, MN; Christopher D. Corcoran, Sc.D., Utah State University, Logan; Yefei Cai, Ph.D., Washington University School of Medicine, St. Louis; Catarina Oliveira, M.D., Ph.D., University of Coimbra, Coimbra, Portugal; Maria Helena Ribeiro, M.S., University of Coimbra, Coimbra, Portugal; Isabel Santana, M.D., Ph.D., University of Coimbra, Coimbra, Portugal; JoAnn T. Tschanz, Ph.D., Utah State University, Logan; J. Raphael Gibbs, B.Sc., UCL Institute of Neurology, London, and National Institute on Aging, National Institutes of Health, Bethesda, MD; Maria C. Norton, Ph.D., Utah State University, Logan; Iwona Kloszewska, Ph.D., Medical University of Lodz, Lodz, Poland; Patrizia Mecocci, M.D., Ph.D., University of Perugia, Perugia, Italy; Hilkka Soininen, M.D., University of Eastern Finland and Kuopio University Hospital, Kuopio, Finland; Magda Tsolaki, M.D., Aristotle University of Thessaloniki, Thessaloniki, Greece; Bruno Vellas, M.D., Ph.D., UMR INSERM 1027, Hôpitaux de Toulouse, Toulouse, France; Ronald G. Munger, Ph.D., M.P.H., Utah State University, Logan; David M.A. Mann, Ph.D., University of Manchester, Manchester, United Kingdom; Stuart Pickering-Brown, Ph.D., University of Manchester, Manchester, United Kingdom; Simon Lovestone, Ph.D., King's College London Institute of Psychiatry, London; Jonathan Beck, B.Sc., MRC Prion Unit and UCL Institute of Neurology, London; Simon Mead, M.D., MRC Prion Unit and UCL Institute of Neurology, London; John Collinge, M.D., MRC Prion Unit and UCL Institute of Neurology, London; Linda Parsons, M.Phil., UCL Institute of Neurology, London; Jennifer Pocock, Ph.D., UCL Institute of Neurology, London; John C. Morris, M.D., Washington University School of Medicine, St. Louis; Tamas Revesz, M.D., UCL Institute of Neurology, London; Tammaryn Lashley, Ph.D., UCL Institute of Neurology, London; Nick C. Fox, M.D., UCL Institute of Neurology, London; Martin N. Rossor, M.D., UCL Institute of Neurology, London; Benjamin Grenier-Boley, M.Sc., INSERM, U744, Institut Pasteur de Lille and Université Lille-Nord de France, Lille, France; Céline Bellenguez, Ph.D., INSERM, U744, Institut Pasteur de Lille and Université Lille-Nord de France, Lille, France; Valentina Moskvina, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, School of Medicine, Cardiff University, Cardiff, United Kingdom; Rebecca Sims, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, United Kingdom; Denise Harold, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, United Kingdom; Julie Williams, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, United Kingdom; Jean-Charles Lambert, Ph.D., INSERM, U744, Institut Pasteur de Lille and Université Lille-Nord de France, Lille, France; Philippe Amouyel, M.D., Ph.D., INSERM, U744, Institut Pasteur de Lille, CHRU de Lille, and Université Lille-Nord de France, Lille, France; Neill Graff-Radford, M.B., B.Ch., Mayo Clinic, Jacksonville, FL; Alison Goate, Ph.D., Washington University School of Medicine, St. Louis; Rosa Rademakers, Ph.D., Mayo Clinic, Jacksonville, FL; Kevin Morgan, Ph.D., School of Molecular Medical Sciences, University of Nottingham, Nottingham, United Kingdom; John Powell, Ph.D., King's College London Institute of Psychiatry, London; Peter St. George-Hyslop, M.D., Tanz Centre, University of Toronto, Toronto, and Cambridge Institute for Medical Research, University of Cambridge, Cambridge, United Kingdom; Andrew Singleton, Ph.D., National Institute on Aging, National Institutes of Health, Bethesda, MD; and John Hardy, Ph.D., UCL Institute of Neurology, London
1,
John Collinge
John Collinge, M.D.
1Rita Guerreiro, Ph.D., University College London (UCL) Institute of Neurology, London, and National Institute on Aging, National Institutes of Health, Bethesda, MD; Aleksandra Wojtas, M.S., Mayo Clinic, Jacksonville, FL; Jose Bras, Ph.D., UCL Institute of Neurology, London; Minerva M. Carrasquillo, Ph.D., Mayo Clinic, Jacksonville, FL; Ekaterina Rogaeva, Ph.D., Tanz Centre, University of Toronto, Toronto; Elisa Majounie, Ph.D., Mayo Clinic, Jacksonville, FL; Carlos Cruchaga, Ph.D., Washington University School of Medicine, St. Louis; Celeste Sassi, M.D., Institute of Neurology, London, and National Institute on Aging, National Institutes of Health, Bethesda, MD; John S.K. Kauwe, Ph.D., Brigham Young University, Provo, Utah; Michelle K. Lupton, Ph.D., King's College London Institute of Psychiatry, London; Mina Ryten, M.D., Ph.D., UCL Institute of Neurology, London; Kristelle Brown, Ph.D., University of Nottingham, Nottingham, United Kingdom; the EADI Consortium, the GERAD Consortium, the UKBE Consortium, James Lowe, D.M., University of Nottingham, Nottingham, United Kingdom; Perry G. Ridge, M.S., Brigham Young University and the ARUP Institute, Salt Lake City; Monia B. Hammer, M.S., National Institute on Aging, National Institutes of Health, Bethesda, MD; Yosuke Wakutani, M.D., Tanz Centre, University of Toronto, Toronto; Lilinaz Hazrati, M.D., Ph.D., Tanz Centre, University of Toronto, Toronto; Petroula Proitsi, Ph.D., King's College London Institute of Psychiatry, London; Stephen Newhouse, Ph.D., King's College London Institute of Psychiatry, London; Ebba Lohmann, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Nihan Erginel-Unaltuna, Ph.D., Institute for Experimental Medical Research, Istanbul University, Istanbul, Turkey; Christopher Medway, Ph.D., University of Nottingham, Nottingham, United Kingdom; Hasmet Hanagasi, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Claire Troakes, Ph.D., King's College London Institute of Psychiatry, London; Hakan Gurvit, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Basar Bilgic, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Safa Al-Sarraj, M.D., King's College London Institute of Psychiatry and King's College Hospital, London; Bruno Benitez, M.D., Washington University School of Medicine, St. Louis; Breanna Cooper, B.S., Washington University School of Medicine, St. Louis; David Carrell, M.S., Washington University School of Medicine, St. Louis; Murat Emre, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Fanggeng Zou, Ph.D., Mayo Clinic, Jacksonville, FL; Li Ma, M.S., Mayo Clinic, Jacksonville, FL; Melissa E. Murray, Ph.D., Mayo Clinic, Jacksonville, FL; Dennis W. Dickson, M.D., Mayo Clinic, Jacksonville, FL; Steven Younkin, M.D., Ph.D., Mayo Clinic, Jacksonville, FL; Ronald C. Petersen, Ph.D., M.D., Mayo Clinic, Rochester, MN; Christopher D. Corcoran, Sc.D., Utah State University, Logan; Yefei Cai, Ph.D., Washington University School of Medicine, St. Louis; Catarina Oliveira, M.D., Ph.D., University of Coimbra, Coimbra, Portugal; Maria Helena Ribeiro, M.S., University of Coimbra, Coimbra, Portugal; Isabel Santana, M.D., Ph.D., University of Coimbra, Coimbra, Portugal; JoAnn T. Tschanz, Ph.D., Utah State University, Logan; J. Raphael Gibbs, B.Sc., UCL Institute of Neurology, London, and National Institute on Aging, National Institutes of Health, Bethesda, MD; Maria C. Norton, Ph.D., Utah State University, Logan; Iwona Kloszewska, Ph.D., Medical University of Lodz, Lodz, Poland; Patrizia Mecocci, M.D., Ph.D., University of Perugia, Perugia, Italy; Hilkka Soininen, M.D., University of Eastern Finland and Kuopio University Hospital, Kuopio, Finland; Magda Tsolaki, M.D., Aristotle University of Thessaloniki, Thessaloniki, Greece; Bruno Vellas, M.D., Ph.D., UMR INSERM 1027, Hôpitaux de Toulouse, Toulouse, France; Ronald G. Munger, Ph.D., M.P.H., Utah State University, Logan; David M.A. Mann, Ph.D., University of Manchester, Manchester, United Kingdom; Stuart Pickering-Brown, Ph.D., University of Manchester, Manchester, United Kingdom; Simon Lovestone, Ph.D., King's College London Institute of Psychiatry, London; Jonathan Beck, B.Sc., MRC Prion Unit and UCL Institute of Neurology, London; Simon Mead, M.D., MRC Prion Unit and UCL Institute of Neurology, London; John Collinge, M.D., MRC Prion Unit and UCL Institute of Neurology, London; Linda Parsons, M.Phil., UCL Institute of Neurology, London; Jennifer Pocock, Ph.D., UCL Institute of Neurology, London; John C. Morris, M.D., Washington University School of Medicine, St. Louis; Tamas Revesz, M.D., UCL Institute of Neurology, London; Tammaryn Lashley, Ph.D., UCL Institute of Neurology, London; Nick C. Fox, M.D., UCL Institute of Neurology, London; Martin N. Rossor, M.D., UCL Institute of Neurology, London; Benjamin Grenier-Boley, M.Sc., INSERM, U744, Institut Pasteur de Lille and Université Lille-Nord de France, Lille, France; Céline Bellenguez, Ph.D., INSERM, U744, Institut Pasteur de Lille and Université Lille-Nord de France, Lille, France; Valentina Moskvina, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, School of Medicine, Cardiff University, Cardiff, United Kingdom; Rebecca Sims, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, United Kingdom; Denise Harold, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, United Kingdom; Julie Williams, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, United Kingdom; Jean-Charles Lambert, Ph.D., INSERM, U744, Institut Pasteur de Lille and Université Lille-Nord de France, Lille, France; Philippe Amouyel, M.D., Ph.D., INSERM, U744, Institut Pasteur de Lille, CHRU de Lille, and Université Lille-Nord de France, Lille, France; Neill Graff-Radford, M.B., B.Ch., Mayo Clinic, Jacksonville, FL; Alison Goate, Ph.D., Washington University School of Medicine, St. Louis; Rosa Rademakers, Ph.D., Mayo Clinic, Jacksonville, FL; Kevin Morgan, Ph.D., School of Molecular Medical Sciences, University of Nottingham, Nottingham, United Kingdom; John Powell, Ph.D., King's College London Institute of Psychiatry, London; Peter St. George-Hyslop, M.D., Tanz Centre, University of Toronto, Toronto, and Cambridge Institute for Medical Research, University of Cambridge, Cambridge, United Kingdom; Andrew Singleton, Ph.D., National Institute on Aging, National Institutes of Health, Bethesda, MD; and John Hardy, Ph.D., UCL Institute of Neurology, London
1,
Jennifer Pocock
Jennifer Pocock, Ph.D.
