Abstract
Small-cell lung cancer and other aggressive neuroendocrine cancers are often associated with early dissemination and frequent metastases. We demonstrate that neurogenic differentiation 1 (NeuroD1) is a regulatory hub securing cross talk among survival and migratory-inducing signaling pathways in neuroendocrine lung carcinomas. We find that NeuroD1 promotes tumor cell survival and metastasis in aggressive neuroendocrine lung tumors through regulation of the receptor tyrosine kinase tropomyosin-related kinase B (TrkB). Like TrkB, the prometastatic signaling molecule neural cell adhesion molecule (NCAM) is a downstream target of NeuroD1, whose impaired expression mirrors loss of NeuroD1. TrkB and NCAM may be therapeutic targets for aggressive neuroendocrine cancers that express NeuroD1.
Keywords: bHLH, SCLC
Based on histology, 15–20% of lung carcinomas are categorized as small-cell lung cancer (SCLC) and 80–85% as non–small-cell lung cancer (NSCLC) (1–4). Lung cancers with neuroendocrine features represent nearly 25% of all lung cancer cases including all SCLC, a subset of NSCLC, and typical and atypical carcinoids. SCLC is the deadliest histological subtype because it is associated with high rates of metastatic disease at time of diagnosis (1, 2). Mixed histological variants containing both SCLC and NSCLC components have previously been linked to a lower overall survival than SCLC alone (5). With neuroendocrine differentiation thought to occur spontaneously in 10–30% of all NSCLC (6), a subset of cells within a tumor may promote poorer prognoses and responses than predicted by original pathology.
The neuronal transcription factor neurogenic differentiation 1 (NeuroD1) is overexpressed in a variety of aggressive neural/neuroendocrine carcinomas. NeuroD1 is important for the development and function of several neural/neuroendocrine tissues, the fate of specific neurons in the central and peripheral nervous system and for insulin gene transcription in adult pancreatic beta cells (7, 8). NeuroD1 mutations can lead to maturity-onset diabetes of the young 6 (MODY6) (Online Mendelian Inheritance in Man 606394), the only setting in which NeuroD1 is thus far known to be critical to disease (9).
To investigate the role of NeuroD1 in tumorigenesis, we focused on neuroendocrine lung cancer cells lines and isogenic normal and tumorigenic immortalized human bronchial epithelial cells (HBEC). We observed that down-regulation of NeuroD1 prevents survival, invasion, and metastasis of several neuroendocrine lung cancer cell lines. Tropomyosin-related kinase B (TrkB) and neural cell adhesion molecule (NCAM) are prometastatic signaling molecules downstream of NeuroD1 and are responsible at least in part for the phenotypic consequences of NeuroD1 expression.
Results
NeuroD1 Is Highly Expressed in Aggressive Neuroendocrine Lung Cancers.
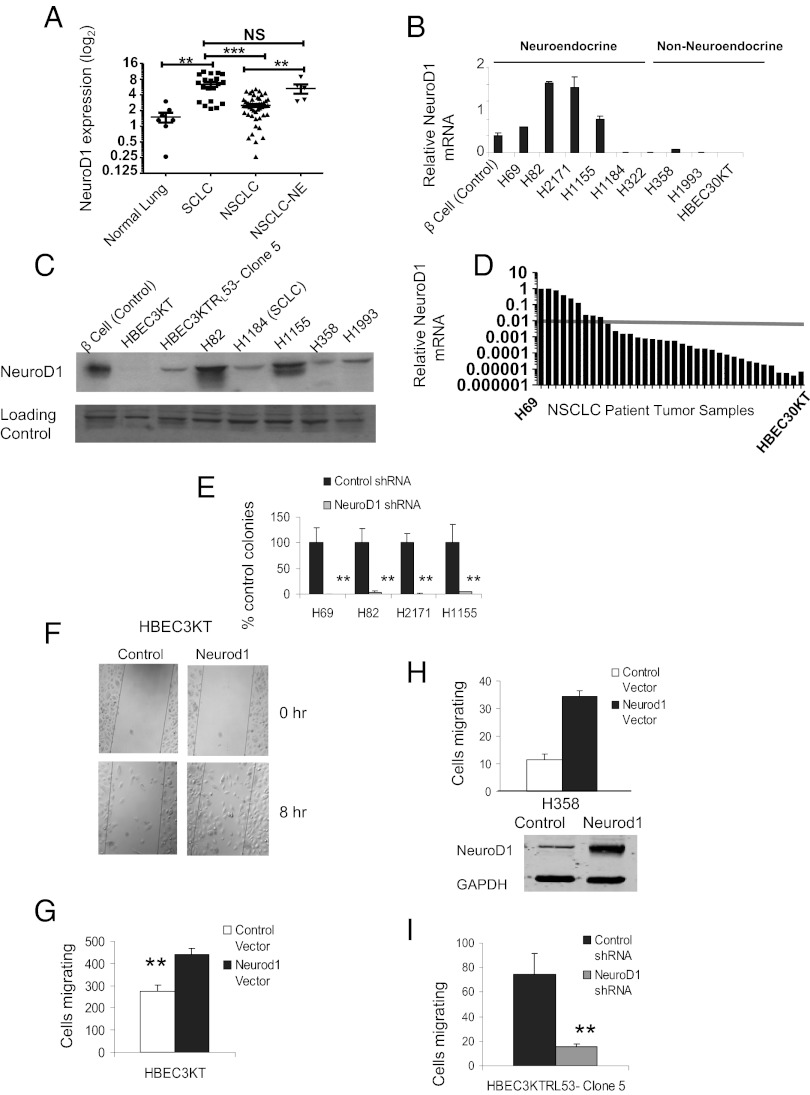
NeuroD1 is overexpressed in several aggressive neural/neuroendocrine cancers including SCLC, medulloblastoma, gastric and prostate cancers, and pituitary adenomas (10–16). To characterize the mechanisms of NeuroD1 action in lung tumor pathogenesis, we analyzed a panel of lung cell lines. HBEC cell lines, assigned a number to distinguish lines from different individuals, are immortalized by overexpression of cyclin-dependent kinase 4, and human telomerase reverse transcriptase (e.g., HBEC3KT) (17). The immortalized HBEC3KT cell line was sequentially transformed by knockdown of the tumor suppressor p53 and expression of K-RasV12 (HBEC3KTRL53) (Table S1) (18, 19). Microarray analysis of lung cell lines revealed that 11 of the 20 SCLC and 3 of the 5 neuroendocrine NSCLC (NSCLC-NE) had significantly higher expression of NEUROD1 compared with HBEC and NSCLC (Fig. 1A and Fig. S1A). We confirmed the neuroendocrine cell lines generally expressed high levels of NeuroD1 compared with HBEC and other lung cancer cell lines (Fig. 1 B and C). We conducted further mechanistic studies in three SCLC cell lines (H69, H82, and H2171) and one NSCLC-NE (H1155).
