Abstract
Yeast cells deleted for the SRO7/SOP1 encoded tumor suppressor homologue show increased sensitivity to NaCl stress. On exposure to growth-inhibiting NaCl concentrations, sro7Δ mutants display a rapid loss in viability that is associated with markers of apoptosis: accumulation of reactive oxygen species, DNA breakage, and nuclear fragmentation. Additional deletion of the yeast metacaspase gene YCA1 prevents the primary fast drop in viability and diminishes nuclear fragmentation and DNA breakage. We also observed that NaCl induced loss in viability of wild-type cells is Yca1p dependent. However, a yeast strain deleted for both SRO7 and its homologue SRO77 exhibits NaCl-induced cell death that is independent on YCA1. Likewise, sro77Δ single mutants do not survive better after additional deletion of the YCA1 gene, and both sro77Δ and sro77Δyca1Δ mutants display apoptotic characteristics when exposed to growth-inhibiting salinity, suggesting that yeast possesses Yca1p-independent pathway(s) for apoptosis-like cell death. The activity of Yca1p increases with increasing NaCl stress and sro7Δ mutants achieve levels that are higher than in wild-type cells. However, mutants lacking SRO77 do not enhance caspase activity when subject to NaCl stress, suggesting that Sro7p and Sro77p exert opposing effects on the cellular activity of Yca1p.
INTRODUCTION
The physiological cell death process of apoptosis is a morphologically distinct form of cellular suicide (Kerr, 2002), designed to remove potentially threatening or undesired cells (Vaux and Korsmeyer, 1999; Beers and McDowell, 2001). In animal cells, apoptosis occurs in an ordered series of event and is associated with activation of the programmed cell death machinery (Wyllie et al., 1980; Joza et al., 2002). Inappropriate regulation of apoptosis is linked to the pathogenesis of many human diseases, such as AIDS, cancer, autoimmune, and neurodegenerative disorders (Uren and Vaux, 1996). The process of programmed cell death can be triggered by a vast array of stimuli, and a complex network of regulators and effectors coordinates the process (Beers and McDowell, 2001; Joza et al., 2002). At the end of almost all apoptotic scenarios, a constellation of morphological characteristics emerge, including chromatin condensation, DNA fragmentation, flipping of phosphatidylserine to the outer leaflet of the plasma membrane, and breakage of the cells to small membrane-enclosed vesicles, so called apoptotic bodies (Wyllie et al., 1980). These structural features constitute the cytological hallmarks of apoptosis.
The finding that cellular suicide occurs in most, if not all, multicellular organisms raises the question on how the mechanisms of apoptosis have evolved. Do these mechanisms have a common origin and have these processes first evolved in single-celled organisms? For unicellular organisms exposed to harsh conditions, cellular suicide may serve to propagate the genome to future generations if some of the members of a population are sacrificed to promote survival of others (Engelberg-Kulka and Glaser, 1999; Matsuyama et al., 1999; Fröhlich and Madeo, 2000). In Saccharomyces cerevisiae, apoptosis-like cell death was first described for a temperature-sensitive cdc48 mutant (Madeo et al., 1997). Cdc48p is involved in vesicle fusion associated with secretion and cell division (Latterich et al., 1995). At nonpermissive temperature, the cdc48 mutant exhibited typical apoptotic markers, i.e., exposition of phosphatidylserine, DNA fragmentation, and chromatin condensation (Madeo et al., 1997). It has also become increasingly clear that there is a connection between ageing and apoptosis-like cell death in yeast, as evidenced by the presence of apoptotic markers in old yeast cells (Fröhlich and Madeo, 2001; Laun et al., 2001). Still, there are arguments against a mechanistic and functional conservation between yeast and mammals, because the yeast genome does not encode obvious homologues to any of the core apoptotic machinery proteins, e.g., the Bcl-2/Bax family of proteins or the caspase family. However, recently, a metacaspase called yeast caspase-1 (Yca1p) was identified in S. cerevisiae, and this protein was required for H2O2 or aging-induced apoptosis in yeast (Madeo et al., 2002). The yeast metacaspase has also been shown to be involved in a telomer-initiated apoptotic pathway that exhibits conserved features with that reported for mammalian cells (Qi et al., 2003). In another recent article, Severin and Hyman (2002) presented evidence that apoptosis in yeast can be induced by a factor that is produced by the yeast cells themselves: the α-factor mating pheromone. The data presented indicate that this mechanism might have evolved to remove cells that fail to mate, suggesting a highly sophisticated death machinery in yeast.
