Abstract
Transmembrane proteins with unknown function 16 (TMEM16A) is a calcium-activated chloride channel (CaCC) important for neuronal, exocrine, and smooth muscle functions. TMEM16A belongs to a family of integral membrane proteins that includes another CaCC, TMEM16B, responsible for controlling action potential waveform and synaptic efficacy, and a small-conductance calcium-activated nonselective cation channel, TMEM16F, linked to Scott syndrome. We find that these channels in the TMEM16 family share a homodimeric architecture facilitated by their cytoplasmic N termini. This dimerization domain is important for channel assembly in eukaryotic cells, and the in vitro association of peptides containing the dimerization domain is consistent with a homotypic protein–protein interaction. Amino acid substitutions in the dimerization domain affect functional TMEM16A-CaCC channel expression, as expected from its critical role in channel subunit assembly.
Recent identification of transmembrane protein with unknown function 16 (TMEM16A) as the calcium-activated chloride channel (CaCC) first described in the frog oocyte has enabled molecular studies of this novel ion channel family (1, 2). Homologs of this channel have been identified in organisms throughout the evolutionary lineage, including yeast, plants, invertebrates, and vertebrates (3). In the mouse, TMEM16A-CaCC regulates smooth muscle contraction and fluid secretion. In addition, TMEM16A and its close homolog TMEM16B contribute to nervous system functions ranging from the modulation of signal transduction in sensory neurons to the control of action potential duration in hippocampal neurons (4, 5). Another member of this family, TMEM16F, is a small-conductance calcium-activated nonselective cation channel that is important for a calcium-activated scramblase activity associated with Scott syndrome’s defects in blood coagulation (6, 7).
The quaternary structure of many ion channels are known to be oligomeric membrane protein complexes assembled from multiple identical or closely related pore-forming subunits. For example, the NMDA-type glutamate receptor (NR) is a tetramer assembled from two obligate NR1 subunits and a choice of two NR2 subunits ranging from NR2A through NR2D (8), and studies of homologous ionotropic glutamate receptors implicate a cytosolic domain at the amino terminus in tetramerization (9). Similarly, the pentameric cys-loop receptors (10), the tetrameric potassium channels (11), and the gap junctions formed by hexameric hemichannels (12) are all protein complexes composed of subunits whose assembly is driven by channel-specific oligomerization domains.
Several groups have recently used biochemical methods to characterize the quaternary structure of TMEM16A channels as homodimers (13, 14). These previous studies on channel stoichiometry have raised the following questions: Is dimerization necessary for channel function? What is the TMEM16A dimerization domain that directs subunit assembly? To address these open questions, we have mapped the TMEM16A dimerization domain to a region within the cytoplasmic N terminus of TMEM16A. We show that this region is necessary and sufficient for dimerization, which is important for functional CaCC expression.
Results
TMEM16 Family Proteins Form Dimeric Protein Complexes.
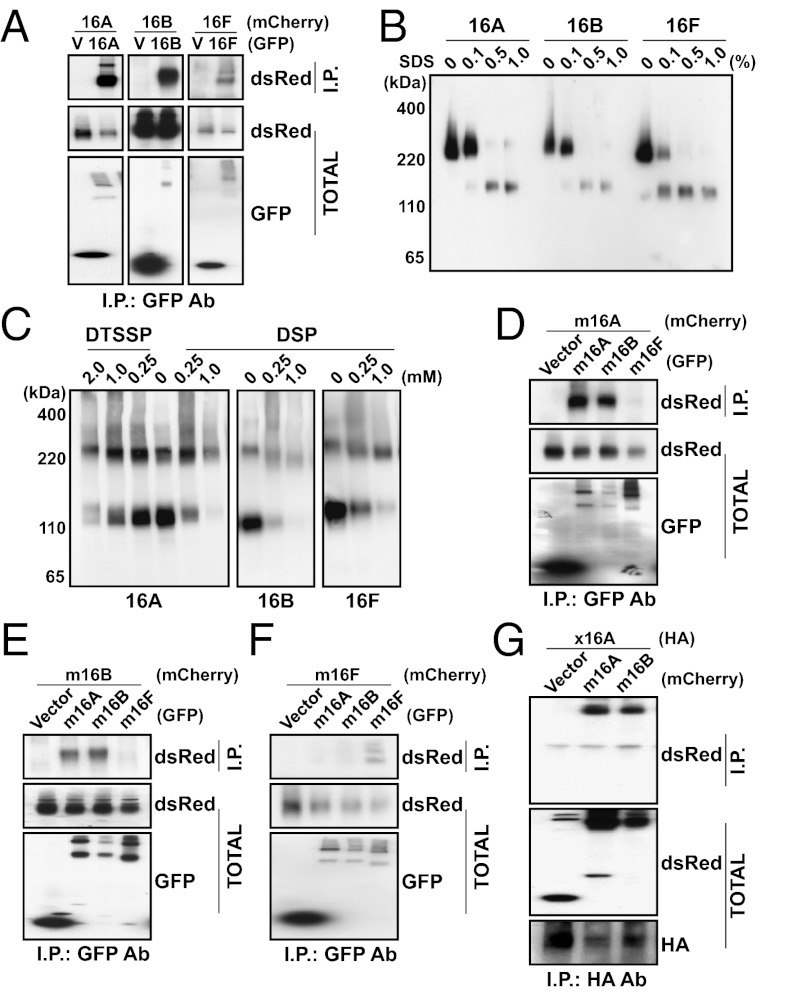
TMEM16A belongs to a family of 10 members in vertebrates (15). Recent studies have characterized TMEM16A, TMEM16B, and TMEM16F as functional calcium-activated ion channels. These channels are closely related, with TMEM16B and TMEM16F sharing 61% and 37% amino acid identity to TMEM16A in the mouse, respectively (Figs. S1 and S2). Because mouse TMEM16A immunoprecipitates TMEM16A and forms homodimers (13, 14) in biochemical studies, including our own (Fig. 1 A–C), we asked whether mouse TMEM16B and TMEM16F are also capable of forming oligomers when expressed in human embryonic kidney (HEK 293) cells. Indeed, GFP-tagged TMEM16B coimmunoprecipitated with mCherry-tagged TMEM16B and GFP-tagged TMEM16F coimmunoprecipitated with mCherry-tagged TMEM16F when each pair was coexpressed in HEK 293 cells (Fig. 1A). Nondenaturing PAGE of GFP-tagged TMEM16B or TMEM16F revealed the presence of dimers that were converted to the monomeric form upon denaturation in increasing concentrations of SDS (Fig. 1B). These dimeric species also appeared on denaturing SDS/PAGE when membrane complexes were stabilized by treating living cells with increasing concentrations of the amine-reactive chemical cross-linkers 3,3′-dithiobis(sulfosuccinimidylpropionate) (DTSSP) or dithiobis(sulfosuccinimidylpropionate) (DSP) (Fig. 1C), suggesting that the dimeric protein is present in physiologically intact cells.
