Abstract
In the fission yeast Schizosaccharomyces pombe, three genes that function in the RNA interference (RNAi) pathway, ago1+, dcr1+, and rdp1+, have recently been shown to be important for timely formation of heterochromatin and accurate chromosome segregation. In the present study, we present evidence that null mutants for ago1+ and dcr1+ but not rdp1+, exhibit abnormal cytokinesis, cell cycle arrest deficiencies, and mating defects. Subsequent analyses showed that ago1+ and dcr1+ are required for regulated hyperphosphorylation of Cdc2 when encountering genotoxic insults. Because rdp1+ is dispensable for this process, the functions of ago1+ and dcr1+ in this pathway are presumably independent of their roles in RNAi-mediated heterochromatin formation and chromosome segregation. This was further supported by the finding that ago1+ is a multicopy suppressor of the S-M checkpoint deficiency and cytokinesis defects associated with loss of Dcr1 function, but not for the chromosome segregation defects of this mutant. Accordingly, we conclude that Dcr1-dependent production of small interfering RNAs is not required for enactment and/or maintenance of certain cell cycle checkpoints and that Ago1 and Dcr1 functionally diverge from Rdp1 to control cell cycle events in fission yeast. Finally, exogenous expression of hGERp95/EIF2C2/hAgo2, a human Ago1 homolog implicated in posttranscriptional gene silencing, compensated for the loss of ago1+ function in S. pombe. This suggests that PPD proteins may also be important for regulation of cell cycle events in higher eukaryotes.
INTRODUCTION
PAZ Piwi Domain (PPD) proteins belong to a family of highly conserved proteins found in all multicellular organisms and in the fission yeast Schizosaccharomyces pombe. They are not present in the budding yeast Saccharomyces cerevisiae or bacteria. Family members are defined by the presence of two signature domains: a 100-amino acid PAZ domain and a 300 amino acid Piwi domain in the N- and C-terminal regions, respectively (Cerutti et al., 2000). The function of the PAZ domain is not known, but recent studies have shown that the Piwi domain (Doi et al., 2003; our unpublished data) is involved in binding to Dicer (Hammond et al., 2000), a ribonuclease that is required for RNA interference (RNAi) (Fire et al., 1998).
Genetic and biochemical studies first linked PPD proteins to signaling processes that affect cellular differentiation and development (Bohmert et al., 1998; Cox et al., 1998; Moussian et al., 1998; Cikaluk et al., 1999; Lynn et al., 1999; Cox et al., 2000; King et al., 2001; Tahbaz et al., 2001). In addition, numerous groups have shown that PPD proteins function as part of the RNAi machinery to mediate posttranscriptional gene and chromatin silencing (Tabara et al., 1999; Fagard et al., 2000; Hammond et al., 2001; Hall et al., 2002; Pal-Bhadra et al., 2002; Volpe et al., 2002). Although it is not known how they work, PPD proteins are hypothesized to serve as effector molecules that provide specificity in gene-silencing pathways (Baulcombe, 2001; Hannon, 2002). Dicer processed micro-RNAs or small interfering RNAs (siRNA) bind PPD proteins to form ribonucleoprotein complexes termed RNA induced silencing complexes (RISCs), which are thought to mediate mRNA degradation, translational repression, or chromatin silencing in a homology-dependent manner (Matzke et al., 2001; Hannon, 2002). Interestingly, mutations in PPD genes that affect either gene silencing or development (but not both processes) have been isolated, demonstrating that these two functions can be genetically uncoupled (Morel et al., 2002).
Because all metazoan genomes encode multiple family members (Carmell et al., 2002), the phenotype caused by a mutation in a given PPD gene may in fact be tempered by the overlapping functions of paralogous genes (Lynn et al., 1999; Grishok et al., 2001; Doi et al., 2003). In this respect, S. pombe is an ideal model organism with which to study PPD gene function because its genome contains a single member of this family, ago1+. The ago1+ gene encodes for Ago1, a protein that is 33% identical and 55% similar to human GERp95/EIF2C2/hAgo2 (Carmell et al., 2002). During the course of this work, a number of studies on the RNAi machinery of S. pombe were reported. First, it was shown that ago1+, dcr1+, and rdp1+ were required for timely formation of centromeric heterochromatin (Hall et al., 2002; Volpe et al., 2002). Moreover, deletion of any one of these genes results in missegregation of chromosomes due to loss of centromeric cohesion and mitotic spindle assembly defects (Provost et al., 2002; Hall et al., 2003; Volpe et al., 2003). A model derived from this work entails that double-strand centromeric transcripts serve as substrates for the RNase Dcr1 (Allshire, 2002; Bailis and Forsburg, 2002; Volpe et al., 2002). Dcr1 cleavage products are then subsequently incorporated into an Ago1-containing RISC that functions to mediate locus-specific chromatin silencing in concert with histone methylation complexes. Although Rdp1 is also required for RNAi-mediated chromatin silencing, it is not clear whether this enzyme is involved mainly in producing double-strand RNA species by using centromeric transcripts as templates and/or in amplification of siRNAs at a later step.
In addition to their well-known roles in RNAi, evidence from Lin and co-workers suggest that PPD proteins regulate cell cycle events. For example in Drosophila, altering the expression level of Piwi leads to changes in the proliferation rates of germline stem cells (Cox et al., 2000). Furthermore, the Piwi signaling network exhibits cross talk with the Hedgehog signaling machinery (King et al., 2001), a developmental patterning system that influences the cell cycle through the actions of Patched1 on cyclin B1 (Barnes et al., 2001; Roy and Ingham, 2002). Presently, it is not clear whether Piwi's role in cell cycle progression is dependent upon siRNAs. In the present study, we demonstrate that ago1+ and dcr1+ are required for regulated hyperphosphorylation of the cyclin-dependent kinase Cdc2. Inhibitory phosphorylation of Cdc2 is required to prevent the onset of mitosis in situations where damaged or unreplicated DNA is present (Enoch and Nurse, 1990; Rhind et al., 1997; Rhind and Russell, 1998). Conversely, dephosphorylation of Cdc2 is required for progression through the cell cycle (Russell and Nurse, 1986; Millar et al., 1991). Because rdp1+ does not seem to be required for regulation of Cdc2 phosphorylation, we suggest that in addition to their roles in chromatin silencing and chromosome segregation, Ago1 and Dcr1 function in a divergent pathway that is not dependent upon Dcr1-derived siRNAs.
MATERIALS AND METHODS
Strains and Media
A list of S. pombe strains used in this study and their corresponding genotypes is shown in Table 1. Unless otherwise indicated, yeast were cultured at 30°C in YES, YPD, YE, or EMM lacking nutritional supplements (Alfa et al., 1993). To induce expression of genes under the control of the no message thiamine promoter, cultures were grown overnight in EMM supplemented with thiamine (5 μg/ml) to repress expression of the gene, washed twice in EMM to remove residual thiamine, and cultured for an additional 48 h to allow expression of the genes of interest. Escherichia coli strain DH5α was used for propagation of plasmids (Table 2).
Table 1.
S. pombe strains used in this study
| Strain | Genotype | Source | Institution |
|---|---|---|---|
| FY254 | h can1-1 leu1-32 ade6-M210 ura4-D18 | Dr. S. Forsburg | Salk Institute |
| FY261 | h− can1-1 leu1-32 ade6-M216 ura4-D18 | Dr. S. Forsburg | Salk Institute |
| JC254K | h− ago1+ :: G418R can1-1 leu1-32 ade6-M210 ura4-D18 | This study | |
| JC261K | h+ ago1+ :: G418R can1-1 leu1-32 ade6-M216 ura4-D18 | This study | |
| FY326 | h+ wee1-50 leu1-32 ura4 ade6-M216 | Dr. S. Forsburg | Salk Institute |
| FY118 | h90 ura4-D18 leu1-32 ade6-M216 | Dr. S. Forsburg | Salk Institute |
| JCD-1 | h−/h′ can1-1 leu1-32 ade6-M210/ade6-M216 ura4-D18 | This study | |
| JCDK | h−/h+ can1-1 leu1-32 ade6-M210/ade6-M216 ura4-D18 ago1′ :: G418R | This study | |
| JC90DK | h90/h+ ago1+ :: G418R ura4-D18 leu1-32 ade6-M216/ade6-M210 can1-1 | This study | |
| JC90K | h90 ago1+ :: G418R ura4-D18 leu1-32 ade6-M216 | This study | |
| TV292 | h− ago1+ :: kanMX6 ura4-D18 DS/E | Dr. T. Volpe | Cold Spring Harbor |
| TV293 | h+ dcr1+ :: kanMX6 ura4-D18 DS/E | Dr. T. Volpe | Cold Spring Harbor |
| TV294 | h− ura4-D18 DS/E | Dr. T. Volpe | Cold Spring Harbor |
| TV296 | h+ rdp1+ :: kanMX6 ura4-D18 DS/E | Dr. T. Volpe | Cold Spring Harbor |
| FY1106 | h+ rad3+ :: ura4+ ura4-D18 leu1-32 ade6-M210 | Dr. S. Forsburg | Salk Institute |
Table 2.