Recently, evidence has also been presented that exposure of yeast to high salinity induces apoptosis (Huh et al., 2002). Several observations suggested that the death was induced by intracellular ion disequilibria rather than by osmotic imbalance. For example, ectopic expression of the antiapoptotic protein Bcl-2 enhanced NaCl tolerance of wild-type cells and a calcineurin-defective cnb1Δ mutant that is defective for ion homeostasis (Huh et al., 2002). However, overexpression of Bcl-2 did not suppress the salt-sensitive phenotype of a yeast hog1Δ mutant, which has decreased capacity to osmoregulate, but seems to maintain ion homeostasis. We have observed that deletion of the SOP1/SRO7 gene (hereafter referred to as SRO7) brings about increased sensitivity to NaCl stress and a defective intracellular ion homeostasis (Larsson et al., 1998). SRO7 is a yeast homologue of the Drosophila l(2)gl tumor suppressor gene. This gene is required for development of epithelial cell polarity (Bilder et al., 2000), and mutations in l(2)gl leads to formation of epithelial derived tumors in fly larvae (Gateff, 1978). The l(2)gl gene is also required for the development of cell polarity associated with asymmetric cell division of neuroblasts (Ohshiro et al., 2000; Peng et al., 2000), and the defective polarity acquisition in l(2)gl mutants may give rise to the neoplastic transformation of optic neuroblasts observed in mutant larvae (Gateff, 1978). The salt-sensitive phenotype of the yeast sro7Δ mutant results from defective targeting of the ENA1 encoded sodium extruding ATPase to the plasma membrane (Wadskog, 2003). Instead, Ena1p is delivered to the vacuole where it is degraded. Deletion of SRO7 together with its iso-gene SRO77 results in hypersensitivity to NaCl stress (Larsson et al., 1998), and a cold-sensitive phenotype (Lehman et al., 1999). At nonpermissive temperature, the sro7Δsro77Δ mutant shows defective secretion and accumulation of post-Golgi vesicles (Lehman et al., 1999). In agreement with these phenotypes, the Sro proteins have been shown to interact physically with Sec9p, a target-soluble N-ethylmaleimide-sensitive factor attachment receptor (t-SNARE) protein for vesicle fusion at the plasma membrane (Lehman et al., 1999). SRO7 also shows genetic interaction with temperature-sensitive alleles of genes for the exocyst complex (Lehman et al., 1999; Wadskog, 2003). Together, these results suggest that the l(2)gl family of proteins assists in the targeting of vesicles to the correct polar destinations in the plasma membrane.
Here, we report the involvement of SRO7 and SRO77 in apoptosis-like cell death in yeast. The studies were initiated for several reasons. First, the two phenotypes associated with sro mutants, salt sensitivity and secretion defects, have both been previously linked to apoptosis in yeast (Madeo et al., 1997; Huh et al., 2002). Second, most of the known tumor suppressor genes are involved in signaling that lead either to cell cycle arrest or apoptosis (Wyllie et al., 1999). Finally, two-hybrid screens have indicated physical interactions between Sro77p and the yeast metacaspase Yca1p (Uetz et al., 2000).
MATERIALS AND METHODS
Yeast Strains and Growth Media
Yeast strains used in this study are listed in Table 1. Rich YPD medium (2% glucose) was used for growth and survival assays, whereas minimal YNB media (2% glucose) plus essential amino acids were used for selective growth. For salt stress conditions, YPD containing 2 M NaCl was added to yeast cultures to obtain final NaCl concentrations.
Table 1.
Yeast strains used in the present study
| Yeast strain | Genotype | Source |
|---|---|---|
| BY4741 | MATa his3Δ leu2Δ met15Δ ura3Δ | Euroscarf |
| BY4742 | MATα his3Δ leu2Δ lys2Δ ura3Δ | Euroscarf |
| Y05451 | BY4741 sro7::kanMX4 | Euroscarf |
| Y13134 | BY4742 sro77::kanMX4 | Euroscarf |
| Y02453 | BY4741 ycal::kanMX4 | Euroscarf |
| Y14387 | BY4742 ckbl::kanMX4 | Euroscarf |
| YIW-1 | BY4741 sro7::kanMX4 sro77::kanMX4 | This study |
| YIW-24 | BY4741 sro7::LEU2 ycal::kanMX4 | This study |
| YIW-25 | BY4741 sro77::HIS3 ycal::kanMX4 | This study |
| YIW-26 | BY4741 sro7::LEU2 sro77::HIS3 ycal::kanMX4 | This study |
| YSH6.142-3A | MATα leu2-3/112 ura3-1 trp1-1 his3-11/15 ade2-1 can1-100 GAL SUC2 mal0 gpdl::TRP1 | (Ansell et al., 1997) |
The yca1Δsro7Δ mutant was constructed using the sro7::LEU2 deletion cassette in pBluescriptKS+ (Larsson et al., 1998). The plasmid was cut with restriction enzymes HindIII and Notl (Roche Diagnostics, Mannheim, Germany) and the ycalΔ mutant transformed with the sro7::LEU2 fragment of 3.5-kb subsequent to gel purification (QIAquick gel extraction kit; QIAGEN, Hilden, Germany). Transformants were selected on YNB medium lacking leucine and subsequently verified by polymerase chain reaction (PCR), by using PCR primers upstream and downstream of the LEU2 insert. The yca1Δsro77Δ mutant was constructed as follows: PCR was used to amplify the sro77Δ::HIS3 fragment of strain WKL-3A (Larsson et al., 1998) by using SRO77 forward primer TTCCGCTTCATAGGAGGAGA (300 base pairs upstream of START codon) and SRO77 backward primer ACGGTTCATCATTCGGAAAA (350 base pairs downstream of STOP codon). PCR products were purified (QIAquick PCR purification kit; QIAGEN) and used to transform the yca1Δ mutant. Transformants were selected on YNB plates lacking histidine and verified by PCR by using primers that bind upstream and downstream of the primers used to amplify the sro77Δ::HIS3 fragment. The yca1Δsro7Δsro77Δ strain was constructed by transforming the yca1Δsro7Δstrain with the sro77::HIS3 fragment described previously. Transformants were selected and verified correspondingly. To construct the BY sro7Δ sro77Δ strain BY sro7Δ Mat a was crossed with BY sro77Δ Mat α. Tetrads were analyzed and potential double mutants checked by PCR. PCR was performed using SRO7 and SRO77 verification primers described above plus additional primers binding within the kanamycin gene.