Fig. 1.
TMEM16 family proteins form dimeric protein complexes. (A) GFP (V), mTMEM16A-GFP, mTMEM16B-GFP, or mTMEM16F-GFP was coexpressed in HEK 293 cells with mTMEM16A-mCherry, mTMEM16B-mCherry, or mTMEM16F-mCherry. Proteins were immunoprecipitated from HEK 293 cell lysates with an anti-GFP antibody. Immunoprecipitated proteins were separated by SDS/PAGE and identified by Western blot analysis with anti-dsRed and anti-GFP antibodies. (B) Lysates from HEK 293 cells transfected with mTMEM16A-GFP, mTMEM16B-GFP, or mTMEM16F-GFP were treated with increasing concentrations of SDS as indicated. Proteins in the lysate were separated by native gel electrophoresis and identified by Western blot analysis with an anti-GFP antibody. (C) HEK 293 cells transfected with mTMEM16A-GFP, mTMEM16B-GFP, or mTMEM16F-GFP were incubated with increasing concentrations of either DTSSP or DSP. Proteins in HEK 293 cell lysates were separated by SDS/PAGE and identified by Western blot analysis with an anti-GFP antibody. (D–F) GFP (Vector), mTMEM16A-GFP, mTMEM16B-GFP, or mTMEM16F-GFP was coexpressed in HEK 293 cells with mTMEM16A-mCherry (D), mTMEM16B-mCherry (E), or mTMEM16F-mCherry (F). Proteins were immunoprecipitated from HEK 293 cell lysates with an anti-GFP antibody. Immunoprecipitated proteins were separated by SDS/PAGE and identified by Western blot analysis with anti-dsRed and anti-GFP antibodies. (G) mCherry (Vector), mTMEM16A-mCherry, or mTMEM16B-mCherry was coexpressed in HEK 293 cells with xTMEM16A-HA. Proteins were immunoprecipitated from HEK 293 cell lysates with an anti-HA antibody. Immunoprecipitated proteins were separated by SDS/PAGE and identified by Western blot analysis with anti-HA and anti-dsRed antibodies.
Because the similar quaternary structure of these proteins may arise from the conservation of a specific dimerization domain of TMEM16 family members, we wondered whether these homologs are able to form heterodimers in vitro. We therefore attempted to coimmunoprecipitate mCherry-labeled mouse TMEM16A with other mouse TMEM16 family members coexpressed in HEK 293 cells. We found that TMEM16A coimmunoprecipitated with the more closely related homolog, TMEM16B, but not the more distant homolog, TMEM16F (Fig. 1D). Similarly, TMEM16B coimmunoprecipitated with TMEM16A but not TMEM16F (Fig. 1E). Moreover, TMEM16F did not coimmunoprecipitate with either TMEM16A or TMEM16B (Fig. 1F). The ability for close homologs to interact extended even across species, as Xenopus TMEM16A was coimmunoprecipitated by both mouse TMEM16A and TMEM16B (Fig. 1G), with which it shares 75% and 61% sequence identity (Fig. S1). Thus, ion channels in the TMEM16 family are dimeric proteins, and the interactions responsible for channel assembly appear to be well-conserved specifically in other CaCCs of the TMEM16 family.
Identification of the TMEM16A Dimerization Domain.
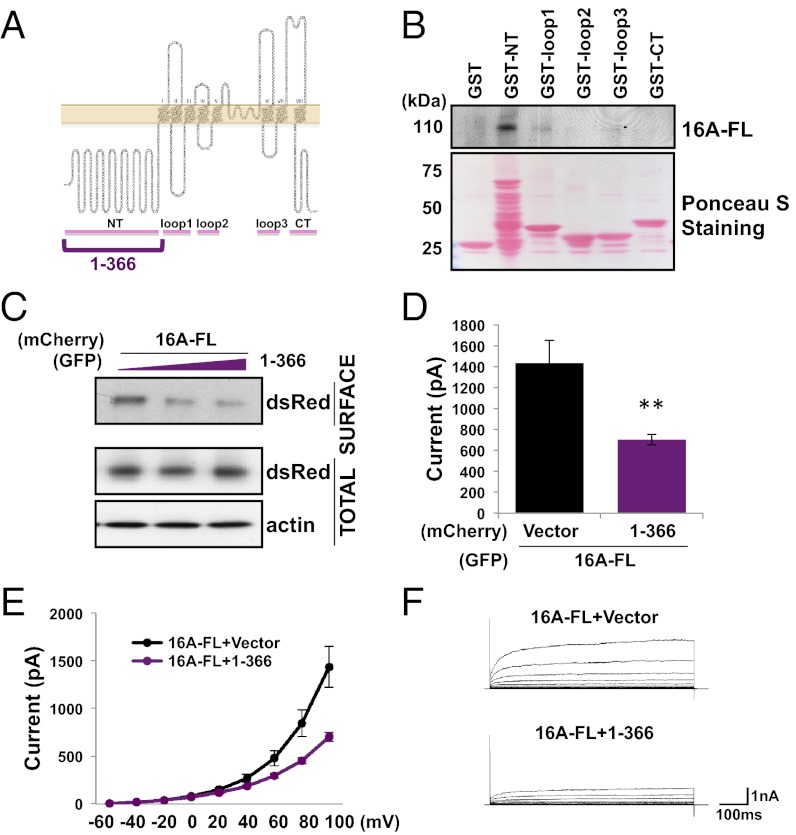
Having found that TMEM16A assembles into dimeric complexes, we next sought to identify the region responsible for this interaction. Because several other channel proteins (9, 11, 12) oligomerize via their cytosolic regions, we began by purifying each of the five predicted cytosolic domains (16) from mouse TMEM16A on glutathione-conjugated beads and mixing them with lysate from HEK 293 cells expressing GFP-tagged TMEM16A (Fig. 2 A and B). Only the GST-tagged peptide containing the very N-terminal domain of TMEM16A was able to pull-down the full-length TMEM16A protein from mammalian cells (Fig. 2B).
Fig. 2.
