Abstract
A posttranslational protein O-mannosylation process resembling that found in fungi and animals has been reported in the major human pathogen Mycobacterium tuberculosis (Mtb) and related actinobacteria. However, the role and incidence of this process, which is essential in eukaryotes, have never been explored in Mtb. We thus analyzed the impact of interrupting O-mannosylation in the nonpathogenic saprophyte Mycobacterium smegmatis and in the human pathogen Mtb by inactivating the respective putative protein mannosyl transferase genes Msmeg_5447 and Rv1002c. Loss of protein O-mannosylation in both mutant strains was unambiguously demonstrated by efficient mass spectrometry-based glycoproteomics analysis. Unexpectedly, although the M. smegmatis phenotype was unaffected by the lack of manno-proteins, the Mtb mutant had severely impacted growth in vitro and in cellulo associated with a strong attenuation of its pathogenicity in immunocompromised mice. These data are unique in providing evidence of the biological significance of protein O-mannosylation in mycobacteria and demonstrate the crucial contribution of this protein posttranslational modification to Mtb virulence in the host.
Keywords: protein glycosylation, O-glycosylation, proteomics
With more than three deaths per minute, Mycobacterium tuberculosis (Mtb) is still a major public health threat, exacerbated by the emergence of multi- or extensively drug-resistant Mtb strains. This situation reflects the weakness of the therapeutic arsenal and highlights the urgent need for a comprehensive understanding of the determinants of Mtb pathogenicity for alternative therapeutics to fight tuberculosis.
We recently described Mtb manno-proteins as an emerging class of bacterial adhesins that contribute to Mtb infectiousness through binding to host innate-immune system receptors, including lung surfactant protein A (SP-A) (1) and DC-SIGN (2), which are considered to be the receptors preferentially used by mycobacteria to enter target cells and evade host defense mechanisms (3). Here, we considered the systemic impact of protein mannosylation on the survival and virulence of Mtb. Indeed the role of the protein glycan chains remains elusive (4), although it has been established that the immunodominant Mtb-secreted alanine and proline-rich antigen (Apa) interacts with the host’s lectin receptors through its mannosyl appendages, which indirectly contribute to the colonization and invasion of the host cell. Moreover, changes in the mannosylation pattern of the Mycobacterium bovis bacillus Calmette–Guérin Apa alter its ability to stimulate CD4+ and CD8+ T lymphocyte responses (5, 6), contributing to the protective properties of bacillus Calmette–Guérin vaccination against tuberculosis (7). In addition, glycosylation (most probably O-mannosylation) of the Mtb lipoprotein LpqH modulates its antigenic processing (8), and glycan decorations of the Mycobacterium leprae lipoprotein LprG have been reported to be indispensable for MHC II-restricted T-cell activation in patients with lepromatous leprosy (9). However, there are still insufficient data to determine clearly the roles and overall influence of mannosylation of mycobacterial proteins on Mtb physiology and infectiousness.
In eukaryotes, protein O-mannosylation is an essential process catalyzed by membrane-associated protein O-mannosyl transferases (PMTs), which transfer mannose residues from lipid carriers (dolichyl-phosphate) to proteins (10). An Mtb genome search for sequence homology with the eukaryotic PMTs identified Rv1002c as the unique putative PMT gene (11). Ectopic expression of Rv1002c in Mycobacterium smegmatis confirmed its mannosyl transferase function and demonstrated that this membrane-associated activity is coupled to the Sec-dependent protein export system, suggesting that O-mannosylation should affect only extracytoplasmic proteins (11). As for the eukaryote PMTs, Rv1002c is categorized as a gene essential for optimal growth of M. tuberculosis in vitro (12, 13). This assumption is corroborated by the high conservation of Rv1002c homologs throughout the mycobacterial genus and, in particular, in the M. leprae genome, which is considered as the minimal set of genes essential for mycobacteria survival (14). On the other hand, PMT knockout mutants of the phylogenetically related Streptomyces coelicolor (15) or Corynebacterium glutamicum (16) are viable and suggest that this posttranslational modification is dispensable in the actinomycetae family. However, because of the proposed similarity of the protein O-mannosylation mechanism in eukaryotes and Mtb, we hypothesized that the conservation in pathogenic mycobacteria of such cell energy-consuming process should result from evolutionary selection of a protein modification required for the parasite to persist in its natural biotope: the infected host. To test this hypothesis we interrupted the putative PMT genes in the nonpathogenic saprophyte M. smegmatis and the human pathogen Mtb, and we verified whether posttranslational O-mannosylation of bacterial proteins is essential for Mtb. Using an efficient and original mass spectrometry-based glycoproteomic approach to tackle the highly challenging molecular analysis of the posttranslational glycosyl protein modifications, we confirmed the interruption of the mannosylation process and analyzed the impact of this interruption on in vitro and ex vivo phenotype and on virulence of Mtb in mice.
Results and Discussion
M. smegmatis Msmeg_5447 Gene Coding for the Putative PMT Is Not Essential.
An in silico search for Mtb Rv1002c gene homologs (Fig. S1B) in the complete genome of the nonpathogenic saprophyte M. smegmatis mc2-155 revealed a unique ORF (Msmeg_5447: 1,551 pb) coding for a putative 516-amino acid membrane protein sharing 72% identity and 81% homology over 489 amino acids with the Mtb PMT sequence (Fig. S1A). An M. smegmatis insertion mutant, ΔM5447, was constructed by disruption of Msmeg_5447 by allelic exchange (Fig. S1 C and D). Phenotypic analysis of the mutant clearly indicated the dispensability of the putative PMT function for M. smegmatis, which contrasts with the essential nature of PMT in eukaryotes. Indeed, there was no significant difference in growth between the WT and the insertion mutant (ΔM5447) (Fig. 1A). Similarly, the relative susceptibility of both strains to fluoroquinolone antibiotic (ciprofloxacin) (Fig. 1B) was similar, suggesting that Msmeg_5447 inactivation had little effect on metabolism or cell wall permeability to drugs. In contrast, tolerance to cell wall stress induced by the SDS detergent was slightly reduced in the ΔM5447 mutant in comparison with the WT (Fig. 1C), which was fully reversed in the complemented mutant strain. The increase in sensitivity to SDS may reflect changes in cell wall organization or composition of the ΔM5447 mutant, weakening its physical and chemical protective properties; however, transmission electron microscopy did not reveal any noticeable differences in the cell wall aspect or cell shape (Fig. 1 D and E) of the WT and the ΔM5447 mutant.