List of plasmids
| Vector | Source | Institution |
|---|---|---|
| pREP3X | Dr. S. Forsburg | Salk Institute |
| pGEM-T | Promega | |
| pFA6a-kanMX6 | Dr. J. Pringle | University of North Carolina |
| pDS473a | Dr. S. Forsburg | Salk Institute |
| pSLF273 | Dr. S. Forsburg | Salk Institute |
Cloning of ago1+
Genomic and cDNA clones of ago1+ (SPCC736.11) were obtained by polymerase chain reaction (PCR) amplification of genomic DNA from FY254 and an S. pombe cDNA library (BD Biosciences Clontech, Palo Alto, CA) with Expand High Fidelity PCR System (Roche Diagnostics, Indianapolis, IN). PCR products were purified from agarose gels by using ELU-QUIK DNA purification kit (Schleicher & Schuell, Keene, NH) and ligated into the pGEM-T vector (Promega, Madison, WI). Individual clones were sequenced using the Department of Cell Biology sequencing facility (University of Alberta, Edmonton, AB, Canada).
Gene Replacement
A G418-resistance cassette was amplified by PCR from pFA6a-kanMX6 (Bahler et al., 1998) and then used to disrupt the ago1+ open reading frame by electroporation (Moreno et al., 1991) in a one-step gene replacement procedure. Replacement of ago1+ was verified by PCR and Southern blot analyses on genomic DNA isolated (Hoffman and Winston, 1987) from transformants that were resistant to G418 (100 μg/ml).
Mating Assays
Heterothallic strains were grown to mid-log phase in liquid YE and equivalent numbers of opposite mating type cells were mixed together and spotted onto malt extract plates. Homothallic (h90) strains were cultured in YE until mid-log phase, washed, and resuspended in media lacking nitrogen or in low glucose/low nitrogen media (Okazaki et al., 1998). Mating assays were conducted over a range of 18-30 h. Mating frequencies were determined by dividing the number of zygotes formed by the total number of cells. Three independent mating assays were performed for each data point with at least 300 cells scored per assay.
Flow Cytometry
Aliquots of 106 cells were collected from liquid cultures, pelleted, and fixed by resuspension in 1 ml of 70% ethanol. Cells were washed in 3 ml of 50 mM sodium citrate, resuspended in 0.5 ml of 50 mM sodium citrate containing 0.1 mg/ml RNaseA and incubated at 37°C for 2 h. Cells were stained by adding 0.5 ml sodium citrate solution containing 2 μM Sytox Green (Molecular Probes, Eugene, OR) and stored at 4°C until processing. Cells were sonicated for 45 s before analyses on an FACScan (BD Biosciences, San Jose, CA).
Drug Treatment and Cdc2 Phosphorylation Assays
Strains were cultured (streaks or serial dilutions) for the indicated time periods at 30°C on YE or EMM plates with indicated hydroxyurea (HU) (3.5-10 mM) or thiabendazole (TBZ) (10 μg/ml). For Cdc2 phosphorylation assays, strains were cultured in YE to OD595 = 0.3 at which time HU (10 mM) was added and incubation continued for various times. For the experiments shown in Figure 3B, 4 h post-HU treatment, cultures were washed, resuspended in YE, and incubations continued at 30°C for the remainder of the experiment. Aliquots were removed at various times, and the proportions of septated cells were determined by microscopic analyses (n = 300).
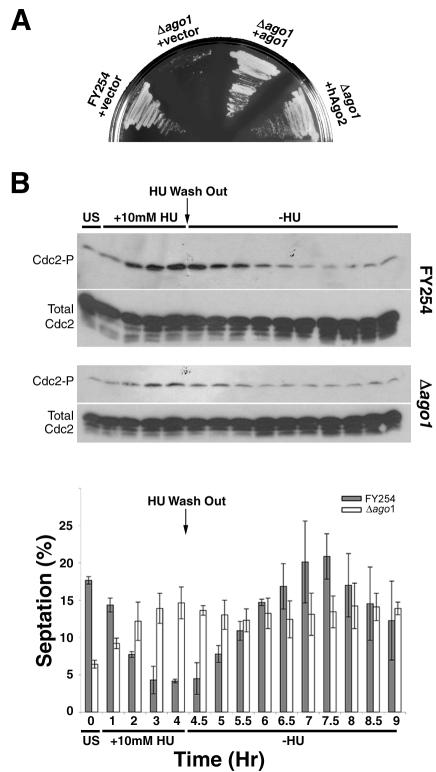
Figure 3.
ago1 null mutants fail to activate the S-M DNA replication checkpoint. (A) FY254 and Δago1 strains transformed with vector (pREP3X), pREP3X +ago1+, or pREP3X +hAgo2 were streaked onto leucine-deficient EMM plates containing 10 mM HU and incubated for 3 d at 30°C. Viability of the Δago1 strain containing pREP3X was confirmed by growth on EMM lacking both leucine and HU (our unpublished data). (B) Immunoblot analyses of total and phosphorylated Cdc2 (Cdc2-P) from FY254 and Δago1 strains in unsynchronized cultures (US), during HU treatment (+10 mM HU) and after removal of HU (Wash Out). Mean septation indices (determined from four independent experiments) are shown for each time point (bar graph).
Whole cell lysates for all Cdc2 phosphorylation assays were prepared by mechanically breaking cells in radioimmunoprecipitation buffer (50 mM Tris-Cl, pH 7.5, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS) with glass beads by using Beadbeater 8 device. After boiling for 3 min, lysates were cleared by centrifugation (1000 × g) for 15 min at 4°C, resolved by SDS-PAGE on 10% gels, and then transferred to Immobilon-P membranes. Total Cdc2 levels and Cdc2 (tyrosine 15) phosphorylation levels were measured by immunoblot analyses by using anti-PSTAIR (Sigma-Aldrich, St. Louis, MO) and anti-phospho-cdc2 (Tyr15) (Cell Signaling Technology, Beverly, MA), respectively, followed by enhanced chemiluminescence detection.
UV Irradiation Assay
Yeast were grown to OD595 = 0.8 in YE, counted, and spread on YE plates at a density of 100-300 cells per plate. The plates were irradiated (0, 75, 150, 225, or 300 J/M2) by using a Hoefer UVC 500 apparatus, wrapped in aluminum foil, and incubated for 24 h at 30°C. The foil was removed and incubation was continued at 30°C for an additional 72 h. Surviving colonies were counted and presented as a percentage of the surviving nonirradiated cells.
Ago1 and Dcr1 Expression Constructs
The cDNAs encoding Ago1 and Dcr1 were subcloned into pREP3X and pREP41HA-N expression vectors (Forsburg and Sherman, 1997), respectively. Yeast were transformed and selected for growth on EMM lacking leucine. Overexpression of Ago1 and HA-Dcr1 were confirmed by immunoblot analysis of whole cell lysates by using rabbit anti-Ago1 or a monoclonal antibody to hemagglutinin (HA), respectively (our unpublished data).
RESULTS
ago1+ and dcr1+ Null Mutants Are Delayed in Cytokinesis
One-step targeted gene disruption was used to replace the entire ago1+ open reading frame with a G418 resistance cassette in the haploid strain FY254. Disruption of the ago1+ locus in the resulting strain JC254K was confirmed by PCR and Southern blot analyses (our unpublished data). In agreement with a recently published study (Volpe et al., 2002), we found that ago1+ was not essential for viability at 25, 30, or 36°C, but it was also clear that deletion of this gene resulted in pronounced growth and morphological defects (see below). Uncondensed and/or mislocalized chromosomes as well as cut phenotypes were also evident in this strain. The latter phenotype is indicative of cytokinesis being uncoupled from DNA segregation. These results are in agreement with those reported in recent studies (Provost et al., 2002; Hall et al., 2003). Backcrossing of the Δago1 strains with FY261 was performed to rule out the possibility that the morphological defects were the result of second site mutations. As well, the morphological defects of ago1 null mutants were suppressed by plasmid-driven expression of Ago1 (our unpublished data).