The TMEM16A N-terminal domain is sufficient for subunit interaction. (A) Hypothetical topology indicating cytosolic regions used in our experiments. (B) GST-tagged cytosolic fragments of TMEM16A were purified on glutathione-conjugated beads and used to affinity capture interacting proteins in cell lysates from HEK 293 cells expressing TMEM16A-GFP. Proteins copurifying with the GST tag were separated by SDS/PAGE, stained with Ponceau S, and identified by Western blot analysis with an anti-GFP antibody. (C) HEK 293 cells stably expressing full-length (FL) TMEM16A-mCherry were transfected with different amounts of a GFP-tagged truncation mutant containing residues 1–366 of TMEM16A. Surface proteins were labeled with a membrane-impermeable biotinylation reagent. Proteins enriched from HEK 293 cell lysates on avidin-conjugated beads were separated by SDS/PAGE and identified by Western blot analysis with anti-dsRed, anti-GFP, and antiactin antibodies. (D) Either mCherry (Vector) or a mCherry-tagged truncation mutant containing residues 1–366 of TMEM16A was coexpressed in HEK 293 cells with wild-type TMEM16A-GFP. Average steady-state whole-cell currents were measured at +100 mV with 300 nM internal free calcium and symmetric chloride. Each bar represents five to seven cells; **P < 0.01 in unpaired t tests with cells coexpressing full-length TMEM16A with only mCherry. Error bars are ± SEM. (E) I-V curve of the cells in D. Plots represent steady-state whole-cell currents recorded with 300 nM internal free calcium and symmetric chloride from −60 mV to +100 mV in +20-mV steps lasting 500 ms. (F) Representative traces of cells illustrated in D and E.
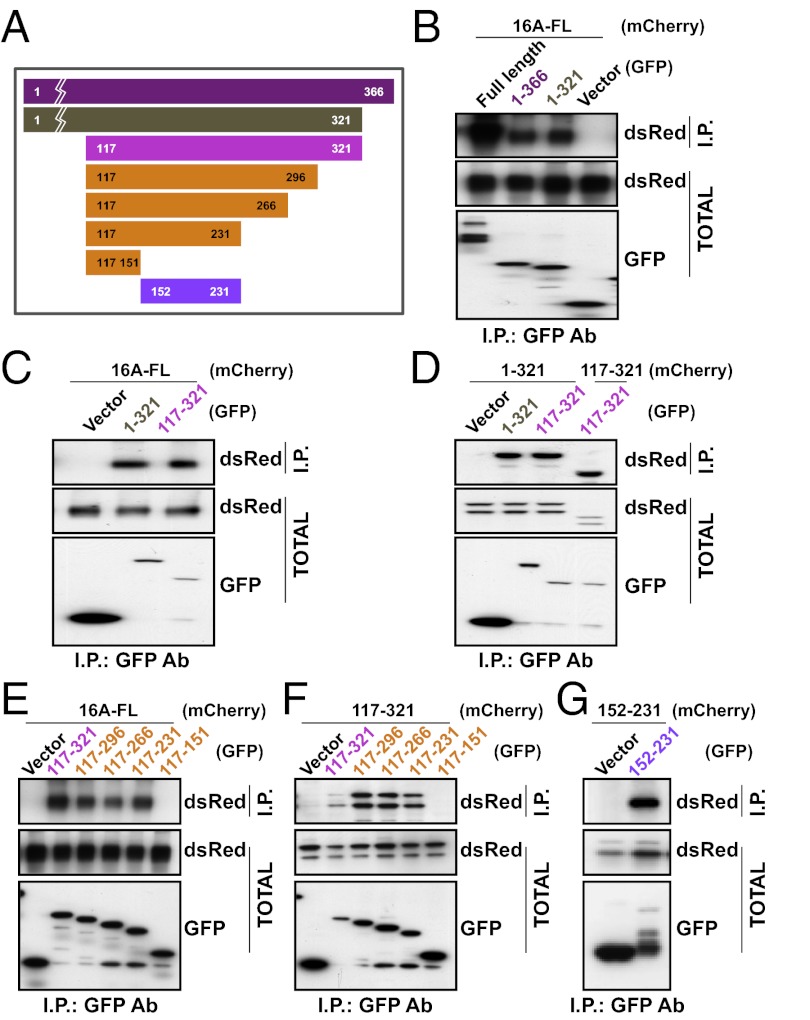
We next verified this N-terminal domain interaction using two further assays. First, hypothesizing that a truncation mutant containing the N-terminal domain and a transmembrane segment would insert into the endoplasmic reticulum (ER) membrane and interfere with the assembly and membrane trafficking of TMEM16A channels, we expressed the first 366 residues containing the entire N-terminal domain along with the first transmembrane segment of TMEM16A in a HEK 293 cell line that stably expressed mCherry-tagged TMEM16A. The population of mCherry-tagged TMEM16A proteins on the cell surface and accessible to labeling by a membrane-impermeable amine-reactive biotin was progressively reduced in cells transfected with increasing concentrations of this truncation mutant (Fig. 2C). This process led to a reduction in functional channel expression as detected in electrophysiological recordings of calcium-activated chloride currents in HEK 293 cells cotransfected with this mutant and wild-type TMEM16A (P < 0.01, unpaired t test) (Fig. 2 D–F). Second, we also tested whether this interaction could be detected using coimmunoprecipitation assays. We found that mCherry-tagged TMEM16A coimmunoprecipitated with not only the truncation mutant containing residues 1–366, but also mutants containing only residues 1–321 without a transmembrane segment (Fig. 3 A and B).
Fig. 3.
A region in the cytoplasmic N terminus is responsible for subunit dimerization. (A) Schematic depiction of TMEM16A N-terminal fragments used in our experiments. (B) GFP (Vector), full-length TMEM16A-GFP, or GFP fused to residues 1–366 or 1–321 was coexpressed in HEK 293 cells with mTMEM16A-mCherry. (C and D) GFP or GFP fused to residues 1–321 or 117–321 was coexpressed in HEK 293 cells with TMEM16A-mCherry (C) or mCherry fused to residues 1–321 or 117–321 (D). (E and F) GFP or GFP fused to TMEM16A truncation mutants containing indicated residues was coexpressed in HEK 293 cells with TMEM16A-mCherry (E) or mCherry fused to residues 117–321 (F). (G) GFP or GFP fused to residues 152–231 was coexpressed in HEK 293 cells with mCherry fused to residues 152–231. (B–G) Proteins were immunoprecipitated from HEK 293 cell lysates with an anti-GFP antibody. Immunoprecipitated proteins were separated by SDS/PAGE and identified by Western blot analysis with anti-dsRed and anti-GFP antibodies.
We next attempted to map the dimerization domain by shortening the truncation mutant further. Because the first 116 residues of the N-terminal domain is not well-conserved between TMEM16A and TMEM16B (Fig. S2), and because Ferrera et al. (17) has reported that the first half of the N-terminal domain is not necessary for TMEM16A function, we hypothesized that the first 116 residues are also not required for protein dimerization. Indeed, when the first 116 residues were removed, residues 117–321 remained sufficient for coimmunoprecipitation with full-length TMEM16A (Fig. 3C). This dimerization domain appears to interact homotypically because residues 117–321 also coimmunoprecipitated with just the N-terminal domain (residues 1–321) and with itself (residues 117–321) (Fig. 3D).