Fig. 1.
NeuroD1 regulates survival and metastasis of neuroendocrine lung cancers. (A) mRNA expression in 86 cell lines, 8 HBEC, 56 NSCLC, and 22 SCLC, was assessed using Affymetrix HG-U133A and B GeneChips. Cell lines were categorized histopathologically and by NEUROD1 expression (**P < 0.001, ***P < 0.0001; two-tailed t test). (B and C) NeuroD1 expression was validated via quantitative RT-PCR (qRT-PCR) and immunoblotting in cell lines from each type noted above; 50 μg of protein was loaded per lane, and NFAT was used as loading control. Pancreatic beta-cell lines are positive controls. (D) Thirty-five adenocarcinoma and squamous patient samples analyzed via qRT-PCR to confirm NEUROD1 expression. Values were normalized to H69 values. The arbitrary line compares expression of NEUROD1 in the normal bronchial epithelial cell line HBEC30KT. (E) Soft agar assays of H69, H82, and H2171 and H1155 infected with shControl or shNeuroD1. Cells were sorted by expression for GFP on the GIPZ plasmids. Plotted are average number of colonies after 2 wk. Error bars represent ±SD of four independent experiments in triplicate (**P < 0.001; one-way ANOVA). (F–H) HBEC3KT and H358 cells were transfected with pCMV-Neurod1, and then subjected to Transwell or wound-healing assay. Graph represents mean ± SD of three independent experiments in duplicate for HBEC3KT (**P < 0.001; one-way ANOVA). (I) Clone 5 cells infected with shNeuroD1 or shControl were subjected to Transwell assay. Graph represents mean ± SD of three independent experiments in triplicate (**P < 0.001; one-way ANOVA).
To investigate the indication that various NSCLC tumors spontaneously undergo neuroendocrine differentiation, microarray analysis was performed on an additional 275 NSCLC primary resected patient samples not previously annotated as neuroendocrine (Fig. S1B). Validation of a subset of the NSCLC tumor samples revealed several had relatively high NeuroD1 expression compared with H69, whereas the majority of tumor samples expressed more NeuroD1 than HBEC (Fig. 1D). As we suspected, several tumors annotated as NSCLC (specifically adenocarcinomas and squamous cell carcinomas) may have undergone neuroendocrine differentiation.
NeuroD1 Regulates Survival and Metastasis of Neuroendocrine Lung Cancers.
To test whether NeuroD1 was essential for tumorigenesis, we established SCLC and NSCLC-NE lines that stably expressed shRNAs against NeuroD1 (Fig. S1C). Depletion of NeuroD1 prevented survival of the neuroendocrine lung cancer cells as measured by reduction of colonies in soft agar (Fig. 1E and Fig. S1D). The residual soft agar colonies were less than 25% of the control colony size, suggesting defects in sustained growth (Fig. S1E). This phenotype could be partially rescued by a shRNA-resistant mouse NeuroD1 plasmid (Fig. S1 F and G). As many patients present with metastases at the time of diagnosis, we investigated the ability of NeuroD1 to regulate migration. Overexpression of NeuroD1 in HBEC3KT resulted in a significant increase in the motility of the cells (Fig. 1 F and G and Fig. S1H), whereas reduction of NeuroD1 in clone 5 resulted in a significant decrease in motility, as well as decreased soft agar colony formation (Fig.1I and Fig. S2A). To test the effect of NeuroD1 on motility in a nonneuroendocrine NSCLC cell line, we overexpressed NeuroD1 in the adenocarcinoma cell line H358 and found a threefold increase in motility (Fig. 1H), suggesting overexpression of NeuroD1 regulates cell migratory potential.
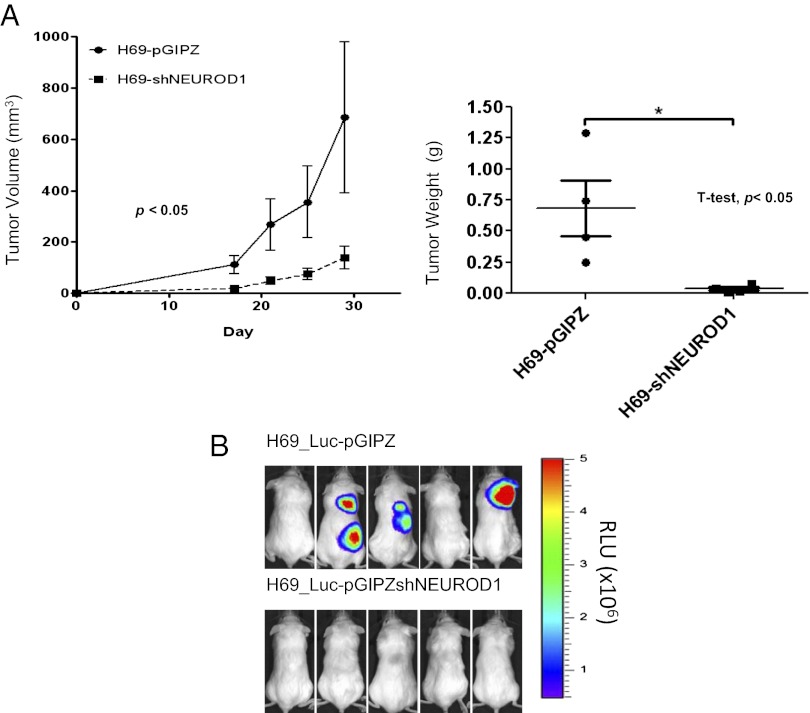
To investigate the tumorigenic role of NeuroD1, neuroendocrine lung cancer cells expressing shNeuroD1 or shControl (pGIPZ) were injected s.c. or i.v. into immune-compromised mice. In the s.c. xenograft model, knockdown of NeuroD1 in the neuroendocrine lung cancer cells resulted in a substantially reduced rate of tumor growth and weight (Fig. 2A and Fig. S2B). We next examined dissemination using H69 stably expressing luciferase (H69-luc). H69-luc expressing shControl colonized to multiple sites, brain, lung, kidney, and lymph nodes, following tail vein injection, whereas knockdown of NeuroD1 prevented colonization and metastasis (Fig. 2B and Fig. S2C). These results provide evidence that NeuroD1 is necessary and sufficient for not only anchorage-independent growth and motility but also tumorigenic and metastatic potential.