Fig. 1.
The Msmeg_5447 gene encoding the putative ortholog of the Mtb Rv1002c mannosyl transferase gene is dispensable for M. smegmatis growth in vitro. Effects of Msmeg_5447 inactivation on the growth (A) and the sensitivity of the M. smegmatis ΔM5447 mutant to the antituberculosis drug Ciprofloxacin (B), and detergent (SDS)-induced cell wall stress (C). Growth and survival were followed by monitoring the OD at 600 nm (A) or the level of MTT reduction by the metabolically active cells at 570 nm (B and C). (D and E) Transmission electron micrographs of negatively stained M. smegmatis wild-type (Wt) and ΔM5447 mutant cells.
Evidence for the Putative PMT Function of the Msmeg_5447 Gene Product.
In the absence of major phenotypic changes associated with inactivation of Msmeg_5447, we analyzed the mutant and WT secreted proteomes to search for potential alterations in protein glycosylation. Comparative SDS/PAGE analysis (Fig. 2A) of the culture filtrates revealed a major difference corresponding to the downsizing of an intense protein gel band from 30 to 25 kDa, consistent with a glycosylation defect in the mutant. Complementation of the mutant strain with Msmeg_5447 led to restoration of the original mobility of the gel band at around 30 kDa, thus confirming that this alteration is related to the Msmeg_5447 gene-product deficiency. Proteomic analysis by nanoLC-MS/MS of the in-gel trypsin digest of the corresponding excised gel bands allowed the identification of several peptides attributable to the predicted M. smegmatis fasciclin domain protein (FasC) (Fig. 2B and Table S1). A recombinant C-terminal histidine tagged FasC (rFasC) was then constructed for expression in M. smegmatis, followed by further purification for precise structural characterization. Carbohydrate content capillary zone electrophoresis analysis (Fig. 2C) and α-exo-mannosidase susceptibility study (Fig. 2D) confirmed that the purified rFasC is glycosylated by α-mannose residues exclusively. Mass spectrometry analysis of the rFasC trypsin digest afforded 42% amino acid sequence coverage and determined the location of the glycosylation (up to four hexose residues) on the N-terminal P30–55 peptide (Fig. 2 E and F, and Tables S1 and S2). Accurate identification of the glycosylated threonine residues was achieved by complementary electron transfer dissociation (ETD) MS/MS analysis (17). Interestingly, we found, that in the rapid growing saprophyte M. smegmatis, as reported in other mycobacteria, the carbohydrate motifs are not randomly distributed along the four potential hydroxylated amino acid acceptors (Thr33, Thr34, Thr35, Thr40) of the P30–55 peptide but are exclusively located on adjacent Thr residues (Thr33-Thr34 doublet) (Fig. S2 and Table S3) (18–20). The identification and the determination of the glycosylation sites of FasC, which is the first ever mannoprotein characterized in M. smegmatis, allowed us to explore the alteration of protein O-mannosylation in the mutant by monitoring specifically the glycosylation status of the FasC P30–55 peptide by mass spectrometry. Searching for the molecular ions of the FasC P30–55 reporter peptide in the ΔM5447 mutant culture filtrate by nanoLC-MS analysis readily revealed the absence of peptide glycosylation (Fig. 2G, Bottom trace), in agreement with the apparent mass reduction of the mutant FasC protein observed by SDS/PAGE (Fig. 2A). Interestingly, the glycosylation defect was reversed by complementation with the Msmeg_5447 gene as well as with the Mtb Rv1002c ortholog (Fig. 2H), providing evidence of functional identity of these genes.
Fig. 2.
Inactivation of the Msmeg_5447 gene interrupts the O-mannosylation of the M. smegmatis FasC protein. (A) Identification of the Msmeg_5447 protein target by silver nitrate-stained SDS/PAGE of the culture filtrate protein extract from M. smegmatis WT, ΔM5447 mutant, and complemented mutant (ΔM5447:M5447) revealing the electrophoretic mobility alteration of a major protein gel band (arrows). (B) Amino acid sequence of the recombinant His-Tagged rFasC (SignalP 3.0 predicted signal peptide is in italic; NetOGlyc 3.1 predicted S and T glycosylation sites are boldface; Histidine tag is into brackets; yellow highlights the amino acid sequence covered by mass spectrometry analysis). (C) Carbohydrate content analysis and (D) Coomassie blue-stained SDS/PAGE of the rFasC, the α-exo-mannosidase–treated rFasC, and the α-exo-mannosidase sham control (Standard: manno-heptose). (E) Identification of the triglycosylated rFasC N-terminal peptide P30–55 by CID MS/MS [(P30–55Hex3+3H)3+ at m/z 1680.23]. Underlined masses correspond to fragment ions resulting from one to three neutral losses of hexose from the parent ion. (F) FasC N-terminal peptide P30–55 sequence reporting the major informative peptide cleavages (b, c, and y peptide fragment ions) observed in the positive CID and ETD MS/MS spectra (Fig. S2). (G) Reconstructed ion chromatograms (mass tolerance: 10 ppm, Sum of [M+3H]3+ [M+2H]2+) of the native FasC P30-55 glycoforms detected in the culture filtrates of the WT and the mannosyl transferase mutants. (H) Cumulated normalized abundances of the native FasC P30–55 glycoforms observed in the mannosyl transferase mutants and their respective complemented strains.