1Rita Guerreiro, Ph.D., University College London (UCL) Institute of Neurology, London, and National Institute on Aging, National Institutes of Health, Bethesda, MD; Aleksandra Wojtas, M.S., Mayo Clinic, Jacksonville, FL; Jose Bras, Ph.D., UCL Institute of Neurology, London; Minerva M. Carrasquillo, Ph.D., Mayo Clinic, Jacksonville, FL; Ekaterina Rogaeva, Ph.D., Tanz Centre, University of Toronto, Toronto; Elisa Majounie, Ph.D., Mayo Clinic, Jacksonville, FL; Carlos Cruchaga, Ph.D., Washington University School of Medicine, St. Louis; Celeste Sassi, M.D., Institute of Neurology, London, and National Institute on Aging, National Institutes of Health, Bethesda, MD; John S.K. Kauwe, Ph.D., Brigham Young University, Provo, Utah; Michelle K. Lupton, Ph.D., King's College London Institute of Psychiatry, London; Mina Ryten, M.D., Ph.D., UCL Institute of Neurology, London; Kristelle Brown, Ph.D., University of Nottingham, Nottingham, United Kingdom; the EADI Consortium, the GERAD Consortium, the UKBE Consortium, James Lowe, D.M., University of Nottingham, Nottingham, United Kingdom; Perry G. Ridge, M.S., Brigham Young University and the ARUP Institute, Salt Lake City; Monia B. Hammer, M.S., National Institute on Aging, National Institutes of Health, Bethesda, MD; Yosuke Wakutani, M.D., Tanz Centre, University of Toronto, Toronto; Lilinaz Hazrati, M.D., Ph.D., Tanz Centre, University of Toronto, Toronto; Petroula Proitsi, Ph.D., King's College London Institute of Psychiatry, London; Stephen Newhouse, Ph.D., King's College London Institute of Psychiatry, London; Ebba Lohmann, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Nihan Erginel-Unaltuna, Ph.D., Institute for Experimental Medical Research, Istanbul University, Istanbul, Turkey; Christopher Medway, Ph.D., University of Nottingham, Nottingham, United Kingdom; Hasmet Hanagasi, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Claire Troakes, Ph.D., King's College London Institute of Psychiatry, London; Hakan Gurvit, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Basar Bilgic, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Safa Al-Sarraj, M.D., King's College London Institute of Psychiatry and King's College Hospital, London; Bruno Benitez, M.D., Washington University School of Medicine, St. Louis; Breanna Cooper, B.S., Washington University School of Medicine, St. Louis; David Carrell, M.S., Washington University School of Medicine, St. Louis; Murat Emre, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Fanggeng Zou, Ph.D., Mayo Clinic, Jacksonville, FL; Li Ma, M.S., Mayo Clinic, Jacksonville, FL; Melissa E. Murray, Ph.D., Mayo Clinic, Jacksonville, FL; Dennis W. Dickson, M.D., Mayo Clinic, Jacksonville, FL; Steven Younkin, M.D., Ph.D., Mayo Clinic, Jacksonville, FL; Ronald C. Petersen, Ph.D., M.D., Mayo Clinic, Rochester, MN; Christopher D. Corcoran, Sc.D., Utah State University, Logan; Yefei Cai, Ph.D., Washington University School of Medicine, St. Louis; Catarina Oliveira, M.D., Ph.D., University of Coimbra, Coimbra, Portugal; Maria Helena Ribeiro, M.S., University of Coimbra, Coimbra, Portugal; Isabel Santana, M.D., Ph.D., University of Coimbra, Coimbra, Portugal; JoAnn T. Tschanz, Ph.D., Utah State University, Logan; J. Raphael Gibbs, B.Sc., UCL Institute of Neurology, London, and National Institute on Aging, National Institutes of Health, Bethesda, MD; Maria C. Norton, Ph.D., Utah State University, Logan; Iwona Kloszewska, Ph.D., Medical University of Lodz, Lodz, Poland; Patrizia Mecocci, M.D., Ph.D., University of Perugia, Perugia, Italy; Hilkka Soininen, M.D., University of Eastern Finland and Kuopio University Hospital, Kuopio, Finland; Magda Tsolaki, M.D., Aristotle University of Thessaloniki, Thessaloniki, Greece; Bruno Vellas, M.D., Ph.D., UMR INSERM 1027, Hôpitaux de Toulouse, Toulouse, France; Ronald G. Munger, Ph.D., M.P.H., Utah State University, Logan; David M.A. Mann, Ph.D., University of Manchester, Manchester, United Kingdom; Stuart Pickering-Brown, Ph.D., University of Manchester, Manchester, United Kingdom; Simon Lovestone, Ph.D., King's College London Institute of Psychiatry, London; Jonathan Beck, B.Sc., MRC Prion Unit and UCL Institute of Neurology, London; Simon Mead, M.D., MRC Prion Unit and UCL Institute of Neurology, London; John Collinge, M.D., MRC Prion Unit and UCL Institute of Neurology, London; Linda Parsons, M.Phil., UCL Institute of Neurology, London; Jennifer Pocock, Ph.D., UCL Institute of Neurology, London; John C. Morris, M.D., Washington University School of Medicine, St. Louis; Tamas Revesz, M.D., UCL Institute of Neurology, London; Tammaryn Lashley, Ph.D., UCL Institute of Neurology, London; Nick C. Fox, M.D., UCL Institute of Neurology, London; Martin N. Rossor, M.D., UCL Institute of Neurology, London; Benjamin Grenier-Boley, M.Sc., INSERM, U744, Institut Pasteur de Lille and Université Lille-Nord de France, Lille, France; Céline Bellenguez, Ph.D., INSERM, U744, Institut Pasteur de Lille and Université Lille-Nord de France, Lille, France; Valentina Moskvina, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, School of Medicine, Cardiff University, Cardiff, United Kingdom; Rebecca Sims, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, United Kingdom; Denise Harold, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, United Kingdom; Julie Williams, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, United Kingdom; Jean-Charles Lambert, Ph.D., INSERM, U744, Institut Pasteur de Lille and Université Lille-Nord de France, Lille, France; Philippe Amouyel, M.D., Ph.D., INSERM, U744, Institut Pasteur de Lille, CHRU de Lille, and Université Lille-Nord de France, Lille, France; Neill Graff-Radford, M.B., B.Ch., Mayo Clinic, Jacksonville, FL; Alison Goate, Ph.D., Washington University School of Medicine, St. Louis; Rosa Rademakers, Ph.D., Mayo Clinic, Jacksonville, FL; Kevin Morgan, Ph.D., School of Molecular Medical Sciences, University of Nottingham, Nottingham, United Kingdom; John Powell, Ph.D., King's College London Institute of Psychiatry, London; Peter St. George-Hyslop, M.D., Tanz Centre, University of Toronto, Toronto, and Cambridge Institute for Medical Research, University of Cambridge, Cambridge, United Kingdom; Andrew Singleton, Ph.D., National Institute on Aging, National Institutes of Health, Bethesda, MD; and John Hardy, Ph.D., UCL Institute of Neurology, London
1,
Tammaryn Lashley
Tammaryn Lashley, Ph.D.
1Rita Guerreiro, Ph.D., University College London (UCL) Institute of Neurology, London, and National Institute on Aging, National Institutes of Health, Bethesda, MD; Aleksandra Wojtas, M.S., Mayo Clinic, Jacksonville, FL; Jose Bras, Ph.D., UCL Institute of Neurology, London; Minerva M. Carrasquillo, Ph.D., Mayo Clinic, Jacksonville, FL; Ekaterina Rogaeva, Ph.D., Tanz Centre, University of Toronto, Toronto; Elisa Majounie, Ph.D., Mayo Clinic, Jacksonville, FL; Carlos Cruchaga, Ph.D., Washington University School of Medicine, St. Louis; Celeste Sassi, M.D., Institute of Neurology, London, and National Institute on Aging, National Institutes of Health, Bethesda, MD; John S.K. Kauwe, Ph.D., Brigham Young University, Provo, Utah; Michelle K. Lupton, Ph.D., King's College London Institute of Psychiatry, London; Mina Ryten, M.D., Ph.D., UCL Institute of Neurology, London; Kristelle Brown, Ph.D., University of Nottingham, Nottingham, United Kingdom; the EADI Consortium, the GERAD Consortium, the UKBE Consortium, James Lowe, D.M., University of Nottingham, Nottingham, United Kingdom; Perry G. Ridge, M.S., Brigham Young University and the ARUP Institute, Salt Lake City; Monia B. Hammer, M.S., National Institute on Aging, National Institutes of Health, Bethesda, MD; Yosuke Wakutani, M.D., Tanz Centre, University of Toronto, Toronto; Lilinaz Hazrati, M.D., Ph.D., Tanz Centre, University of Toronto, Toronto; Petroula Proitsi, Ph.D., King's College London Institute of Psychiatry, London; Stephen Newhouse, Ph.D., King's College London Institute of Psychiatry, London; Ebba Lohmann, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Nihan Erginel-Unaltuna, Ph.D., Institute for Experimental Medical Research, Istanbul University, Istanbul, Turkey; Christopher Medway, Ph.D., University of Nottingham, Nottingham, United Kingdom; Hasmet Hanagasi, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Claire Troakes, Ph.D., King's College London Institute of Psychiatry, London; Hakan Gurvit, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Basar Bilgic, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Safa Al-Sarraj, M.D., King's College London Institute of Psychiatry and King's College Hospital, London; Bruno Benitez, M.D., Washington University School of Medicine, St. Louis; Breanna Cooper, B.S., Washington University School of Medicine, St. Louis; David Carrell, M.S., Washington University School of Medicine, St. Louis; Murat Emre, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Fanggeng Zou, Ph.D., Mayo Clinic, Jacksonville, FL; Li Ma, M.S., Mayo Clinic, Jacksonville, FL; Melissa E. Murray, Ph.D., Mayo Clinic, Jacksonville, FL; Dennis W. Dickson, M.D., Mayo Clinic, Jacksonville, FL; Steven Younkin, M.D., Ph.D., Mayo Clinic, Jacksonville, FL; Ronald C. Petersen, Ph.D., M.D., Mayo Clinic, Rochester, MN; Christopher D. Corcoran, Sc.D., Utah State University, Logan; Yefei Cai, Ph.D., Washington University School of Medicine, St. Louis; Catarina Oliveira, M.D., Ph.D., University of Coimbra, Coimbra, Portugal; Maria Helena Ribeiro, M.S., University of Coimbra, Coimbra, Portugal; Isabel Santana, M.D., Ph.D., University of Coimbra, Coimbra, Portugal; JoAnn T. Tschanz, Ph.D., Utah State University, Logan; J. Raphael Gibbs, B.Sc., UCL Institute of Neurology, London, and National Institute on Aging, National Institutes of Health, Bethesda, MD; Maria C. Norton, Ph.D., Utah State University, Logan; Iwona Kloszewska, Ph.D., Medical University of Lodz, Lodz, Poland; Patrizia Mecocci, M.D., Ph.D., University of Perugia, Perugia, Italy; Hilkka Soininen, M.D., University of Eastern Finland and Kuopio University Hospital, Kuopio, Finland; Magda Tsolaki, M.D., Aristotle University of Thessaloniki, Thessaloniki, Greece; Bruno Vellas, M.D., Ph.D., UMR INSERM 1027, Hôpitaux de Toulouse, Toulouse, France; Ronald G. Munger, Ph.D., M.P.H., Utah State University, Logan; David M.A. Mann, Ph.D., University of Manchester, Manchester, United Kingdom; Stuart Pickering-Brown, Ph.D., University of Manchester, Manchester, United Kingdom; Simon Lovestone, Ph.D., King's College London Institute of Psychiatry, London; Jonathan Beck, B.Sc., MRC Prion Unit and UCL Institute of Neurology, London; Simon Mead, M.D., MRC Prion Unit and UCL Institute of Neurology, London; John Collinge, M.D., MRC Prion Unit and UCL Institute of Neurology, London; Linda Parsons, M.Phil., UCL Institute of Neurology, London; Jennifer Pocock, Ph.D., UCL Institute of Neurology, London; John C. Morris, M.D., Washington University School of Medicine, St. Louis; Tamas Revesz, M.D., UCL Institute of Neurology, London; Tammaryn Lashley, Ph.D., UCL Institute of Neurology, London; Nick C. Fox, M.D., UCL Institute of Neurology, London; Martin N. Rossor, M.D., UCL Institute of Neurology, London; Benjamin Grenier-Boley, M.Sc., INSERM, U744, Institut Pasteur de Lille and Université Lille-Nord de France, Lille, France; Céline Bellenguez, Ph.D., INSERM, U744, Institut Pasteur de Lille and Université Lille-Nord de France, Lille, France; Valentina Moskvina, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, School of Medicine, Cardiff University, Cardiff, United Kingdom; Rebecca Sims, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, United Kingdom; Denise Harold, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, United Kingdom; Julie Williams, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, United Kingdom; Jean-Charles Lambert, Ph.D., INSERM, U744, Institut Pasteur de Lille and Université Lille-Nord de France, Lille, France; Philippe Amouyel, M.D., Ph.D., INSERM, U744, Institut Pasteur de Lille, CHRU de Lille, and Université Lille-Nord de France, Lille, France; Neill Graff-Radford, M.B., B.Ch., Mayo Clinic, Jacksonville, FL; Alison Goate, Ph.D., Washington University School of Medicine, St. Louis; Rosa Rademakers, Ph.D., Mayo Clinic, Jacksonville, FL; Kevin Morgan, Ph.D., School of Molecular Medical Sciences, University of Nottingham, Nottingham, United Kingdom; John Powell, Ph.D., King's College London Institute of Psychiatry, London; Peter St. George-Hyslop, M.D., Tanz Centre, University of Toronto, Toronto, and Cambridge Institute for Medical Research, University of Cambridge, Cambridge, United Kingdom; Andrew Singleton, Ph.D., National Institute on Aging, National Institutes of Health, Bethesda, MD; and John Hardy, Ph.D., UCL Institute of Neurology, London
1,
Julie Williams
Julie Williams, Ph.D.