Fig. 2.
NeuroD1 is required for tumorigenesis and metastasis in neuroendocrine lung. (A) NOD/SCID female mice were injected with 106 H69 cells infected with shNeuroD1 or shControl. Tumors were measured every 3–5 d until maximum tumor burden was reached. (n = 10, 5 mice per group). P values were computed by linear regression (of slopes) for volume measurements and Student t test for weights. Means are ±SEM. (B) The H69-luc cell line was infected with shcontrol or shNeuroD1. A total of 106 cells was injected into the tail vein of mice and monitored for metastases via bioluminescence imaging.
NeuroD1 Downstream Targets, TrkB and NCAM, Regulate Survival and Migration in Neuroendocrine Lung Cancer.
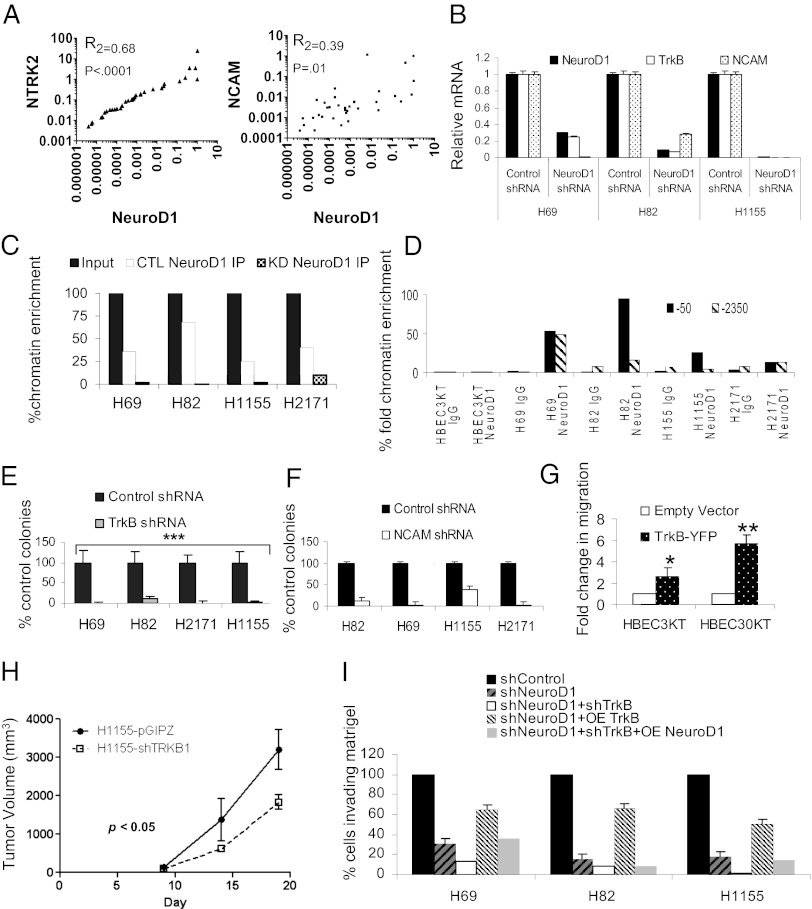
Expression of both NCAM and TrkB correlates with and is potentially regulated by NeuroD1 during neuronal/neuroendocrine differentiation (20–24). TrkB was identified in an unbiased screen for genes that overcame anoikis and is implicated in metastases of several cancers (25–27). NCAM, like TrkB, regulates neuronal differentiation, cell survival, neurite outgrowth, and migration, and also has been implicated in metastasis (28–31). NCAM and TrkB both exhibited higher expression in neuroendocrine cancer cells than in HBEC or NSCLC (Fig. S3A). Analysis of the NSCLC lung cancer patient samples confirmed a significant correlation between expression of NeuroD1 and both NCAM and TrkB (TrkB) (Fig. 3A). In neuroendocrine lung cancer cells, expression of NCAM and TrkB was reduced following knockdown of NeuroD1 (Fig. 3B and Fig. S3 B and C).
Fig. 3.
TrkB and NCAM are downstream targets of NeuroD1 that phenocopy loss of NeuroD1. (A) XY scatter plots of 35 adenocarcinoma and squamous patient samples examining correlation between NEUROD1, NRTK2 (TrkB), and NCAM. P values and R2 values were obtained by Pearson’s test. (B) qRT-PCR analysis of NRTK2, NCAM, and NEUROD1 in lung cancer cells with stable knockdown of NeuroD1. (C) ChIP of NeuroD1 on NTRK2 promoter in cell lines expressing shNeuroD1 or shcontrol (GIPZ). One of four independent experiments in duplicate. (D) ChIP of NeuroD1 on two E boxes in the NCAM promoter with NeuroD1 consensus binding sites in HBEC3KT, three SCLCs, and a NSCLC-NE. NeuroD1 immunoprecipitation values were compared with input, and then plotted as percentage chromatin fold enrichment normalized to HBEC-3KT. (E) Soft agar assay of SCLC and NSCLC-NE lines infected with shControl or shTrkB. The average numbers of colonies after 2 wk are shown. Error bars indicate ±SD from the mean of four independent experiments in triplicate (***P < 0.001; one-way ANOVA). (F) Cell lines were infected with shControl or shNCAM and subjected to soft agar assay. The average numbers of colonies after two weeks are shown. Error bars indicate ±SD from the mean of two independent experiments in triplicate. (G) HBEC3KT and HBEC30KT cells were transfected with a plasmid encoding human TrkB, and then subjected to Transwell assay. Graph represents fold mean ± SD of four and three independent experiments, respectively (**P < 0.005, *P < 0.05; one-way ANOVA). (H) Mice were injected with 106 H1155 cells infected with shNeuroD1 or shControl. Tumors were measured until maximum tumor burden was reached (n = 10, 5 mice per group). P values were computed by linear regression (of slopes) for volume measurements. Means are ±SEM. (I) H69, H82, and H1155 were cell lines were subjected to knockdown of NeuroD1 and/or NeuroD1/TrkB. Knockdown cells were then subjected to overexpression of either NeuroD1 or TrkB. Cells were then embedded in growth factor-reduced Matrigel, for Transwell migration assays as described in Experimental Procedures.
