Abstract
Next-generation sequencing of antibody transcripts from HIV-1–infected individuals with broadly neutralizing antibodies could provide an efficient means for identifying somatic variants and characterizing their lineages. Here, we used 454 pyrosequencing and identity/divergence grid sampling to analyze heavy- and light-chain sequences from donor N152, the source of the broadly neutralizing antibody 10E8. We identified variants with up to 28% difference in amino acid sequence. Heavy- and light-chain phylogenetic trees of identified 10E8 variants displayed similar architectures, and 10E8 variants reconstituted from matched and unmatched phylogenetic branches displayed significantly lower autoreactivity when matched. To test the generality of phylogenetic pairing, we analyzed donor International AIDS Vaccine Initiative 84, the source of antibodies PGT141–145. Heavy- and light-chain phylogenetic trees of PGT141–145 somatic variants also displayed remarkably similar architectures; in this case, branch pairings could be anchored by known PGT141–145 antibodies. Altogether, our findings suggest that phylogenetic matching of heavy and light chains can provide a means to approximate natural pairings.
Keywords: antibody-affinity maturation, antibodyomics, B-cell ontogeny, DNA sequencing, immunological tolerance
Approximately 20% of HIV-1–infected individuals develop antibodies capable of neutralizing diverse isolates of HIV-1 (1–3), and monoclonal antibodies identified from these individuals are revolutionizing our understanding of how the human immune system can recognize highly variable antigens (reviewed in refs. 4 and 5). Currently, such identification is occurring primarily through the sequencing of antibody heavy and light chains from individually sorted B cells selected by antigen-specific probes (6, 7) or by direct assessment of neutralization from secreted IgG (8–10). Highly effective monoclonal neutralizers have now been identified by these techniques from more than 20 donors (reviewed in ref. 11). Generally, only a few monoclonal antibodies from each donor have been identified, although it is possible to identify substantially more (6, 12). Indeed, next-generation sequencing technologies (13–18) seem to offer an efficient means for identifying thousands of somatic variants (19). The massively parallel sequencing that is used by such technologies, however, leads to the loss of information on individual pairings of heavy and light chain and as such, has made it a challenge to discern native antibodies (with naturally paired heavy and light chains) in next-generation sequencing data.
We investigated the identification and functional pairing of heavy and light chains determined by 454 pyrosequencing, which currently allows ∼1,000,000 sequences of 400–500 bp from parallel sequencing on a single chip (19, 20). Beginning from a single HIV-1 neutralizing antibody (10E8), we identified clonal variants of heavy and light chain that we assessed for function by pairing with the WT 10E8 complementary chain. Phylogenetic trees of heavy and light chains revealed similar branch topologies relative to WT 10E8 sequences, thereby allowing branches of the heavy- and light-chain phylogenetic trees to be matched based on their relative distances from 10E8. By assessing a matrix of antibodies reconstituted from matched and mismatched branches for neutralization of HIV-1 and reactivity with self-antigens, we could quantify the use of phylogenetic pairing on function. Lastly, to establish the generality of phylogenetic pairing, we examined B-cell transcripts from donor International AIDS Vaccine Initiative (IAVI) 84, the source of the broadly neutralizing antibodies PGT141–145. As with donor N152, the heavy- and light-chain phylogenetic trees were remarkably similar. Bioinformatics coupled to functional assessment of next-generation sequencing-determined antibody transcripts can thus furnish a genetic record for clonal families of broadly neutralizing antibodies, including effective HIV-1 neutralizers, with phylogenetic matching of heavy and light chains providing a means to approximate natural pairings.
Results
Next-Generation Sequencing of B-Cell Transcripts from Donor N152.
The broadly neutralizing antibody 10E8 was recently identified in the HIV-1–infected donor N152 (10). 10E8 recognizes a helix-turn-helix in the membrane-proximal external region (MPER) of the transmembrane-spanning HIV-1 gp41 glycoprotein and neutralizes 98% of diverse HIV-1 isolates at a geometric mean IC50 of 0.22 μg/mL (10). Unlike other HIV-1–neutralizing antibodies that target the MPER (21), the 10E8 antibody does not react with self-antigens (10). Its extraordinary breadth and lack of autoreactivity have raised interest in using it as an anti–HIV-1 prophylactic and in understanding its lineage for use as a vaccine template. The heavy chain of antibody 10E8 derives from IgHV3-15 and IgHJ1, has a third complementarity-determining region (CDR H3) of 22 aa, and displays a somatic mutation level of 21% (10). The light chain of antibody 10E8 derives from IgVL3-19 and IgLJ3, has a CDR L3 of 12 aa, and displays a somatic mutation level of 14% (10). We performed next-generation sequencing of donor B-cell transcripts using PCR to amplify IgG heavy-chain sequences from the IgHV3 family and amplify IgG light-chain sequences from the IgVL3 family. mRNA from an estimated 5 million peripheral blood mononuclear cells (PBMCs) per sequencing reaction was used for reverse transcription to produce template cDNA (19), and in both cases, we used primers (12) that were upstream from the start of the V-gene leader sequences and downstream from the end of the J chain (Table S1); although there should be ∼500,000 B cells per sequencing reaction (10% of PBMCs), the total number of B-cell transcripts is less clear, and oversampling may occur (SI Materials and Methods, Potential Oversampling of B-Cell Transcripts). Overall, Roche 454 pyrosequencing (20) provided 843,084 heavy-chain reads and 1,219,214 light-chain reads for donor N152.