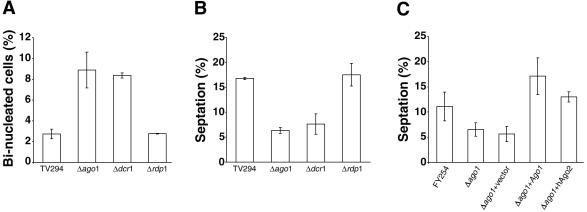
A striking feature of the ago1 null mutants when examined by light microscopy was the relative abundance of binucleated cells. This apparent cytokinesis defect was first noticed in the Δago1 strain JC254K (our unpublished data). Because dcr1+ and rdp1+ are known to function together with ago1+ in a chromatin silencing pathway (Volpe et al., 2002), we investigated whether cytokinesis would also be affected in these null strains. The proportions of binucleated cells were significantly higher in a Δdcr1 strain and a second Δago1 strain (TV292), compared with the parental (TV294) strain, whereas no change was observed in a Δrdp1 strain (Figure 1A). Binucleates, which comprised ∼3% of the parental and Δrdp1 populations, increased by threefold to ∼9% of cells in the Δago1 and Δdcr1 strains under the same culture conditions.
Figure 1.
ago1+ and dcr1+ are both required for normal cytokinesis. All strains were cultured at 30°C in YE to mid-log phase (OD595 = 0.6-0.9), stained with 4,6-diamidino-2-phenylindole and placed onto microscope slides coated with poly l-lysine (5 mg/ml). Samples were examined by fluorescence and differential interference contrast microscopy. Representative fields were photographed and scored for nuclei or septation. For each sample, at least 500 cells were scored. (A) Quantitation of binucleated cells in parental strain TV294, Δago1 (TV292), Δdcr1 (TV293), and Δrdp1 (TV296) strains. (B) Quantitation of septation indices of TV294, Δago1 (TV292), Δdcr1 (TV293), and Δrdp1 (TV296). (C) Quantitation of septation indices in FY254 and Δago1 (JC254K) strains transformed with vector (pREP3X), pREP3X +ago1+, or pREP3X +hAgo2.
Cytokinesis is preceded by septum formation in fission yeast and therefore the septation index can be used as an indicator of cell division (Alfa et al., 1993). As shown in Figure 1B, the proportions of septated cells were significantly reduced when ago1+ or dcr1+ gene function was ablated. However, deletion of rdp1+ had no effect on this process. Plasmid-driven overexpression of Ago1 or the human PPD protein hAgo2 corrected the cytokinesis defect in the ago1+ mutant (Figure 1C). Although it cannot be ruled out that loss of ago1+ or dcr1+ results in accelerated mitosis, these results suggest that Ago1 and Dcr1 may function together to regulate septum formation and subsequent cytokinesis in S. pombe.
ago1+ and dcr1+, but Not rdp1+ Are Required for G1 Arrest and Mating in Response to Nitrogen Starvation
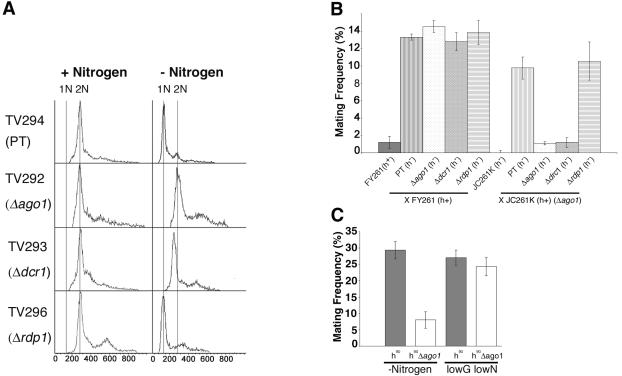
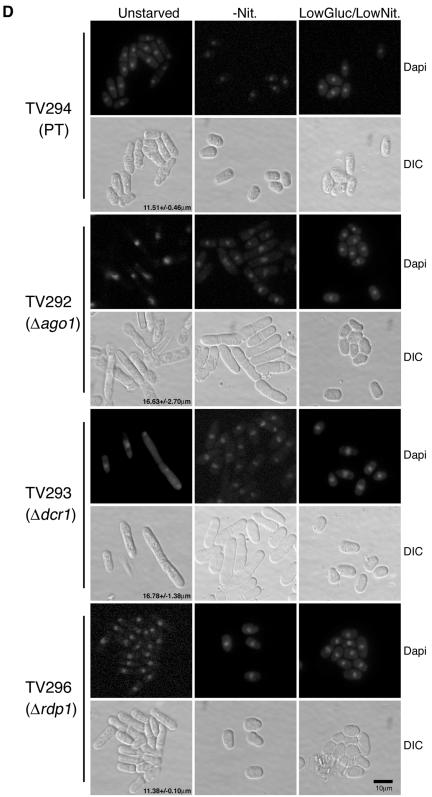
To enable survival when starved of nitrogen, S. pombe cells normally arrest in G1 and if a partner of opposite mating type is in proximity such that pheromones are sensed, commit to sexual differentiation and subsequent meiosis (Egel and Egel-Mitani, 1974; Crandall et al., 1977; Breeding et al., 1998). Alternatively, cells enter a quiescent state (G0) when mating partners are absent. These processes are mediated by the Wis1 MAPK pathway (Shiozaki and Russell, 1996). Fluorescence-activated cell sorting (FACS) analyses of JC254K demonstrated that ago1+ was required for G1 arrest during nitrogen depletion (our unpublished data). The inability to arrest in G1 was also observed in another Δago1 strain (TV292) as well as in a Δdcr1 (TV293) strain (Figure 2A). Moreover, the characteristic shortening and rounding of cells (Alfa et al., 1993; Su et al., 1996) in response to nitrogen depletion was not evident in the nitrogen-starved Δago1 and Δdcr1 strains (Figure 2D). In contrast, the control parental strain (TV294) and the Δrdp1 strain (TV296) both arrested in G1 in response to nitrogen limitation.
Figure 2.
ago1 and dcr1 mutants are defective for G1 arrest and mating after nitrogen limitation. (A) FACS analyses of log phase (+Nitrogen) and nitrogen starved (-Nitrogen) TV294, Δago1 (TV292), Δdcr1 (TV293), and Δrdp1 (TV296) cultures. Insets at the right show the morphologies of yeast cells under nitrogen-deficient conditions. (B and C) Mating mixtures were incubated at 30°C for 18-30 h on malt extract plates and scored for zygote production microscopically. (B) Mating frequencies of TV294, Δago1 (TV292), Δdcr1 (TV293), and Δrdp1 (TV296) crossed with FY261 (parental type) or JC261K (Δago1). The low level of self-mating in strains FY261 and JC261K presumably occurs due to mating type switching. (C) Mating frequencies for homothallic wild-type (h90) or homothallic ago1 null (h90Δago1) strains under nitrogen-deficient or low glucose/low nitrogen condition. (D) 4,6-diamidino-2-phenylindole-stained and corresponding differential interference contrast (DIC) images of TV294, Δago1, Δdcr1, and Δrdp1 strains cultured under normal growth (unstarved), nitrogen starvation (-Nit.) and low glucose/low nitrogen (LowGluc./LowNit.) conditions. Average cell lengths for each strain (unstarved condition) are shown at the bottoms of the DIC panels.