We confirmed that endogenous NeuroD1 bound to the expected E box of the TrkB promoter in the neuroendocrine cancer cell lines (Fig. 3C). Because the correlation between NCAM and NeuroD1 expression has been made via overexpression data and promoter binding predicted in silico, we investigated whether endogenous NeuroD1 directly bound NCAM promoter elements (24, 32–34). We found 10 consensus E-box CANNTG binding sites upstream of the transcriptional start site in the NCAM promoter. Two of the E boxes (located at −50 and −2350) were NeuroD1-preferred binding sites (24). Enhanced NeuroD1 binding was observed on both sites in the neuroendocrine lung cancer cell lines, whereas no binding was observed in HBEC3KT (Fig. 3D).
Next, we inquired whether loss of NCAM or TrkB would phenocopy loss of NeuroD1. Reduction in expression of either TrkB or NCAM also led to a decreased ability of neuroendocrine cell lines to form colonies in soft agar (Fig. 3 E and F and Fig. S3 D and E). Additionally, as with NeuroD1, overexpression of TrkB in HBEC3KT and HBEC30KT led to twofold and fivefold increases in motility, respectively, compared with controls (Fig. 3G and Fig. S3F). Loss of TrkB also led to a decrease in the sustained rate of tumor growth (Fig. 3H). To further explore the relationship between NeuroD1 and TrkB, we knocked down either NeuroD1 alone or NeuroD1 in conjunction with TrkB. Loss of NeuroD1 alone or together with TrkB decreased the ability of the cells to invade through Matrigel (Fig. 3I and Fig. S3G). The loss of invasion caused solely by NeuroD1 depletion was substantially restored by overexpression of TrkB. However, complementation with mouse Neurod1 did not restore invasion caused by depletion of both proteins (Fig. 3I and Fig. S3G), confirming that NeuroD1 acts mostly through TrkB.
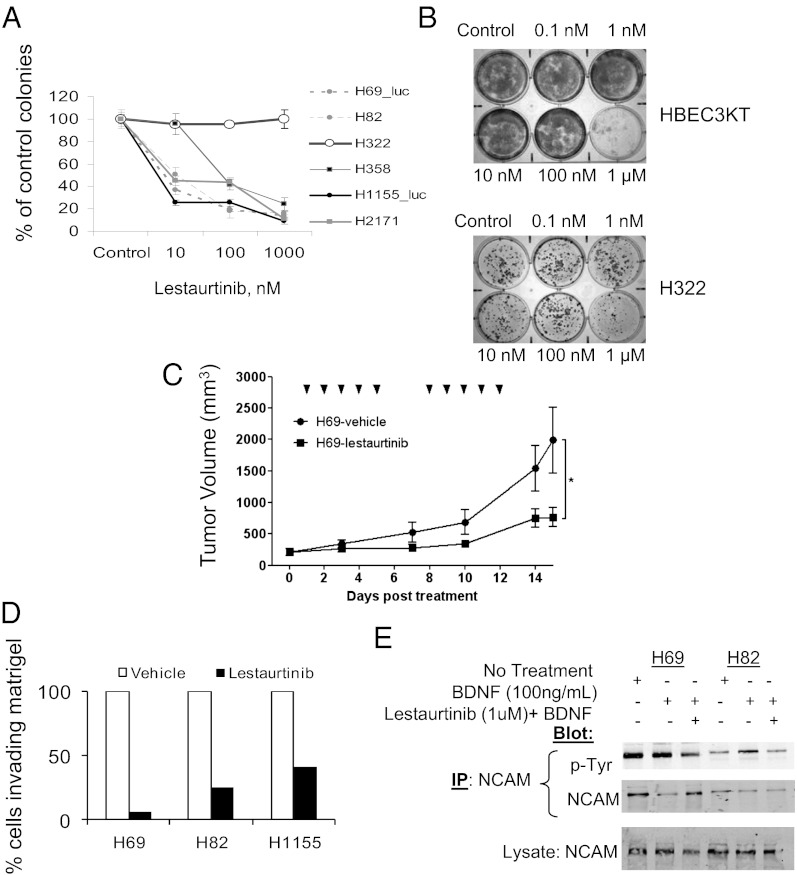
Lestaurtinib, an inhibitor of Trk and certain other tyrosine kinases, has progressed to phase II clinical trials for the treatment of acute myelogenous leukemia (35, 36). Subnanomolar concentrations of lestaurtinib reduced the ability of NeuroD1-expressing cell lines to form colonies in soft agar (Fig. 4A and Fig. S4A). In comparison, 1,000-fold higher drug concentrations were required to perturb colony formation by HBEC3KT or NSCLC with little NeuroD1 (Fig. 4 A and B and Fig. S4B), suggesting the potential for a therapeutic window to treat properly selected neuroendocrine cancers in a clinical setting. Treatment with lestaurtinib resulted in a significant reduction in the rate of tumor growth in xenografts, consistent with a reduction in phosphorylated TrkB (Fig. 4C and Fig. S4C). We also found that lestaurtinib significantly decreased the ability of the cells to invade Matrigel, indicating the importance of TrkB activity in neuroendocrine lung cancer invasion (Fig. 4D).
Fig. 4.
Lestaurtinib regulates survival and invasion. (A) Soft agar assays of SCLC and NSCLC-NE cells treated with 10, 100, and 1,000 nM lestaurtinib. (B) Liquid colony assays of HBEC3KT and the NSCLC cell line H322 exposed to increasing concentrations of lestaurtinib. (C) A total of 106 H69-Luc cells was s.c. injected into the flank of mice and monitored every 2–3 d. Lestaurtinib or vehicle treatment commenced once tumor volume reached ∼200 mm3. n = 10 for each group. The tick marks represent treatment days (*P ≤ 0.05; Student t test). (D) Transwell assays of H69, H82, and H1155 embedded in a 1-mm-thick layer of growth factor-reduced Matrigel with or without 100 μM lestaurtinib. (E) SCLC were serum starved for 8 h, and then treated with 100 ng/mL BDNF without or with 1 μM lestaurtinib for 30 min. NCAM was immunoprecipitated and blotted with anti-phosphotyrosine. Representative of four independent experiments.