1Rita Guerreiro, Ph.D., University College London (UCL) Institute of Neurology, London, and National Institute on Aging, National Institutes of Health, Bethesda, MD; Aleksandra Wojtas, M.S., Mayo Clinic, Jacksonville, FL; Jose Bras, Ph.D., UCL Institute of Neurology, London; Minerva M. Carrasquillo, Ph.D., Mayo Clinic, Jacksonville, FL; Ekaterina Rogaeva, Ph.D., Tanz Centre, University of Toronto, Toronto; Elisa Majounie, Ph.D., Mayo Clinic, Jacksonville, FL; Carlos Cruchaga, Ph.D., Washington University School of Medicine, St. Louis; Celeste Sassi, M.D., Institute of Neurology, London, and National Institute on Aging, National Institutes of Health, Bethesda, MD; John S.K. Kauwe, Ph.D., Brigham Young University, Provo, Utah; Michelle K. Lupton, Ph.D., King's College London Institute of Psychiatry, London; Mina Ryten, M.D., Ph.D., UCL Institute of Neurology, London; Kristelle Brown, Ph.D., University of Nottingham, Nottingham, United Kingdom; the EADI Consortium, the GERAD Consortium, the UKBE Consortium, James Lowe, D.M., University of Nottingham, Nottingham, United Kingdom; Perry G. Ridge, M.S., Brigham Young University and the ARUP Institute, Salt Lake City; Monia B. Hammer, M.S., National Institute on Aging, National Institutes of Health, Bethesda, MD; Yosuke Wakutani, M.D., Tanz Centre, University of Toronto, Toronto; Lilinaz Hazrati, M.D., Ph.D., Tanz Centre, University of Toronto, Toronto; Petroula Proitsi, Ph.D., King's College London Institute of Psychiatry, London; Stephen Newhouse, Ph.D., King's College London Institute of Psychiatry, London; Ebba Lohmann, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Nihan Erginel-Unaltuna, Ph.D., Institute for Experimental Medical Research, Istanbul University, Istanbul, Turkey; Christopher Medway, Ph.D., University of Nottingham, Nottingham, United Kingdom; Hasmet Hanagasi, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Claire Troakes, Ph.D., King's College London Institute of Psychiatry, London; Hakan Gurvit, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Basar Bilgic, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Safa Al-Sarraj, M.D., King's College London Institute of Psychiatry and King's College Hospital, London; Bruno Benitez, M.D., Washington University School of Medicine, St. Louis; Breanna Cooper, B.S., Washington University School of Medicine, St. Louis; David Carrell, M.S., Washington University School of Medicine, St. Louis; Murat Emre, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Fanggeng Zou, Ph.D., Mayo Clinic, Jacksonville, FL; Li Ma, M.S., Mayo Clinic, Jacksonville, FL; Melissa E. Murray, Ph.D., Mayo Clinic, Jacksonville, FL; Dennis W. Dickson, M.D., Mayo Clinic, Jacksonville, FL; Steven Younkin, M.D., Ph.D., Mayo Clinic, Jacksonville, FL; Ronald C. Petersen, Ph.D., M.D., Mayo Clinic, Rochester, MN; Christopher D. Corcoran, Sc.D., Utah State University, Logan; Yefei Cai, Ph.D., Washington University School of Medicine, St. Louis; Catarina Oliveira, M.D., Ph.D., University of Coimbra, Coimbra, Portugal; Maria Helena Ribeiro, M.S., University of Coimbra, Coimbra, Portugal; Isabel Santana, M.D., Ph.D., University of Coimbra, Coimbra, Portugal; JoAnn T. Tschanz, Ph.D., Utah State University, Logan; J. Raphael Gibbs, B.Sc., UCL Institute of Neurology, London, and National Institute on Aging, National Institutes of Health, Bethesda, MD; Maria C. Norton, Ph.D., Utah State University, Logan; Iwona Kloszewska, Ph.D., Medical University of Lodz, Lodz, Poland; Patrizia Mecocci, M.D., Ph.D., University of Perugia, Perugia, Italy; Hilkka Soininen, M.D., University of Eastern Finland and Kuopio University Hospital, Kuopio, Finland; Magda Tsolaki, M.D., Aristotle University of Thessaloniki, Thessaloniki, Greece; Bruno Vellas, M.D., Ph.D., UMR INSERM 1027, Hôpitaux de Toulouse, Toulouse, France; Ronald G. Munger, Ph.D., M.P.H., Utah State University, Logan; David M.A. Mann, Ph.D., University of Manchester, Manchester, United Kingdom; Stuart Pickering-Brown, Ph.D., University of Manchester, Manchester, United Kingdom; Simon Lovestone, Ph.D., King's College London Institute of Psychiatry, London; Jonathan Beck, B.Sc., MRC Prion Unit and UCL Institute of Neurology, London; Simon Mead, M.D., MRC Prion Unit and UCL Institute of Neurology, London; John Collinge, M.D., MRC Prion Unit and UCL Institute of Neurology, London; Linda Parsons, M.Phil., UCL Institute of Neurology, London; Jennifer Pocock, Ph.D., UCL Institute of Neurology, London; John C. Morris, M.D., Washington University School of Medicine, St. Louis; Tamas Revesz, M.D., UCL Institute of Neurology, London; Tammaryn Lashley, Ph.D., UCL Institute of Neurology, London; Nick C. Fox, M.D., UCL Institute of Neurology, London; Martin N. Rossor, M.D., UCL Institute of Neurology, London; Benjamin Grenier-Boley, M.Sc., INSERM, U744, Institut Pasteur de Lille and Université Lille-Nord de France, Lille, France; Céline Bellenguez, Ph.D., INSERM, U744, Institut Pasteur de Lille and Université Lille-Nord de France, Lille, France; Valentina Moskvina, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, School of Medicine, Cardiff University, Cardiff, United Kingdom; Rebecca Sims, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, United Kingdom; Denise Harold, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, United Kingdom; Julie Williams, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, United Kingdom; Jean-Charles Lambert, Ph.D., INSERM, U744, Institut Pasteur de Lille and Université Lille-Nord de France, Lille, France; Philippe Amouyel, M.D., Ph.D., INSERM, U744, Institut Pasteur de Lille, CHRU de Lille, and Université Lille-Nord de France, Lille, France; Neill Graff-Radford, M.B., B.Ch., Mayo Clinic, Jacksonville, FL; Alison Goate, Ph.D., Washington University School of Medicine, St. Louis; Rosa Rademakers, Ph.D., Mayo Clinic, Jacksonville, FL; Kevin Morgan, Ph.D., School of Molecular Medical Sciences, University of Nottingham, Nottingham, United Kingdom; John Powell, Ph.D., King's College London Institute of Psychiatry, London; Peter St. George-Hyslop, M.D., Tanz Centre, University of Toronto, Toronto, and Cambridge Institute for Medical Research, University of Cambridge, Cambridge, United Kingdom; Andrew Singleton, Ph.D., National Institute on Aging, National Institutes of Health, Bethesda, MD; and John Hardy, Ph.D., UCL Institute of Neurology, London
1,
Jean-Charles Lambert
Jean-Charles Lambert, Ph.D.
1Rita Guerreiro, Ph.D., University College London (UCL) Institute of Neurology, London, and National Institute on Aging, National Institutes of Health, Bethesda, MD; Aleksandra Wojtas, M.S., Mayo Clinic, Jacksonville, FL; Jose Bras, Ph.D., UCL Institute of Neurology, London; Minerva M. Carrasquillo, Ph.D., Mayo Clinic, Jacksonville, FL; Ekaterina Rogaeva, Ph.D., Tanz Centre, University of Toronto, Toronto; Elisa Majounie, Ph.D., Mayo Clinic, Jacksonville, FL; Carlos Cruchaga, Ph.D., Washington University School of Medicine, St. Louis; Celeste Sassi, M.D., Institute of Neurology, London, and National Institute on Aging, National Institutes of Health, Bethesda, MD; John S.K. Kauwe, Ph.D., Brigham Young University, Provo, Utah; Michelle K. Lupton, Ph.D., King's College London Institute of Psychiatry, London; Mina Ryten, M.D., Ph.D., UCL Institute of Neurology, London; Kristelle Brown, Ph.D., University of Nottingham, Nottingham, United Kingdom; the EADI Consortium, the GERAD Consortium, the UKBE Consortium, James Lowe, D.M., University of Nottingham, Nottingham, United Kingdom; Perry G. Ridge, M.S., Brigham Young University and the ARUP Institute, Salt Lake City; Monia B. Hammer, M.S., National Institute on Aging, National Institutes of Health, Bethesda, MD; Yosuke Wakutani, M.D., Tanz Centre, University of Toronto, Toronto; Lilinaz Hazrati, M.D., Ph.D., Tanz Centre, University of Toronto, Toronto; Petroula Proitsi, Ph.D., King's College London Institute of Psychiatry, London; Stephen Newhouse, Ph.D., King's College London Institute of Psychiatry, London; Ebba Lohmann, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Nihan Erginel-Unaltuna, Ph.D., Institute for Experimental Medical Research, Istanbul University, Istanbul, Turkey; Christopher Medway, Ph.D., University of Nottingham, Nottingham, United Kingdom; Hasmet Hanagasi, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Claire Troakes, Ph.D., King's College London Institute of Psychiatry, London; Hakan Gurvit, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Basar Bilgic, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Safa Al-Sarraj, M.D., King's College London Institute of Psychiatry and King's College Hospital, London; Bruno Benitez, M.D., Washington University School of Medicine, St. Louis; Breanna Cooper, B.S., Washington University School of Medicine, St. Louis; David Carrell, M.S., Washington University School of Medicine, St. Louis; Murat Emre, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Fanggeng Zou, Ph.D., Mayo Clinic, Jacksonville, FL; Li Ma, M.S., Mayo Clinic, Jacksonville, FL; Melissa E. Murray, Ph.D., Mayo Clinic, Jacksonville, FL; Dennis W. Dickson, M.D., Mayo Clinic, Jacksonville, FL; Steven Younkin, M.D., Ph.D., Mayo Clinic, Jacksonville, FL; Ronald C. Petersen, Ph.D., M.D., Mayo Clinic, Rochester, MN; Christopher D. Corcoran, Sc.D., Utah State University, Logan; Yefei Cai, Ph.D., Washington University School of Medicine, St. Louis; Catarina Oliveira, M.D., Ph.D., University of Coimbra, Coimbra, Portugal; Maria Helena Ribeiro, M.S., University of Coimbra, Coimbra, Portugal; Isabel Santana, M.D., Ph.D., University of Coimbra, Coimbra, Portugal; JoAnn T. Tschanz, Ph.D., Utah State University, Logan; J. Raphael Gibbs, B.Sc., UCL Institute of Neurology, London, and National Institute on Aging, National Institutes of Health, Bethesda, MD; Maria C. Norton, Ph.D., Utah State University, Logan; Iwona Kloszewska, Ph.D., Medical University of Lodz, Lodz, Poland; Patrizia Mecocci, M.D., Ph.D., University of Perugia, Perugia, Italy; Hilkka Soininen, M.D., University of Eastern Finland and Kuopio University Hospital, Kuopio, Finland; Magda Tsolaki, M.D., Aristotle University of Thessaloniki, Thessaloniki, Greece; Bruno Vellas, M.D., Ph.D., UMR INSERM 1027, Hôpitaux de Toulouse, Toulouse, France; Ronald G. Munger, Ph.D., M.P.H., Utah State University, Logan; David M.A. Mann, Ph.D., University of Manchester, Manchester, United Kingdom; Stuart Pickering-Brown, Ph.D., University of Manchester, Manchester, United Kingdom; Simon Lovestone, Ph.D., King's College London Institute of Psychiatry, London; Jonathan Beck, B.Sc., MRC Prion Unit and UCL Institute of Neurology, London; Simon Mead, M.D., MRC Prion Unit and UCL Institute of Neurology, London; John Collinge, M.D., MRC Prion Unit and UCL Institute of Neurology, London; Linda Parsons, M.Phil., UCL Institute of Neurology, London; Jennifer Pocock, Ph.D., UCL Institute of Neurology, London; John C. Morris, M.D., Washington University School of Medicine, St. Louis; Tamas Revesz, M.D., UCL Institute of Neurology, London; Tammaryn Lashley, Ph.D., UCL Institute of Neurology, London; Nick C. Fox, M.D., UCL Institute of Neurology, London; Martin N. Rossor, M.D., UCL Institute of Neurology, London; Benjamin Grenier-Boley, M.Sc., INSERM, U744, Institut Pasteur de Lille and Université Lille-Nord de France, Lille, France; Céline Bellenguez, Ph.D., INSERM, U744, Institut Pasteur de Lille and Université Lille-Nord de France, Lille, France; Valentina Moskvina, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, School of Medicine, Cardiff University, Cardiff, United Kingdom; Rebecca Sims, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, United Kingdom; Denise Harold, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, United Kingdom; Julie Williams, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, United Kingdom; Jean-Charles Lambert, Ph.D., INSERM, U744, Institut Pasteur de Lille and Université Lille-Nord de France, Lille, France; Philippe Amouyel, M.D., Ph.D., INSERM, U744, Institut Pasteur de Lille, CHRU de Lille, and Université Lille-Nord de France, Lille, France; Neill Graff-Radford, M.B., B.Ch., Mayo Clinic, Jacksonville, FL; Alison Goate, Ph.D., Washington University School of Medicine, St. Louis; Rosa Rademakers, Ph.D., Mayo Clinic, Jacksonville, FL; Kevin Morgan, Ph.D., School of Molecular Medical Sciences, University of Nottingham, Nottingham, United Kingdom; John Powell, Ph.D., King's College London Institute of Psychiatry, London; Peter St. George-Hyslop, M.D., Tanz Centre, University of Toronto, Toronto, and Cambridge Institute for Medical Research, University of Cambridge, Cambridge, United Kingdom; Andrew Singleton, Ph.D., National Institute on Aging, National Institutes of Health, Bethesda, MD; and John Hardy, Ph.D., UCL Institute of Neurology, London
1,
Philippe Amouyel
Philippe Amouyel, M.D., Ph.D.