Because they cannot effectively block in G1 under nitrogen limiting condition, we anticipated that the Δago1 and Δdcr1 strains would be compromised in their abilities to undergo sexual differentiation. Indeed, heterothallic Δago1 (TV292) and Δdcr1 (TV293) strains were unable to mate efficiently with a Δago1 (JC261K) strain of opposite mating type (Figure 2B). In contrast, crossing null ago1 and dcr1 strains with FY261 resulted in formation of zygotes at the same level as TV294/FY261 crosses or with the rdp1 null strain. Because mating is dependent upon sexual partners being in proximity to each other, any mating defect associated with the ago1 null phenotype would be exaggerated by using two different mating strains. To circumvent this problem, we constructed an h90 strain deleted for the ago1+ gene and tested the ability of this strain to form zygotes. The h90 ago1 null strain (h90Δ ago1) mated at a greatly reduced frequency compared with the parental strain (h90) under nitrogen-deficient conditions (Figure 2C). Interestingly, when subjected to low glucose/low nitrogen conditions, the h90Δago1 strain showed the characteristic morphological change indicative of a G1 arrest (Figure 2D). Moreover, under the same conditions, FACS analyses (our unpublished data) were used to confirm that this strain had efficiently blocked in G1. As expected, the h90Δago1 strain mated with the same efficiency as the parental h90 strain under low glucose/low nitrogen conditions (Figure 3C). These results indicate that ago1+ and dcr1+ but not rdp1+, are required for mating through the facilitation of a nitrogen-specific block in G1.
ago1 and dcr1 Null Mutants Fail to Enact and/or Maintain the S-M Checkpoint
The above-mentioned results demonstrate that yeast lacking Ago1 function share a number of similarities to Spc1/StyI mutants (Warbrick and Fantes, 1991; Stettler et al., 1996). Specifically, cells are elongated, do not block in G1 in response to nitrogen deprivation, and mating ability is compromised. It has also been shown that strains deficient in Spc1/Sty1 are highly sensitive to deoxyribonucleotide depletion (Taricani et al., 2001) and radiation-induced DNA damage (Degols and Russell, 1997). In response to the ribo-nucleotide reductase inhibitor HU, checkpoint kinases ultimately mediate enactment of the S-M checkpoint through inhibitory phosphorylation of tyrosine 15 on Cdc2 (al-Khodairy and Carr, 1992; Enoch et al., 1992; Furnari et al., 1997; Rhind and Russell, 1998). Cdc2 is a cyclin-dependent kinase and is the central regulator of the cell cycle that controls passage through S-phase and into mitosis (Nurse, 1990; Fisher and Nurse, 1996). Experimental results shown in Figure 3A offered the first clue that ago1+ may be important for enactment of the S-M checkpoint. JC254K (Δago1) and the parent strain FY254 transformed with vector pREP3X, pREP3X +ago1+, or pREP3X +hAgo2 were streaked onto plates containing 10 mM HU to test their abilities to activate the DNA replication checkpoint. After 3 d, Δago1 cells transformed with vector alone were not able to grow under these conditions, indicating that they were unable to activate, maintain or recover from activation of this checkpoint (Figure 3A). In comparison, the Δago1 strain ectopically expressing Ago1 or hAgo2 grew well on HU-containing media.
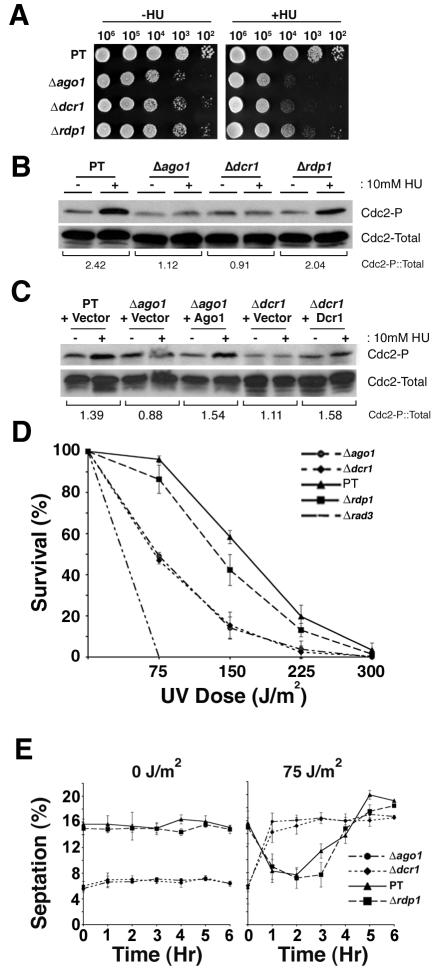
Because hyperphosphorylation of Cdc2 is required for initiation of the S-M checkpoint, we predicted that phosphorylation of this kinase would be decreased in the Δago1 strain. As expected in response to HU treatment, the levels of phosphorylated Cdc2 in the parent strain FY254 increased, coinciding with a decrease in the septation index (Figure 3B). After removal of HU, the levels of phosphorylated Cdc2 decreased and were accompanied by a concomitant increase in the number of septated cells. In contrast, Cdc2 hyperphosphorylation was delayed in Δago1 cultures, and peak phosphorylation levels were substantially lower than those seen in FY254 (Figure 3B). Consequently, the septation index of Δago1 cultures continued to increase rather than decrease in the presence of HU (Figure 3B). The Δdcr1 strain was also hypersensitive to HU, whereas the Δrdp1 mutant was significantly less sensitive to this drug (Figure 4A). Acting on the premise that the HU sensitivity of the Δdcr1 strain resulted from an inability to activate/maintain the S-M checkpoint, we verified that this mutant was indeed unable to hyperphosphorylate Cdc2 in response to HU treatment (Figure 4B). On the contrary, HU-induced hyperphosphorylation of Cdc2 was not affected by loss of Rdp1 function. Overexpression of Ago1 or Dcr1 in their cognate null mutants, restored the ability of these strains to hyperphosphorylate Cdc2 in response to HU treatment (Figure 4C).
Figure 4.
ago1+ and dcr1+ are both required for hyperphosphorylation of Cdc2 and enactment of the DNA damage and replication checkpoints. (A) Serial dilutions of TV294 and corresponding Δago1, Δdcr1, and Δrdp1 strains were spotted on YE agar or YE agar containing 3.5 mM HU and were cultured for 3 and 5 d at 30°C, respectively. (B) TV294, Δago1, Δdcr1, and Δrdp1 liquid cultures were grown at 30°C to an OD595 = 0.5, and aliquots were removed before (-) or after (+) a 4-h HU treatment. Whole cell lysates were prepared and separated by SDS-PAGE before immunoblot analyses of total and phosphorylated Cdc2. The normalized levels of HU-induced Cdc2 phosphorylation are shown below the immunoblots in B and C. (C) Levels of total and phosphorylated Cdc2 were analyzed in strains transformed with vector alone or vector encoding Ago1 or Dcr1. (D) TV294, Δago1, Δdcr1 Δrdp1, and Δrad3 liquid cultures were grown at 30°C to an OD595 = 0.8, spread at a density of 100-300 cells per plate and exposed to UV radiation (0, 75, 150, 225, or 300 J/M2). Cell survival is shown as a percentage of the nonirradiated control cell survival. (E) TV294, Δago1, Δdcr1, and Δrdp1 were subjected to mock (0 J/m2) treatment and UV irradiation (75 J/m2), and the septation index for each strain was determined every hour postirradiation (n = 3, 300 cells counted/time point). Similar results were obtained when these strains were subjected to a 100 J/m2 dose (our unpublished data).
ago1 and dcr1 Null Mutants Are Defective for the DNA Damage Checkpoint
The degree of overlap between components of the DNA replication and DNA damage checkpoints is considerable (al-Khodairy and Carr, 1992; Rowley et al., 1992; Furnari et al., 1997; Rhind et al., 1997; Rhind and Russell, 1998; Raleigh and O'Connell, 2000). In both cases, activation of the checkpoints after exposure to genotoxic insults requires hyperphosphorylation of Cdc2. Accordingly, we predicted that Δago1 and Δdcr1 strains would be hypersensitive to UV irradiation, a commonly used inducer of the DNA damage checkpoint. Indeed, Δago1 and Δdcr1 strains showed increased sensitivity to UV radiation compared with the rdp1 null and the parental strain TV294, but were less sensitive than the control checkpoint mutant Δrad3 (Jimenez et al., 1992) (Figure 4D). To confirm that the sensitivity of ago1 and dcr1 null mutants to UV light was the result of a failure to activate the DNA damage checkpoint, strains were irradiated and the septation indices were determined over a 6-h period (Figure 4E). As expected the parental and Δrdp1 strains showed decreased septation after exposure to UV light (Francesconi et al., 2002), an indication that the DNA damage checkpoint was functioning in these strains. In contrast, ago1 and dcr1 null mutants did not exhibit decreased septation after exposure to UV radiation. These results suggest that ago1+ and dcr1+ are both required for survival when encountering DNA-damaging conditions, possibly through Cdc2-dependent activation of the DNA damage checkpoint.