Bioinformatics Identification of 10E8-Related Transcripts.
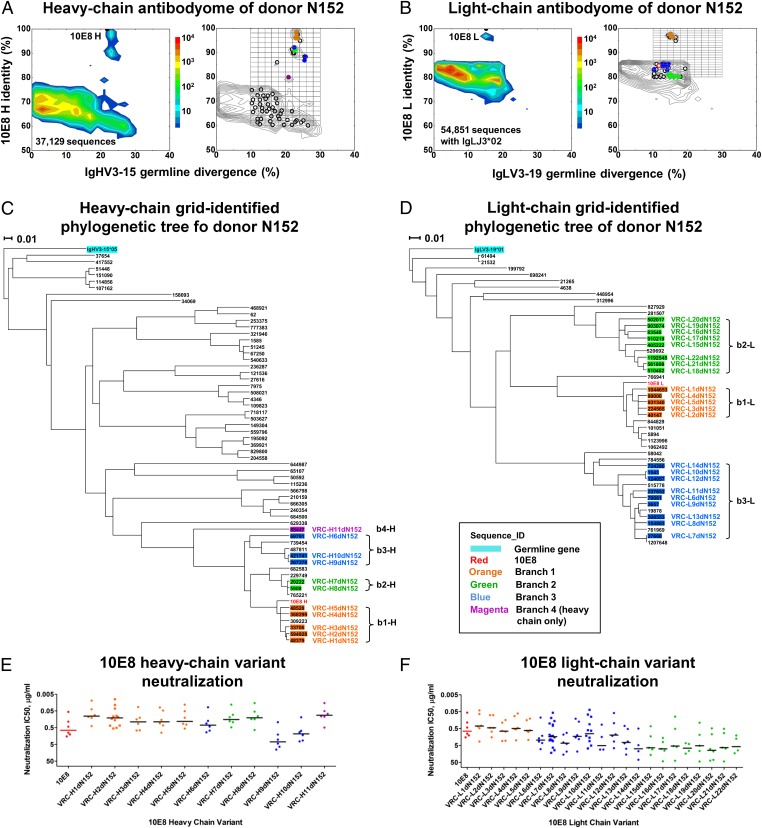
After primary analysis using a previously described bioinformatics pipeline (10, 19) (Figs. S1 and S2), 37,129 full-length heavy-chain sequences were assigned to the IgHV3-15 allelic family, and 54,851 full-length light-chain sequences were assigned to the IgVL3-IgJ3 allelic families. Processed sequences were analyzed for identity to 10E8 and divergence from the unmutated V genes, and their frequencies were displayed on identity/divergence plots (10, 19) (Fig. 1 A, Left and B, Left). For the heavy chain, several well-separated islands of high identity and about 25% divergence were observed, and for the light chain, a single well-separated island of high identity and about 15% divergence were observed. Grid-based sampling (19) of the high identity–divergence region initially selected 60 heavy-chain sequences and 48 light-chain sequences. After manual inspection to remove erroneous or redundant sequences, 59 heavy chains and 45 light chains remained, which were analyzed for their phylogenetic relationship to 10E8 (Fig. 1 C and D).
Fig. 1.