Growth and Survival Tests
Growth was monitored by plate assays. Yeast strains were grown overnight in YPD medium, adjusted to identical OD610 and diluted 100, 10-1, 10-2, 10-3, and 10-4. Each dilution was spotted in aliquots of 5 μl onto YPD plates with or without NaCl supplementation. For cell survival experiments, yeast cells were grown until they reached exponential phase. NaCl was added to the desired concentration, and samples containing a defined number of cells were plated onto YPD plates after various periods of salt exposure. The number of cells was determined (∼20 cells/μl, 1:1000 dilution) for each culture by using a Helber counting chamber and a total of 600 cells, divided into three aliquots, were spread onto YPD plates. The number of surviving colonies was determined after 2 d of incubation at 30°C.
Cytological Analysis
For determination of reactive oxygen species (ROS) accumulation, cells were grown into exponential phase (OD610 ∼0.5) and exposed to NaCl for l h. Dihydrorhodamine 123 (Sigma-Aldrich, St. Louis, MO) was added (5 μg/ml cell culture) and cultivation continued for another 2 h. Stained cells were viewed using a Leica DMRXA fluorescence microscope equipped with a rhodamine optical filter. To determine the frequency of stained cells, at least 500 cells of three independent experiments were evaluated.
Analysis of nuclear fragmentation (4,6-diamidino-2-phenylindole [DAPI] staining) and DNA strand breaks (terminal deoxynucleotidyl transferase dUTP nick-end labeling [TUNEL] analysis) was performed on fixed cells. Exponentially growing cells were fixed by the addition of 1:10 volumes of formaldehyde (leading to a final concentration of 3.7%) followed by incubation for 1 h in 30°C. The cells were washed twice in apoptosis buffer 1 (35 mM potassium phosphate and 0.5 mM MgC12, pH 6.8), resuspended in apoptosis buffer 2 (apoptosis buffer 1 + 1.2 M sorbitol), and treated with lyticase (50 U/ml) for 30 min at 30°C. After careful washing in apoptosis buffer 2 sphaeroplasts were bound on poly-lysine-coated slides. For DAPI staining, cells were incubated with diaminophenylindole (1 μg/ml; Sigma-Aldrich) for 2 min, washed twice with apoptosis buffer 2, and examined under a Leica DMRXA fluorescence microscope. TUNEL analysis was performed as described previously (Madeo et al., 1997, 1999).
Caspase Activity Measurements
Caspase activity was measured by using fluorescein isothiocyanate (FITC)VAD-fmk (CaspACE; Promega, Madison, WI), which binds at the catalytic center of active caspases. NaCl was added to stationary overnight cultures of wild-type and yca1Δ cells at various concentration, and the cultures were incubated for 1 or 4 h at 28°C. After incubation, cells were harvested (5 × 106 cells, washed in 1 ml phosphate-buffered saline, and stained for 20 min in 200 μl FITC-VAD-fmk diluted 1:1000 in the same buffer. For flow cytometric analysis, stained cells were counted using FACSCalibur (BD Biosciences, San Jose, CA) and CellQuest analysis software with excitation and emission settings of 488 nm and 525-550 nm (filter FL1), respectively.
RESULTS
NaCl Stress Reduces Survival of sro7Δ and sro7Δsro77Δ Mutants and Induces Intracellular Accumulation of Reactive Oxygen Species
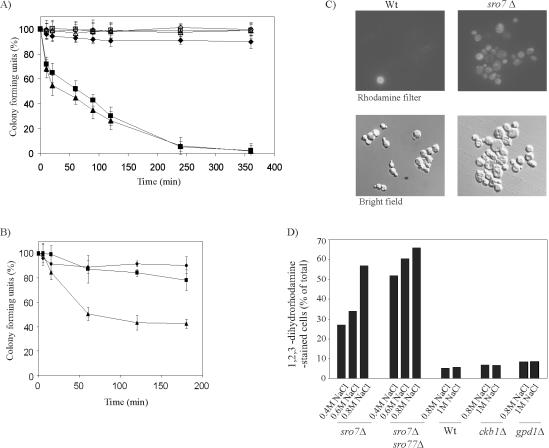
Survival tests demonstrated that sro7Δ and sro7Δsro77Δ mutants are highly sensitive to NaCl stress. Already, a 10-min exposure to 0.7 M NaCl killed ∼30% of either population (Figure 1A), and <5% remained viable after 6 h. Equivalent treatment of wild-type cells resulted in <5% loss in viability. These results demonstrate that sro7Δ and sro7Δsro77Δ mutants not only exhibit reduced ability to grow at moderate salt stress, as has been reported previously (Larsson et al., 1998) but also that the cells are actually killed by the stress. The observations prompted us to analyze the cells for the presence of ROS, which are considered a key stimulus for celldeathbyapoptosis(Madeoet al.,1999).Usingdihydrorhodamine 123 as ROS indicator, accumulation of ROS was observed in ∼60% of sro7Δ or sro7Δsro77Δ mutants exposed to 0.8 M NaCl (Figure 1, C and D). Control wild-type cells showed little dihydrorhodamine staining, even when exposed to 1 M NaCl (Figure 1D). At lower salinities (0.4 and 0.6 M NaCl), the proportion of stained sro7Δ cells decreased to become about one-half of that of the sro7Δsro77Δ mutants. In agreement with this pattern the double mutant is more efficiently killed by moderate salt stress than the sro7Δ single mutant (Figure 1B). We also examined ROS mediated staining of two other yeast mutants, gpd1Δ and ckb1Δ, that exhibit a salt-sensitive growth phenotype similar to that of sro7Δ. Interestingly, the accumulation of ROS in the gpd1Δ and the ckb1Δ mutants remained similar to that of wild-type cells, even at 1 M NaCl (Figure 1D).