Ago1 May Function Downstream of Dcr1 to Regulate Cell Cycle Events
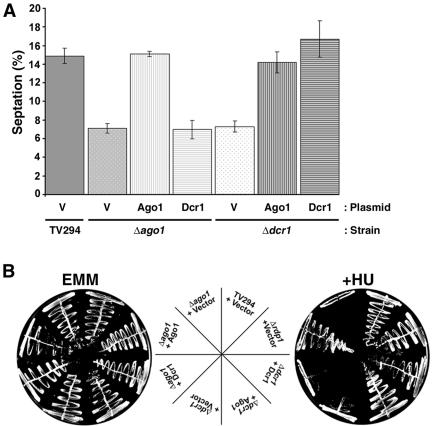
To gain insight into how Ago1 and Dcr1 interact genetically to enact the S-phase replication checkpoint, we determined whether overexpression of Ago1 could compensate for lack of Dcr1 function and vice versa. We noticed that Δdcr1 strains overexpressing Ago1 exhibited normal morphology compared with Δdcr1 strains expressing vector only (our unpublished data). To investigate this finding further, we assayed whether Ago1 overexpression could correct the cytokinesis defect associated with loss of Dcr1 function. Data shown in Figure 5A clearly show that high level expression of Ago1 restores the septation index of Δdcr1 cells to normal levels. In contrast, overexpression of Dcr1 did not compliment the cytokinesis defect of Δago1 strains. Moreover, ectopic expression of Ago1 complemented that S-M checkpoint deficiency of both Δago1 and Δdcr1 strains, whereas Dcr1 overexpression corrected the checkpoint deficiency in Δdcr1 strains only (Figure 5B). These results indicate that the requirement for Ago1 in regulation of cytokinesis and enactment of cell cycle checkpoints is downstream of Dcr1 and independent of Dcr1-derived siRNAs.
Figure 5.
ago1+ functions downstream of dcr1+ to regulate cell cycle events. Yeast strains were transformed with plasmids encoding Ago1, HA-Dcr1, or vector alone. (A) Yeast strains grown in EMM-leu to log phase were examined by phase contrast microscopy, and septated cells were counted and expressed as a percentage of total cells. (B) Yeast strains were streaked onto EMM lacking leucine (EMM) in the presence or absence of 7.5 mM HU (+HU) and were then cultured at 30°C for 3 to 5 d.
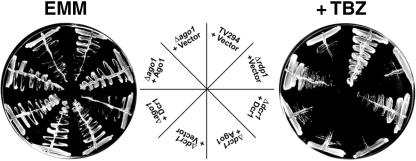
Recent studies have shown that ago1+, dcr1+, and rdp1+ are required for accurate segregation of chromosomes and consequently, null mutations in any one of these genes results in hypersensitivity to the microtubule-destabilizing drug TBZ (Provost et al., 2002; Hall et al., 2003; Volpe et al., 2003). Because Rdp1 and Dcr1 are both required for efficient chromosome segregation, we assume that production and targeting of siRNAs are also required for this process. Accordingly, we predicted that ectopic expression of Ago1 would not complement the TBZ sensitivity of the Δdcr1 strains. Overexpression of Ago1 or Dcr1 in the ago1 or dcr1 mutants, respectively, resulted in decreased sensitivity to TBZ (Figure 6). However, overexpression of Ago1 in Δdcr1 or Dcr1 in Δago1 strains did not alleviate sensitivity to TBZ. Together, these results suggest that the roles of Ago1 and Dcr1 in centromere function and chromosome segregation are distinct from their functions in cytokinesis and cell cycle checkpoints.
Figure 6.
Ectopic overexpression of Ago1 does not correct the chromosome segregation defect of dcr1 nulls. Yeast strains were transformed with plasmids encoding Ago1, HA-Dcr1 or vector alone. Strains were streaked onto EMM lacking leucine in the presence (+TBZ) or absence (EMM) of 10 μg/ml TBZ and were then cultured at 30°C for 3 to 5 d.
DISCUSSION
Depending upon the organism, metazoan genomes encode between five and 24 PPD genes (Carmell et al., 2002). Not surprisingly, some paralogs have overlapping functions (Lynn et al., 1999; Doi et al., 2003), a situation that complicates the genetic and biochemical analyses of this family of proteins. In contrast, the S. pombe genome encodes for a single PPD gene ago1+. This organism also encodes homologues of dicer and RNA-dependent RNA polymerase that together with ago1+ function in chromatin silencing/segregation (Provost et al., 2002; Volpe et al., 2002; Hall et al., 2003). Accordingly, the fission yeast is an excellent experimental system with which to study the molecular functions of PPD proteins.
Targeted gene deletion demonstrated that ago1+ was not essential for growth in haploid strains under a variety of conditions. Our results are consistent with recent studies that showed none of the genes that encode core RNAi components, ago1+, dcr1+, or rdp1+, are essential for viability in S. pombe (Provost et al., 2002; Volpe et al., 2002). However, these genes were shown to be necessary for timely formation of centromeric heterochromatin (Hall et al., 2002; Volpe et al., 2002), a process that is not only important for epigenetic silencing in S. pombe but also for sister chromosome segregation (reviewed in Karpen and Allshire, 1997; Lee and Orr-Weaver, 2001). Heterochromatin formation at centromeric sites, results in enrichment of cohesion molecules that mediate sister chromosome attachment and kinetochore orientation, which in turn facilitates microtubule binding and proper chromosome segregation (Bernard et al., 2001; Nonaka et al., 2002). In keeping with recent work by others (Hall et al., 2003), we also found that assembly and or localization of mitotic spindles was abnormal in the Δago1 strain JC254K (our unpublished data). Similar phenotypes were reported for S. pombe strains devoid of Dcr1 function (Provost et al., 2002; Hall et al., 2003). However, a finding not previously reported for Δago1 and Δdcr1 strains is the fact that these mutants are impaired for cytokinesis. Interestingly, the rdp1 null mutant did not exhibit this phenotype, providing the first indication that Ago1 and Dcr1 may function independently of Rdp1 in a pathway distinct from that required for efficient heterochromatin formation and chromosome segregation.
PPD Proteins and Dicer Function to Regulate Cell Cycle Events
In S. pombe, cell division is preceded by the establishment of a septum in the medial portion of a dividing cell (Le Goff et al., 1999; Balasubramanian et al., 2000). This process is regulated by a signaling network known as the septation initiation network (McCollum and Gould, 2001). The absence of Ago1 or Dcr1 activity was associated with a dramatic reduction in septation and subsequent cytokinesis. However, these cell cycle defects are apparently not related to the inability to efficiently form heterochromatin or segregate chromomsomes because septation and cytokinesis were normal in rdp1 mutants.
Of relevance to the present study is the previous finding that the Piwi signaling network exhibits cross talk with the hedgehog signaling pathway (King et al., 2001). The latter pathway is known to influence the regulation of various cell cycle events (reviewed in Barnes et al., 2001). With respect to Piwi, ectopic overexpression of this protein increases the division rate of germline stem cells, whereas mutations that compromise Piwi function seem to have the opposite effect (Cox et al., 2000). We also found that plasmid-driven overexpression of Ago1 increased the growth rate of parental yeast strains such as FY254, whereas growth is impaired in ago1 null strains (our unpublished data). At this point, it is unknown whether siRNAs are required for Piwi function in signaling pathways that regulate the cell cycle.
Our analyses showed that ago1+ and dcr1+ are both required for establishment and/or maintenance of G1 arrest in response to nitrogen-limiting conditions. Accordingly, Δago1 and Δdcr1 strains are severely impaired for mating, a hallmark phenotype for PPD and Dicer mutations in metazoans (Cox et al., 2000; Smardon et al., 2000; Grishok et al., 2001; Knight and Bass, 2001; Parrish and Fire, 2001). The results in this study are in apparent contrast to those of a previous study (using the same strains) where no obvious mating defects were reported (Volpe et al., 2002). However, the discrepancy in findings could potentially be explained if different mating assays were used in the two studies. For example, ago1+ and dcr1+-dependent mating defects were only observed in our hands when assays were conducted under nitrogen-deficient conditions. In contrast, when mating assays were performed under low glucose/low nitrogen conditions, these mutants mated with the same frequencies as parental and Δrdp1 strains. Importantly, these data suggest that the mating defects associated with the ago1 and dcr1 null mutants are not due to global defects stemming from lack of a functioning RNAi pathway or missegregation of chromosomes, but rather a failure to block in G1 when nitrogen is absent.