Identification of somatic variants of antibody 10E8 by next-generation sequencing and grid sampling. (A) 10E8 identity/divergence plots of donor N152 heavy-chain antibodyome (Left) with grid sampling (Right). Identity to 10E8 is shown on the vertical axis, and divergence from germ-line V gene origin is plotted on the horizontal axis, with frequency of antibodies shown as a heat map. Grid sampling is shown, with selected antibodies that either did not express or bind to MPER as open circles and selected antibodies that did bind as solid circles colored according to their phylogenetic distance from 10E8 in C. (B) 10E8 identity/divergence plots of donor N152 light-chain antibodyome (Left) with grid sampling (Right). Axes and coloring are the same as in A except for the open red circle, which represents an antibody that failed to express at the 250-mL scale. (C and D) Phylogenetic trees of grid-identified variants for heavy chain in C and light chain in D. (E and F) 10E8 and 10E8 variant neutralization of six HIV-1 isolates assessed in duplicate for heavy-chain variants, E, and light-chain variants, F. The average IC50 values of gVRC-H1dN152:10E8L and gVRC-H11dN152:10E8L were roughly sixfold improved over the original template 10E8. Variants are arranged and named by their genetic distance from 10E8, and colored relative to their phylogenetic segregation. Horizontal bars represent median IC50 values for each variant. Bars representing 0.01 changes per nucleotide site are shown.
Functional Assessment of Grid-Selected Transcripts.
Grid-selected transcripts were synthesized and reconstituted with their complementary WT 10E8 chain in a 96-well format transient transfection expression system. ELISAs of the expressed 10E8 variants identified 11 heavy chains and 22 light chains, which when paired with the partner WT 10E8 chains bound to a peptide corresponding to the entire MPER of HIV-1 gp41 (Fig. 1 A, Right and B, Right and Fig. S3). Expression was then scaled to 250 mL, and all but one light-chain sequence provided sufficient antibody to allow neutralization to be assessed. On a panel of six HIV-1 isolates, up to approximately fivefold increases in neutralization potency relative to 10E8 were observed (Fig. 1 E and F and Table S2).
Maturation Patterns in 10E8-Related Transcripts.
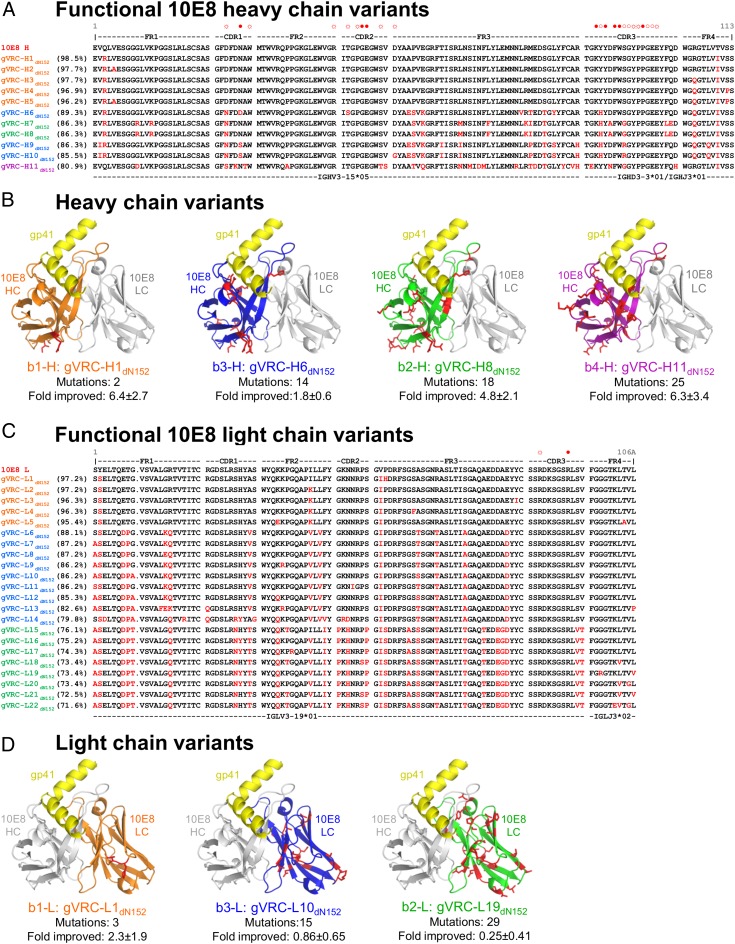
Functional 10E8-like heavy chains were derived from three distinct islands on the identity/divergence plots (Fig. 1A) and exhibited sequences and mutational patterns consistent with a common clonal origin (Fig. 2A). Mutations clustered in CDR H1 and H3 and also, the first, third, and fourth framework regions (FRs; FR-1, -3, and -4). The most divergent sequence, gVRC-H11dN152, had 25 amino acid changes, corresponding to 19.1% difference from the WT 10E8 heavy chain. Often, mutations were distal from the MPER-interacting region but still affected neutralization; for example, in gVRC-H1dN152 (Fig. 2B), mutations were observed at the distal portion of the heavy chain variable (VH) domain, but neutralization improved by 6.4 ± 2.7-fold. Functional 10E8-like light chains derived from several regions of the identity/divergence plots, including a single distinct island and several regions overlapping the primary light-chain population (Fig. 1B). Like heavy chain, functional light chains exhibited mutational patterns consistent with a common clonal origin (Fig. 2C). Mutations clustered in CDR L1 and L2 regions and all of the FRs. The most divergent sequence, gVRC-L22dN152, had 31 amino acid changes, corresponding to 28.4% difference from the WT 10E8 light chain. As with heavy chain, light-chain mutations were often distal from the MPER-contacting region (Fig. 2D). Unlike the heavy chain, where several variants showed improved potency, most light-chain variants showed reduced neutralization potency.