1Rita Guerreiro, Ph.D., University College London (UCL) Institute of Neurology, London, and National Institute on Aging, National Institutes of Health, Bethesda, MD; Aleksandra Wojtas, M.S., Mayo Clinic, Jacksonville, FL; Jose Bras, Ph.D., UCL Institute of Neurology, London; Minerva M. Carrasquillo, Ph.D., Mayo Clinic, Jacksonville, FL; Ekaterina Rogaeva, Ph.D., Tanz Centre, University of Toronto, Toronto; Elisa Majounie, Ph.D., Mayo Clinic, Jacksonville, FL; Carlos Cruchaga, Ph.D., Washington University School of Medicine, St. Louis; Celeste Sassi, M.D., Institute of Neurology, London, and National Institute on Aging, National Institutes of Health, Bethesda, MD; John S.K. Kauwe, Ph.D., Brigham Young University, Provo, Utah; Michelle K. Lupton, Ph.D., King's College London Institute of Psychiatry, London; Mina Ryten, M.D., Ph.D., UCL Institute of Neurology, London; Kristelle Brown, Ph.D., University of Nottingham, Nottingham, United Kingdom; the EADI Consortium, the GERAD Consortium, the UKBE Consortium, James Lowe, D.M., University of Nottingham, Nottingham, United Kingdom; Perry G. Ridge, M.S., Brigham Young University and the ARUP Institute, Salt Lake City; Monia B. Hammer, M.S., National Institute on Aging, National Institutes of Health, Bethesda, MD; Yosuke Wakutani, M.D., Tanz Centre, University of Toronto, Toronto; Lilinaz Hazrati, M.D., Ph.D., Tanz Centre, University of Toronto, Toronto; Petroula Proitsi, Ph.D., King's College London Institute of Psychiatry, London; Stephen Newhouse, Ph.D., King's College London Institute of Psychiatry, London; Ebba Lohmann, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Nihan Erginel-Unaltuna, Ph.D., Institute for Experimental Medical Research, Istanbul University, Istanbul, Turkey; Christopher Medway, Ph.D., University of Nottingham, Nottingham, United Kingdom; Hasmet Hanagasi, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Claire Troakes, Ph.D., King's College London Institute of Psychiatry, London; Hakan Gurvit, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Basar Bilgic, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Safa Al-Sarraj, M.D., King's College London Institute of Psychiatry and King's College Hospital, London; Bruno Benitez, M.D., Washington University School of Medicine, St. Louis; Breanna Cooper, B.S., Washington University School of Medicine, St. Louis; David Carrell, M.S., Washington University School of Medicine, St. Louis; Murat Emre, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Fanggeng Zou, Ph.D., Mayo Clinic, Jacksonville, FL; Li Ma, M.S., Mayo Clinic, Jacksonville, FL; Melissa E. Murray, Ph.D., Mayo Clinic, Jacksonville, FL; Dennis W. Dickson, M.D., Mayo Clinic, Jacksonville, FL; Steven Younkin, M.D., Ph.D., Mayo Clinic, Jacksonville, FL; Ronald C. Petersen, Ph.D., M.D., Mayo Clinic, Rochester, MN; Christopher D. Corcoran, Sc.D., Utah State University, Logan; Yefei Cai, Ph.D., Washington University School of Medicine, St. Louis; Catarina Oliveira, M.D., Ph.D., University of Coimbra, Coimbra, Portugal; Maria Helena Ribeiro, M.S., University of Coimbra, Coimbra, Portugal; Isabel Santana, M.D., Ph.D., University of Coimbra, Coimbra, Portugal; JoAnn T. Tschanz, Ph.D., Utah State University, Logan; J. Raphael Gibbs, B.Sc., UCL Institute of Neurology, London, and National Institute on Aging, National Institutes of Health, Bethesda, MD; Maria C. Norton, Ph.D., Utah State University, Logan; Iwona Kloszewska, Ph.D., Medical University of Lodz, Lodz, Poland; Patrizia Mecocci, M.D., Ph.D., University of Perugia, Perugia, Italy; Hilkka Soininen, M.D., University of Eastern Finland and Kuopio University Hospital, Kuopio, Finland; Magda Tsolaki, M.D., Aristotle University of Thessaloniki, Thessaloniki, Greece; Bruno Vellas, M.D., Ph.D., UMR INSERM 1027, Hôpitaux de Toulouse, Toulouse, France; Ronald G. Munger, Ph.D., M.P.H., Utah State University, Logan; David M.A. Mann, Ph.D., University of Manchester, Manchester, United Kingdom; Stuart Pickering-Brown, Ph.D., University of Manchester, Manchester, United Kingdom; Simon Lovestone, Ph.D., King's College London Institute of Psychiatry, London; Jonathan Beck, B.Sc., MRC Prion Unit and UCL Institute of Neurology, London; Simon Mead, M.D., MRC Prion Unit and UCL Institute of Neurology, London; John Collinge, M.D., MRC Prion Unit and UCL Institute of Neurology, London; Linda Parsons, M.Phil., UCL Institute of Neurology, London; Jennifer Pocock, Ph.D., UCL Institute of Neurology, London; John C. Morris, M.D., Washington University School of Medicine, St. Louis; Tamas Revesz, M.D., UCL Institute of Neurology, London; Tammaryn Lashley, Ph.D., UCL Institute of Neurology, London; Nick C. Fox, M.D., UCL Institute of Neurology, London; Martin N. Rossor, M.D., UCL Institute of Neurology, London; Benjamin Grenier-Boley, M.Sc., INSERM, U744, Institut Pasteur de Lille and Université Lille-Nord de France, Lille, France; Céline Bellenguez, Ph.D., INSERM, U744, Institut Pasteur de Lille and Université Lille-Nord de France, Lille, France; Valentina Moskvina, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, School of Medicine, Cardiff University, Cardiff, United Kingdom; Rebecca Sims, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, United Kingdom; Denise Harold, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, United Kingdom; Julie Williams, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, United Kingdom; Jean-Charles Lambert, Ph.D., INSERM, U744, Institut Pasteur de Lille and Université Lille-Nord de France, Lille, France; Philippe Amouyel, M.D., Ph.D., INSERM, U744, Institut Pasteur de Lille, CHRU de Lille, and Université Lille-Nord de France, Lille, France; Neill Graff-Radford, M.B., B.Ch., Mayo Clinic, Jacksonville, FL; Alison Goate, Ph.D., Washington University School of Medicine, St. Louis; Rosa Rademakers, Ph.D., Mayo Clinic, Jacksonville, FL; Kevin Morgan, Ph.D., School of Molecular Medical Sciences, University of Nottingham, Nottingham, United Kingdom; John Powell, Ph.D., King's College London Institute of Psychiatry, London; Peter St. George-Hyslop, M.D., Tanz Centre, University of Toronto, Toronto, and Cambridge Institute for Medical Research, University of Cambridge, Cambridge, United Kingdom; Andrew Singleton, Ph.D., National Institute on Aging, National Institutes of Health, Bethesda, MD; and John Hardy, Ph.D., UCL Institute of Neurology, London
1,
Alison Goate
Alison Goate, Ph.D.