NCAM signaling was initially thought to occur via tyrosine phosphorylation by the fibroblast growth factor receptor (10, 37, 38) and recently by TrkB (39). We investigated whether changing TrkB signaling would alter NCAM modification in SCLC. Treatment with the TrkB ligand, brain-derived neurotrophic factor (BDNF), increased tyrosine phosphorylation, whereas treatment with lestaurtinib decreased its tyrosine phosphorylation, consistent with the evidence that TrkB can phosphorylate NCAM in neuroendocrine lung cancers (Fig. 4E). Taken together, these data suggest this interaction may rely on the initiation or maintenance of NCAM and TrkB expression by NeuroD1.
Discussion
NeuroD1 and other factors such as the lineage-restricted oncogene Achaete-scute complex homolog 1 (ASCL1) have been shown to be anomalously expressed in a several aggressive neuroendocrine tumors (15). Initial examination of microarray data revealed that subsets of aggressive SCLCs and certain neuroendocrine NSCLCs have high expression of NeuroD1 compared with HBECs and other NSCLCs. Mechanism of action of NeuroD1 is carried out by downstream targets, which include the signaling molecules, the tyrosine kinase TrkB and NCAM. Neuroendocrine differentiation of tumors has become a topic of interest as differentiation of these tumors from epithelial cells has been hypothesized to be involved in acquisition of invasive/metastatic phenotypes (12, 14, 16). NeuroD1 expression has recently been speculated to contribute to the transformation of epithelial cells to neuronal-like cells (12); this transformation may be the onset or termination neuroendocrine differentiation of prostate and other neuroendocrine cancers. We find that depletion of NeuroD1 reverses or suppresses neuroendocrine characteristics, for example, a reduction in NCAM expression.
TrkB induces neuronal migration and, similar to NeuroD1, neuronal differentiation (20, 40, 41). TrkB has also recently been implicated in invasion and metastasis of pancreatic, prostate, and colorectal cancers (26, 42). TrkB is mutated or overexpressed in certain NSCLCs and ovarian, prostate, pancreatic, and gastric cancers, and can suppress cell death caused by loss of cell-substratum contacts. NCAM, like TrkB, also regulates survival, differentiation, and migration of neurons (10, 31, 39, 43). NCAM is highly expressed in neuroendocrine lung tumors and immunotherapy using an antibody targeting NCAM linked to the microtubule-depolymerizing agent BB10901 has entered phase II clinical trials for SCLC (44). We hypothesize that overexpression of NeuroD1 may contribute to the development and metastasis of extremely aggressive SCLC, via regulation of each of these downstream factors involved in differentiation, cell survival, and invasiveness.
Experimental Procedures
Plasmids, Primers, and Luciferase Assays.
Stable shNeuroD1 and shTrkB cells lines were generated via infection of human pGIPZ lentiviral shRNA plasmids created by the RNAi Consortium. These were purchased by University of Texas Southwestern as a library [TRC-Hs1.0 (human)] from Open Biosystems. NeuroD1 short hairpins V2LHS_152218 (shRNA-1), V2LHS_152220 (shRNA-2), TrkB short hairpins, V2LHS_63731, and NCAM short hairpins V2LHS_111710 were used (sequences are available online at Open Biosystems website). SCLC cell lines were selected in puromycin (<2 mg/mL) for 6 d. Plasmids were transfected into 293T cells for viral production using FUGENE 6. Oligonucleotides used were as follows: NeuroD1-1, sense, CGAAUUUGGUGUGGCUGUA, and antisense, UACAGCCACACCAAAUUCG (QIAGEN); NeuroD1-AB, sense, GGAUCAAUCUUCUCAGGCA, and antisense, UGCCUGAGAAGAUUGAUCC (Ambion); NTRK2-5, sense, GACGAGUUUGUCUAGGAAA, and antisense, UUUCCUAGACAAACUCGUC (QIAGEN). For p53 experiments, cells were transfected with pCMV5, SV40 (internal control), pcDNA.1-p53, or pGL3-NeuroD1 constructs using Fugene HD. Luciferase assays used the Promega dual luciferase kit according to manufacturer’s protocol.
Quantitative Real-Time PCR.
Total RNA from xenograft tumors and cell lines was isolated with TRI Reagent. RNA from tumor samples was from M. D. Anderson Cancer Center. cDNA was synthesized using iSCRIPT cDNA Synthesis Kit (Bio-Rad). RNAs for mouse and human NeuroD1, TrkB, NCAM, and 18s ribosomal RNA were quantified by RT-PCR with iTaq (Bio-Rad) master mix using TaqMan probes (Applied Biosystems) on an ABI 7500 thermocycler. Relative transcript levels were normalized to 18s rRNA. Transcript amounts in knockdown cells were plotted as fold change relative to control. Data were analyzed using ABI 7500 system software.
Statistical Analyses.
Student t test, one-way analysis of variance (ANOVA), Pearson’s test, and linear regression were used to determine statistical significance. Statistical significance for all tests, assessed by calculating the P values, was defined as <0.05.
Chromatin Immunoprecipitation.
ChIP was performed as previously described (45). TrkB primers were as described (22). NCAM E-box primers were as follows: −2350 forward, GGGGAGAGAGGTCCAGTGA; −2350 reverse, TTCTAGAATGCTGCCCCAGT; −50 forward, ATCAAAATATGCAAACTGCTGATTA; and −50 reverse, CGAACATCAAGGAGGTAAGAGA.
Colony Formation and in Vivo Assays.
Soft agar and liquid colony assays were as described previously (18). As indicated, lestaurtinib was added once for 24 h 1 d after seeding. In vivo experiments were performed in 6- to 8-wk-old nonobese diabetic (NOD)/SCID female mice. Viable cells from subconfluent cultures in normal growth medium were counted using Trypan Blue (Invitrogen). Subcutaneous tumors were elicited by injecting 1.0 × 106 cells in 0.2 mL of PBS into the flank of mice and monitored every 2–3 d. Tumor size was assessed with digital calipers; tumor volume was taken to be equal to the width × length2 × π/6. The effect of lestaurtinib (LC Laboratories) was determined on tumors grown as above. Lestaurtinib or vehicle treatment commenced once tumor volume reached ∼200 mm3. Lestaurtinib was dissolved in 40% (wt/vol) polyethylene glycol, 10% (wt/vol) polyvinylpyrrolidone, and 2% (vol/vol) benzyl alcohol (all from Sigma-Aldrich) in distilled water and administered s.c. at 20 mg/kg once daily, 5 d a week. Tail vein injections of 1.0 × 106 cells in 0.1 mL of PBS were used to compare the metastatic capacity of H69 cells expressing luciferase driven by the CMV promoter (H69-luc). Tumor growth was monitored every week with bioluminescence imaging following s.c. injection of 450 mg/kg D-luciferin offline (Biosynth) in PBS into anesthetized mice (46). Images were captured 10 min after D-luciferin injection with a 60-s exposure using a CCD camera (Caliper Xenogen). Animal care was in accord with University of Texas Southwestern Medical Center guidelines and approved Institutional Animal Care and Use Committee protocols.