Fig. 2.
Sequences and modeled structures of 10E8 variants that neutralize HIV-1. (A) Heavy-chain sequences. Sequences are arranged by genetic distance from 10E8 and colored according to their phylogenetic segregation, with sequence changes from 10E8 highlighted in red. Framework and CDR residues are labeled along with residues that interact with the gp41 MPER epitope (open circle, main chain interactions; open circle with rays, side chain interactions; solid circle, both main chain and side chain interactions). (B) Modeled structures of heavy-chain variants in complex with gp41 epitope. The most potent neutralizers from each 10E8 phylogenetic heavy chain subgroup (with heavy chains colored according to phylogenetic segregation as in Fig. 1B) were threaded onto the structure of WT 10E8 in complex with the MPER region of HIV-1 gp41 (yellow). Structures are displayed as Cα-ribbons, with amino acid side chains that vary from WT 10E8 highlighted in red stick representation. (C) Light-chain sequences. Sequences are arranged by genetic distance from 10E8 and colored according to their phylogenetic segregation, with sequence changes from 10E8 highlighted in red. Framework and CDR residues are labeled along with residues that interact with the gp41 MPER epitope (as described in A). (D) Modeled structures of light-chain variants in complex with gp41 epitope. The most potent neutralizers from each 10E8 phylogenetic light-chain subgroup (with light chains colored according to phylogenetic segregation as in Fig. 1D) were threaded onto the structure of WT 10E8 in complex with the MPER region of HIV-1 gp41 (yellow). Structures are displayed as Cα-ribbons, with amino acid side chains that vary from 10E8 highlighted in red stick representation.
Phylogeny-Based Pairing of Antibody Heavy and Light Chains.
Although functional, the 10E8 variants reconstituted with 10E8 WT complementary chains do not represent natural pairs. As described above, a drawback of the next-generation sequencing approach to antibody identification is that the parallel unlinked sequencing of heavy and light chains loses critical information related to specific pairings of heavy and light chains. We therefore asked whether an evolution-based analysis could provide sufficient information to recapitulate approximate natural pairings. In principle, the maturation/evolution of heavy and light chains should be linked because of their physical association as proteins, the presence of their evolving genes in the same cells subject to the same enzymatic mutation processes, and the requirement for cooperative structural change in response to the same immunogen. Furthermore, the sampling of paired heavy and light chains in a single cDNA library of mRNA population of antibody transcripts should be highly correlated, because they originate from the same cells. In bioinformatics analyses, such coevolution and similarity in sampling should lead to correlations in both topologies and frequencies of corresponding heavy- and light-chain branches of phylogenetic trees.
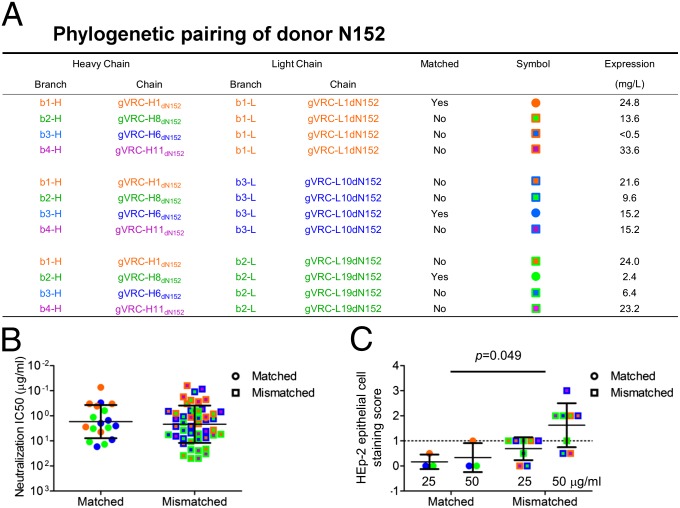
Phylogenetic analysis of the grid-selected experimentally tested 10E8 heavy and light chains found most of the neutralizing antibodies to populate three branches close to and including template 10E8 (Fig. 1 C and D). Branch b1-H for the heavy chain (or b1-L for the light chain) contained 10E8, branch b2-H (b2-L) was the closest branch to b1-H, and branch b3-H (b3-L) was the next closest branch. A single neutralizing sequence (gVRC-H11dN152) occupied a more distant branch (b4-H). Because 454 pyrosequencing produces, on average, about five errors per variable antibody domain (10, 22), we chose the most potent antibody from each branch as representative; therefore, the most functional antibody would likely have the least 454 error-related impairment. We reconstituted 12 antibodies, comprising a complete matrix of heavy-/light-chain pairs from the four heavy-chain branches and the three light-chain branches (Fig. 3A); 11 of 12 reconstituted antibodies expressed sufficient levels of IgG to assess neutralization, which was performed on the same panel of six HIV-1 isolates used to test the grid-identified 10E8 variants (Table S3). All 11 expressed antibodies were neutralizing. Heavy- and light-chain pairings that matched phylogenetic distance from 10E8 (e.g., b1-H to b1-L, b2-H to b2-L, and b3-H to b3-L) were slightly more potent, on average, than mismatched pairings, but the difference was not statistically significant (Fig. 3B).