Initiation and Elongation of the FasC Mannosyl Appendages Requires Msmeg_5447 PMT and PimE.
O-glycosylation in bacteria has been described to proceed either by en bloc transfer (of a lipid-linked oligosaccharide to the protein) or by a stepwise processing initiated by attachment of an initial glycosyl residue to the target serine or threonine and continued by elongation of the oligosaccharide chain by successive glycosylation. By analogy with the eukaryote PMT-mediated process, protein O-mannosylation in Mtb has been considered to proceed according to the latter mechanism (11), which is the exclusive rule for O-glycosylation in eukaryotes. However, the mycobacterial enzymes involved in the elongation process are not known. The oligomannosyl chains of the Mtb proteins have been found elongated through α1–2 interglycosidic linkages (18, 20), as in the polar phosphatidyl inositol mannosides (PIM6) or the mannose branches of the lipomannan and lipoarabinomannan core (21, 22). Thus, mutants of the α1–2 mannosyl transferases Msmeg_5149 (pimE) (23) and Msmeg_4247, respectively responsible for the elongation of PIMs and lipoglycans in M. smegmatis, were constructed to verify the possible involvement of these enzymes in the elongation of the FasC mannosyl chain. As revealed by the reporter FasC P30–55 glycoforms detected by nanoLC-MS in the culture filtrate of the ΔM4247 mutant (Fig. 2G), disruption of Msmeg_4247 failed to induce any significant modification of the distribution of the reporter glycopeptide, suggesting that this α1–2 mannosyl transferase does not contribute to the elongation of FasC mannosyl chains. In contrast, inactivation of the PimE mannosyl transferase resulted as expected in the loss of the polar PIMs (Fig. S3B), but also in the total extinction of the triglycosylated form of the FasC P30–55 peptide with the presence of trace amounts (<1.5%) of the diglycosylated isoform (Fig. 2G). This glycosylation defect was reversed by complementation with the Mtb pimE (Rv1159) gene (Fig. 2H), which confirms the involvement of the Msmeg_5149 mannosyl transferase in the elongation of FasC P30–55 oligomannosyl appendages and the functional identity between Msmeg_5149 and Rv1159. These results show that, protein mannosylation in M. smegmatis occurs through a stepwise process initiated by the transfer of the primary mannose residue onto protein by the PMT encoded by Msmeg_5447 followed by the elongation of the mannosyl chain through α1–2 linkages by PimE. It is noteworthy that the pleiotropic function of the M. smegmatis PimE evokes the shared glycosylation machinery for glycolipids and glycoproteins reported in the Gram-negative pathogens (24). However, unlike the Gram-negative bacteria oligosaccharidyl-transferase, which catalyzes the oligosaccharide transfer en bloc from glycolipids to acceptor glycoproteins, PimE catalyzes the stepwise oligomannosyl chain elongation indifferently on manno-proteins and manno-lipids, proving a remarkable relaxed specificity. In contrast, the PMT is protein-specific and unlikely to contribute to the PIM biosynthesis (Fig. S3C).
Absence of Phenotype for the ΔM5447 Mutant Is Not Because of PMT Functional Complementation.
To determine whether the absence of a phenotype in the ΔM5447 mutant was a result of the presence of homologous PMT with nonoverlapping target specificity, as seen in eukaryotes, we attempted to evaluate the extent of the extinction of the protein mannosylation process resulting from Msmeg_5447 disruption. This attempt was achieved by implementing a dedicated original nanoLC-MS/MS–based glycopeptidomic strategy to search specifically for glycosylated peptides in a complex proteome digest containing thousands of nonglycosylated peptides. The approach relies on the widely reported observation that collision-induced dissociation (CID) mass spectra of O-glycosylated peptides are dominated by intense fragment ions resulting from loss of the carbohydrates as neutral fragments from the parent ion (25). An automatic filter was thus designed to extract selectively the CID MS/MS spectra exhibiting such a characteristic signature for the major carbohydrate residue types encountered in mycobacteria [i.e., pentose (arabinose), amino-hexose (amino-galactose), and hexose (galactose, glucose, mannose)]. This specific MS/MS data analysis was applied to the secreted proteomes of M. smegmatis WT and mutant strains after protein fractionation by SDS/PAGE into 12 bands and nanoLC-MS/MS analysis of the corresponding trypsin digests. Among more than 2 × 105 MS/MS spectra acquired for the WT M. smegmatis secreted proteome, fewer than 2,000 MS/MS spectra corresponding to about 300 precursor ions were selected. The selected spectra exclusively exhibited “hexose loss signatures” suggesting the absence of any other type of protein glycosylation in M. smegmatis; however, only a minor proportion (40) of these spectra could be attributed with confidence to glycopeptides, allowing the characterization of at least 10 M. smegmatis unique glycoproteins experimentally confirmed by mass spectrometry (Table 1). Interestingly, comparative glycoproteomic analysis of the ΔM5447 mutant confirmed that none of the peptides identified above were found as glycosylated in the mutant, although they were all present in the complemented strain culture filtrate. Taken together, these data provide evidence that O-glycosylation of the M. smegmatis proteins is exclusively dependent on Msmeg_5447 and confirm the dispensability of this process for M. smegmatis, as in other saprophytic actinomycetae (4).
Table 1.