Figure 1.
Cells lacking SRO7 accumulate ROS and exhibit reduced survival in saline environments. (A) Survival of wild-type, sro7Δ, and sro7Δsro77Δ mutants on exposure to 0.7 M NaCl for various periods. Data represent the mean of four different experiments. Wild-type (♦), sro7Δ (▪), and sro7Δsro77Δ (▴). Open symbols represent 0 M NaCl. (B) Survival of wild-type, sro7Δ, and sro7Δsro77Δ mutants on exposure to 0.4 M NaCl for various periods. Data represent the mean of four different experiments. Wild-type (♦), sro7Δ (▪), and sro7Δsro77Δ (▴). (C) Bright field (bottom row) and fluorescence (top row) photomicrographs of dihydrorhodamine 123 stained wild-type (Wt) and sro7Δ cells exposed to 0.8 M NaCl. (D) Diagram showing the percentage of dihydrorhodamine-stained cells of sro7Δ, sro7Δsro77Δ, Wt, ckb1Δ, and gpd1Δ strains after exposure to various concentrations of NaCl. Each data bar is based on the evaluation of 500 cells in several independent experiments. For further details, see MATERIAL AND METHODS.
sro7Δ and sro7Δsro77Δ Mutants Display Chromatin Condensation and DNA Fragmentation
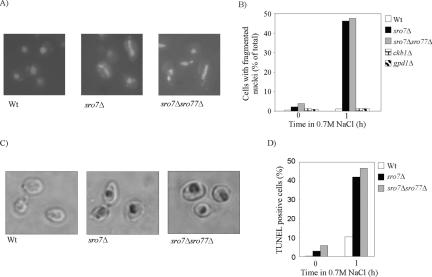
Having established increased ROS occurrence in salt-stressed sro mutants, we wanted to examine the cells for nuclear fragmentation, a well-established cytological hallmark of apoptosis. For the sro7Δ mutant, cells with irregularly shaped and fragmented nuclei were evident 30 min after exposure to 0.7 M NaCl (Figure 2A). In contrast, virtually no cells of the similarly treated wild-type strain showed abnormal nuclei. After 1 h at 0.7 M NaCl, about one-half of the sro7Δ and sro7Δsro77Δ mutants displayed fragmented nuclei (Figure 2B). At lower salinities, the proportion of aberrant nuclei decreased, and this tendency was, as expected, more pronounced for sro7Δ mutants than for sro7Δsro77Δ mutants (our unpublished data). The salt-sensitive gpd1Δ and ckb1Δ, mutants showed little chromatin condensation and fragmentation on exposure to NaCl stress similar to wild-type cells (Figure 2B). These mutants show that sensitivity to NaCl is not generally associated with apoptotic features such as ROS accumulation (Figure 1D) and nuclear fragmentation.
Figure 2.
sro7Δ and sro7Δsro77Δ mutants display fragmented nuclei and DNA strand breaks after NaCl treatment. (A) Photomicrograph of DAPI-stained wild-type (Wt), sro7Δ, and sro7Δsro77Δ cells exposed to YPD containing 0.7 M NaCl for 1 h. (B) Percentage of wild-type (Wt), sro7Δ, sro7Δsro77Δ, ckb1Δ, and gpd1Δ cells containing fragmented nuclei after 0 and 1 h in YPD containing 0.7 M NaCl. (C) TUNEL staining of wild-type, sro7Δ, and sro7Δsro77Δ cells after 30 min of incubation in YPD containing 0.7 M NaCl. (D) Quantification of TUNEL-positive staining of wild-type (Wt), sro7Δ, and sro7Δsro77Δ after 1-h exposure to 0.7 M NaCl.
Another familiar apoptotic feature is fragmentation of DNA in the nucleus. DNA strand breaks can be visualized by TUNEL tests, in which fluorescent nucleotides are added to the 3′-OH end of the DNA fragment, making the phenomenon visible by fluorescent microscopy. Both sro7Δ and sro7Δsro77Δ mutants display TUNEL-positive nuclei after 30 min of exposure to 0.7 M NaCl (Figure 2C), whereas the nuclei of control wild-type cells remained unstained or only slightly stained. After 1 h at 0.7 M NaCl, about one-half of the sro7Δ and sro7Δsro77Δ population showed a clear TUNEL-positive staining, whereas ∼10% of the wild-type cells exhibited this characteristic (Figure 2D). We conclude that NaCl stress that does not markedly affect viability of wild-type cells, induces cell death in sro7Δ and sro7Δ sro77Δ mutants that bears the structural attributes of apoptosis with respect to ROS accumulation, nuclear fragmentation, and DNA strand breakage.