Additional evidence for the functioning of ago1+ and dcr1+ in an rdp1+-independent pathway came from the finding that the former two genes are essential for initiation and/or maintenance of the DNA replication and damage checkpoints. Significantly, enactment of these two checkpoints requires a common set of genes (al-Khodairy and Carr, 1992; Rowley et al., 1992; Furnari et al., 1997; Rhind et al., 1997; Rhind and Russell, 1998; Raleigh and O'Connell, 2000) whose collective functions ultimately determine the activity level of the master cell cycle regulator Cdc2. Inhibitory phosphorylation of tyrosine 15 in Cdc2 leads to a block in the cell cycle until replication inhibition or DNA damage is relieved. With respect to the ago1 and dcr1 null mutants, failure to enact these checkpoints likely results from their inability to hyperphosphorylate Cdc2 when exposed to genotoxic insults. Moreover, the inability to inactivate Cdc2 by inhibitory phosphorylation could also be the underlying cause of the cytokinesis and mating defects in the ago1 and dcr1 nulls. Activation of the septation initiation network and subsequent septation as well as sexual mating in yeast, requires a reduction in Cdc2 activity before these processes can occur (He et al., 1997; Wu and Russell, 1997; Stern and Nurse, 1998; Guertin et al., 2000; Chang et al., 2001). Intriguingly, a human Ago1 homolog, hAgo2, was able to fully complement the cytokinesis defect and DNA replication checkpoint deficiencies of ago1 mutants, suggesting that PPD proteins may regulate cell cycle events in mammalian cells, a function not reported previously. In contrast, human dicer was found to only partially complement the chromosomal segregation defects in a dcr1 knockout strain of S. pombe (Provost et al., 2002).
The phosphorylation status and activity of Cdc2 is controlled primarily by the opposing actions of the phosphatase Cdc25 and two kinases, Wee1 and to a lesser extent, Mik1 (Russell and Nurse,1986, 1987; Gould and Nurse, 1989; Lundgren et al., 1991; McGowan and Russell, 1993; Lee et al., 1994). Accordingly, the decreased Cdc2 phosphorylation in ago1 and dcr1 mutants may result from inhibition of kinases that target Cdc2 and/or through increased Cdc25 phosphatase activity. Our repeated attempts to cross Δago1 strains with a wee1-50 mutant strain (Nurse, 1975) at the permissive temperature were unsuccessful. However, given the mating defects associated with wee1 (Wu and Russell, 1997) and ago1 mutants, successful conjugation between these two strain types may indeed be a rare event.
Ago1 and Dcr1 Function Independently of the Classical RNAi Pathway to Regulate Cell Cycle Events
Of course, it must be considered that the cell cycle defects associated with ago1 and dcr1 mutants are the result of an inability to enact transcriptional and/or posttranscriptional silencing of genes that modulate the cell cycle. However, if this were the case, it is difficult to explain why rdp1+ is not required for these processes. Furthermore, expression profile analyses indicated that only two S. pombe genes require dcr1+ for silencing (Provost et al., 2002) and neither of these genes has recognized functions in cell cycle regulation.
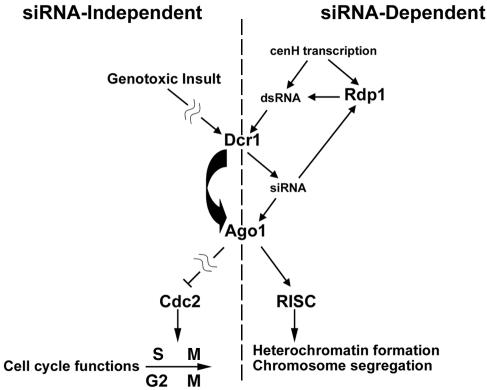
Rather, we propose that Ago1 and Dcr1 function independently of Rdp1 and the classical RNAi pathway to regulate Cdc2-dependent cell cycle events (Figure 7). This model, which also proposes that Ago1 acts downstream of Dcr1, is based upon the following experimental observations: 1) Rdp1 function is dispensable for regulated hyperphosphorylation of Cdc2. 2) Overexpression of Ago1 results in complementation of the S-phase check-point deficiency in a dcr1 null strain. In contrast, dcr1+ does not act as a multicopy suppressor to correct the S-phase checkpoint deficiency of Δago1 mutants. 3) Overexpression of Ago1 in a dcr1 null background does not complement the chromosome segregation defects associated with this mutant. Furthermore, a search of the S. pombe genome revealed that no other Dcr1-like or Rdp1-like sequences are present, and therefore it seems likely that Dcr1-dependent production of siRNAs is not required for Ago1-mediated regulation of cell cycle checkpoints. Together, these findings suggest that Ago1 and Dcr1 function in at least two pathways, one of which may be independent of siRNAs.
Figure 7.
Model for RNAi-independent functions of Ago1 and Dcr1. The right side of the model depicts the classical RNAi pathway in which Rdp1 is required for production/amplification of centromere-derived double-strand transcripts (cenH) that serve as substrates for Dcr1. The resulting siRNAs are incorporated into Ago1-containing RISCs that facilitate heterochromatin formation, an event that is required for attachment of mitotic spindles to kinetochores and subsequent orderly chromosome segregation. The left side of the model depicts a pathway in which Dcr1 and Ago1 in response to genotoxic insults such as DNA damage or HU, mediate downstream events that lead to inhibitory phosphorylation of Cdc2. This process is proposed to be independent of siRNAs and Rdp1.
Presently, it is unknown how Dcr1 and Ago1 function to regulate Cdc2 phosphorylation, but given that neither of these proteins is predicted to have kinase or phosphatase activity, it seems likely that their actions are indirect. Consequently, important next steps are to understand how ago1+ and dcr1+ interact genetically with genes that regulate the phosphorylation status of Cdc2 and to determine whether PPD proteins in higher eukaryotes regulate cell cycle events by affecting the activity of this kinase.
Acknowledgments
We are indebted to Dr. Susan Forsburg for the gifts of yeast strains, plasmids, and invaluable advice. We thank Drs. Rob Martienssen, John Pringle, and Tom Volpe for gifts of plasmids and strains. We also thank Dr. Dave Stuart for timely and insightful advice and Dr. Henry Parker for comments on the manuscript. This work was funded by grants from the Canadian Institutes of Health Research and the Natural Sciences and Engineering Research Council. T.C.H. is the recipient of a Senior Medical Scholarship from the Alberta Heritage Foundation for Medical Research.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03-06-0433. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-06-0433.