M. smegmatis glycoproteins identified by glycopeptidomics analysis of the WT strain culture filtrate
| Protein description | Access no.* | Gene no. | MM† | Peptide sequence | Hexose no. |
| Fasciclin domain protein | A0R2Q4 | 5196 | 3020.3 | (A)EPETTTEAEPTVEIPDPQGPGC‡DDFK(K) | 1–3 Hex |
| 3148.4 | (A)EPETTTEAEPTVEIPDPQGPGC‡DDFKK(A) | 1–3 Hex | |||
| ABC-type amino acid transport system secreted component |
A0QXB0 | 3235 | 1867.8 | (S)GEGGGESSPESTQAASGAK(V) | 1–2 Hex |
| 1954.8 | (A)SGEGGGESSPESTQAASGAK(V) | 1–2 Hex | |||
| 2025.9 | (C)ASGEGGGESSPESTQAASGAK(V) | 1–3Hex | |||
| Putative protein | A0R6M4 | 6601 | 2574.3 | (K)STTPTTPLPPPLPAEVGGSAAK(A) | 3–5 Hex |
| Putative uncharacterized protein |
A0QYI9 | 3674 | 2777.3 | (K)VILMLDC§TEAAAQQAQDTAVSGGPR(V) | 1 Hex |
| 3068.5 | (R)VGSN¶GVTTVTPTPVVPIAGAPGAGTPPPA(-) | 3–4 Hex | |||
| Putative uncharacterized protein |
A0QRP2 | 1188 | 1876.9 | (R)AQPVTPTQSDTVPGR(Y) | 2 Hex |
| 3702.9 | (R)YLPPVPHEPDPVALPVTVLTPPPAQPVAPNTR(T) | 2 Hex | |||
| Immunogenic protein MPT63 | A0R3B5 | 5412 | 1904.0 | (T)TTVPAPAPTTAPVSHGPAA(-) | 1 Hex |
| 2389.2 | (K)DLLVWDKPAASATAPSGTGQSR(P) | 1 Hex | |||
| 4227.9 | (R)ATDLPAATAAEAETEATATDAETTPVTEETPAAPAPEGTTPV(S) | 1 Hex | |||
| Putative protein | A0QSU1 | 1600 | 1994.0 | (K)LQTLTTTVATVNDEAQK(Q) | 1 Hex |
| Immunogenic protein MPT63 | A0QQP4 | 828 | 1492.7 | (R)PAATGSGTSASTPAR(T) | 1 Hex |
| Putative protein | A0QVP4 | 2645 | 3127.5 | (K)WPVIETTDPKPFDPC‡NDIPIDVIER(I) | 1 Hex |
| Alanine and proline-rich secreted protein apa | A0QYD3 | 3618 | 2119.0 | (R)PGVGVPVPVTDAPPEMMPPA(-) | 1 Hex |
Boldface T and S correspond to potential glycosylation sites according NetOGlyc (v3.1) predictions.
UniProtKB accession number.
Experimental mass of the glycosylated peptide of lower glycosylation degree.
Carboxymethylated cysteine.
Cysteine acrylamide adduct.
Deamidation.
Construction and Phenotype of the Mtb ΔRv1002c Mutant.
Because the Mtb PMT mutant has never been reported, we next sought to use the same strategy to analyze the impact of a protein mannosylation defect on the biology of the human pathogen Mtb. The Msmeg_5447 ortholog in Mtb, Rv1002c, was predicted to be essential for Mtb viability, but we succeeded in disrupting it by homologous recombination (Fig. S4). Although the ΔRv1002c mutant was viable, it had profoundly impaired growth in vitro. The mutant exhibited a slight growth delay in Albumin-Dextrose-Catalase–enriched 7H9 broth (Fig. 3A) but was almost completely unable to grow in 7H9 supplemented with dextrose only (Fig. 3A, Inset). This inability to grow in the absence of supplemented proteins was readily reversed by complementation with the WT gene, demonstrating that inactivation of Rv1002c is the sole cause of the observed phenotype. A severe growth defect was also observed on solid medium. Indeed, compared with the WT or the complemented mutant, the ΔRv1002c mutant showed a 103-fold attenuation in its capacity to form colonies on albumin-enriched 7H11 solid medium (Fig. 3B). Similar diminished ability of Mtb mutants to form colonies on solid medium has been previously attributed to disruption of cell wall integrity, resulting in an increased susceptibility to chaotropic or bacteriostatic agents (such as Malachite green, ordinarily incorporated in solid media for mycobacterial culture) (26). However, the persistence of the deficiency to form colony on Malachite green-free agar medium (Fig. S5A), together with the absence of any noticeable increase in sensitivity to SDS or ciprofloxacin (Fig. S5 B and C), fails to support any functional alteration of cell wall integrity in the ΔRv1002c mutant.
Fig. 3.
Inactivation of the M. tuberculosis Rv1002c gene impairs Apa protein O-mannosylation and impacts on Mtb growth. (A) Impact of the Rv1002c inactivation on the Mtb ΔRv1002c mutant growth in Middlebrook 7H9 broth with Albumin-Dextrose-Catalase or dextrose alone (Inset). (B) Growth impairment of the mutant on solid Middlebrook 7H11 agar medium. Serial dilution of cultures containing 108 bacteria per milliliter were plated and colony sizes were compared after 4 wk of growth at 37 °C. (C, Right) Relative abundances of the Apa peptides detected in the culture filtrates of wild-type (Wt), ΔRv1002c (Mt), and complemented ΔRv1002c:Rv1002c (Cp) mutant (sum of the abundances are set at 100% for each strain). (Left) Weighted deviation from the mean relative abundance of each peptide in the three strains. (D) LC-MS reconstructed ion chromatograms of the P278–321, P285–321, and P287–321 Apa C-terminal peptides and their monoglycosylated forms (*) detected in the different Mtb strains; (reconstructed ion chromatograms parameters as in Fig. 2).
ΔRv1002c Mutant Apa Protein Is Devoid of Mannosylation.