Fig. 3.
Pairing of phylogenetic branches of heavy- and light-chain variants of 10E8. (A) Phylogenetic branch matching. From the phylogenetic trees of grid-identified antibodies (Fig. 1 C and D), branches were named based on phylogenetic distance from 10E8 (b1-H for heavy and b1-L for light for the branch containing 10E8) and in descending order [b2-H (b2-L), b3-H (b3-L), and b4-H for the farthest branch]. The variant from each branch that displayed the most potent neutralization (lowest median IC50) with a 10E8 WT partner (Fig. 1 E and F) was chosen, and a full matrix of 12 antibodies was reconstituted. (B) HIV-1 neutralization. Neutralization was assessed on six isolates for 10E8 variants from matched and mismatched branch pairings. (C) HEp-2 epithelial cell staining. Autoreactivity was assessed with HEp-2 epithelial cell staining for 10E8 variants from matched and mismatched branch pairings. The dotted line represents the threshold for autoreactivity; HEp-2 epithelial cell staining scores below 1.0 are not considered autoreactive. Measurements were made at antibody concentrations of 25 and 50 μg/mL, as indicated. P value, 0.049 in this case, based on comparison of autoreactivity between matched and mismatched antibodies when both 25- and 50-μg/mL data are used in a two-way ANOVA.
Antibody Pairing and Autoreactivity.
We next tested matched and mismatched antibody pairings for reactivity with self antigens (Tables S4 and S5). Notably, the matched pairings showed significantly lower HEp-2 epithelial cell staining (P = 0.049) (Figs. 3C and Fig. S4). Assessment of reactivity with other self antigens, including cardiolipin and a panel of anti-nuclear antigens (23–25), revealed that matched antibodies trended to lower mean reactivity (in 6/6 antibody doses for cardiolipin and 35/36 antibody doses for anti-nuclear antigens) but did not reach statistical significance, likely because mismatched antibodies exhibited a broad range of reactivities (Fig. S5 and Tables S4 and S5).
Together, the results show that with 10E8 and donor N152, (i) identity/divergence grid sampling can be used to identify somatic variants, (ii) phylogenetic tree architectures and distances can be used to approximate natural pairings, and (iii) antibodies paired by phylogenetic matching exhibit less autoreactivity. Such reduced autoreactivity is likely related to in vivo selection that natural antibodies undergo (26), observed here within a single clonal lineage. With an antibody like 10E8, which may mechanistically be more prone to autoreactivity than other antibodies (10, 27), recapitulation of natural antibody-chain pairings may be critical for isolating potential therapeutic antibodies.
Next-Generation Sequencing and Phylogenetic Pairing of B-Cell Transcripts from Donor IAVI 84.