1Rita Guerreiro, Ph.D., University College London (UCL) Institute of Neurology, London, and National Institute on Aging, National Institutes of Health, Bethesda, MD; Aleksandra Wojtas, M.S., Mayo Clinic, Jacksonville, FL; Jose Bras, Ph.D., UCL Institute of Neurology, London; Minerva M. Carrasquillo, Ph.D., Mayo Clinic, Jacksonville, FL; Ekaterina Rogaeva, Ph.D., Tanz Centre, University of Toronto, Toronto; Elisa Majounie, Ph.D., Mayo Clinic, Jacksonville, FL; Carlos Cruchaga, Ph.D., Washington University School of Medicine, St. Louis; Celeste Sassi, M.D., Institute of Neurology, London, and National Institute on Aging, National Institutes of Health, Bethesda, MD; John S.K. Kauwe, Ph.D., Brigham Young University, Provo, Utah; Michelle K. Lupton, Ph.D., King's College London Institute of Psychiatry, London; Mina Ryten, M.D., Ph.D., UCL Institute of Neurology, London; Kristelle Brown, Ph.D., University of Nottingham, Nottingham, United Kingdom; the EADI Consortium, the GERAD Consortium, the UKBE Consortium, James Lowe, D.M., University of Nottingham, Nottingham, United Kingdom; Perry G. Ridge, M.S., Brigham Young University and the ARUP Institute, Salt Lake City; Monia B. Hammer, M.S., National Institute on Aging, National Institutes of Health, Bethesda, MD; Yosuke Wakutani, M.D., Tanz Centre, University of Toronto, Toronto; Lilinaz Hazrati, M.D., Ph.D., Tanz Centre, University of Toronto, Toronto; Petroula Proitsi, Ph.D., King's College London Institute of Psychiatry, London; Stephen Newhouse, Ph.D., King's College London Institute of Psychiatry, London; Ebba Lohmann, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Nihan Erginel-Unaltuna, Ph.D., Institute for Experimental Medical Research, Istanbul University, Istanbul, Turkey; Christopher Medway, Ph.D., University of Nottingham, Nottingham, United Kingdom; Hasmet Hanagasi, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Claire Troakes, Ph.D., King's College London Institute of Psychiatry, London; Hakan Gurvit, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Basar Bilgic, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Safa Al-Sarraj, M.D., King's College London Institute of Psychiatry and King's College Hospital, London; Bruno Benitez, M.D., Washington University School of Medicine, St. Louis; Breanna Cooper, B.S., Washington University School of Medicine, St. Louis; David Carrell, M.S., Washington University School of Medicine, St. Louis; Murat Emre, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Fanggeng Zou, Ph.D., Mayo Clinic, Jacksonville, FL; Li Ma, M.S., Mayo Clinic, Jacksonville, FL; Melissa E. Murray, Ph.D., Mayo Clinic, Jacksonville, FL; Dennis W. Dickson, M.D., Mayo Clinic, Jacksonville, FL; Steven Younkin, M.D., Ph.D., Mayo Clinic, Jacksonville, FL; Ronald C. Petersen, Ph.D., M.D., Mayo Clinic, Rochester, MN; Christopher D. Corcoran, Sc.D., Utah State University, Logan; Yefei Cai, Ph.D., Washington University School of Medicine, St. Louis; Catarina Oliveira, M.D., Ph.D., University of Coimbra, Coimbra, Portugal; Maria Helena Ribeiro, M.S., University of Coimbra, Coimbra, Portugal; Isabel Santana, M.D., Ph.D., University of Coimbra, Coimbra, Portugal; JoAnn T. Tschanz, Ph.D., Utah State University, Logan; J. Raphael Gibbs, B.Sc., UCL Institute of Neurology, London, and National Institute on Aging, National Institutes of Health, Bethesda, MD; Maria C. Norton, Ph.D., Utah State University, Logan; Iwona Kloszewska, Ph.D., Medical University of Lodz, Lodz, Poland; Patrizia Mecocci, M.D., Ph.D., University of Perugia, Perugia, Italy; Hilkka Soininen, M.D., University of Eastern Finland and Kuopio University Hospital, Kuopio, Finland; Magda Tsolaki, M.D., Aristotle University of Thessaloniki, Thessaloniki, Greece; Bruno Vellas, M.D., Ph.D., UMR INSERM 1027, Hôpitaux de Toulouse, Toulouse, France; Ronald G. Munger, Ph.D., M.P.H., Utah State University, Logan; David M.A. Mann, Ph.D., University of Manchester, Manchester, United Kingdom; Stuart Pickering-Brown, Ph.D., University of Manchester, Manchester, United Kingdom; Simon Lovestone, Ph.D., King's College London Institute of Psychiatry, London; Jonathan Beck, B.Sc., MRC Prion Unit and UCL Institute of Neurology, London; Simon Mead, M.D., MRC Prion Unit and UCL Institute of Neurology, London; John Collinge, M.D., MRC Prion Unit and UCL Institute of Neurology, London; Linda Parsons, M.Phil., UCL Institute of Neurology, London; Jennifer Pocock, Ph.D., UCL Institute of Neurology, London; John C. Morris, M.D., Washington University School of Medicine, St. Louis; Tamas Revesz, M.D., UCL Institute of Neurology, London; Tammaryn Lashley, Ph.D., UCL Institute of Neurology, London; Nick C. Fox, M.D., UCL Institute of Neurology, London; Martin N. Rossor, M.D., UCL Institute of Neurology, London; Benjamin Grenier-Boley, M.Sc., INSERM, U744, Institut Pasteur de Lille and Université Lille-Nord de France, Lille, France; Céline Bellenguez, Ph.D., INSERM, U744, Institut Pasteur de Lille and Université Lille-Nord de France, Lille, France; Valentina Moskvina, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, School of Medicine, Cardiff University, Cardiff, United Kingdom; Rebecca Sims, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, United Kingdom; Denise Harold, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, United Kingdom; Julie Williams, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, United Kingdom; Jean-Charles Lambert, Ph.D., INSERM, U744, Institut Pasteur de Lille and Université Lille-Nord de France, Lille, France; Philippe Amouyel, M.D., Ph.D., INSERM, U744, Institut Pasteur de Lille, CHRU de Lille, and Université Lille-Nord de France, Lille, France; Neill Graff-Radford, M.B., B.Ch., Mayo Clinic, Jacksonville, FL; Alison Goate, Ph.D., Washington University School of Medicine, St. Louis; Rosa Rademakers, Ph.D., Mayo Clinic, Jacksonville, FL; Kevin Morgan, Ph.D., School of Molecular Medical Sciences, University of Nottingham, Nottingham, United Kingdom; John Powell, Ph.D., King's College London Institute of Psychiatry, London; Peter St. George-Hyslop, M.D., Tanz Centre, University of Toronto, Toronto, and Cambridge Institute for Medical Research, University of Cambridge, Cambridge, United Kingdom; Andrew Singleton, Ph.D., National Institute on Aging, National Institutes of Health, Bethesda, MD; and John Hardy, Ph.D., UCL Institute of Neurology, London
1,
Rosa Rademakers
Rosa Rademakers, Ph.D.
1Rita Guerreiro, Ph.D., University College London (UCL) Institute of Neurology, London, and National Institute on Aging, National Institutes of Health, Bethesda, MD; Aleksandra Wojtas, M.S., Mayo Clinic, Jacksonville, FL; Jose Bras, Ph.D., UCL Institute of Neurology, London; Minerva M. Carrasquillo, Ph.D., Mayo Clinic, Jacksonville, FL; Ekaterina Rogaeva, Ph.D., Tanz Centre, University of Toronto, Toronto; Elisa Majounie, Ph.D., Mayo Clinic, Jacksonville, FL; Carlos Cruchaga, Ph.D., Washington University School of Medicine, St. Louis; Celeste Sassi, M.D., Institute of Neurology, London, and National Institute on Aging, National Institutes of Health, Bethesda, MD; John S.K. Kauwe, Ph.D., Brigham Young University, Provo, Utah; Michelle K. Lupton, Ph.D., King's College London Institute of Psychiatry, London; Mina Ryten, M.D., Ph.D., UCL Institute of Neurology, London; Kristelle Brown, Ph.D., University of Nottingham, Nottingham, United Kingdom; the EADI Consortium, the GERAD Consortium, the UKBE Consortium, James Lowe, D.M., University of Nottingham, Nottingham, United Kingdom; Perry G. Ridge, M.S., Brigham Young University and the ARUP Institute, Salt Lake City; Monia B. Hammer, M.S., National Institute on Aging, National Institutes of Health, Bethesda, MD; Yosuke Wakutani, M.D., Tanz Centre, University of Toronto, Toronto; Lilinaz Hazrati, M.D., Ph.D., Tanz Centre, University of Toronto, Toronto; Petroula Proitsi, Ph.D., King's College London Institute of Psychiatry, London; Stephen Newhouse, Ph.D., King's College London Institute of Psychiatry, London; Ebba Lohmann, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Nihan Erginel-Unaltuna, Ph.D., Institute for Experimental Medical Research, Istanbul University, Istanbul, Turkey; Christopher Medway, Ph.D., University of Nottingham, Nottingham, United Kingdom; Hasmet Hanagasi, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Claire Troakes, Ph.D., King's College London Institute of Psychiatry, London; Hakan Gurvit, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Basar Bilgic, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Safa Al-Sarraj, M.D., King's College London Institute of Psychiatry and King's College Hospital, London; Bruno Benitez, M.D., Washington University School of Medicine, St. Louis; Breanna Cooper, B.S., Washington University School of Medicine, St. Louis; David Carrell, M.S., Washington University School of Medicine, St. Louis; Murat Emre, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Fanggeng Zou, Ph.D., Mayo Clinic, Jacksonville, FL; Li Ma, M.S., Mayo Clinic, Jacksonville, FL; Melissa E. Murray, Ph.D., Mayo Clinic, Jacksonville, FL; Dennis W. Dickson, M.D., Mayo Clinic, Jacksonville, FL; Steven Younkin, M.D., Ph.D., Mayo Clinic, Jacksonville, FL; Ronald C. Petersen, Ph.D., M.D., Mayo Clinic, Rochester, MN; Christopher D. Corcoran, Sc.D., Utah State University, Logan; Yefei Cai, Ph.D., Washington University School of Medicine, St. Louis; Catarina Oliveira, M.D., Ph.D., University of Coimbra, Coimbra, Portugal; Maria Helena Ribeiro, M.S., University of Coimbra, Coimbra, Portugal; Isabel Santana, M.D., Ph.D., University of Coimbra, Coimbra, Portugal; JoAnn T. Tschanz, Ph.D., Utah State University, Logan; J. Raphael Gibbs, B.Sc., UCL Institute of Neurology, London, and National Institute on Aging, National Institutes of Health, Bethesda, MD; Maria C. Norton, Ph.D., Utah State University, Logan; Iwona Kloszewska, Ph.D., Medical University of Lodz, Lodz, Poland; Patrizia Mecocci, M.D., Ph.D., University of Perugia, Perugia, Italy; Hilkka Soininen, M.D., University of Eastern Finland and Kuopio University Hospital, Kuopio, Finland; Magda Tsolaki, M.D., Aristotle University of Thessaloniki, Thessaloniki, Greece; Bruno Vellas, M.D., Ph.D., UMR INSERM 1027, Hôpitaux de Toulouse, Toulouse, France; Ronald G. Munger, Ph.D., M.P.H., Utah State University, Logan; David M.A. Mann, Ph.D., University of Manchester, Manchester, United Kingdom; Stuart Pickering-Brown, Ph.D., University of Manchester, Manchester, United Kingdom; Simon Lovestone, Ph.D., King's College London Institute of Psychiatry, London; Jonathan Beck, B.Sc., MRC Prion Unit and UCL Institute of Neurology, London; Simon Mead, M.D., MRC Prion Unit and UCL Institute of Neurology, London; John Collinge, M.D., MRC Prion Unit and UCL Institute of Neurology, London; Linda Parsons, M.Phil., UCL Institute of Neurology, London; Jennifer Pocock, Ph.D., UCL Institute of Neurology, London; John C. Morris, M.D., Washington University School of Medicine, St. Louis; Tamas Revesz, M.D., UCL Institute of Neurology, London; Tammaryn Lashley, Ph.D., UCL Institute of Neurology, London; Nick C. Fox, M.D., UCL Institute of Neurology, London; Martin N. Rossor, M.D., UCL Institute of Neurology, London; Benjamin Grenier-Boley, M.Sc., INSERM, U744, Institut Pasteur de Lille and Université Lille-Nord de France, Lille, France; Céline Bellenguez, Ph.D., INSERM, U744, Institut Pasteur de Lille and Université Lille-Nord de France, Lille, France; Valentina Moskvina, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, School of Medicine, Cardiff University, Cardiff, United Kingdom; Rebecca Sims, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, United Kingdom; Denise Harold, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, United Kingdom; Julie Williams, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, United Kingdom; Jean-Charles Lambert, Ph.D., INSERM, U744, Institut Pasteur de Lille and Université Lille-Nord de France, Lille, France; Philippe Amouyel, M.D., Ph.D., INSERM, U744, Institut Pasteur de Lille, CHRU de Lille, and Université Lille-Nord de France, Lille, France; Neill Graff-Radford, M.B., B.Ch., Mayo Clinic, Jacksonville, FL; Alison Goate, Ph.D., Washington University School of Medicine, St. Louis; Rosa Rademakers, Ph.D., Mayo Clinic, Jacksonville, FL; Kevin Morgan, Ph.D., School of Molecular Medical Sciences, University of Nottingham, Nottingham, United Kingdom; John Powell, Ph.D., King's College London Institute of Psychiatry, London; Peter St. George-Hyslop, M.D., Tanz Centre, University of Toronto, Toronto, and Cambridge Institute for Medical Research, University of Cambridge, Cambridge, United Kingdom; Andrew Singleton, Ph.D., National Institute on Aging, National Institutes of Health, Bethesda, MD; and John Hardy, Ph.D., UCL Institute of Neurology, London
1,
Kevin Morgan
Kevin Morgan, Ph.D.