We sought to characterize the molecular phenotype of the Mtb ΔRv1002c mutant. Unfortunately, the global glycopeptidomics approach used to verify the interruption of protein O-mannosylation in M. smegmatis could not be used here because of the prohibitive concentration of albumin (2.5 g/L) required in the culture medium for the mutant to grow, which was more than 10-fold the concentration of bacterial proteins (∼100–200 mg/L). Thus, to confirm the abrogation of the protein O-mannosylation process resulting from the Rv1002c disruption, we specifically monitored the glycosylation defects in the reporter protein Apa known to be glycosylated and secreted by Mtb (27, 28). To this end, the Mtb WT, mutant (ΔRv1002c), and complemented (ΔRv1002c:Rv1002c) mutant culture filtrates were analyzed by shotgun proteomics. Comparative analysis of the Apa peptides identified by nanoLC-MS/MS revealed relatively low interstrain variability in the abundance (estimated from the area under the curve of the selected ion chromatogram) of the individual peptides except for the C-terminal peptides P278–321, P285–321, and P287–321 and their monoglycosylated counterparts G1P278–321, G1P285–321, and G1P287–321 (Fig. 3C). The corresponding peptide ion reconstructed chromatograms confirmed that, in the WT and complemented strains, both glycosylated and nonglycosylated forms were observed for the peptides P278–321, P285–321, and P287–321 (Fig. 3D), whereas only the unglycosylated peptides were observed in the mutant strain (Fig. 3D). Further normalization of the peptide glycoform abundances (relative to the closely eluted prototypic P174–184 Apa peptide used as internal standard) supported this observation by showing that the mannosylation defect was correlated to a relative increase in the abundances of the unmodified peptides (Fig. 3C, Left). These results clearly demonstrate that disruption of Rv1002c in Mtb abolishes protein mannosylation, similar to the situation in the model strain M. smegmatis.
Maintenance of the ΔRv1002c Mutant in Macrophages Is Compromised.
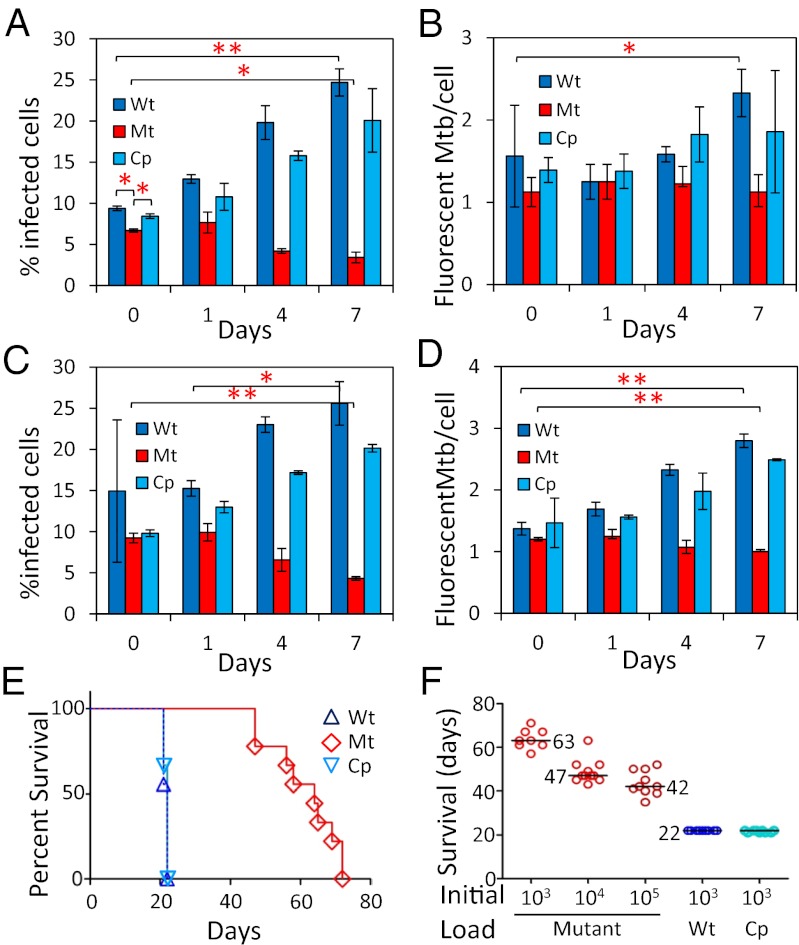
We next investigated the impact of the deficiency in Rv1002c-dependent protein O-mannosylation on the interaction of Mtb with the host. The growth defect on solid medium induced by the inactivation of Rv1002c clearly impaired the use of the conventional counting of colony forming units to monitor the intracellular survival of the mutant. Fluorescent Mtb strains were thus generated by expressing the gfp gene from a replicative plasmid in the parental WT, the ΔRv1002c mutant, and the complemented mutant to evaluate the consequences of the inactivation of the Rv1002c gene on the ability of the mutant to infect and proliferate in host cells. Resting cultures of murine alveolar macrophage cell line and primary human monocyte-derived macrophages were infected for 1 h at a multiplicity of infection (MOI) of one bacterium per macrophage. In the murine macrophages, the initial rate of infection is slightly lower for the mutant than for the WT or complemented strains (Fig. 4A). This finding is consistent with the view that surface-exposed glycoproteins such Apa may act as bacterial adhesins for the phagocyte membrane C-type lectin receptors (1). However, the absence of similar statistical difference with the human macrophages (Fig. 4C) not only reveals subtle differences in the impact of the surface manno-proteins on the initial uptake by mouse or human phagocytes, but also suggests that the overall divergence observed between the mutant and the WT intracellular behavior is not formally related to an altered initial capacity of the mutant to interact with the host cell. Indeed, the main difference between the ΔRv1002c mutant and the Mtb WT or complemented mutant strains was the almost total incapacity of the mutant to proliferate within the phagocytes. The Mtb WT strain proliferated in either macrophage cell culture, as shown by both the number of infected macrophages and the bacterial load values, which increased perceptibly as early as 48 h postinfection and had almost doubled at day 7 (Fig. 4). On the other hand, the number of ΔRv1002c mutant fluorescent cells per infected macrophage remained almost stable, and the number of macrophages infected by the mutant cells declined by about 50% over the 7-d observation period. These findings indicate a possible block in the capacity of the mutant to multiply within the macrophage. This phenotype was nearly completely reversed in the complemented strain, confirming that the functional disruption of the putative PMT encoded by Rv1002c dramatically compromises the mutant ability to survive and multiply in phagocytes.