Reagents, Antibodies, Immunoblotting.
Immunoblot analyses were as previously described using equal amounts of protein from each sample (45). The following antibodies were used for blotting, immunoprecipitation, and ChIP: goat NeuroD1 (N-19), rabbit pan-phospho-Trk (E-6), synaptophysin (H-8), mouse pTyr (PY20), p53 (DO-1), GAPDH (FL-335) (Santa Cruz); mouse ASCL1, mouse N-cadherin, mouse E-cadherin (BD Biosciences); rabbit TrkB (Chemicon), mouse CD56/NCAM rabbit β-catenin (Cell Signaling), and α-tubulin hybridoma (The Hybridoma Bank Studies at University of Iowa) were purchased. Lestaurtinib was purchased from LC Laboratories, and BDNF, from R&D Systems. Band intensities were quantified using LI-COR Odyssey Infrared Imaging System.
Immunoprecipitation of NCAM.
SCLC cells were lysed in 0.1% SDS, 1% Nonidet P-40, and 1% sodium deoxycholate with phosphatase inhibitor mixture (Sigma). NCAM was immunoprecipitated from lysates with anti-NCAM overnight.
Cell Culture.
Min6 cells were grown in Dulbecco’s modified Eagle medium with 10% FBS. SCLC and NSCLC lines were from the Hamon Cancer Center Collection (University of Texas Southwestern). SCLC, NSCLC-NE, HBEC3KTRL53-clone 5 were cultured in RPMI 1640 with 10% FBS. Immortalized HBECs (except HBEC3KTRL53-clone 5) (18) were cultured in keratinocyte serum-free medium (KSFM) (Invitrogen) with 5 ng/mL epidermal growth factor and 50 μg/mL bovine pituitary extract. The lung cancer cell lines were DNA fingerprinted using the PowerPlex 1.2 kit (Promega) and confirmed to be the same as the DNA fingerprint library maintained either by ATCC or the Hamon Cancer Center. The lines were also tested to be free of mycoplasma by e-Myco kit (Boca Scientific).
Migration Assays.
For migration assays, cells were seeded 48 h following either transient expression of mouse NeuroD1 (pCMV-Neurod1) or human TrkB-YFP in HBEC3KT and HBEC30KT or transient knockdown of NeuroD1 (shNeuroD1) in clone 5. Transwell migration was assayed in a 10-well Boyden chamber (Neuro Probe) or Transwell permeable supports (Corning; no. 3422). Clone 5, H358, and HBEC3KT cells were seeded in the top chamber in either RPMI with 1% FBS or KSFM without FBS, respectively, and allowed to migrate along a concentration gradient through a polycarbonate membrane with 8-μm pores to the bottom chamber containing medium with 10% FBS. For TrkB studies, HBECs transfected with TrkB or empty vector were treated with 100 ng/mL BDNF. After 24 h, cells were fixed, stained (with hematoxylin and eosin stain), and counted. For invasion assays, 1.5 × 105 cells were imbedded in growth factor-reduced Matrigel in the presence or absence of 100 nM lestaurtinib in Transwell permeable supports. Cells were allowed to migrate for 48 h across membranes with a gradient of 10% serum in the bottom chamber. In the wound-healing assays, 2 × 105 HBEC3KT cells were seeded in six-well dishes in KSFM and grown to confluence for 24–48 h. A wound was created in confluent monolayers using a sterile pipette tip; cell migration was quantified using ImageJ software after 8 h.
Microarray Analysis.
RNA from tumor samples was from M. D. Anderson Cancer Center. RNA was prepared using the RNeasy Midi kit (Qiagen) and analyzed for quality on RNA 6000 Nano kit (Agilent Technologies) with Agilent Bioanalyzer software. Five micrograms of total RNA were labeled and hybridized to Affymetrix GeneChips HG-U133A and B according to the manufacturer’s protocol (www.affymetrix.com), whereas 0.5 μg of total RNA was used for Illumina BeadChip HumanWG-6 V3 (www.illumina.com). These data are available in the Gene Expression Omnibus database (accession nos. GSE4824 and GSE32036). Array data were preprocessed with MAS5 (Affymetrix algorithm for probe summarization) or MBCB [Illumina algorithm for background subtraction (47)], quantile-normalized, and log-transformed.
Supplementary Material
Acknowledgments
We thank A-Young Lee, Elma Zaganjor, Andres Lorente-Rodriguez (M.H.C. lab), Rolf Brekken [Department of Surgery, UT Southwestern (UTSW)], and Joseph Albanesi (Department of Pharmacology, UTSW) and Banu Eskiocak (Department of Cell Biology, UTSW) for comments on the manuscript, and Dionne Ware for administrative assistance. We thank Luis Parada (Department of Developmental Biology, UTSW) for TrkB-YFP. This work was supported by National Institutes of Health Grants R01DK55310 (to M.H.C.) and P50CA70907 (to J.D.M.), grants from the Cancer Prevention and Research Institute of Texas (to M.H.C. and J.D.M.), and Department of Defense PROSPECT and The Longenbaugh Foundation (J.D.M.). J.K.O. is supported by National Institute of General Medical Sciences Pharmacological Sciences Training Grant 5-T32 GM007062. J.E.L. is supported by National Health and Medical Research Council Biomedical Fellowship 494511 and Transplantation Society of Australia and New Zealand/Allen and Hanburys Respiratory Research Fellowship. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession nos. GSE4824 and GSE32036).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1303932110/-/DCSupplemental.