1Rita Guerreiro, Ph.D., University College London (UCL) Institute of Neurology, London, and National Institute on Aging, National Institutes of Health, Bethesda, MD; Aleksandra Wojtas, M.S., Mayo Clinic, Jacksonville, FL; Jose Bras, Ph.D., UCL Institute of Neurology, London; Minerva M. Carrasquillo, Ph.D., Mayo Clinic, Jacksonville, FL; Ekaterina Rogaeva, Ph.D., Tanz Centre, University of Toronto, Toronto; Elisa Majounie, Ph.D., Mayo Clinic, Jacksonville, FL; Carlos Cruchaga, Ph.D., Washington University School of Medicine, St. Louis; Celeste Sassi, M.D., Institute of Neurology, London, and National Institute on Aging, National Institutes of Health, Bethesda, MD; John S.K. Kauwe, Ph.D., Brigham Young University, Provo, Utah; Michelle K. Lupton, Ph.D., King's College London Institute of Psychiatry, London; Mina Ryten, M.D., Ph.D., UCL Institute of Neurology, London; Kristelle Brown, Ph.D., University of Nottingham, Nottingham, United Kingdom; the EADI Consortium, the GERAD Consortium, the UKBE Consortium, James Lowe, D.M., University of Nottingham, Nottingham, United Kingdom; Perry G. Ridge, M.S., Brigham Young University and the ARUP Institute, Salt Lake City; Monia B. Hammer, M.S., National Institute on Aging, National Institutes of Health, Bethesda, MD; Yosuke Wakutani, M.D., Tanz Centre, University of Toronto, Toronto; Lilinaz Hazrati, M.D., Ph.D., Tanz Centre, University of Toronto, Toronto; Petroula Proitsi, Ph.D., King's College London Institute of Psychiatry, London; Stephen Newhouse, Ph.D., King's College London Institute of Psychiatry, London; Ebba Lohmann, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Nihan Erginel-Unaltuna, Ph.D., Institute for Experimental Medical Research, Istanbul University, Istanbul, Turkey; Christopher Medway, Ph.D., University of Nottingham, Nottingham, United Kingdom; Hasmet Hanagasi, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Claire Troakes, Ph.D., King's College London Institute of Psychiatry, London; Hakan Gurvit, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Basar Bilgic, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Safa Al-Sarraj, M.D., King's College London Institute of Psychiatry and King's College Hospital, London; Bruno Benitez, M.D., Washington University School of Medicine, St. Louis; Breanna Cooper, B.S., Washington University School of Medicine, St. Louis; David Carrell, M.S., Washington University School of Medicine, St. Louis; Murat Emre, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Fanggeng Zou, Ph.D., Mayo Clinic, Jacksonville, FL; Li Ma, M.S., Mayo Clinic, Jacksonville, FL; Melissa E. Murray, Ph.D., Mayo Clinic, Jacksonville, FL; Dennis W. Dickson, M.D., Mayo Clinic, Jacksonville, FL; Steven Younkin, M.D., Ph.D., Mayo Clinic, Jacksonville, FL; Ronald C. Petersen, Ph.D., M.D., Mayo Clinic, Rochester, MN; Christopher D. Corcoran, Sc.D., Utah State University, Logan; Yefei Cai, Ph.D., Washington University School of Medicine, St. Louis; Catarina Oliveira, M.D., Ph.D., University of Coimbra, Coimbra, Portugal; Maria Helena Ribeiro, M.S., University of Coimbra, Coimbra, Portugal; Isabel Santana, M.D., Ph.D., University of Coimbra, Coimbra, Portugal; JoAnn T. Tschanz, Ph.D., Utah State University, Logan; J. Raphael Gibbs, B.Sc., UCL Institute of Neurology, London, and National Institute on Aging, National Institutes of Health, Bethesda, MD; Maria C. Norton, Ph.D., Utah State University, Logan; Iwona Kloszewska, Ph.D., Medical University of Lodz, Lodz, Poland; Patrizia Mecocci, M.D., Ph.D., University of Perugia, Perugia, Italy; Hilkka Soininen, M.D., University of Eastern Finland and Kuopio University Hospital, Kuopio, Finland; Magda Tsolaki, M.D., Aristotle University of Thessaloniki, Thessaloniki, Greece; Bruno Vellas, M.D., Ph.D., UMR INSERM 1027, Hôpitaux de Toulouse, Toulouse, France; Ronald G. Munger, Ph.D., M.P.H., Utah State University, Logan; David M.A. Mann, Ph.D., University of Manchester, Manchester, United Kingdom; Stuart Pickering-Brown, Ph.D., University of Manchester, Manchester, United Kingdom; Simon Lovestone, Ph.D., King's College London Institute of Psychiatry, London; Jonathan Beck, B.Sc., MRC Prion Unit and UCL Institute of Neurology, London; Simon Mead, M.D., MRC Prion Unit and UCL Institute of Neurology, London; John Collinge, M.D., MRC Prion Unit and UCL Institute of Neurology, London; Linda Parsons, M.Phil., UCL Institute of Neurology, London; Jennifer Pocock, Ph.D., UCL Institute of Neurology, London; John C. Morris, M.D., Washington University School of Medicine, St. Louis; Tamas Revesz, M.D., UCL Institute of Neurology, London; Tammaryn Lashley, Ph.D., UCL Institute of Neurology, London; Nick C. Fox, M.D., UCL Institute of Neurology, London; Martin N. Rossor, M.D., UCL Institute of Neurology, London; Benjamin Grenier-Boley, M.Sc., INSERM, U744, Institut Pasteur de Lille and Université Lille-Nord de France, Lille, France; Céline Bellenguez, Ph.D., INSERM, U744, Institut Pasteur de Lille and Université Lille-Nord de France, Lille, France; Valentina Moskvina, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, School of Medicine, Cardiff University, Cardiff, United Kingdom; Rebecca Sims, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, United Kingdom; Denise Harold, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, United Kingdom; Julie Williams, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, United Kingdom; Jean-Charles Lambert, Ph.D., INSERM, U744, Institut Pasteur de Lille and Université Lille-Nord de France, Lille, France; Philippe Amouyel, M.D., Ph.D., INSERM, U744, Institut Pasteur de Lille, CHRU de Lille, and Université Lille-Nord de France, Lille, France; Neill Graff-Radford, M.B., B.Ch., Mayo Clinic, Jacksonville, FL; Alison Goate, Ph.D., Washington University School of Medicine, St. Louis; Rosa Rademakers, Ph.D., Mayo Clinic, Jacksonville, FL; Kevin Morgan, Ph.D., School of Molecular Medical Sciences, University of Nottingham, Nottingham, United Kingdom; John Powell, Ph.D., King's College London Institute of Psychiatry, London; Peter St. George-Hyslop, M.D., Tanz Centre, University of Toronto, Toronto, and Cambridge Institute for Medical Research, University of Cambridge, Cambridge, United Kingdom; Andrew Singleton, Ph.D., National Institute on Aging, National Institutes of Health, Bethesda, MD; and John Hardy, Ph.D., UCL Institute of Neurology, London
1,
John Powell
John Powell, Ph.D.
1Rita Guerreiro, Ph.D., University College London (UCL) Institute of Neurology, London, and National Institute on Aging, National Institutes of Health, Bethesda, MD; Aleksandra Wojtas, M.S., Mayo Clinic, Jacksonville, FL; Jose Bras, Ph.D., UCL Institute of Neurology, London; Minerva M. Carrasquillo, Ph.D., Mayo Clinic, Jacksonville, FL; Ekaterina Rogaeva, Ph.D., Tanz Centre, University of Toronto, Toronto; Elisa Majounie, Ph.D., Mayo Clinic, Jacksonville, FL; Carlos Cruchaga, Ph.D., Washington University School of Medicine, St. Louis; Celeste Sassi, M.D., Institute of Neurology, London, and National Institute on Aging, National Institutes of Health, Bethesda, MD; John S.K. Kauwe, Ph.D., Brigham Young University, Provo, Utah; Michelle K. Lupton, Ph.D., King's College London Institute of Psychiatry, London; Mina Ryten, M.D., Ph.D., UCL Institute of Neurology, London; Kristelle Brown, Ph.D., University of Nottingham, Nottingham, United Kingdom; the EADI Consortium, the GERAD Consortium, the UKBE Consortium, James Lowe, D.M., University of Nottingham, Nottingham, United Kingdom; Perry G. Ridge, M.S., Brigham Young University and the ARUP Institute, Salt Lake City; Monia B. Hammer, M.S., National Institute on Aging, National Institutes of Health, Bethesda, MD; Yosuke Wakutani, M.D., Tanz Centre, University of Toronto, Toronto; Lilinaz Hazrati, M.D., Ph.D., Tanz Centre, University of Toronto, Toronto; Petroula Proitsi, Ph.D., King's College London Institute of Psychiatry, London; Stephen Newhouse, Ph.D., King's College London Institute of Psychiatry, London; Ebba Lohmann, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Nihan Erginel-Unaltuna, Ph.D., Institute for Experimental Medical Research, Istanbul University, Istanbul, Turkey; Christopher Medway, Ph.D., University of Nottingham, Nottingham, United Kingdom; Hasmet Hanagasi, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Claire Troakes, Ph.D., King's College London Institute of Psychiatry, London; Hakan Gurvit, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Basar Bilgic, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Safa Al-Sarraj, M.D., King's College London Institute of Psychiatry and King's College Hospital, London; Bruno Benitez, M.D., Washington University School of Medicine, St. Louis; Breanna Cooper, B.S., Washington University School of Medicine, St. Louis; David Carrell, M.S., Washington University School of Medicine, St. Louis; Murat Emre, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Fanggeng Zou, Ph.D., Mayo Clinic, Jacksonville, FL; Li Ma, M.S., Mayo Clinic, Jacksonville, FL; Melissa E. Murray, Ph.D., Mayo Clinic, Jacksonville, FL; Dennis W. Dickson, M.D., Mayo Clinic, Jacksonville, FL; Steven Younkin, M.D., Ph.D., Mayo Clinic, Jacksonville, FL; Ronald C. Petersen, Ph.D., M.D., Mayo Clinic, Rochester, MN; Christopher D. Corcoran, Sc.D., Utah State University, Logan; Yefei Cai, Ph.D., Washington University School of Medicine, St. Louis; Catarina Oliveira, M.D., Ph.D., University of Coimbra, Coimbra, Portugal; Maria Helena Ribeiro, M.S., University of Coimbra, Coimbra, Portugal; Isabel Santana, M.D., Ph.D., University of Coimbra, Coimbra, Portugal; JoAnn T. Tschanz, Ph.D., Utah State University, Logan; J. Raphael Gibbs, B.Sc., UCL Institute of Neurology, London, and National Institute on Aging, National Institutes of Health, Bethesda, MD; Maria C. Norton, Ph.D., Utah State University, Logan; Iwona Kloszewska, Ph.D., Medical University of Lodz, Lodz, Poland; Patrizia Mecocci, M.D., Ph.D., University of Perugia, Perugia, Italy; Hilkka Soininen, M.D., University of Eastern Finland and Kuopio University Hospital, Kuopio, Finland; Magda Tsolaki, M.D., Aristotle University of Thessaloniki, Thessaloniki, Greece; Bruno Vellas, M.D., Ph.D., UMR INSERM 1027, Hôpitaux de Toulouse, Toulouse, France; Ronald G. Munger, Ph.D., M.P.H., Utah State University, Logan; David M.A. Mann, Ph.D., University of Manchester, Manchester, United Kingdom; Stuart Pickering-Brown, Ph.D., University of Manchester, Manchester, United Kingdom; Simon Lovestone, Ph.D., King's College London Institute of Psychiatry, London; Jonathan Beck, B.Sc., MRC Prion Unit and UCL Institute of Neurology, London; Simon Mead, M.D., MRC Prion Unit and UCL Institute of Neurology, London; John Collinge, M.D., MRC Prion Unit and UCL Institute of Neurology, London; Linda Parsons, M.Phil., UCL Institute of Neurology, London; Jennifer Pocock, Ph.D., UCL Institute of Neurology, London; John C. Morris, M.D., Washington University School of Medicine, St. Louis; Tamas Revesz, M.D., UCL Institute of Neurology, London; Tammaryn Lashley, Ph.D., UCL Institute of Neurology, London; Nick C. Fox, M.D., UCL Institute of Neurology, London; Martin N. Rossor, M.D., UCL Institute of Neurology, London; Benjamin Grenier-Boley, M.Sc., INSERM, U744, Institut Pasteur de Lille and Université Lille-Nord de France, Lille, France; Céline Bellenguez, Ph.D., INSERM, U744, Institut Pasteur de Lille and Université Lille-Nord de France, Lille, France; Valentina Moskvina, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, School of Medicine, Cardiff University, Cardiff, United Kingdom; Rebecca Sims, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, United Kingdom; Denise Harold, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, United Kingdom; Julie Williams, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, United Kingdom; Jean-Charles Lambert, Ph.D., INSERM, U744, Institut Pasteur de Lille and Université Lille-Nord de France, Lille, France; Philippe Amouyel, M.D., Ph.D., INSERM, U744, Institut Pasteur de Lille, CHRU de Lille, and Université Lille-Nord de France, Lille, France; Neill Graff-Radford, M.B., B.Ch., Mayo Clinic, Jacksonville, FL; Alison Goate, Ph.D., Washington University School of Medicine, St. Louis; Rosa Rademakers, Ph.D., Mayo Clinic, Jacksonville, FL; Kevin Morgan, Ph.D., School of Molecular Medical Sciences, University of Nottingham, Nottingham, United Kingdom; John Powell, Ph.D., King's College London Institute of Psychiatry, London; Peter St. George-Hyslop, M.D., Tanz Centre, University of Toronto, Toronto, and Cambridge Institute for Medical Research, University of Cambridge, Cambridge, United Kingdom; Andrew Singleton, Ph.D., National Institute on Aging, National Institutes of Health, Bethesda, MD; and John Hardy, Ph.D., UCL Institute of Neurology, London
1,
Peter St George-Hyslop
Peter St George-Hyslop, M.D.