Fig. 4.
Rv1002c-dependent protein O-mannosylation is required for Mtb intracellular persistence and proliferation and for full virulence in SCID mice. (A–C) Persistence and (B–D) intracellular proliferation of GFP-tagged Mtb wild-type (Wt), ΔRv1002c mutant (Mt), and complemented mutant (Cp) in the mouse alveolar macrophage cell line (A and B) and primary human blood monocyte-derived macrophages (C and D). [Data (Table S5) were analyzed by one-way Anova tests: *P < 0.05, **P < 0.01]. Survival of immunocompromised SCID mice infected intranasally with 103 Mtb WT, ΔRv1002c mutant, or complemented mutant cells (E), or with 103, 104, or 105 ΔRv1002c mutant cells or 103 Mtb WT and complemented mutant cells (F). Numbers correspond to the median survival time.
Virulence of the ΔRv1002c Mutant Is Attenuated in SCID Mice.
Given the incapacity of the mutant to proliferate in macrophages, we predicted a severe loss of pathogenicity in vivo, even in the absence of host-acquired immunity. To test this theory, virulence assays were carried out by infecting immunocompromised SCID mice via the intranasal route with 103 cells of the Mtb parental WT, the ΔRv1002c mutant, or the complemented mutant. Control mice infected with the virulent WT Mtb strain did not survive for more than 22 d, whereas no death was observed until day 47 (median survival time = 63 d, n = 9) in the mice infected with the mutant strain (Fig. 4E). By increasing the dose of ΔRv1002c mutant, it was clear that survival time was dependent on the initial bacterial load used to infect the mice (Fig. 4F). A rough estimate of the relative pathogenicity of the mutant, obtained by extrapolating the median survival time as a function of the initial bacterial load, indicated a virulence attenuation of about 10−3 to 10−4 [103- to 104-fold more mutant bacteria are required to give a median survival time (∼22 d) equivalent to that observed with 103 WT Mtb cells]. Interestingly, complementation of the mutant with the Rv1002c gene readily restored mutant pathogenicity, as demonstrated by the death of all of the mice infected with the complemented strain by day 22 (Fig. 4E). This finding clearly confirms that the attenuation of the mutant virulence is related to the inactivation of Rv1002c gene coding for the Mtb PMT.
Conclusion and Future Prospects.
Taken together, our findings reveal that although mycobacterial protein O-mannosylation occurs by a similar mechanism to that in eukaryotes (11, 29), it strongly differs in its biological impact. Unlike in eukaryotes, this process is dispensable, at least in laboratory culture conditions, for nonpathogenic saprophyte and pathogenic parasite mycobacteria. On the other hand, we demonstrate in vivo that interruption of protein O-mannosylation crucially attenuates Mtb virulence in an immunocompromised mouse model. Therefore, we speculate that decorations of the Mtb surface and secreted proteins with α1–2 linked mannobioses mimicking the high mannose branch motifs of the eukaryotic intracellular glycoproteins are not fortuitous and may be crucial for Mtb to establish effectively parasitic relationships with its eukaryotic hosts. Further explorations need to be undertaken to determine the molecular basis of the phenotypic alterations of the ΔRv1002c mutant and to decipher the relationships between the interruption of Rv1002c-dependent protein O-mannosylation, the colony-forming deficiency, and the virulence attenuation. Our findings are unique in providing evidence for the crucial influence of mycobacterial protein O-mannosylation on Mtb physiology and virulence, and open the door for the exhaustive exploration of the PMT target proteome and analysis of the roles of mannosylated proteins in tuberculosis pathophysiology. Such holistic consideration of protein O-mannosylation–dependent Mtb persistence and proliferation should provide new perspectives in the search for original strategies to fight tuberculosis.
Materials and Methods
The M. smegmatis Msmeg_5447 and M. tuberculosis Rv1002c mutants (Figs. S1 and S4, and Table S4) were complemented with pMV361 derivative carrying an HygR cassette and the Msmeg_5447 or Rv1002c genes under the control of the pBlaF* promoter. The FasC Msmeg_5196 gene was fused to a polyhistidine tag within Hygr-pMIP12 derivative under the control of pBlaF* promoter. M. smegmatis mc2155 and M. tuberculosis H37Rv cells were grown for 5–7 d and 20–30 d (OD600 = 0.5–0.6) (30). Mtb culture mediums were doubly filtred on 0.22-µm membrane and concentrated on Vivaspin 5 kDa. rFasC was purified by FPLC before carbohydrate analysis and α-exomannosidase treatment (1). NanoLC-MS/MS analyses were performed on an Ultimate3000 system (Dionex) coupled to an LTQ-Orbitrap XL or an ETD-enabled LTQ Orbitrap Velos mass spectrometer (Thermo Fisher Scientific), respectively, for CID or ETD experiments. Data analysis and database searches were performed according to journal guidelines using Mascot Daemon against actinobacteria entries in Sprot-Trembl_20090707 database and the in-house developed software MFPaQ v4.0.0 (31, 32). The detection of peak signatures corresponding to carbohydrate neutral loss in CID MS/MS spectra has been performed using an in-house developed Perl script. Human peripheral blood mononuclear cells (hPBMC) were prepared from nontuberculous control donor blood (33). hPBMC and murine macrophages (34) were infected by GFP- mycobacteria at a MOI = 1 for 1 h at 37 °C. Cells collected at 0, 24, 96 h, and 7 d were fixed with paraformaldehyde and green fluorescent intracellular mycobacteria were quantified by fluorescence microscopy (Table S5). Animal studies were approved by the Comité d’Ethique Midi-Pyrénées and conducted according the Centre National de la Recherche Scientifique housing and care guidelines for laboratory animals. Groups of nine female SCID mice were infected intranasally with 103 cfu (otherwise stated) of either M. tuberculosis H37Rv strain and monitored daily. Animals showing signs of illness were killed.