References
- 1.Fischer B, Arcaro A. Current status of clinical trials for small cell lung cancer. Rev Recent Clin Trials. 2008;3(1):40–61. doi: 10.2174/157488708783330503. [DOI] [PubMed] [Google Scholar]
- 2.Jackman DM, Johnson BE. Small-cell lung cancer. Lancet. 2005;366(9494):1385–1396. doi: 10.1016/S0140-6736(05)67569-1. [DOI] [PubMed] [Google Scholar]
- 3.Sato M, Shames DS, Gazdar AF, Minna JD. A translational view of the molecular pathogenesis of lung cancer. J Thorac Oncol. 2007;2(4):327–343. doi: 10.1097/01.JTO.0000263718.69320.4c. [DOI] [PubMed] [Google Scholar]
- 4.Sun S, Schiller JH, Spinola M, Minna JD. New molecularly targeted therapies for lung cancer. J Clin Invest. 2007;117(10):2740–2750. doi: 10.1172/JCI31809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Radice PA, et al. The clinical behavior of “mixed” small cell/large cell bronchogenic carcinoma compared to “pure” small cell subtypes. Cancer. 1982;50(12):2894–2902. doi: 10.1002/1097-0142(19821215)50:12<2894::aid-cncr2820501232>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 6.Ionescu DN, et al. Nonsmall cell lung carcinoma with neuroendocrine differentiation—an entity of no clinical or prognostic significance. Am J Surg Pathol. 2007;31(1):26–32. doi: 10.1097/01.pas.0000213319.04919.97. [DOI] [PubMed] [Google Scholar]
- 7.Miyata T, Maeda T, Lee JE. NeuroD is required for differentiation of the granule cells in the cerebellum and hippocampus. Genes Dev. 1999;13(13):1647–1652. doi: 10.1101/gad.13.13.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Naya FJ, Stellrecht CM, Tsai MJ. Tissue-specific regulation of the insulin gene by a novel basic helix-loop-helix transcription factor. Genes Dev. 1995;9(8):1009–1019. doi: 10.1101/gad.9.8.1009. [DOI] [PubMed] [Google Scholar]
- 9.Malecki MT, et al. Mutations in NEUROD1 are associated with the development of type 2 diabetes mellitus. Nat Genet. 1999;23(3):323–328. doi: 10.1038/15500. [DOI] [PubMed] [Google Scholar]
- 10.Abrahamsson PA. Neuroendocrine differentiation in prostatic carcinoma. Prostate. 1999;39(2):135–148. doi: 10.1002/(sici)1097-0045(19990501)39:2<135::aid-pros9>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 11.Cantile M, et al. cAMP induced modifications of HOX D gene expression in prostate cells allow the identification of a chromosomal area involved in vivo with neuroendocrine differentiation of human advanced prostate cancers. J Cell Physiol. 2005;205(2):202–210. doi: 10.1002/jcp.20384. [DOI] [PubMed] [Google Scholar]
- 12.Cindolo L, et al. NeuroD1 expression in human prostate cancer: Can it contribute to neuroendocrine differentiation comprehension? Eur Urol. 2007;52(5):1365–1373. doi: 10.1016/j.eururo.2006.11.030. [DOI] [PubMed] [Google Scholar]
- 13.Fratticci A, et al. Differential expression of neurogenins and NeuroD1 in human pituitary tumours. J Endocrinol. 2007;194(3):475–484. doi: 10.1677/JOE-07-0020. [DOI] [PubMed] [Google Scholar]
- 14.Gupta A, et al. Neuroendocrine differentiation in the 12T-10 transgenic prostate mouse model mimics endocrine differentiation of pancreatic beta cells. Prostate. 2008;68(1):50–60. doi: 10.1002/pros.20650. [DOI] [PubMed] [Google Scholar]
- 15.Rostomily RC, et al. Expression of neurogenic basic helix-loop-helix genes in primitive neuroectodermal tumors. Cancer Res. 1997;57(16):3526–3531. [PubMed] [Google Scholar]
- 16.Syder AJ, et al. A transgenic mouse model of metastatic carcinoma involving transdifferentiation of a gastric epithelial lineage progenitor to a neuroendocrine phenotype. Proc Natl Acad Sci USA. 2004;101(13):4471–4476. doi: 10.1073/pnas.0307983101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramirez RD, et al. Immortalization of human bronchial epithelial cells in the absence of viral oncoproteins. Cancer Res. 2004;64(24):9027–9034. doi: 10.1158/0008-5472.CAN-04-3703. [DOI] [PubMed] [Google Scholar]
- 18.Sato M, et al. Multiple oncogenic changes (K-RAS(V12), p53 knockdown, mutant EGFRs, p16 bypass, telomerase) are not sufficient to confer a full malignant phenotype on human bronchial epithelial cells. Cancer Res. 2006;66(4):2116–2128. doi: 10.1158/0008-5472.CAN-05-2521. [DOI] [PubMed] [Google Scholar]
- 19.Sato M, et al. Human lung epithelial cells progressed to malignancy through specific oncogenic manipulations. Molec Cancer Res. 2013 doi: 10.1158/1541-7786.MCR-12-0634-T. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaplan DR, Matsumoto K, Lucarelli E, Thiele CJ. Eukaryotic Signal Transduction Group Induction of TrkB by retinoic acid mediates biologic responsiveness to BDNF and differentiation of human neuroblastoma cells. Neuron. 1993;11(2):321–331. doi: 10.1016/0896-6273(93)90187-v. [DOI] [PubMed] [Google Scholar]
- 21.Liu K, et al. MiR-124 regulates early neurogenesis in the optic vesicle and forebrain, targeting NeuroD1. Nucleic Acids Res. 2011;39(7):2869–2879. doi: 10.1093/nar/gkq904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Y, Encinas M, Comella JX, Aldea M, Gallego C. Basic helix-loop-helix proteins bind to TrkB and p21(Cip1) promoters linking differentiation and cell cycle arrest in neuroblastoma cells. Mol Cell Biol. 2004;24(7):2662–2672. doi: 10.1128/MCB.24.7.2662-2672.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seki T. Expression patterns of immature neuronal markers PSA-NCAM, CRMP-4 and NeuroD in the hippocampus of young adult and aged rodents. J Neurosci Res. 2002;70(3):327–334. doi: 10.1002/jnr.10387. [DOI] [PubMed] [Google Scholar]