References
- al-Khodairy, F., and Carr, A.M. (1992). DNA repair mutants defining G2 checkpoint pathways in Schizosaccharomyces pombe. EMBO J. 11, 1343-1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfa, C., Fantes, P., Hyams, J., McLeod, M., and Warbrick, E. (1993). Experiments with Fission Yeast: A Laboratory Course Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Allshire, R. (2002). Molecular biology. RNAi and heterochromatin - a hushed-up affair. Science 297, 1818-1819. [DOI] [PubMed] [Google Scholar]
- Bahler, J., Wu, J.Q., Longtine, M.S., Shah, N.G., McKenzie, A., 3rd, Steever, A.B., Wach, A., Philippsen, P., and Pringle, J.R. (1998). Heterologous modules for efficient and versatile P.C.R-based gene targeting in Schizosaccharomyces pombe. Yeast 14, 943-951. [DOI] [PubMed] [Google Scholar]
- Bailis, J.M., and Forsburg, S.L. (2002). RNAi hushes heterochromatin. Genome Biol. 3, REVIEWS1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian, M.K., McCollum, D., and Surana, U. (2000). Tying the knot: linking cytokinesis to the nuclear cycle. J. Cell Sci. 113, 1503-1513. [DOI] [PubMed] [Google Scholar]
- Barnes, E.A., Kong, M., Ollendorff, V., and Donoghue, D.J. (2001). Patched1 interacts with cyclin B1 to regulate cell cycle progression. EMBO J. 20, 2214-2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baulcombe, D. (2001). RNA silencing. Diced defence. Nature 409, 295-296. [DOI] [PubMed] [Google Scholar]
- Bernard, P., Maure, J.F., Partridge, J.F., Genier, S., Javerzat, J.P., and Allshire, R.C. (2001). Requirement of heterochromatin for cohesion at centromeres. Science 294, 2539-2542. [DOI] [PubMed] [Google Scholar]
- Bohmert, K., Camus, I., Bellini, C., Bouchez, D., Caboche, M., and Benning, C. (1998). AGO1 defines a novel locus of Arabidopsis controlling leaf development. EMBO J. 17, 170-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breeding, C.S., Hudson, J., Balasubramanian, M.K., Hemmingsen, S.M., Young, P.G., and Gould, K.L. (1998). The cdr2(+) gene encodes a regulator of G2/M progression and cytokinesis in Schizosaccharomyces pombe. Mol. Biol. Cell 9, 3399-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmell, M., Xuan, Z., Zhang, M., and Hannon, G. (2002). The Argonaute family: tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes Dev. 16, 2733-2742. [DOI] [PubMed] [Google Scholar]
- Cerutti, L., Mian, N., and Bateman, A. (2000). Domains in gene silencing and cell differentiation proteins: the novel PAZ domain and redefinition of the Piwi domain. Trends Biochem. Sci. 25, 481-482. [DOI] [PubMed] [Google Scholar]
- Chang, L., Morrell, J.L., Feoktistova, A., and Gould, K.L. (2001). Study of cyclin proteolysis in anaphase-promoting complex (APC) mutant cells reveals the requirement for APC function in the final steps of the fission yeast septation initiation network. Mol. Cell Biol. 21, 6681-6694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cikaluk, D.E., Tahbaz, N., Hendricks, L.C., DiMattia, G.E., Hansen, D., Pilgrim, D., and Hobman, T.C. (1999). GERp95, a membrane-associated protein that belongs to a family of proteins involved in stem cell differentiation. Mol. Biol. Cell 10, 3357-3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox, D.N., Chao, A., Baker, J., Chang, L., Qiao, D., and Lin, H. (1998). A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev. 12, 3715-3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox, D.N., Chao, A., and Lin, H. (2000). piwi encodes a nucleoplasmic factor whose activity modulates the number and division rate of germline stem cells. Development 127, 503-514. [DOI] [PubMed] [Google Scholar]
- Crandall, M., Egel, R., and Mackay, V.L. (1977). Physiology of mating in three yeasts. Adv. Microbiol. Physiol. 15, 307-398. [DOI] [PubMed] [Google Scholar]
- Degols, G., and Russell, P. (1997). Discrete roles of the Spc1 kinase and the Atf1 transcription factor in the UV response of Schizosaccharomyces pombe. Mol. Cell Biol. 17, 3356-3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi, N., Zenno, S., Ueda, R., Ohki-Hamazaki, H., UiTei, K., and Saigo, K. (2003). Short-interfering-RNA-mediated gene silencing in mammalian cells requires Dicer and eIF2C translation initiation factors. Curr. Biol. 13, 41-46. [DOI] [PubMed] [Google Scholar]
- Egel, R., and Egel-Mitani, M. (1974). Premeiotic DNA synthesis in fission yeast. Exp. Cell Res. 88, 127-134. [DOI] [PubMed] [Google Scholar]
- Enoch, T., Carr, A.M., and Nurse, P. (1992). Fission yeast genes involved in coupling mitosis to completion of DNA replication. Genes Dev. 6, 2035-2046. [DOI] [PubMed] [Google Scholar]
- Enoch, T., and Nurse, P. (1990). Mutation of fission yeast cell cycle control genes abolishes dependence of mitosis on DNA replication. Cell 60, 665-673. [DOI] [PubMed] [Google Scholar]
- Fagard, M., Boutet, S., Morel, J.B., Bellini, C., and Vaucheret, H. (2000). AGO1, QDE-2, and RDE-1 are related proteins required for post-transcriptional gene silencing in plants, quelling in fungi, and RNA interference in animals [In Process Citation]. Proc. Natl. Acad. Sci. USA 97, 11650-11654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire, A., Xu, S., Montgomery, M.K., Kostas, S.A., Driver, S.E., and Mello, C.C. (1998). Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806-811. [DOI] [PubMed] [Google Scholar]
- Fisher, D.L., and Nurse, P. (1996). A single fission yeast mitotic cyclin B p34cdc2 kinase promotes both S-phase and mitosis in the absence of G1 cyclins. EMBO J. 15, 850-860. [PMC free article] [PubMed] [Google Scholar]
- Forsburg, S.L., and Sherman, D.A. (1997). General purpose tagging vectors for fission yeast. Gene 191, 191-195. [DOI] [PubMed] [Google Scholar]
- Francesconi, S., Smeets, M., Grenon, M., Tillit, J., Blaisonneau, J., and Baldacci, G. (2002). Fission yeast chk1 mutants show distinct responses to different types of DNA damaging treatments. Genes Cells 7, 663-673. [DOI] [PubMed] [Google Scholar]
- Furnari, B., Rhind, N., and Russell, P. (1997). Cdc25 mitotic inducer targeted by chk1 DNA damage checkpoint kinase. Science 277, 1495-1497. [DOI] [PubMed] [Google Scholar]
- Gould, K.L., and Nurse, P. (1989). Tyrosine phosphorylation of the fission yeast cdc2+ protein kinase regulates entry into mitosis. Nature 342, 39-45. [DOI] [PubMed] [Google Scholar]
- Grishok, A., Pasquinelli, A.E., Conte, D., Li, N., Parrish, S., Ha, I., Baillie, D.L., Fire, A., Ruvkun, G., and Mello, C.C. (2001). Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106, 23-34. [DOI] [PubMed] [Google Scholar]
- Guertin, D.A., Chang, L., Irshad, F., Gould, K.L., and McCollum, D. (2000). The role of the sid1p kinase and cdc14p in regulating the onset of cytokinesis in fission yeast. EMBO J. 19, 1803-1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, I., Noma, K., and Grewal, S.I. (2003). RNA interference machinery regulates chromosome dynamics during mitosis and meiosis in fission yeast. Proc. Natl. Acad. Sci. USA 100, 193-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, I.M., Shankaranarayana, G.D., Noma, K., Ayoub, N., Cohen, A., and Grewal, S.I. (2002). Establishment and maintenance of a heterochromatin domain. Science 297, 2232-2237. [DOI] [PubMed] [Google Scholar]
- Hammond, S.M., Bernstein, E., Beach, D., and Hannon, G.J. (2000). An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404, 293-296. [DOI] [PubMed] [Google Scholar]
- Hammond, S.M., Boettcher, S., Caudy, A.A., Kobayashi, R., and Hannon, G.J. (2001). Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 293, 1146-1150. [DOI] [PubMed] [Google Scholar]
- Hannon, G.J. (2002). RNA interference. Nature 418, 244-251. [DOI] [PubMed] [Google Scholar]
- He, X., Patterson, T.E., and Sazer, S. (1997). The Schizosaccharomyces pombe spindle checkpoint protein mad2p blocks anaphase and genetically interacts with the anaphase-promoting complex. Proc. Natl. Acad. Sci. USA 94, 7965-7970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman, C.S., and Winston, F. (1987). A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene 57, 267-272. [DOI] [PubMed] [Google Scholar]
- Jimenez, G., Yucel, J., Rowley, R., and Subramani, S. (1992). The rad3+ gene of Schizosaccharomyces pombe is involved in multiple checkpoint functions and in DNA repair. Proc. Natl. Acad. Sci. USA 89, 4952-4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpen, G.H., and Allshire, R.C. (1997). The case for epigenetic effects on centromere identity and function. Trends Genet. 13, 489-496. [DOI] [PubMed] [Google Scholar]