Supplementary Material
Acknowledgments
We thank Dr. P. Constant (MTT micro-assay), F. Moreau and L. Lepourry (animal experiments), C. Froment (electron transfer dissociation mass spectrometry), Dr. M. Gilleron (phosphatidyl inositol mannoside analysis), and Dr. G. Larrouy-Maumus (Institut de Pharmacologie et de Biologie Structurale, Toulouse, France), the microscopy facilities IFR109 (Institut de Biologie Cellulaire et de Génétique, Toulouse, France), and Dr. D. Kaur (ΔM4247 and ΔM5149 mutants) (Colorado State University). This work was supported by the Agence Nationale de la Recherche Grant 11BSVB6016 (to M.R.); the National Institute of Allergy and Infectious Diseases/National Institutes of Health Grant AI064798 (to M.C.J.); the French Ministery of Research (Investissements d'avenir, Proteomics French Infrastructure) and the Fonds Européens de Développement Régional and the Région Midi-Pyrénées (O.B.-S.); the Centre National de la Recherche Scientifique (M.B.); the Région Midi-Pyrénées (L.T.); and the National Science Council, Taiwan NSC98-2917-I-564-173 (to C.-F.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1219704110/-/DCSupplemental.
References
- 1.Ragas A, Roussel L, Puzo G, Rivière M. The Mycobacterium tuberculosis cell-surface glycoprotein apa as a potential adhesin to colonize target cells via the innate immune system pulmonary C-type lectin surfactant protein A. J Biol Chem. 2007;282(8):5133–5142. doi: 10.1074/jbc.M610183200. [DOI] [PubMed] [Google Scholar]
- 2.Pitarque S, et al. Deciphering the molecular bases of Mycobacterium tuberculosis binding to the lectin DC-SIGN reveals an underestimated complexity. Biochem J. 2005;392(Pt 3):615–624. doi: 10.1042/BJ20050709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nguyen L, Pieters J. The Trojan horse: Survival tactics of pathogenic mycobacteria in macrophages. Trends Cell Biol. 2005;15(5):269–276. doi: 10.1016/j.tcb.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 4.Espitia C, Servín-González L, Mancilla R. New insights into protein O-mannosylation in actinomycetes. Mol Biosyst. 2010;6(5):775–781. doi: 10.1039/b916394h. [DOI] [PubMed] [Google Scholar]
- 5.Romain F, et al. Deglycosylation of the 45/47-kilodalton antigen complex of Mycobacterium tuberculosis decreases its capacity to elicit in vivo or in vitro cellular immune responses. Infect Immun. 1999;67(11):5567–5572. doi: 10.1128/iai.67.11.5567-5572.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Horn C, et al. Decreased capacity of recombinant 45/47-kDa molecules (Apa) of Mycobacterium tuberculosis to stimulate T lymphocyte responses related to changes in their mannosylation pattern. J Biol Chem. 1999;274(45):32023–32030. doi: 10.1074/jbc.274.45.32023. [DOI] [PubMed] [Google Scholar]
- 7.Kumar P, Amara RR, Challu VK, Chadda VK, Satchidanandam V. The Apa protein of Mycobacterium tuberculosis stimulates gamma interferon-secreting CD4+ and CD8+ T cells from purified protein derivative-positive individuals and affords protection in a guinea pig model. Infect Immun. 2003;71(4):1929–1937. doi: 10.1128/IAI.71.4.1929-1937.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Herrmann JL, O’Gaora P, Gallagher A, Thole JE, Young DB. Bacterial glycoproteins: A link between glycosylation and proteolytic cleavage of a 19 kDa antigen from Mycobacterium tuberculosis. EMBO J. 1996;15(14):3547–3554. [PMC free article] [PubMed] [Google Scholar]
- 9.Sieling PA, et al. Conserved mycobacterial lipoglycoproteins activate TLR2 but also require glycosylation for MHC class II-restricted T cell activation. J Immunol. 2008;180(9):5833–5842. doi: 10.4049/jimmunol.180.9.5833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Strahl-Bolsinger S, Gentzsch M, Tanner W. Protein O-mannosylation. Biochim Biophys Acta. 1999;1426(2):297–307. doi: 10.1016/s0304-4165(98)00131-7. [DOI] [PubMed] [Google Scholar]
- 11.VanderVen BC, Harder JD, Crick DC, Belisle JT. Export-mediated assembly of mycobacterial glycoproteins parallels eukaryotic pathways. Science. 2005;309(5736):941–943. doi: 10.1126/science.1114347. [DOI] [PubMed] [Google Scholar]
- 12.Griffin JE, et al. High-resolution phenotypic profiling defines genes essential for mycobacterial growth and cholesterol catabolism. PLoS Pathog. 2011;7(9):e1002251. doi: 10.1371/journal.ppat.1002251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sassetti CM, Rubin EJ. Genetic requirements for mycobacterial survival during infection. Proc Natl Acad Sci USA. 2003;100(22):12989–12994. doi: 10.1073/pnas.2134250100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vissa VD, Brennan PJ. The genome of Mycobacterium leprae: A minimal mycobacterial gene set. Genome Biol. 2001;2(8):S1023. doi: 10.1186/gb-2001-2-8-reviews1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wehmeier S, et al. Glycosylation of the phosphate binding protein, PstS, in Streptomyces coelicolor by a pathway that resembles protein O-mannosylation in eukaryotes. Mol Microbiol. 2009;71(2):421–433. doi: 10.1111/j.1365-2958.2008.06536.x. [DOI] [PubMed] [Google Scholar]