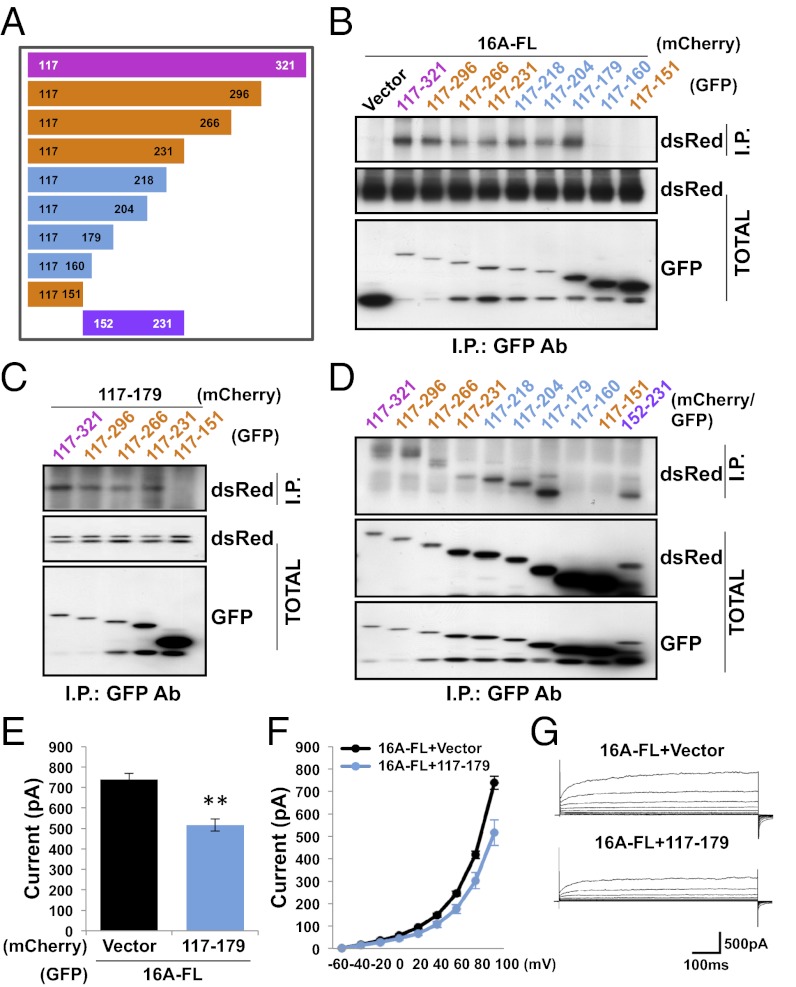
To further define the dimerization domain, we made a series of smaller truncation mutants based on the predicted secondary structure (18–20) and the sequence conservation in the N-terminal domains of the TMEM16A and TMEM16B homologs (Fig. 3A). We tested each with coimmunoprecipitation experiments to identify the minimally necessary region for protein–protein interaction. Constructs spanning residues 117–296, residues 117–266, and residues 117–231 were able to interact with both the full-length TMEM16A protein (Fig. 3E) and residues 117–321 (Fig. 3F), but residues 117–151 could not, suggesting that the 78 amino acids corresponding to residues 152–231 are critical for TMEM16A dimerization. In fact, not only are these 78 residues necessary for dimer interaction, they are also sufficient, because peptides containing these residues tagged with GFP or mCherry were able to coimmunoprecipitate (Fig. 3G). Focusing on this critical region, we made another series of truncations (Fig. 4A) and found that although residues 117–218, residues 117–204, and residues 117–179 were able to interact with full-length TMEM16A, constructs containing only residues 117–160 could not (Fig. 4B). The interaction in this minimal region appears to be homotypic because residues 117–179 not only bound to all other truncation mutants containing residues 117–179 (Fig. 4C), it also bound to itself (Fig. 4D).
Fig. 4.
Residues 117–179 are sufficient for a homotypic dimer interaction. (A) Schematic depiction of TMEM16A N-terminal fragments used in our experiments. (B) GFP (Vector) or GFP fused to TMEM16A truncation mutants containing indicated residues was coexpressed in HEK 293 cells with TMEM16A-mCherry. (C) GFP fused to TMEM16A truncation mutants was coexpressed in HEK 293 cells with mCherry fused to residues 117–179. (D) GFP-tagged TMEM16A truncation mutants were coexpressed in HEK 293 cells with mCherry-tagged versions of the same mutants. (B–D) Proteins were immunoprecipitated from HEK 293 cell lysates with an anti-GFP antibody. Immunoprecipitated proteins were separated by SDS/PAGE and identified by Western blot analysis with anti-dsRed and anti-GFP antibodies. (E) Either mCherry (Vector) or mCherry fused to residues 117–179 was coexpressed in HEK 293 cells with wild-type TMEM16A-GFP. Average steady-state whole-cell currents were measured at +100 mV with 300 nM internal free calcium, and symmetric chloride. Each bar represents five to seven cells; **P < 0.01 in unpaired t tests with cells coexpressing full-length TMEM16A with only mCherry. Error bars are ± SEM. (F) I-V curve of the cells in E. Plots represent steady-state whole-cell currents recorded with 300 nM internal free calcium and symmetric chloride from −60 mV to +100 mV in +20-mV steps lasting 500 ms. (G) Representative traces of cells illustrated in E and F.
Because residues 117–179 are sufficient to interact with wild-type TMEM16A channels in coimmunoprecipitation assays, we wondered whether overexpression of this fragment would also act as a dominant-negative in electrophysiological assays of channel function. When wild-type TMEM16A was cotransfected into HEK 293 cells with a construct containing only residues 117–179, functional channel expression was decreased relative to cells cotransfected with an empty vector (P < 0.01, unpaired t test) (Fig. 4 E–G). The decrease in whole-cell current was smaller than that observed when residues 1–399 were cotransfected (Fig. 2D), possibly because a fragment containing only residues 117–179 is expected to be cytosolic and would therefore interact with wild-type TMEM16A subunits less efficiently than constructs containing a transmembrane segment.