- 24.Seo S, Lim JW, Yellajoshyula D, Chang LW, Kroll KL. Neurogenin and NeuroD direct transcriptional targets and their regulatory enhancers. EMBO J. 2007;26(24):5093–5108. doi: 10.1038/sj.emboj.7601923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Au CW, et al. Tyrosine kinase B receptor and BDNF expression in ovarian cancers—effect on cell migration, angiogenesis and clinical outcome. Cancer Lett. 2009;281(2):151–161. doi: 10.1016/j.canlet.2009.02.025. [DOI] [PubMed] [Google Scholar]
- 26.Douma S, et al. Suppression of anoikis and induction of metastasis by the neurotrophic receptor TrkB. Nature. 2004;430(7003):1034–1039. doi: 10.1038/nature02765. [DOI] [PubMed] [Google Scholar]
- 27.Kupferman ME, et al. TrkB induces EMT and has a key role in invasion of head and neck squamous cell carcinoma. Oncogene. 2010;29(14):2047–2059. doi: 10.1038/onc.2009.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cavallaro U, Niedermeyer J, Fuxa M, Christofori G. N-CAM modulates tumour-cell adhesion to matrix by inducing FGF-receptor signalling. Nat Cell Biol. 2001;3(7):650–657. doi: 10.1038/35083041. [DOI] [PubMed] [Google Scholar]
- 29.Daniloff JK, Chuong CM, Levi G, Edelman GM. Differential distribution of cell adhesion molecules during histogenesis of the chick nervous system. J Neurosci. 1986;6(3):739–758. doi: 10.1523/JNEUROSCI.06-03-00739.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoffman S, Friedlander DR, Chuong CM, Grumet M, Edelman GM. Differential contributions of Ng-CAM and N-CAM to cell adhesion in different neural regions. J Cell Biol. 1986;103(1):145–158. doi: 10.1083/jcb.103.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vutskits L, et al. PSA-NCAM modulates BDNF-dependent survival and differentiation of cortical neurons. Eur J Neurosci. 2001;13(7):1391–1402. doi: 10.1046/j.0953-816x.2001.01516.x. [DOI] [PubMed] [Google Scholar]
- 32.Neptune ER, et al. Targeted disruption of NeuroD, a proneural basic helix-loop-helix factor, impairs distal lung formation and neuroendocrine morphology in the neonatal lung. J Biol Chem. 2008;283(30):21160–21169. doi: 10.1074/jbc.M708692200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee JE, et al. Conversion of Xenopus ectoderm into neurons by NeuroD, a basic helix-loop-helix protein. Science. 1995;268(5212):836–844. doi: 10.1126/science.7754368. [DOI] [PubMed] [Google Scholar]
- 34.Kashiwagi K, et al. Differences of molecular expression mechanisms among neural cell adhesion molecule 1, synaptophysin, and chromogranin A in lung cancer cells. Pathol Int. 2012;62(4):232–245. doi: 10.1111/j.1440-1827.2011.02781.x. [DOI] [PubMed] [Google Scholar]
- 35.Knapper S, et al. A phase 2 trial of the FLT3 inhibitor lestaurtinib (CEP701) as first-line treatment for older patients with acute myeloid leukemia not considered fit for intensive chemotherapy. Blood. 2006;108(10):3262–3270. doi: 10.1182/blood-2006-04-015560. [DOI] [PubMed] [Google Scholar]
- 36.Thiele CJ, Li Z, McKee AE. On Trk—the TrkB signal transduction pathway is an increasingly important target in cancer biology. Clin Cancer Res. 2009;15(19):5962–5967. doi: 10.1158/1078-0432.CCR-08-0651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kiselyov VV, et al. Structural basis for a direct interaction between FGFR1 and NCAM and evidence for a regulatory role of ATP. Structure. 2003;11(6):691–701. doi: 10.1016/s0969-2126(03)00096-0. [DOI] [PubMed] [Google Scholar]
- 38.Soroka V, et al. Structure and interactions of NCAM Ig1-2-3 suggest a novel zipper mechanism for homophilic adhesion. Structure. 2003;11(10):1291–1301. doi: 10.1016/j.str.2003.09.006. [DOI] [PubMed] [Google Scholar]
- 39.Cassens C, et al. Binding of the receptor tyrosine kinase TrkB to the neural cell adhesion molecule (NCAM) regulates phosphorylation of NCAM and NCAM-dependent neurite outgrowth. J Biol Chem. 2010;285(37):28959–28967. doi: 10.1074/jbc.M110.114835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu ZZ, Zhu LQ, Eide FF. Critical role of TrkB and brain-derived neurotrophic factor in the differentiation and survival of retinal pigment epithelium. J Neurosci. 1997;17(22):8749–8755. doi: 10.1523/JNEUROSCI.17-22-08749.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luikart BW, et al. Neurotrophin-dependent dendritic filopodial motility: A convergence on PI3K signaling. J Neurosci. 2008;28(27):7006–7012. doi: 10.1523/JNEUROSCI.0195-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sclabas GM, et al. Overexpression of tropomysin-related kinase B in metastatic human pancreatic cancer cells. Clin Cancer Res. 2005;11(2 Pt 1):440–449. [PubMed] [Google Scholar]
- 43.Ditlevsen DK, Povlsen GK, Berezin V, Bock E. NCAM-induced intracellular signaling revisited. J Neurosci Res. 2008;86(4):727–743. doi: 10.1002/jnr.21551. [DOI] [PubMed] [Google Scholar]
- 44.Blackhall FH, Shepherd FA. Small cell lung cancer and targeted therapies. Curr Opin Oncol. 2007;19(2):103–108. doi: 10.1097/CCO.0b013e328011bec3. [DOI] [PubMed] [Google Scholar]
- 45.Lawrence MC, McGlynn K, Park BH, Cobb MH. ERK1/2-dependent activation of transcription factors required for acute and chronic effects of glucose on the insulin gene promoter. J Biol Chem. 2005;280(29):26751–26759. doi: 10.1074/jbc.M503158200. [DOI] [PubMed] [Google Scholar]
- 46.Paroo Z, et al. Validating bioluminescence imaging as a high-throughput, quantitative modality for assessing tumor burden. Mol Imaging. 2004;3(2):117–124. doi: 10.1162/15353500200403172. [DOI] [PubMed] [Google Scholar]
- 47.Ding LH, Xie Y, Park S, Xiao G, Story MD. Enhanced identification and biological validation of differential gene expression via Illumina whole-genome expression arrays through the use of the model-based background correction methodology. Nucleic Acids Res. 2008;36(10):e58. doi: 10.1093/nar/gkn234. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.