- 16.Mahne M, Tauch A, Pühler A, Kalinowski J. The Corynebacterium glutamicum gene pmt encoding a glycosyltransferase related to eukaryotic protein-O-mannosyltransferases is essential for glycosylation of the resuscitation promoting factor (Rpf2) and other secreted proteins. FEMS Microbiol Lett. 2006;259(2):226–233. doi: 10.1111/j.1574-6968.2006.00269.x. [DOI] [PubMed] [Google Scholar]
- 17.Zubarev RA, et al. Electron capture dissociation for structural characterization of multiply charged protein cations. Anal Chem. 2000;72(3):563–573. doi: 10.1021/ac990811p. [DOI] [PubMed] [Google Scholar]
- 18.Dobos KM, Khoo KH, Swiderek KM, Brennan PJ, Belisle JT. Definition of the full extent of glycosylation of the 45-kilodalton glycoprotein of Mycobacterium tuberculosis. J Bacteriol. 1996;178(9):2498–2506. doi: 10.1128/jb.178.9.2498-2506.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Michell SL, et al. The MPB83 antigen from Mycobacterium bovis contains O-linked mannose and (1→3)-mannobiose moieties. J Biol Chem. 2003;278(18):16423–16432. doi: 10.1074/jbc.M207959200. [DOI] [PubMed] [Google Scholar]
- 20.Sartain MJ, Belisle JT. N-Terminal clustering of the O-glycosylation sites in the Mycobacterium tuberculosis lipoprotein SodC. Glycobiology. 2009;19(1):38–51. doi: 10.1093/glycob/cwn102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chatterjee D, Lowell K, Rivoire B, McNeil MR, Brennan PJ. Lipoarabinomannan of Mycobacterium tuberculosis. Capping with mannosyl residues in some strains. J Biol Chem. 1992;267(9):6234–6239. [PubMed] [Google Scholar]
- 22.Hunter SW, Brennan PJ. Evidence for the presence of a phosphatidylinositol anchor on the lipoarabinomannan and lipomannan of Mycobacterium tuberculosis. J Biol Chem. 1990;265(16):9272–9279. [PubMed] [Google Scholar]
- 23.Morita YS, et al. PimE is a polyprenol-phosphate-mannose-dependent mannosyltransferase that transfers the fifth mannose of phosphatidylinositol mannoside in mycobacteria. J Biol Chem. 2006;281(35):25143–25155. doi: 10.1074/jbc.M604214200. [DOI] [PubMed] [Google Scholar]
- 24.Nothaft H, Szymanski CM. Protein glycosylation in bacteria: Sweeter than ever. Nat Rev Microbiol. 2010;8(11):765–778. doi: 10.1038/nrmicro2383. [DOI] [PubMed] [Google Scholar]
- 25.Zaia J. Mass spectrometry and glycomics. OMICS. 2010;14(4):401–418. doi: 10.1089/omi.2009.0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Banaei N, et al. Lipoprotein processing is essential for resistance of Mycobacterium tuberculosis to malachite green. Antimicrob Agents Chemother. 2009;53(9):3799–3802. doi: 10.1128/AAC.00647-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Espitia C, Mancilla R. Identification, isolation and partial characterization of Mycobacterium tuberculosis glycoprotein antigens. Clin Exp Immunol. 1989;77(3):378–383. [PMC free article] [PubMed] [Google Scholar]
- 28.Dobos KM, Swiderek K, Khoo KH, Brennan PJ, Belisle JT. Evidence for glycosylation sites on the 45-kilodalton glycoprotein of Mycobacterium tuberculosis. Infect Immun. 1995;63(8):2846–2853. doi: 10.1128/iai.63.8.2846-2853.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lommel M, Strahl S. Protein O-mannosylation: Conserved from bacteria to humans. Glycobiology. 2009;19(8):816–828. doi: 10.1093/glycob/cwp066. [DOI] [PubMed] [Google Scholar]
- 30.Le Dantec C, Winter N, Gicquel B, Vincent V, Picardeau M. Genomic sequence and transcriptional analysis of a 23-kilobase mycobacterial linear plasmid: Evidence for horizontal transfer and identification of plasmid maintenance systems. J Bacteriol. 2001;183(7):2157–2164. doi: 10.1128/JB.183.7.2157-2164.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mouton-Barbosa E, et al. In-depth exploration of cerebrospinal fluid by combining peptide ligand library treatment and label-free protein quantification. Mol Cell Proteomics. 2010;9(5):1006–1021. doi: 10.1074/mcp.M900513-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bouyssié D, et al. Mascot file parsing and quantification (MFPaQ), a new software to parse, validate, and quantify proteomics data generated by ICAT and SILAC mass spectrometric analyses: Application to the proteomics study of membrane proteins from primary human endothelial cells. Mol Cell Proteomics. 2007;6(9):1621–1637. doi: 10.1074/mcp.T600069-MCP200. [DOI] [PubMed] [Google Scholar]
- 33.Astarie-Dequeker C, et al. The mannose receptor mediates uptake of pathogenic and nonpathogenic mycobacteria and bypasses bactericidal responses in human macrophages. Infect Immun. 1999;67(2):469–477. doi: 10.1128/iai.67.2.469-477.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu CF, Rivere M, Huang HJ, Puzo G, Wang JY. Surfactant protein D inhibits mite-induced alveolar macrophage and dendritic cell activations through TLR signalling and DC-SIGN expression. Clin Exp Allergy. 2010;40(1):111–122. doi: 10.1111/j.1365-2222.2009.03367.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.