Because this region contains a cysteine at residue 166 (Fig. S3A), we performed further mutagenesis experiments to test the role of C166 and disulfide bond formation in subunit dimerization. Mutant N-terminal domains with triple alanine substitutions scanning the region including C166 were still able to coimmunoprecipitate with full-length TMEM16A proteins as well as truncation mutants containing only residues 117–204 (Fig. S3 B–C). This result is consistent with the location of the dimerization domain in the cytoplasmic N terminus where it is exposed to the reducing intracellular environment, and thus unlikely to form disulfide bonds. In addition, C166 is poorly conserved across TMEM16A/B/F homologs, suggesting that it is dispensable for channel oligomerization.
TMEM16A Dimerization Domain Is Responsible for Specific Subunit Interactions.
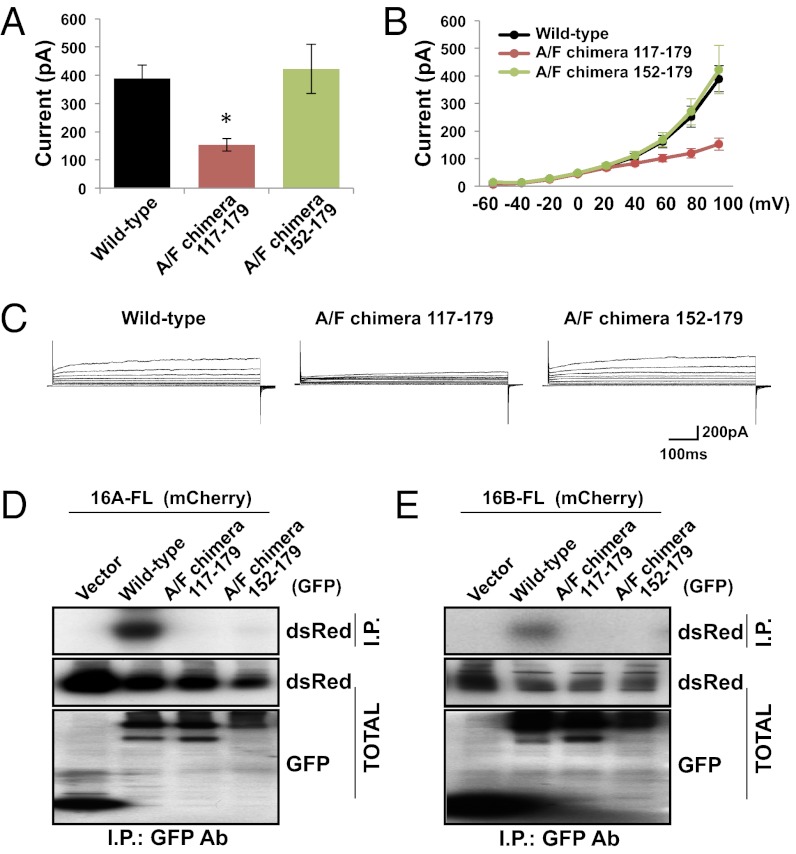
Having observed that TMEM16F subunits normally form homodimers even though TMEM16A subunits do not heterodimerize with TMEM16F subunits (Fig. 1 D and F), we wondered whether the TMEM16F dimerization domain is also in its N-terminal region and whether the dimerization domain is responsible for this specificity in subunit interactions. To test these questions, we engineered chimera mutants by substituting residues 117–179 in TMEM16A’s dimerization domain with the homologous sequence from TMEM16F (A/F chimera 117–179) (Fig. S4). The dimerization domain in these mutants appeared to be sufficient for homodimerization in the context of the TMEM16A protein and facilitated functional channel assembly when expressed in HEK 293 cells (Fig. 5 A–C). However, the whole-cell calcium-activated chloride currents generated by these channels were smaller than wild-type currents (P < 0.05, Dunn’s test) (Fig. 5A).
Fig. 5.
Replacement of the TMEM16A dimerization domain with that of TMEM16F yields functional channels. (A) GFP-tagged wild-type TMEM16A, A/F chimera 117–179, or A/F chimera 152–179 was expressed in HEK 293 cells. Average steady-state whole-cell currents were measured at +100 mV with 300 nM internal free calcium and symmetric chloride. Each bar represents five to six cells; *P < 0.05 in Dunn’s test with cells expressing only wild-type TMEM16A. Error bars are ± SEM. (B) I-V curve of the cells in A. Plots represent steady-state whole-cell currents recorded with 300 nM internal free calcium and symmetric chloride from −60 mV to +100 mV in +20-mV steps lasting 500 ms. (C) Representative traces of cells illustrated in A and B. (D and E) GFP (Vector) or GFP-tagged wild-type TMEM16A, A/F chimera 117–179, or A/F chimera 152–179 was coexpressed in HEK 293 cells with TMEM16A-mCherry (D) or TMEM16B-mCherry (E). Proteins were immunoprecipitated from HEK 293 cell lysates with an anti-GFP antibody. Immunoprecipitating proteins were separated by SDS/PAGE and identified by Western blot analysis with anti-dsRed and anti-GFP antibodies.
Hypothesizing that the reduction in current may be the result of inefficient protein folding due to the mutation of such a large region within the N-terminal domain of TMEM16A, we engineered a second chimera protein targeting a smaller region for mutagenesis. Because truncation mutants containing residues 117–179 and residues 152–231 were able to form homodimers in vitro (Fig. 4D), we reasoned that residues 152–179 could be a critical region sufficient for protein–protein interaction. We therefore replaced TMEM16A residues 152–179 with the homologous region from TMEM16F (A/F chimera 152–179) (Fig. S4). This smaller mutation was able to efficiently form homodimers when expressed in HEK 293 cells and produced whole-cell calcium-activated chloride currents with amplitudes similar to those observed with wild-type TMEM16A channels (Fig. 5 A–C).
Having found that both chimera mutants were able to form homodimers when expressed in HEK 293 cells, we next tested the ability of these mutants to form heterodimers. As expected, neither chimera mutant coimmunoprecipitated with wild-type TMEM16A or TMEM16B subunits (Fig. 5 D and E), suggesting that the dimerization domains of TMEM16F and TMEM16A are not compatible for heterodimeric interactions despite their homology (Fig. S4). Thus, we have identified a dimerization domain within the TMEM16A protein that facilitates specific intersubunit assembly among TMEM16 family members.
Conserved Peptide of 19 Residues Within the Dimerization Domain Is Necessary for TMEM16A Assembly.