Yca1p Is Involved in the NaCl-induced Lethality of sro7Δ Mutants
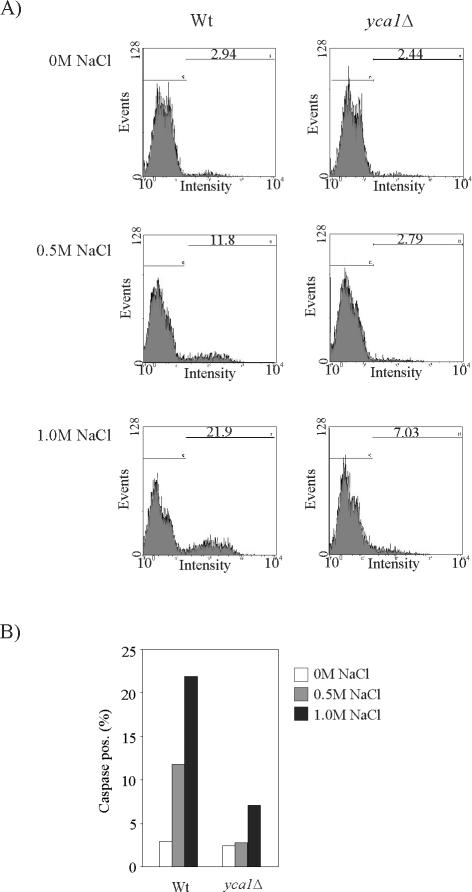
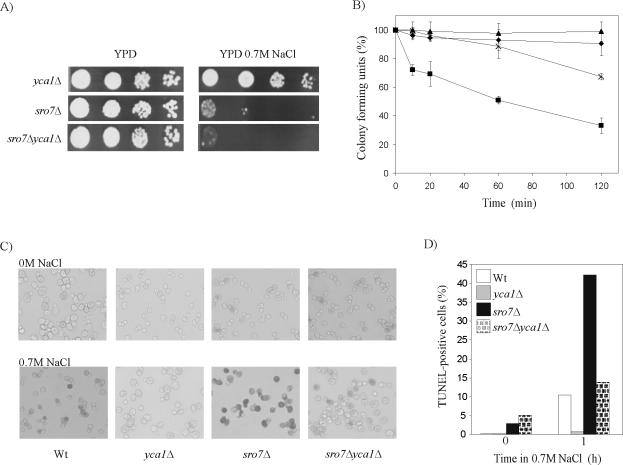
To investigate whether the yeast metacaspase Yca1p has a role in NaCl-mediated cell death, we first examined whether salt stressed cells show increased caspase activity. To this end, cells were incubated with the FITC-labeled pan-caspase inhibitor VAD-fmk that binds to the active site of the caspase, allowing for flow cytometric determination of cells with active enzyme (Madeo et al., 2002). This assay revealed a significant increase in the proportion of cells with active caspase after exposure to NaCl stress for 4 h (Figure 3). Compared with nonstressed conditions, wild-type cells exposed to 1.0 M NaCl increase their caspase activity more than sevenfold. Cells lacking YCA1 exhibit basal level caspase activity both at 0 and 0.5 M NaCl, whereas a modest increase was apparent at 1 M NaCl. We next examined the involvement of Yca1p in the NaCl induced lethality by deleting the YCA1 gene in the sro7Δ background. If the salt sensitivity of the sro7Δ strain is caused by Yca1p-mediated cell death, the sro7Δyca1Δ double mutant would display higher salt tolerance than the sro7Δ single mutant. This simple prediction was, however, not supported by testing serial dilutions for growth on YPD plates (Figure 4A), which showed that the sensitivity of sro7Δ mutants to 0.7 M NaCl is very similar to that of sro7Δyca1Δ mutants. In fact, the two strains grew equally well at all NaCl concentrations tested, showing inhibited growth by salt concentrations of ≥0.5 M NaCl (our unpublished data). Similar result was obtained with the sro7Δsro77Δ double mutant. This mutant experience growth inhibition at ≥0.2 M NaCl, and additional deletion of YCA1 did not improve NaCl tolerance as revealed by growth tests on YPD plates containing various salt concentrations (our unpublished data). However, similar growth inhibition in the presence of NaCl does not necessarily mean that the strains survive equally well on exposure to salt stress. Actually, there was a significant difference between the survival traits of sro7Δ and sro7Δyca1Δ mutants (Figure 4B). The sro7Δ mutants exhibit an immediate decrease in viability after exposure to 0.7 M NaCl, leading to a rapid elimination of ∼30% of the population. Similar stress exposure of the sro7Δyca1Δ mutants resulted only in a slight decrease in cell viability (∼5%). However, after the rapid phase of cell demise, the death rate for the sro7Δ population changed to a pace that was similar to that shown by the sro7Δ yca1Δ mutants (Figure 4B). These results suggest that Yca1p is required for the primary, fast drop in viability of the sro7Δ mutants. The better survival of the sro7Δ yca1Δ strain is consistent with markedly decreased DNA breakage (Figure 4, C and D). A similar tendency was apparent for the control yca1Δ and wild-type strains, although the salt concentration is too low to produce strong effects in these strains.
Figure 3.
Salt-stressed cells exhibit increased caspase activity. (A) Caspase activity measurement by flow cytometric analysis of wild-type and yca1Δ mutant cells exposed to NaCl (0, 0.5, or 1.0 M) for 4 h. (B) Percentage of FITC-VAD-fmk-positive cells in fluorescence-activated cell sorting measurements of wild-type (Wt) and yca1Δ mutant strains exposed to different NaCl concentrations.