1Rita Guerreiro, Ph.D., University College London (UCL) Institute of Neurology, London, and National Institute on Aging, National Institutes of Health, Bethesda, MD; Aleksandra Wojtas, M.S., Mayo Clinic, Jacksonville, FL; Jose Bras, Ph.D., UCL Institute of Neurology, London; Minerva M. Carrasquillo, Ph.D., Mayo Clinic, Jacksonville, FL; Ekaterina Rogaeva, Ph.D., Tanz Centre, University of Toronto, Toronto; Elisa Majounie, Ph.D., Mayo Clinic, Jacksonville, FL; Carlos Cruchaga, Ph.D., Washington University School of Medicine, St. Louis; Celeste Sassi, M.D., Institute of Neurology, London, and National Institute on Aging, National Institutes of Health, Bethesda, MD; John S.K. Kauwe, Ph.D., Brigham Young University, Provo, Utah; Michelle K. Lupton, Ph.D., King's College London Institute of Psychiatry, London; Mina Ryten, M.D., Ph.D., UCL Institute of Neurology, London; Kristelle Brown, Ph.D., University of Nottingham, Nottingham, United Kingdom; the EADI Consortium, the GERAD Consortium, the UKBE Consortium, James Lowe, D.M., University of Nottingham, Nottingham, United Kingdom; Perry G. Ridge, M.S., Brigham Young University and the ARUP Institute, Salt Lake City; Monia B. Hammer, M.S., National Institute on Aging, National Institutes of Health, Bethesda, MD; Yosuke Wakutani, M.D., Tanz Centre, University of Toronto, Toronto; Lilinaz Hazrati, M.D., Ph.D., Tanz Centre, University of Toronto, Toronto; Petroula Proitsi, Ph.D., King's College London Institute of Psychiatry, London; Stephen Newhouse, Ph.D., King's College London Institute of Psychiatry, London; Ebba Lohmann, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Nihan Erginel-Unaltuna, Ph.D., Institute for Experimental Medical Research, Istanbul University, Istanbul, Turkey; Christopher Medway, Ph.D., University of Nottingham, Nottingham, United Kingdom; Hasmet Hanagasi, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Claire Troakes, Ph.D., King's College London Institute of Psychiatry, London; Hakan Gurvit, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Basar Bilgic, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Safa Al-Sarraj, M.D., King's College London Institute of Psychiatry and King's College Hospital, London; Bruno Benitez, M.D., Washington University School of Medicine, St. Louis; Breanna Cooper, B.S., Washington University School of Medicine, St. Louis; David Carrell, M.S., Washington University School of Medicine, St. Louis; Murat Emre, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Fanggeng Zou, Ph.D., Mayo Clinic, Jacksonville, FL; Li Ma, M.S., Mayo Clinic, Jacksonville, FL; Melissa E. Murray, Ph.D., Mayo Clinic, Jacksonville, FL; Dennis W. Dickson, M.D., Mayo Clinic, Jacksonville, FL; Steven Younkin, M.D., Ph.D., Mayo Clinic, Jacksonville, FL; Ronald C. Petersen, Ph.D., M.D., Mayo Clinic, Rochester, MN; Christopher D. Corcoran, Sc.D., Utah State University, Logan; Yefei Cai, Ph.D., Washington University School of Medicine, St. Louis; Catarina Oliveira, M.D., Ph.D., University of Coimbra, Coimbra, Portugal; Maria Helena Ribeiro, M.S., University of Coimbra, Coimbra, Portugal; Isabel Santana, M.D., Ph.D., University of Coimbra, Coimbra, Portugal; JoAnn T. Tschanz, Ph.D., Utah State University, Logan; J. Raphael Gibbs, B.Sc., UCL Institute of Neurology, London, and National Institute on Aging, National Institutes of Health, Bethesda, MD; Maria C. Norton, Ph.D., Utah State University, Logan; Iwona Kloszewska, Ph.D., Medical University of Lodz, Lodz, Poland; Patrizia Mecocci, M.D., Ph.D., University of Perugia, Perugia, Italy; Hilkka Soininen, M.D., University of Eastern Finland and Kuopio University Hospital, Kuopio, Finland; Magda Tsolaki, M.D., Aristotle University of Thessaloniki, Thessaloniki, Greece; Bruno Vellas, M.D., Ph.D., UMR INSERM 1027, Hôpitaux de Toulouse, Toulouse, France; Ronald G. Munger, Ph.D., M.P.H., Utah State University, Logan; David M.A. Mann, Ph.D., University of Manchester, Manchester, United Kingdom; Stuart Pickering-Brown, Ph.D., University of Manchester, Manchester, United Kingdom; Simon Lovestone, Ph.D., King's College London Institute of Psychiatry, London; Jonathan Beck, B.Sc., MRC Prion Unit and UCL Institute of Neurology, London; Simon Mead, M.D., MRC Prion Unit and UCL Institute of Neurology, London; John Collinge, M.D., MRC Prion Unit and UCL Institute of Neurology, London; Linda Parsons, M.Phil., UCL Institute of Neurology, London; Jennifer Pocock, Ph.D., UCL Institute of Neurology, London; John C. Morris, M.D., Washington University School of Medicine, St. Louis; Tamas Revesz, M.D., UCL Institute of Neurology, London; Tammaryn Lashley, Ph.D., UCL Institute of Neurology, London; Nick C. Fox, M.D., UCL Institute of Neurology, London; Martin N. Rossor, M.D., UCL Institute of Neurology, London; Benjamin Grenier-Boley, M.Sc., INSERM, U744, Institut Pasteur de Lille and Université Lille-Nord de France, Lille, France; Céline Bellenguez, Ph.D., INSERM, U744, Institut Pasteur de Lille and Université Lille-Nord de France, Lille, France; Valentina Moskvina, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, School of Medicine, Cardiff University, Cardiff, United Kingdom; Rebecca Sims, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, United Kingdom; Denise Harold, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, United Kingdom; Julie Williams, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, United Kingdom; Jean-Charles Lambert, Ph.D., INSERM, U744, Institut Pasteur de Lille and Université Lille-Nord de France, Lille, France; Philippe Amouyel, M.D., Ph.D., INSERM, U744, Institut Pasteur de Lille, CHRU de Lille, and Université Lille-Nord de France, Lille, France; Neill Graff-Radford, M.B., B.Ch., Mayo Clinic, Jacksonville, FL; Alison Goate, Ph.D., Washington University School of Medicine, St. Louis; Rosa Rademakers, Ph.D., Mayo Clinic, Jacksonville, FL; Kevin Morgan, Ph.D., School of Molecular Medical Sciences, University of Nottingham, Nottingham, United Kingdom; John Powell, Ph.D., King's College London Institute of Psychiatry, London; Peter St. George-Hyslop, M.D., Tanz Centre, University of Toronto, Toronto, and Cambridge Institute for Medical Research, University of Cambridge, Cambridge, United Kingdom; Andrew Singleton, Ph.D., National Institute on Aging, National Institutes of Health, Bethesda, MD; and John Hardy, Ph.D., UCL Institute of Neurology, London
1,
Andrew Singleton
Andrew Singleton, Ph.D.