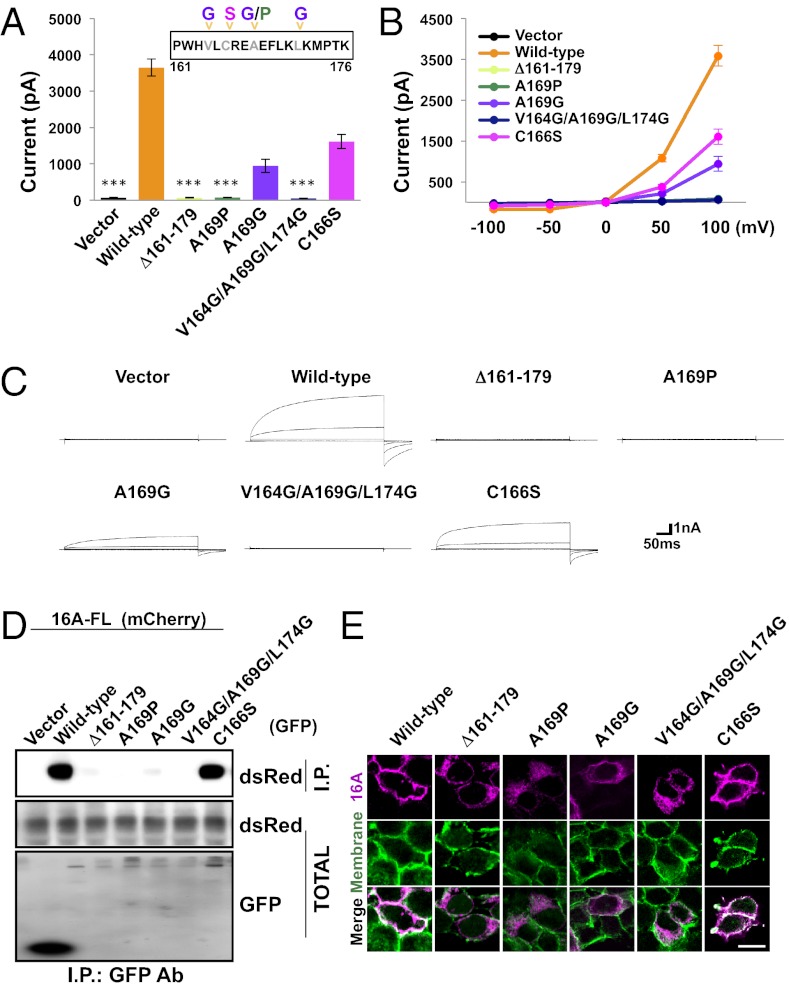
We noticed that the region spanning residues 161–179 in the dimerization domain is predicted to form α-helical structures in solution. Hypothesizing that this region might form the core structure of the dimerization domain, we wondered whether disruption of this segment might impede TMEM16A-CaCC channel assembly and activity. Transfection of TMEM16A mutants where residues 161–179 (Δ161–179) have been deleted did not result in the expression of calcium-activated chloride currents in HEK 293 cells (P < 0.001, Dunn’s test with respect to wild-type channels) (Fig. 6 A–C), suggesting that TMEM16A is unable to assemble into functional channels without this segment of its dimerization domain. The loss of functional channel expression is likely caused by the failure of TMEM16A subunits to properly assemble into quaternary structures because mutants lacking residues 161–179 failed to bind to other TMEM16A subunits in coimmunoprecipitation assays (Fig. 6D) and also mislocalized from the plasma membrane to the ER (Fig. 6E and Fig. S5). This phenomenon has been observed in previous studies of transmembrane proteins (21, 22) and may be attributed to quality-control mechanisms in the ER that prevent the export of proteins until they can be folded into a native conformation and fully assembled or tagged for degradation (23).
Fig. 6.
A segment of 19 residues is necessary for TMEM16A function. (A) GFP (Vector), wild-type TMEM16A-GFP, or GFP-tagged TMEM16A mutants was expressed in HEK 293 cells as indicated. Average steady-state whole-cell currents were measured at +100 mV with 300 nM internal free calcium and symmetric chloride. Each bar represents 8 to 10 cells; ***P < 0.001 in Dunn’s tests with cells expressing wild-type TMEM16A-GFP. Error bars are ± SEM. (B) I-V curve of the cells in A. Plots represent steady-state whole-cell currents recorded with 300 nM internal free calcium and symmetric chloride from −100 mV to +100 mV in +50-mV steps lasting 300 ms. (C) Representative traces of cells illustrated in A and B. (D) GFP, wild-type TMEM16A-GFP, or GFP-tagged TMEM16A mutants was coexpressed in HEK 293 cells with TMEM16A-mCherry. Proteins were immunoprecipitated from HEK 293 cell lysates with an anti-GFP antibody. Immunoprecipitating proteins were separated by SDS/PAGE and identified by Western blot analysis with anti-dsRed and anti-GFP antibodies. (E) Wild-type TMEM16A-mCherry or mCherry-tagged TMEM16A mutants was expressed in HEK 293 cells. Surface proteins were labeled with a membrane-impermeable biotinylation reagent. Immunostaining was performed using a ds-Red antibody (magenta, Top) and biotin was detected with an avidin-conjugated Alexa-fluor (green, Middle). Colocalization of dsRed and membrane biotin shown in white (Bottom). (Scale bar, 20 μm.)
Because the deletion of 19 residues is a large mutation that may cause major conformational rearrangements and protein misfolding, we also tested several smaller mutations to preserve as much of the native structure as possible while still disrupting the dimerization domain’s intermolecular associations. Because the peptide of 19 residues within the dimerization domain is predicted to be α-helical, we mutated A169 at the center of this peptide to proline (A169P), an amino acid known to have low helix-forming propensities (24). Substitution of alanine 169 with proline completely abolished calcium-activated chloride currents (P < 0.001, Dunn’s test with respect to wild-type channels) (Fig. 6 A–C), eliminated protein–protein interaction (Fig. 6D), and blocked channel trafficking to the plasma membrane (Fig. 6E and Fig. S5), similar to what we observed for the Δ161–179 mutant.
Another helix-disrupting mutation of A169, namely substitution with glycine (A160G), appears to be more benign, as the mutation tended to reduce the observed current by only 74% (Fig. 6 A–C). This is likely caused by a disruption in channel assembly and trafficking as the A169G mutant protein showed a reduced ability to interact with wild-type TMEM16A (Fig. 6D) and to localize to the cell surface (Fig. 6E). Further disruptions of this domain by the introduction of glycine substitutions at V164 and L174 (V164G/A169G/L174G) eliminated the remainder of the chloride current (P < 0.001, Dunn’s test with respect to wild-type channels) (Fig. 4 A–C), protein–protein interactions (Fig. 6D), and trafficking to the cell membrane (Fig. 6E and Fig. S5).