To test the generality of phylogenetic pairing, we performed next-generation sequencing on B-cell transcripts from donor IAVI 84, the source of the broadly neutralizing antibodies PGT141–145 (8). The clonally related PGT141–145 recognize an N-linked glycan at residue 160 in the V1/V2 region of the HIV-1 gp120 envelope glycoprotein, with residues in strand C of V1/V2 contributing important contacts (28, 29). The most potent of these five antibodies, PGT145, neutralizes ∼80% of diverse HIV-1 isolates at a median IC50 of 0.29 μg/mL, with PGT141–144 neutralizing 30–50% of HIV-1 at median IC50 values of 0.21–2.06 μg/mL (8). The heavy chains of PGT141–145 derive from IgHV1–8, have CDR H3s of 31–32 aa, and display somatic mutation levels of 16–17%, whereas their light chains derive from IgKV2–28, have CDR L3s of 9 aa, and display somatic mutation levels of 13–17% (8). As with donor N152, we performed 454 pyrosequencing of donor B-cell transcripts using PCR to amplify IgG heavy-chain sequences from the IgHV1 family and amplify IgG light-chain sequences from the IgKV2 family (Table S6). In total, 31,324 full-length heavy-chain sequences with IgHV1–8 origin and 72,397 full-length light-chain sequences with IgKV2–28 origin were identified (Figs. S6 and S7).
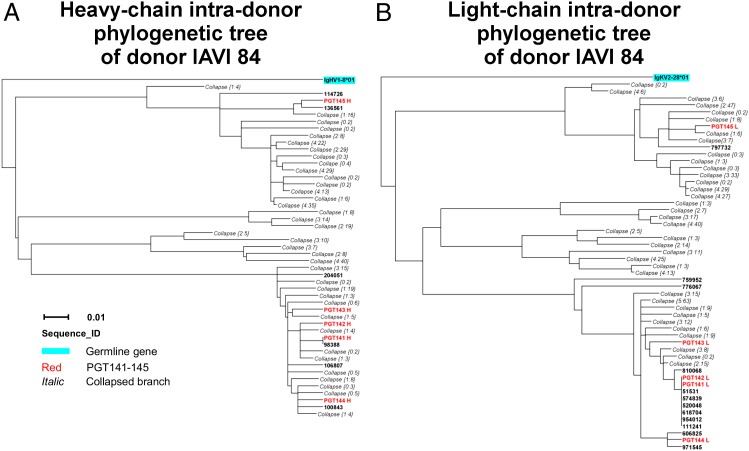
To identify heavy or light chains related to the known PGT141–145 antibodies, we performed intradonor phylogenetic analysis (20). Intradonor phylogenetic analysis uses the same procedure as cross-donor phylogenetic analysis, except that the template antibodies are from the same donor (intradonor) rather than added exogenously (cross-donor) (19). In total, this procedure identified 377 heavy-chain members and 481 light-chain members of the PGT141–145 clonal lineage (SI Materials and Methods and Fig. S8). We used these sequences to construct phylogenetic trees for the variable domains of heavy and light chains of PGT141–145 (Fig. 4).
Fig. 4.
Phylogenetic trees of PGT141–145 somatic variants from donor IAVI 84. Maximum likelihood trees of sequences identified by intradonor phylogenetic analysis from donor IAVI 84, along with five known antibodies from this donor (PGT141–145), are rooted by their respective germ-line genes for both heavy chains (A) and light chains (B). Bars representing 0.01 changes per nucleotide site are shown.
In the heavy-chain dendrogram, antibodies PGT141–144 were positioned on closely related branches, whereas antibody PGT145 was positioned on a separate distant branch (Fig. 4A). In the light-chain dendrogram, antibodies PGT141–144 were also positioned on closely related branches, whereas antibody PGT145 was also positioned on a separate distant branch (Fig. 4B), illustrating the expected correlation in evolution between paired heavy and light chains. The numbers of sequences associated with these distinct heavy- and light-chain dendrogram branches were also approximately correlated: the PGT141–144 branch was populated in heavy- and light-chain trees with 1.14 and 1.62 times the number of sequences as the PGT145 branch. The positions of inferred maturation intermediates—at nodal points of heavy- and light-chain phylogenetic trees—also seemed to be correlated, suggestive of similarities throughout the phylogenetic tree architecture.
Discussion
A key aspect of this study is the bioinformatics prediction of heavy- and light-chain pairing based on the architectures of phylogenetic trees of heavy- and light-chain sequences within a clonal family. Although natural pairings are lost with current next-generation sequencing, our work suggests that it is possible to approximate them by using topological similarities between antibody heavy- and light-chain phylogenetic trees. We note that branch topology is sensitive to small changes in sequence. In the case of PGT141–145, the five naturally paired antibodies provided anchors with which to pair architectures of heavy and light phylogenetic trees. In the case of 10E8, natural pairings did not seem to yield enhanced neutralization potency relative to the total combinatorial possibilities of all heavy- and light-chain pairings, but other functional properties, such as autoreactivity, seemed to be dependent on matching phylogenetic branches. It will be interesting to see if antibodies are selected in vivo less by differences in HIV-1 Env affinity and neutralization potency than reductions in autoreactivity.