- King, F.J., Szakmary, A., Cox, D.N., and Lin, H. (2001). Yb modulates the divisions of both germline and somatic stem cells through piwi- and hh-mediated mechanisms in the Drosophila ovary. Mol. Cell 7, 497-508. [DOI] [PubMed] [Google Scholar]
- Knight, S.W., and Bass, B.L. (2001). A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science 293, 2269-2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Goff, X., Utzig, S., and Simanis, V. (1999). Controlling septation in fission yeast: finding the middle, and timing it right. Curr. Genet. 35, 571-584. [DOI] [PubMed] [Google Scholar]
- Lee, J.Y., and Orr-Weaver, T.L. (2001). The molecular basis of sister-chromatid cohesion. Annu. Rev. Cell Dev. Biol. 17, 753-777. [DOI] [PubMed] [Google Scholar]
- Lee, M.S., Enoch, T., and Piwnica-Worms, H. (1994). mik1+ encodes a tyrosine kinase that phosphorylates p34cdc2 on tyrosine 15. J. Biol. Chem. 269, 30530-30537. [PubMed] [Google Scholar]
- Lundgren, K., Walworth, N., Booher, R., Dembski, M., Kirschner, M., and Beach, D. (1991). mik1 and wee1 cooperate in the inhibitory tyrosine phosphorylation of cdc2. Cell 64, 1111-1122. [DOI] [PubMed] [Google Scholar]
- Lynn, K., Fernandez, A., Aida, M., Sedbrook, J., Tasaka, M., Masson, P., and Barton, M.K. (1999). The PINHEAD/ZWILLE gene acts pleiotropically in Arabidopsis development and has overlapping functions with the ARGONAUTE1 gene. Development 126, 469-481. [DOI] [PubMed] [Google Scholar]
- Matzke, M., Matzke, A.J., and Kooter, J.M. (2001). RNA: guiding gene silencing. Science 293, 1080-1083. [DOI] [PubMed] [Google Scholar]
- McCollum, D., and Gould, K.L. (2001). Timing is everything: regulation of mitotic exit and cytokinesis by the MEN and SIN. Trends Cell Biol. 11, 89-95. [DOI] [PubMed] [Google Scholar]
- McGowan, C.H., and Russell, P. (1993). Human Wee1 kinase inhibits cell division by phosphorylating p34cdc2 exclusively on Tyr15. EMBO J. 12, 75-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar, J.B., McGowan, C.H., Lenaers, G., Jones, R., and Russell, P. (1991). p80cdc25 mitotic inducer is the tyrosine phosphatase that activates p34cdc2 kinase in fission yeast. EMBO J. 10, 4301-4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel, J.B., Godon, C., Mourrain, P., Beclin, C., Boutet, S., Feuerbach, F., Proux, F., and Vaucheret, H. (2002). Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance. Plant Cell 14, 629-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno, S., Klar, A., and Nurse, P. (1991). Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194, 795-823. [DOI] [PubMed] [Google Scholar]
- Moussian, B., Schoof, H., Haecker, A., Juergens, G., and Laux, T. (1998). Role of the ZWILLE gene in the regulation of central shoot meristem cell fate during Arabidopsis embryogenesis. EMBO J. 17, 1799-1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka, N., Kitajima, T., Yokobayashi, S., Xiao, G., Yamamoto, M., Grewal, S.I., and Watanabe, Y. (2002). Recruitment of cohesin to heterochromatic regions by Swi6/HP1 in fission yeast. Nat. Cell Biol. 4, 89-93. [DOI] [PubMed] [Google Scholar]
- Nurse, P. (1975). Genetic control of cell size at cell division in yeast. Nature 256, 547-551. [DOI] [PubMed] [Google Scholar]
- Nurse, P. (1990). Universal control mechanism regulating onset of M-phase. Nature 344, 503-508. [DOI] [PubMed] [Google Scholar]
- Okazaki, N., Okazaki, K., Watanabe, Y., Kato-Hayashi, M., Yamamoto, M., and Okayama, H. (1998). Novel factor highly conserved among eukaryotes controls sexual development in fission yeast. Mol. Cell Biol. 18, 887-895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal-Bhadra, M., Bhadra, U., and Birchler, J.A. (2002). RNAi related mechanisms affect both transcriptional and posttranscriptional transgene silencing in Drosophila. Mol. Cell 9, 315-327. [DOI] [PubMed] [Google Scholar]
- Parrish, S., and Fire, A. (2001). Distinct roles for RDE-1 and RDE-4 during RNA interference in Caenorhabditis elegans. RNA 7, 1397-1402. [PMC free article] [PubMed] [Google Scholar]
- Provost, P., Silverstein, R., Dishart, D., Walfridsson, J., Djupedal, I., Kniola, B., Wright, A., Samuelsson, B., Radmark, O., and Ekwall, K. (2002). Dicer is required for chromosome segregation and gene silencing in fission yeast cells. Proc. Natl. Acad. Sci. USA 99, 16648-16653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raleigh, J.M., and O'Connell, M.J. (2000). The G(2) DNA damage checkpoint targets both Wee1 and Cdc25. J. Cell Sci. 113, 1727-1736. [DOI] [PubMed] [Google Scholar]
- Rhind, N., Furnari, B., and Russell, P. (1997). Cdc2 tyrosine phosphorylation is required for the DNA damage checkpoint in fission yeast. Genes Dev. 11, 504-511. [DOI] [PubMed] [Google Scholar]
- Rhind, N., and Russell, P. (1998). Tyrosine phosphorylation of cdc2 is required for the replication checkpoint in Schizosaccharomyces pombe. Mol. Cell Biol. 18, 3782-3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley, R., Subramani, S., and Young, P.G. (1992). Checkpoint controls in Schizosaccharomyces pombe: rad1. EMBO J. 11, 1335-1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy, S., and Ingham, P.W. (2002). Hedgehogs tryst with the cell cycle. J. Cell Sci. 115, 4393-4397. [DOI] [PubMed] [Google Scholar]
- Russell, P., and Nurse, P. (1986). cdc25+ functions as an inducer in the mitotic control of fission yeast. Cell 45, 145-153. [DOI] [PubMed] [Google Scholar]
- Russell, P., and Nurse, P. (1987). Negative regulation of mitosis by wee1+, a gene encoding a protein kinase homolog. Cell 49, 559-567. [DOI] [PubMed] [Google Scholar]
- Shiozaki, K., and Russell, P. (1996). Conjugation, meiosis, and the osmotic stress response are regulated by Spc1 kinase through Atf1 transcription factor in fission yeast. Genes Dev. 10, 2276-2288. [DOI] [PubMed] [Google Scholar]
- Smardon, A., Spoerke, J.M., Stacey, S.C., Klein, M.E., Mackin, N., and Maine, E.M. (2000). EGO-1 is related to RNA-directed RNA polymerase and functions in germ-line development and RNA interference in C. elegans. Curr. Biol. 10, 169-178. [DOI] [PubMed] [Google Scholar]
- Stern, B., and Nurse, P. (1998). Cyclin B proteolysis and the cyclin-dependent kinase inhibitor rum1p are required for pheromone-induced G1 arrest in fission yeast. Mol. Biol. Cell 9, 1309-1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stettler, S., Warbrick, E., Prochnik, S., Mackie, S., and Fantes, P. (1996). The wis1 signal transduction pathway is required for expression of cAMP-repressed genes in fission yeast. J. Cell Sci. 109, 1927-1935. [DOI] [PubMed] [Google Scholar]
- Su, S.S., Tanaka, Y., Samejima, I., Tanaka, K., and Yanagida, M. (1996). A nitrogen starvation-induced dormant G0 state in fission yeast: the establishment from uncommitted G1 state and its delay for return to proliferation. J. Cell Sci. 109, 1347-1357. [DOI] [PubMed] [Google Scholar]
- Tabara, H., Sarkissian, M., Kelly, W.G., Fleenor, J., Grishok, A., Timmons, L., Fire, A., and Mello, C.C. (1999). The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell 99, 123-132. [DOI] [PubMed] [Google Scholar]
- Tahbaz, N., Carmichael, J.B., and Hobman, T.C. (2001). GERp95 belongs to a family of signal-transducing proteins and requires Hsp90 activity for stability and Golgi localization. J. Biol. Chem. 276, 43294-43299. [DOI] [PubMed] [Google Scholar]
- Taricani, L., Feilotter, H.E., Weaver, C., and Young, P.G. (2001). Expression of hsp16 in response to nucleotide depletion is regulated via the spc1 MAPK pathway in Schizosaccharomyces pombe. Nucleic Acids Res. 29, 3030-3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe, T., Schramke, V., Hamilton, G.L., White, S.A., Teng, G., Martienssen, R.A., and Allshire, R.C. (2003). RNA interference is required for normal centromere function in fission yeast. Chromosome Res. 11, 137-146. [DOI] [PubMed] [Google Scholar]
- Volpe, T.A., Kidner, C., Hall, I.M., Teng, G., Grewal, S.I., and Martienssen, R.A. (2002). Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297, 1833-1837. [DOI] [PubMed] [Google Scholar]
- Warbrick, E., and Fantes, P.A. (1991). The wis1 protein kinase is a dosage-dependent regulator of mitosis in Schizosaccharomyces pombe. EMBO J. 10, 4291-4299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, L., and Russell, P. (1997). Roles of Wee1 and Nim1 protein kinases in regulating the switch from mitotic division to sexual development in Schizosaccharomyces pombe. Mol. Cell Biol. 17, 10-17. [DOI] [PMC free article] [PubMed] [Google Scholar]