Figure 4.
Rate of killing of sro7Δ cells after exposure to salt stress is YCA1 dependent, and the level of salt induced double stranded DNA breaks is reduced in the sro7Δyca1Δ mutant. (A) Growth of sro7Δ, yca1Δ, and sro7Δyca1Δ mutants at high salinity monitored by drop assays. Serial 10-fold dilutions of cell suspensions were spotted onto YPD medium with and without 0.7 M NaCl. (B) Survival curves for wild-type (♦), sro7Δ (▪), yca1Δ (▴), and sro7Δyca1Δ (X) mutants after incubation with 0.7 M NaCl. Samples were taken after incubation with 0.7 M NaCl for various periods of time and plated onto YPD medium. Each data point is based the plating of 200 cells in three independent experiments. (C) Photographs of TUNEL staining showing DNA breakage in the sro7Δ mutant after 1 h of incubation in YPD medium containing 0.7 M NaCl. (D) Quantification of TUNEL-positive cells subsequent to 1 h 0.7 M NaCl exposure. To determine the frequency of TUNEL-positive cells, at least 1000 cells were evaluated for each strain and each condition.
Deletion of YCA1 Improves Survival of Wild-Type Cells but Has Little Effect on Survival of sro77Δ Mutants
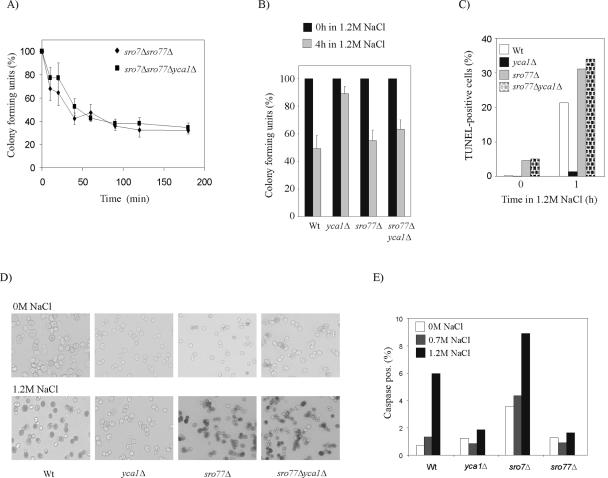
Yca1p has previously been shown to mediate apoptosis-like cell death in aged yeast cells and in yeasts treated with H2O2 or acetic acid (Madeo et al., 2002). For example, cells lacking YCA1 survive high levels of H2O2 much longer than wild-type cells. Yca1p obviously also has a role in mediating demise of wild-type cells exposed to high salinity. About 50% of wild-type yeast survives exposure to 1.2 M NaCl for 4 h, whereas ∼85% of the cells survives the same treatment if deleted for the YCA1 gene (Figure 5B). This observation agrees with the decreased DNA cleavage seen for the yca1Δ mutant (Figure 5, C and D). Hence, survival assays and caspase activity measurements demonstrate that the apoptotic-like response seen for wild-type cells during salt stress is strongly dependent on Yca1p activity.
Figure 5.
Survival at high salinity is YCA1dependent for wild-type cells but not for sro77Δ mutants. (A) Survival kinetics of sro7Δsro77Δ and sro7Δsro77Δyca1Δ cells in YPD containing 0.4 M NaCl. (B) Survival of wild-type, yca1Δ, sro77Δ, and sro77Δyca1Δ after incubation for 4 h in YPD containing 1.2 M NaCl. (C) Percentage of TUNEL-positive cells after 1-h exposure to 1.2 M NaCl. For quantification, at least 1000 cells were evaluated for each strain and each condition. (D) Photographs showing examples of TUNEL staining of wild-type (Wt), yca1Δ, sro77Δ, and sro77Δyca1Δ cells before and after 1-h exposure to1.2 M NaCl. (E) Caspase activity, shown as percentage of FITC-VAD-positive cells by fluorescence-activated cell sorting analysis of wild-type, yca1Δ, sro7Δ, and sro77Δ strains, grown overnight, and exposed to NaCl stress for 1 h.
The sro77Δ mutants show a similar tolerance to 1.2 M NaCl as wild-type cells. Interestingly, however, deletion of YCA1 in sro77Δ background does not significantly improve the viability at high salinity, as it does for wild-type cells (Figure 5B). In accordance with these results, the sro7Δsro77Δ and the sro7Δsro77Δyca1Δ mutants also display similar survival curves after exposure to 0.4 M NaCl (Figure 5A). The observation that NaCl induced lethality is independent of Yca1p in strains lacking SRO77, is in agreement with the finding that DNA breakage (Figure 5, C and D) and nuclear fragmentation (our unpublished data) is very similar for sro77Δ and sro77Δyca1Δ cells. Similarly, nuclear fragmentation and the proportion of TUNEL-positive cells are very similar for NaCl exposed sro7Δsro77Δ and the sro7Δsro77Δyca1Δ mutants (our unpublished data). These results imply a genetic interaction between SRO77 and YCA1, which encode proteins that have been previously shown to interact in two-hybrid screens (Uetz et al., 2000; Drees et al., 2001). To further study the effect of Sro7p and Sro77p on Yca1p, we examined the proportion of cells with active caspase in wild-type, yca1Δ, sro7Δ, and sro77Δ cultures exposed to NaCl stress for 1 h (Figure 5E). The observed caspase activity remained unaffected by salt stress in sro77Δ mutants, similar to what was noted for yca1Δ cells, whereas mutants lacking SRO7 exhibited a generally increased casapase activity, which consistently was kept higher than that of wild-type cells. These results indicate that Sro7p and Sro77p have opposing effects on Yca1p; Sro77p being required for the salt induced activation of Yca1p, whereas Sro7p seems to moderate the activity of the caspase. Furthermore, the observation that sro77Δyca1Δ mutants show NaCl induced nuclear fragmentation and strong DNA strand breakage (Figure 5, C and D) suggests the existence of an Yca1p independent apoptotic pathway in yeast.