Our results also provide examples of how to populate phylogenetic trees from next-generation sequencing data by either grid searching coupled to antibody synthesis and experimental assessment (which was done to identify 10E8 somatic variants) or an exclusively computational means, such as lineage analysis or phylogenetic sieving (which was done to identify PGT141–145 somatic variants). We previously used similar methods to identify somatic variants for the CD4 binding site antibody VRC-PG04 and the N332-directed antibodies PGT135–137 (19, 20). In the current experiment, grid sampling did not identify a light-chain branch corresponding to the heavy-chain b4-H branch (likely because the b4-L light-chain branch did not form a separate resolved island and the sampling grid did not extend to low enough identity relative to 10E8) (Fig. 1B). A combination of grid searching, computational analysis, and functional assessment may prove to be a superior means of identifying somatic variants.
Overall, our work shows how—from a single founder sequence, such as for 10E8, or a handful of antibodies, such as for PGT141–145—phylogeny-based bioinformatics analyses of antibody-transcript sequences obtained by next-generation sequencing techniques can both populate a clonal lineage and approximate heavy/light-chain natural pairings (Figs. 1 C and D and 4). It remains to be seen whether such phylogenetic analyses from cross-sectional data are sufficient to reveal the initial recombinant and chronological order of somatic mutations that produced a broad HIV-1–neutralizing antibody. With both 10E8 and PGT141–145, next-generation sequencing-inferred lineages extended less than halfway to the initial recombinant, suggesting either substantially greater coverage (e.g., starting with 500 million PBMCs) or longitudinal sampling (e.g., monthly from time of infection) will be required.
Materials and Methods
Appropriate informed consent and institutional review board approval were obtained for the use of Donors N152 and IAVI 84 samples. A cDNA library of B-cell transcripts was prepared from 33 million PBMCs. V gene-specific primers were used to amplify 10E8-related transcripts, which were subjected to 454 pyrosequencing and analyzed with the Antibodyomics1.0 pipeline. The Antibodyomics1.0 pipeline is available upon request from J.Z., L.S., or P.D.K. Similar methods were followed with IAVI 84. Transcripts were synthesized and expressed by transient transfection of 293F cells in either 96-well microplate or 250-mL formats. Functional analysis used ELISA assessment of MPER–peptide binding, HIV-1 neutralization, and autoreactivity assays. Detailed materials and methods and complete references can be found in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank H. Coleman, M. Park, B. Schmidt, and A. Young for 454 pyrosequencing at the National Institutes of Health Intramural Sequencing Center (NISC); J. Huang, L. Laub, and M. Connors for donor N152 materials and sequence of 10E8; J. Stuckey for assistance with figures; and Rahul Kohli and members of the Structural Biology Section and Structural Bioinformatics Core, Vaccine Research Center, for discussions or comments on the manuscript. Support for this work was provided by the Intramural Research Program of the Vaccine Research Center, National Institute of Allergy and Infectious Diseases; the National Human Genome Research Institute, National Institutes of Health; the International AIDS Vaccine Initiative; and Center for HIV/AIDS Vaccine Immunology-Immunogen Design Grant UM1 AI100645 (to B.F.H.) from the Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. KC754704-KC754736). The data reported in this paper have been deposited in the NCBI Sequence Read Archive (SRA) (accession no. SRP018335).
2A complete list of the National Institutes of Health Intramural Sequencing Center (NISC) Comparative Sequencing Program can be found in SI Materials and Methods.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1219320110/-/DCSupplemental.