1Rita Guerreiro, Ph.D., University College London (UCL) Institute of Neurology, London, and National Institute on Aging, National Institutes of Health, Bethesda, MD; Aleksandra Wojtas, M.S., Mayo Clinic, Jacksonville, FL; Jose Bras, Ph.D., UCL Institute of Neurology, London; Minerva M. Carrasquillo, Ph.D., Mayo Clinic, Jacksonville, FL; Ekaterina Rogaeva, Ph.D., Tanz Centre, University of Toronto, Toronto; Elisa Majounie, Ph.D., Mayo Clinic, Jacksonville, FL; Carlos Cruchaga, Ph.D., Washington University School of Medicine, St. Louis; Celeste Sassi, M.D., Institute of Neurology, London, and National Institute on Aging, National Institutes of Health, Bethesda, MD; John S.K. Kauwe, Ph.D., Brigham Young University, Provo, Utah; Michelle K. Lupton, Ph.D., King's College London Institute of Psychiatry, London; Mina Ryten, M.D., Ph.D., UCL Institute of Neurology, London; Kristelle Brown, Ph.D., University of Nottingham, Nottingham, United Kingdom; the EADI Consortium, the GERAD Consortium, the UKBE Consortium, James Lowe, D.M., University of Nottingham, Nottingham, United Kingdom; Perry G. Ridge, M.S., Brigham Young University and the ARUP Institute, Salt Lake City; Monia B. Hammer, M.S., National Institute on Aging, National Institutes of Health, Bethesda, MD; Yosuke Wakutani, M.D., Tanz Centre, University of Toronto, Toronto; Lilinaz Hazrati, M.D., Ph.D., Tanz Centre, University of Toronto, Toronto; Petroula Proitsi, Ph.D., King's College London Institute of Psychiatry, London; Stephen Newhouse, Ph.D., King's College London Institute of Psychiatry, London; Ebba Lohmann, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Nihan Erginel-Unaltuna, Ph.D., Institute for Experimental Medical Research, Istanbul University, Istanbul, Turkey; Christopher Medway, Ph.D., University of Nottingham, Nottingham, United Kingdom; Hasmet Hanagasi, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Claire Troakes, Ph.D., King's College London Institute of Psychiatry, London; Hakan Gurvit, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Basar Bilgic, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Safa Al-Sarraj, M.D., King's College London Institute of Psychiatry and King's College Hospital, London; Bruno Benitez, M.D., Washington University School of Medicine, St. Louis; Breanna Cooper, B.S., Washington University School of Medicine, St. Louis; David Carrell, M.S., Washington University School of Medicine, St. Louis; Murat Emre, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Fanggeng Zou, Ph.D., Mayo Clinic, Jacksonville, FL; Li Ma, M.S., Mayo Clinic, Jacksonville, FL; Melissa E. Murray, Ph.D., Mayo Clinic, Jacksonville, FL; Dennis W. Dickson, M.D., Mayo Clinic, Jacksonville, FL; Steven Younkin, M.D., Ph.D., Mayo Clinic, Jacksonville, FL; Ronald C. Petersen, Ph.D., M.D., Mayo Clinic, Rochester, MN; Christopher D. Corcoran, Sc.D., Utah State University, Logan; Yefei Cai, Ph.D., Washington University School of Medicine, St. Louis; Catarina Oliveira, M.D., Ph.D., University of Coimbra, Coimbra, Portugal; Maria Helena Ribeiro, M.S., University of Coimbra, Coimbra, Portugal; Isabel Santana, M.D., Ph.D., University of Coimbra, Coimbra, Portugal; JoAnn T. Tschanz, Ph.D., Utah State University, Logan; J. Raphael Gibbs, B.Sc., UCL Institute of Neurology, London, and National Institute on Aging, National Institutes of Health, Bethesda, MD; Maria C. Norton, Ph.D., Utah State University, Logan; Iwona Kloszewska, Ph.D., Medical University of Lodz, Lodz, Poland; Patrizia Mecocci, M.D., Ph.D., University of Perugia, Perugia, Italy; Hilkka Soininen, M.D., University of Eastern Finland and Kuopio University Hospital, Kuopio, Finland; Magda Tsolaki, M.D., Aristotle University of Thessaloniki, Thessaloniki, Greece; Bruno Vellas, M.D., Ph.D., UMR INSERM 1027, Hôpitaux de Toulouse, Toulouse, France; Ronald G. Munger, Ph.D., M.P.H., Utah State University, Logan; David M.A. Mann, Ph.D., University of Manchester, Manchester, United Kingdom; Stuart Pickering-Brown, Ph.D., University of Manchester, Manchester, United Kingdom; Simon Lovestone, Ph.D., King's College London Institute of Psychiatry, London; Jonathan Beck, B.Sc., MRC Prion Unit and UCL Institute of Neurology, London; Simon Mead, M.D., MRC Prion Unit and UCL Institute of Neurology, London; John Collinge, M.D., MRC Prion Unit and UCL Institute of Neurology, London; Linda Parsons, M.Phil., UCL Institute of Neurology, London; Jennifer Pocock, Ph.D., UCL Institute of Neurology, London; John C. Morris, M.D., Washington University School of Medicine, St. Louis; Tamas Revesz, M.D., UCL Institute of Neurology, London; Tammaryn Lashley, Ph.D., UCL Institute of Neurology, London; Nick C. Fox, M.D., UCL Institute of Neurology, London; Martin N. Rossor, M.D., UCL Institute of Neurology, London; Benjamin Grenier-Boley, M.Sc., INSERM, U744, Institut Pasteur de Lille and Université Lille-Nord de France, Lille, France; Céline Bellenguez, Ph.D., INSERM, U744, Institut Pasteur de Lille and Université Lille-Nord de France, Lille, France; Valentina Moskvina, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, School of Medicine, Cardiff University, Cardiff, United Kingdom; Rebecca Sims, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, United Kingdom; Denise Harold, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, United Kingdom; Julie Williams, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, United Kingdom; Jean-Charles Lambert, Ph.D., INSERM, U744, Institut Pasteur de Lille and Université Lille-Nord de France, Lille, France; Philippe Amouyel, M.D., Ph.D., INSERM, U744, Institut Pasteur de Lille, CHRU de Lille, and Université Lille-Nord de France, Lille, France; Neill Graff-Radford, M.B., B.Ch., Mayo Clinic, Jacksonville, FL; Alison Goate, Ph.D., Washington University School of Medicine, St. Louis; Rosa Rademakers, Ph.D., Mayo Clinic, Jacksonville, FL; Kevin Morgan, Ph.D., School of Molecular Medical Sciences, University of Nottingham, Nottingham, United Kingdom; John Powell, Ph.D., King's College London Institute of Psychiatry, London; Peter St. George-Hyslop, M.D., Tanz Centre, University of Toronto, Toronto, and Cambridge Institute for Medical Research, University of Cambridge, Cambridge, United Kingdom; Andrew Singleton, Ph.D., National Institute on Aging, National Institutes of Health, Bethesda, MD; and John Hardy, Ph.D., UCL Institute of Neurology, London
1,
John Hardy
John Hardy, Ph.D.
1Rita Guerreiro, Ph.D., University College London (UCL) Institute of Neurology, London, and National Institute on Aging, National Institutes of Health, Bethesda, MD; Aleksandra Wojtas, M.S., Mayo Clinic, Jacksonville, FL; Jose Bras, Ph.D., UCL Institute of Neurology, London; Minerva M. Carrasquillo, Ph.D., Mayo Clinic, Jacksonville, FL; Ekaterina Rogaeva, Ph.D., Tanz Centre, University of Toronto, Toronto; Elisa Majounie, Ph.D., Mayo Clinic, Jacksonville, FL; Carlos Cruchaga, Ph.D., Washington University School of Medicine, St. Louis; Celeste Sassi, M.D., Institute of Neurology, London, and National Institute on Aging, National Institutes of Health, Bethesda, MD; John S.K. Kauwe, Ph.D., Brigham Young University, Provo, Utah; Michelle K. Lupton, Ph.D., King's College London Institute of Psychiatry, London; Mina Ryten, M.D., Ph.D., UCL Institute of Neurology, London; Kristelle Brown, Ph.D., University of Nottingham, Nottingham, United Kingdom; the EADI Consortium, the GERAD Consortium, the UKBE Consortium, James Lowe, D.M., University of Nottingham, Nottingham, United Kingdom; Perry G. Ridge, M.S., Brigham Young University and the ARUP Institute, Salt Lake City; Monia B. Hammer, M.S., National Institute on Aging, National Institutes of Health, Bethesda, MD; Yosuke Wakutani, M.D., Tanz Centre, University of Toronto, Toronto; Lilinaz Hazrati, M.D., Ph.D., Tanz Centre, University of Toronto, Toronto; Petroula Proitsi, Ph.D., King's College London Institute of Psychiatry, London; Stephen Newhouse, Ph.D., King's College London Institute of Psychiatry, London; Ebba Lohmann, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Nihan Erginel-Unaltuna, Ph.D., Institute for Experimental Medical Research, Istanbul University, Istanbul, Turkey; Christopher Medway, Ph.D., University of Nottingham, Nottingham, United Kingdom; Hasmet Hanagasi, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Claire Troakes, Ph.D., King's College London Institute of Psychiatry, London; Hakan Gurvit, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Basar Bilgic, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Safa Al-Sarraj, M.D., King's College London Institute of Psychiatry and King's College Hospital, London; Bruno Benitez, M.D., Washington University School of Medicine, St. Louis; Breanna Cooper, B.S., Washington University School of Medicine, St. Louis; David Carrell, M.S., Washington University School of Medicine, St. Louis; Murat Emre, M.D., Faculty of Medicine, Istanbul University, Istanbul, Turkey; Fanggeng Zou, Ph.D., Mayo Clinic, Jacksonville, FL; Li Ma, M.S., Mayo Clinic, Jacksonville, FL; Melissa E. Murray, Ph.D., Mayo Clinic, Jacksonville, FL; Dennis W. Dickson, M.D., Mayo Clinic, Jacksonville, FL; Steven Younkin, M.D., Ph.D., Mayo Clinic, Jacksonville, FL; Ronald C. Petersen, Ph.D., M.D., Mayo Clinic, Rochester, MN; Christopher D. Corcoran, Sc.D., Utah State University, Logan; Yefei Cai, Ph.D., Washington University School of Medicine, St. Louis; Catarina Oliveira, M.D., Ph.D., University of Coimbra, Coimbra, Portugal; Maria Helena Ribeiro, M.S., University of Coimbra, Coimbra, Portugal; Isabel Santana, M.D., Ph.D., University of Coimbra, Coimbra, Portugal; JoAnn T. Tschanz, Ph.D., Utah State University, Logan; J. Raphael Gibbs, B.Sc., UCL Institute of Neurology, London, and National Institute on Aging, National Institutes of Health, Bethesda, MD; Maria C. Norton, Ph.D., Utah State University, Logan; Iwona Kloszewska, Ph.D., Medical University of Lodz, Lodz, Poland; Patrizia Mecocci, M.D., Ph.D., University of Perugia, Perugia, Italy; Hilkka Soininen, M.D., University of Eastern Finland and Kuopio University Hospital, Kuopio, Finland; Magda Tsolaki, M.D., Aristotle University of Thessaloniki, Thessaloniki, Greece; Bruno Vellas, M.D., Ph.D., UMR INSERM 1027, Hôpitaux de Toulouse, Toulouse, France; Ronald G. Munger, Ph.D., M.P.H., Utah State University, Logan; David M.A. Mann, Ph.D., University of Manchester, Manchester, United Kingdom; Stuart Pickering-Brown, Ph.D., University of Manchester, Manchester, United Kingdom; Simon Lovestone, Ph.D., King's College London Institute of Psychiatry, London; Jonathan Beck, B.Sc., MRC Prion Unit and UCL Institute of Neurology, London; Simon Mead, M.D., MRC Prion Unit and UCL Institute of Neurology, London; John Collinge, M.D., MRC Prion Unit and UCL Institute of Neurology, London; Linda Parsons, M.Phil., UCL Institute of Neurology, London; Jennifer Pocock, Ph.D., UCL Institute of Neurology, London; John C. Morris, M.D., Washington University School of Medicine, St. Louis; Tamas Revesz, M.D., UCL Institute of Neurology, London; Tammaryn Lashley, Ph.D., UCL Institute of Neurology, London; Nick C. Fox, M.D., UCL Institute of Neurology, London; Martin N. Rossor, M.D., UCL Institute of Neurology, London; Benjamin Grenier-Boley, M.Sc., INSERM, U744, Institut Pasteur de Lille and Université Lille-Nord de France, Lille, France; Céline Bellenguez, Ph.D., INSERM, U744, Institut Pasteur de Lille and Université Lille-Nord de France, Lille, France; Valentina Moskvina, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, School of Medicine, Cardiff University, Cardiff, United Kingdom; Rebecca Sims, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, United Kingdom; Denise Harold, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, United Kingdom; Julie Williams, Ph.D., MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, United Kingdom; Jean-Charles Lambert, Ph.D., INSERM, U744, Institut Pasteur de Lille and Université Lille-Nord de France, Lille, France; Philippe Amouyel, M.D., Ph.D., INSERM, U744, Institut Pasteur de Lille, CHRU de Lille, and Université Lille-Nord de France, Lille, France; Neill Graff-Radford, M.B., B.Ch., Mayo Clinic, Jacksonville, FL; Alison Goate, Ph.D., Washington University School of Medicine, St. Louis; Rosa Rademakers, Ph.D., Mayo Clinic, Jacksonville, FL; Kevin Morgan, Ph.D., School of Molecular Medical Sciences, University of Nottingham, Nottingham, United Kingdom; John Powell, Ph.D., King's College London Institute of Psychiatry, London; Peter St. George-Hyslop, M.D., Tanz Centre, University of Toronto, Toronto, and Cambridge Institute for Medical Research, University of Cambridge, Cambridge, United Kingdom; Andrew Singleton, Ph.D., National Institute on Aging, National Institutes of Health, Bethesda, MD; and John Hardy, Ph.D., UCL Institute of Neurology, London
1;
The Alzheimer Genetic Analysis Group1,*