DISCUSSION
Exposed to Salt Stress, the sro7Δ Mutant Exhibits Hallmarks of Apoptosis
Cells committing active suicide by apoptosis are subject to a series of stereotyped morphological changes that differentiate apoptotic cells from healthy or necrotic cells (Webb et al., 1997; Kerr, 2002). The most typical structural changes occur within the nucleus, where chromatin condenses and aggregates along the margins of the nuclear membrane and DNA is degraded, first into large fragments and later into short oligomers of ∼180-200 base pairs. Coincident with these changes are alterations of the nuclear structural framework leading to fragmentation of the nucleus. Here, we demonstrate NaCl-induced cell death associated with the typical nuclear features of apoptosis for yeast cells deleted for the tumor suppressor homologue SRO7. NaCl stress is one of the environmental factors that has been demonstrated previously to induce apoptosis-like cell death in yeast and plants (Katsuhara, 1997; Huh et al., 2002). Wild-type cells of S. cerevisiae were shown to exhibit apoptotic features after treatment with 1.2 M NaCl (Katsuhara, 1997; Huh et al., 2002). The sro7Δ mutant displays a similar response at NaCl concentrations beginning at 0.5 M NaCl. Because the inducing salinity correlates with the NaCl concentration for strong growth inhibition of each strain, a simple interpretation would be that severely growth-inhibiting concentrations suffice to initiate apoptosis-like cell death. In other words, the lack of SRO7 may simply lower the salt concentration needed for apoptotic induction. However, two other yeast mutants that display NaCl sensitivity similar to that of sro7Δ, namely, ckb1Δ (de Nadal et al., 1999) and gpd1Δ (Larsson et al., 1993; Albertyn et al., 1994), do not show nuclear fragmentation (Figure 2B) and do not seem to accumulate reactive oxygen species (Figure 1D) on exposure to growth preventing NaCl concentrations. The salt sensitivity of the gpd1Δ mutant is due to a defective production and intracellular accumulation of glycerol, the primary yeast osmoregulator (Larsson et al., 1993). The reason for the salt-sensitive phenotype of the ckb1Δ mutant, which lacks a functional casein kinase II, is less clear, although the mutant seems to maintain intracellular ion homeostasis under salt stress (de Nadal et al., 1999). Hence, the different display of apoptotic features in response to growth inhibiting NaCl concentrations by sro7Δ mutants, on the one hand, and gpd1Δ and ckb1Δ mutants on the other, reinforces the observations by Huh et al. (2002) that disruption of inorganic ion homeostasis seems to be the primary reason for salt-induced cell death of yeast cells. Due to the defective targeting and subsequent degradation of the Ena1p sodium transporter in sro7Δ mutants, these cells experience increased intracellular Na+/K+ ratios when exposed to NaCl stress (Larsson et al., 1998; Wadskog, 2003). The ENA1 gene encodes a P-type ATPase that uses ATP to drive sodium ions out of the cell, and this pump has a primary role in maintaining cation homeostasis in NaCl-stressed yeast (Haro et al., 1991; Wieland et al., 1995). Interestingly, expression of the ENA1 gene is subject to a highly complex regulation, being controlled by a number of distinct regulatory pathways (Serrano et al., 1999; Wadskog and Adler, 2003). It is tempting to speculate that this elaborate control of ENA1 expression reflects a strict need to fine-tune ion homeostasis to prevent creating an intracellular environment that favors the activation of a cell death program. An additional indication for a role of ENA1 in this context stems from the observation that the human antiapoptotic protein Bcl-2 promoted transcriptional activation of ENA1 when expressed in a salt-sensitive cnb1Δ mutant (Huh et al., 2002).
The Yca1p Metacaspase Promotes Salt-induced Cell Death
The YCA1 gene in S. cerevisiae encodes a metacaspase that is activated by H2O2 and aging and is required for apoptotic demise of yeast (Madeo et al., 2002). Yca1p undergoes proteolytic processing similar to mammalian caspases and cleaves petidyl-caspase substrates, giving direct support for the existence of a specific effector of apoptosis in yeast. To investigate whether the apoptotic traits of NaCl-exposed cells were dependent on Yca1p, we compared wild-type and sro7Δ mutants with the corresponding strains lacking YCA1. For wild-type cells, deletion of YCA1 significantly improved survival rate at high salinity (Figure 4B), which suggests a general role for Yca1p in salt-induced apoptosis. The marked increase in caspase activity of cells exposed to harsh NaCl stress (Figure 3) gives further support to this view. Two additional observations provide evidence that Yca1p serves as an effector of the salt-induced cell death in yeast. Yca1p was more strongly activated in the salt sensitive sro7Δ mutants than in the more tolerant wild-type cells (Figure 5E), and deletion of YCA1 markedly affected both survival rate and DNA cleavage of the NaCl stressed strains (Figures 4, B-D, and 5, B-D). These results suggest that Yca1p mediates an apoptosis-like cell death, which is rapid and efficient. However, only a fraction of the cells seems to be susceptible to the Yca1p-mediated cell death. The first dramatic decrease of cell viability of sro7Δ cells was followed by a more moderate rate of killing that seemed independent of Yca1p. This decrease in viability may result from a more general effect of Na+ toxicity on the sro7Δ mutant and explain why the sro7Δyca1Δ mutant displays a salt-sensitive phenotype on plate assays.