References
- 1.Binley JM, et al. Profiling the specificity of neutralizing antibodies in a large panel of plasmas from patients chronically infected with human immunodeficiency virus type 1 subtypes B and C. J Virol. 2008;82(23):11651–11668. doi: 10.1128/JVI.01762-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li Y, et al. Broad HIV-1 neutralization mediated by CD4-binding site antibodies. Nat Med. 2007;13(9):1032–1034. doi: 10.1038/nm1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Simek MD, et al. Human immunodeficiency virus type 1 elite neutralizers: Individuals with broad and potent neutralizing activity identified by using a high-throughput neutralization assay together with an analytical selection algorithm. J Virol. 2009;83(14):7337–7348. doi: 10.1128/JVI.00110-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kwong PD, Wilson IA. HIV-1 and influenza antibodies: Seeing antigens in new ways. Nat Immunol. 2009;10(6):573–578. doi: 10.1038/ni.1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burton DR, Poignard P, Stanfield RL, Wilson IA. Broadly neutralizing antibodies present new prospects to counter highly antigenically diverse viruses. Science. 2012;337(6091):183–186. doi: 10.1126/science.1225416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scheid JF, et al. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature. 2009;458(7238):636–640. doi: 10.1038/nature07930. [DOI] [PubMed] [Google Scholar]
- 7.Wu X, et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science. 2010;329(5993):856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walker LM, et al. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011;477(7365):466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walker LM, et al. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326(5950):285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang J, et al. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature. 2012;491(7424):406–412. doi: 10.1038/nature11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kwong PD, Mascola JR. Human antibodies that neutralize HIV-1: Identification, structures, and B cell ontogenies. Immunity. 2012;37(3):412–425. doi: 10.1016/j.immuni.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scheid JF, et al. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science. 2011;333(6049):1633–1637. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weinstein JA, Jiang N, White RA, 3rd, Fisher DS, Quake SR. High-throughput sequencing of the zebrafish antibody repertoire. Science. 2009;324(5928):807–810. doi: 10.1126/science.1170020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reddy ST, et al. Monoclonal antibodies isolated without screening by analyzing the variable-gene repertoire of plasma cells. Nat Biotechnol. 2010;28(9):965–969. doi: 10.1038/nbt.1673. [DOI] [PubMed] [Google Scholar]
- 15.Ravn U, et al. By-passing in vitro screening—next generation sequencing technologies applied to antibody display and in silico candidate selection. Nucleic Acids Res. 2010;38(21):e193. doi: 10.1093/nar/gkq789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheung WC, et al. A proteomics approach for the identification and cloning of monoclonal antibodies from serum. Nat Biotechnol. 2012;30(5):447–452. doi: 10.1038/nbt.2167. [DOI] [PubMed] [Google Scholar]
- 17.Reddy ST, Georgiou G. Systems analysis of adaptive immunity by utilization of high-throughput technologies. Curr Opin Biotechnol. 2011;22(4):584–589. doi: 10.1016/j.copbio.2011.04.015. [DOI] [PubMed] [Google Scholar]
- 18.Fischer N. Sequencing antibody repertoires: The next generation. MAbs. 2011;3(1):17–20. doi: 10.4161/mabs.3.1.14169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu X, et al. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science. 2011;333(6049):1593–1602. doi: 10.1126/science.1207532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu J, et al. (2012) Somatic populations of PGT135-137 HIV-1-neutralizing antibodies identified by 454 pyrosequencing and bioinformatics. Front Microbiol. 3(2012):315. [DOI] [PMC free article] [PubMed]
- 21.Haynes BF, et al. Cardiolipin polyspecific autoreactivity in two broadly neutralizing HIV-1 antibodies. Science. 2005;308(5730):1906–1908. doi: 10.1126/science.1111781. [DOI] [PubMed] [Google Scholar]
- 22.Prabakaran P, Streaker E, Chen W, Dimitrov DS. (2011) 454 antibody sequencing—error characterization and correction. BMC Res Notes. 4(2011):404. [DOI] [PMC free article] [PubMed]
- 23.Ichikawa K, Khamashta MA, Koike T, Matsuura E, Hughes GR. beta 2-Glycoprotein I reactivity of monoclonal anticardiolipin antibodies from patients with the antiphospholipid syndrome. Arthritis Rheum. 1994;37(10):1453–1461. doi: 10.1002/art.1780371008. [DOI] [PubMed] [Google Scholar]
- 24.Vismara A, et al. Relationship between anti-cardiolipin and anti-endothelial cell antibodies in systemic lupus erythematosus. Clin Exp Immunol. 1988;74(2):247–253. [PMC free article] [PubMed] [Google Scholar]
- 25.Lafer EM, et al. Polyspecific monoclonal lupus autoantibodies reactive with both polynucleotides and phospholipids. J Exp Med. 1981;153(4):897–909. doi: 10.1084/jem.153.4.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goodnow CC, et al. Altered immunoglobulin expression and functional silencing of self-reactive B lymphocytes in transgenic mice. Nature. 1988;334(6184):676–682. doi: 10.1038/334676a0. [DOI] [PubMed] [Google Scholar]
- 27.Haynes BF, Moody MA, Verkoczy L, Kelsoe G, Alam SM. Antibody polyspecificity and neutralization of HIV-1: A hypothesis. Hum Antibodies. 2005;14(3–4):59–67. [PMC free article] [PubMed] [Google Scholar]
- 28.McLellan JS, et al. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature. 2011;480(7377):336–343. doi: 10.1038/nature10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Doria-Rose NA, et al. (2012) A short segment of the HIV-1 gp120 V1/V2 region is a major determinant of resistance to V1/V2 neutralizing antibodies. J Virol. 86(15):8319–8323. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.