The predicted helical domain within the dimerization domain also contains the cysteine residue at position 166. Because our immunoprecipitation experiments showed that C166 is dispensable for TMEM16A dimerization (Fig. S3), we wondered whether small perturbations of this region, such as substituting this cysteine with serine, would have any effect on channel assembly or function. The C166S mutation of the dimerization domain tended to reduce the whole cell calcium-activated chloride current in HEK 293 cells (Fig. 6 A–C). However, this reduction in channel activity was not statistically significant, consistent with the observations that the mutant channel still coimmunoprecipitated with other TMEM16A subunits (Fig. 6D) and transported to the plasma membrane (Fig. 6E and Fig. S5). Thus, C166 does not appear to be required for channel assembly and substitution of C166 with either serine or alanine (Fig. S3) does not compromise protein interactions in the N-terminal dimerization domain.
Discussion
Many ion channels are assembled as modular complexes of identical or closely homologous pore-forming subunits. In addition to interactions of the membrane-spanning regions of these channel subunits, their cytosolic regions often harbor interaction domains that are important for channel assembly. Our study of the TMEM16A-CaCC extends this trend with a dimerization domain in the cytosolic N-terminal region crucial for channel assembly and function. We identify a region that is necessary and sufficient for homotypic protein–protein interactions in biochemical and electrophysiological assays. Perturbation of this domain interferes with channel subunit assembly and disrupts channel function. Replacement of this segment with homologous residues from TMEM16F alters the specificity of subunit interactions.
In TMEM16A-CaCC, this domain resides in a region highly conserved among the TMEM16 homologs, with the predicted helical segment (residues 161–179 of mTMEM16A) varying at just three residues in species that are as evolutionary divergent as rats, turkeys, chimpanzees, and zebrafish. The domain that we have identified is similar in size compared with the homotypic interaction domains found in other proteins. The T1 domain of KV1 channels is ∼87-residues long (25) and the A-domain tail of the KV7 channels is ∼41 residues long (26). Despite its short length, this region of the TMEM16A protein contains a segment that is predicted with high probability to be in an α-helical conformation (18–20), possibly forming a core for protein–protein interactions. It is conceivable that these interactions are further stabilized by other parts of the protein.
Even though our data show that this N-terminal domain is necessary and sufficient for dimerization, it is unlikely to be the only intersubunit contact present in the TMEM16A-CaCC channel. The interaction between TMEM16A subunits are probably additionally facilitated by the high effective concentration of proteins embedded in the lipid bilayer, the favorable energetic of side-chain packing in the transmembrane domains, as well as the other protein–protein interfaces present in other portions of the channel (27). These disparate sources of interaction are well established in the channel structure literature and each channel family seems to adopt a unique strategy for protein oligomerization (11, 26, 28, 29). Because the dimerization domain identified in our studies exhibits no homology to any other known protein according to protein BLAST algorithms, further investigation of this channel will likely reveal unique oligomerization strategies that may not have been evident from these previously described models.
TMEM16A plays an important physiological role in many different cell types (2). In addition, its close homologs TMEM16B and TMEM16F have been implicated in sensory transduction and modulation of central neuronal signaling (4, 5) and blood coagulation diseases (6, 7), respectively. To gain mechanistic insights to the function of this family of calcium-activated ion channels, it is important to probe the structural features of these unique proteins. Our study continues this effort by identifying a region critical for directing proper channel assembly. These results will have implications for understanding the molecular interactions that regulate ion conductance through the channel as well as for developing chemical modulators of this channel.
Materials and Methods
Immunoprecipitation.
Transfected HEK 293 cells were lysed by homogenization in PBS (1.5 mM KH2PO4, 4.3 mM Na2HPO4, 2.7 mM KCl, 137 mM NaCl at pH 7.4) supplemented with 1% cholic acid, 1% Triton X-100, and 1× Complete protease inhibitor (Roche) with brief sonication. Insoluble matter was removed by centrifuging at 20,000 × g for 40 min at 4 °C. Supernantants were incubated with mouse monoclonal antibodies raised against GFP (Neuromab) overnight at 4 °C and then pulled-down with protein A-conjugated Sepharose beads (Invitrogen) for 2 h at 4 °C. Beads were washed three times with lysis buffer and once with PBS before elution with SDS/PAGE running buffer.
Western Blot Analysis.
Proteins denatured by heating for 5–10 min at 95 °C in Laemmli sample buffer (30) were separated by SDS/PAGE and transferred to nitrocellulose membranes. After blocking in TTBS buffer [10 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.05% Tween-20, and 5% (wt/vol) skim milk powder], the membranes were incubated with primary antibodies overnight at 4 °C followed by horseradish peroxidase (HRP)-conjugated secondary antibodies for 1 h at room temperature. Detection was performed using enhanced chemiluminescence kit and hyperfilm MP (Amersham).
Electrophysiology.
HEK 293 cells transiently transfected for 14–20 h were plated onto poly-L-lysine–coated coverslips 2–3 h before recording. Whole-cell recordings were performed at room temperature on transfected cells using an Axopatch 200B patch-clamp amplifier and pClamp9 software (Molecular Devices) following conditions modified from Schroeder et al. (31). External solutions contained 140 mM NaCl, 5 mM KCl, 2 mM CaCl2, 1 mM MgCl2, and 10 mM Hepes and 300 nM free Ca2+ internal solutions contained 140 mM NaCl, 10 mM Hepes, 7.4 mM CaCl2, and 12 mM EGTA. Free calcium concentrations were determined using Maxchelator software (http://maxchelator.stanford.edu) and the pH-metric method (32). All solutions were titrated with NaOH to pH 7.2.
Miscellaneous.
Additional information can be found in SI Materials and Methods. For a list of primers used in this study, see Table S1.
Supplementary Material
Acknowledgments
We thank Dr. Sung Ho Ryu and Dr. Jae Yoon Kim of Pohang University of Science and Technology (POSTECH), Pohang, Korea for GST-tagged protein purification protocols. This work was supported by the National Institutes of Health Grants NS069229 (to L.Y.J.) and DC007664 (to D.L.M.), and the American Asthma Foundation 09-0051 (to D.L.M.). L.Y.J. and Y.N.J. are investigators of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1303672110/-/DCSupplemental.
References
- 1.Hartzell C, Putzier I, Arreola J. Calcium-activated chloride channels. Annu Rev Physiol. 2005;67:719–758. doi: 10.1146/annurev.physiol.67.032003.154341. [DOI] [PubMed] [Google Scholar]
- 2.Huang F, Wong X, Jan LY. International Union of Basic and Clinical Pharmacology. LXXXV: Calcium-activated chloride channels. Pharmacol Rev. 2012;64(1):1–15. doi: 10.1124/pr.111.005009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berg J, Yang H, Jan LY. Ca2+-activated Cl- channels at a glance. J Cell Sci. 2012;125(Pt 6):1367–1371. doi: 10.1242/jcs.093260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stöhr H, et al. TMEM16B, a novel protein with calcium-dependent chloride channel activity, associates with a presynaptic protein complex in photoreceptor terminals. J Neurosci. 2009;29(21):6809–6818. doi: 10.1523/JNEUROSCI.5546-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang WC, et al. Calcium-activated chloride channels (CaCCs) regulate action potential and synaptic response in hippocampal neurons. Neuron. 2012;74(1):179–192. doi: 10.1016/j.neuron.2012.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Suzuki J, Umeda M, Sims PJ, Nagata S. Calcium-dependent phospholipid scrambling by TMEM16F. Nature. 2010;468(7325):834–838. doi: 10.1038/nature09583. [DOI] [PubMed] [Google Scholar]
- 7.Yang H, et al. TMEM16F forms a Ca2+-activated cation channel required for lipid scrambling in platelets during blood coagulation. Cell. 2012;151(1):111–122. doi: 10.1016/j.cell.2012.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cull-Candy SG, Leszkiewicz DN. Role of distinct NMDA receptor subtypes at central synapses. Sci STKE. 2004;2004(255):re16. doi: 10.1126/stke.2552004re16. [DOI] [PubMed] [Google Scholar]
- 9.Madden DR. The structure and function of glutamate receptor ion channels. Nat Rev Neurosci. 2002;3(2):91–101. doi: 10.1038/nrn725. [DOI] [PubMed] [Google Scholar]
- 10.Yu XM, Hall ZW. Extracellular domains mediating epsilon subunit interactions of muscle acetylcholine receptor. Nature. 1991;352(6330):64–67. doi: 10.1038/352064a0. [DOI] [PubMed] [Google Scholar]
- 11.Li M, Jan YN, Jan LY. Specification of subunit assembly by the hydrophilic amino-terminal domain of the Shaker potassium channel. Science. 1992;257(5074):1225–1230. doi: 10.1126/science.1519059. [DOI] [PubMed] [Google Scholar]
- 12.Lagree V, et al. Specific amino-acid residues in the N-terminus and TM3 implicated in channel function and oligomerization compatibility of connexin43. J Cell Sci. 2003;116(Pt 15):3189–3201. doi: 10.1242/jcs.00604. [DOI] [PubMed] [Google Scholar]
- 13.Fallah G, et al. TMEM16A(a)/anoctamin-1 shares a homodimeric architecture with CLC chloride channels. Mol Cell Proteomics. 2011;10(2) doi: 10.1074/mcp.M110.004697. M110.004697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sheridan JT, et al. Characterization of the oligomeric structure of the Ca(2+)-activated Cl- channel Ano1/TMEM16A. J Biol Chem. 2011;286(2):1381–1388. doi: 10.1074/jbc.M110.174847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Milenkovic VM, Brockmann M, Stöhr H, Weber BH, Strauss O. Evolution and functional divergence of the anoctamin family of membrane proteins. BMC Evol Biol. 2010;10:319. doi: 10.1186/1471-2148-10-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Das S, et al. NGEP, a prostate-specific plasma membrane protein that promotes the association of LNCaP cells. Cancer Res. 2007;67(4):1594–1601. doi: 10.1158/0008-5472.CAN-06-2673. [DOI] [PubMed] [Google Scholar]
- 17.Ferrera L, et al. A minimal isoform of the TMEM16A protein associated with chloride channel activity. Biochim Biophys Acta. 2011;1808(9):2214–2223. doi: 10.1016/j.bbamem.2011.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cole C, Barber JD, Barton GJ. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 2008;36(Web Server issue):W197–W201. doi: 10.1093/nar/gkn238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Combet C, Blanchet C, Geourjon C, Deléage G. NPS@: Network protein sequence analysis. Trends Biochem Sci. 2000;25(3):147–150. doi: 10.1016/s0968-0004(99)01540-6. [DOI] [PubMed] [Google Scholar]
- 20.Cheng J, Randall AZ, Sweredoski MJ, Baldi P. SCRATCH: A protein structure and structural feature prediction server. Nucleic Acids Res. 2005;33(Web Server issue):W72–W76. doi: 10.1093/nar/gki396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zerangue N, Jan YN, Jan LY. An artificial tetramerization domain restores efficient assembly of functional Shaker channels lacking T1. Proc Natl Acad Sci USA. 2000;97(7):3591–3595. doi: 10.1073/pnas.060016797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.VanSlyke JK, Deschenes SM, Musil LS. Intracellular transport, assembly, and degradation of wild-type and disease-linked mutant gap junction proteins. Mol Biol Cell. 2000;11(6):1933–1946. doi: 10.1091/mbc.11.6.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ellgaard L, Helenius A. Quality control in the endoplasmic reticulum. Nat Rev Mol Cell Biol. 2003;4(3):181–191. doi: 10.1038/nrm1052. [DOI] [PubMed] [Google Scholar]
- 24.O’Neil KT, DeGrado WF. A thermodynamic scale for the helix-forming tendencies of the commonly occurring amino acids. Science. 1990;250(4981):646–651. doi: 10.1126/science.2237415. [DOI] [PubMed] [Google Scholar]
- 25.Kreusch A, Pfaffinger PJ, Stevens CF, Choe S. Crystal structure of the tetramerization domain of the Shaker potassium channel. Nature. 1998;392(6679):945–948. doi: 10.1038/31978. [DOI] [PubMed] [Google Scholar]
- 26.Howard RJ, Clark KA, Holton JM, Minor DL., Jr Structural insight into KCNQ (Kv7) channel assembly and channelopathy. Neuron. 2007;53(5):663–675. doi: 10.1016/j.neuron.2007.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adam G, Delbruck M. Reduction of Dimensionality. In: Rich A, Davidson N, editors. Biological Diffusion Processes. Structural Chemistry and Molecular Biology. San Francisco: W. H. Freeman; 1968. pp. 198–215. [Google Scholar]
- 28.Martínez AD, et al. Different domains are critical for oligomerization compatibility of different connexins. Biochem J. 2011;436(1):35–43. doi: 10.1042/BJ20110008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ayalon G, Stern-Bach Y. Functional assembly of AMPA and kainate receptors is mediated by several discrete protein-protein interactions. Neuron. 2001;31(1):103–113. doi: 10.1016/s0896-6273(01)00333-6. [DOI] [PubMed] [Google Scholar]
- 30.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 31.Schroeder BC, Cheng T, Jan YN, Jan LY. Expression cloning of TMEM16A as a calcium-activated chloride channel subunit. Cell. 2008;134(6):1019–1029. doi: 10.1016/j.cell.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsien R, Pozzan T. Measurement of cytosolic free Ca2+ with quin2. Methods Enzymol. 1989;172:230–262. doi: 10.1016/s0076-6879(89)72017-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.