Role of SRO7 and SRO77 in Apoptosis-like Cell Death
Because interactions between Sro77p and Yca1p are suggested by two-hybrid screens (Uetz et al., 2000; Drees et al., 2001), we wanted to investigate the role of SRO77 in salt-induced apoptosis-like cell death. Interestingly, deletion of YCA1 in sro7Δsro77Δ or sro77Δ backgrounds did not improve viability on NaCl treatment (Figure 5, A and B), indicating that the function of Yca1p is dependent on SRO77. Indeed, our measurement of salt-induced Yca1p activation shows that the caspase activity does not increase in mutants lacking SRO77, but remains similar to that of cells having a deleted YCA1 gene. The SRO-encoded proteins have been implicated in a late step in exocytosis by their ability to physically interact with Sec9p, a t-soluble N-ethylmaleimide-sensitive factor attachment (t-SNARE) protein receptor involved in the fusion of post-Golgi vesicles with the plasma membrane (Lehman et al., 1999). Given the proposed role in vesicle targeting, the opposing consequences of loss of function of SRO7 and SRO77 on the susceptibility of yeast to NaCl-induced cell death, suggest that the exocytosis machinery might sensitively attune the responsiveness of the cells to the exogenous apoptotic stimuli. The observed hyperactivation of the caspase in sro7Δ strain (Figure 5E) and the fact that these mutants show signs of DNA strand breakage (Figure 2C), even before exposure to NaCl stress, indicates that loss of SRO7 promotes a generally increased disposition for apoptosis-like cell death. The precise mechanistic relationship between Yca1p and the Sro proteins remains, however, to be determined. Because both sro77Δyca1Δ (Figure 5, C and D) and sro7Δsro77Δyca1Δ (our unpublished data) cells exhibit diagnostic markers of apoptosis when exposed to sufficiently high NaCl concentration, our findings also indicate the existence of a Yca1p-independent pathway(s) for cellular suicide in S. cerevisiae. This pathway(s) may be under negative control that is dependent on Sro77p.
Are There Links to the Function of the Authentic Tumor Suppressor?
Whether the observed sensitivity of sro7Δ and sro7Δsro77Δ mutants to NaCl-induced cell death bears any relevance to tumor development in Drosophila larvae lacking the authentic tumor suppressor l(2)gl is as yet speculative. Homozygous l(2)gl mutants show phenotypes involving loss of epithelial cell polarity and neoplastic overgrowth of tissues (Wodarz, 2000). Recent studies have indicated that loss of epithelial cell polarity may confer increased susceptibility to apoptosis (Weaver et al., 2002). This sensitivity has been suggested to counterbalance the proliferative advantage that is associated with nonpolarized cells, thereby reducing the possibility for malignant transformation (Humbert et al., 2003). Interestingly, De Lorenzo et al. (1999) have reported that a mutant form of l(2)gl induces apoptosis of the germ line during oogenesis, indicating that a defective tumor suppressor may promote apoptosis. In most epithelial cells the Na+, K+ ATPase is selectively sorted to the basolateral membrane (Dunbar and Caplan, 2001). Whether l(2)gl mutants exhibit defective epithelial distribution of this sodium transporter and suffer from a consequent disruption of inorganic ion homeostasis remain to be determined. Additional studies of the relationship between ion homeostasis and epithelial tumor development/apoptosis may further our understanding of the signals controlling growth, survival, and death of cells.
Acknowledgments
We thank Thomas Nyström for helpful discussions. This work was supported by grants from the Swedish Research Council and Magn. Bergvalls Stiftelse.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03-02-0114. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-02-0114.
References
- Albertyn, J., Hohmann, S., Thevelein, J.M., and Prior, B.A. (1994). GPD1, which encodes glycerol-3-phosphate dehydrogenase is essential for growth under osmotic stress in Saccharomyces cerevisiae and its expression is regulated by the high-osmolarity glycerol response pathway. Mol. Cell. Biol. 14, 4135-4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansell, R., Granath, K., Hohmann, S., Thevelein, J., and Adler, L. (1997). The two isoenzymes for yeast NAD-dependent glycerol 3-phosphate dehydrogenase encoded by GPD1 and GPD2, have distinct roles in osmoadaption and redox regulation. EMBO J. 16, 2179-2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beers, E.P., and McDowell, J.M. (2001). Regulation and execution of programmed cell death in response to pathogens, stress and developmental cues. Curr. Opin. Plant Biol. 4, 561-567. [DOI] [PubMed] [Google Scholar]
- Bilder, D., Li, M., and Perrimon, N. (2000). Cooperative regulation of cell polarity and growth by Drosophila tumor suppressors. Science 289, 113-116. [DOI] [PubMed] [Google Scholar]
- De Lorenzo, C., Strand, D., and Mechler, B.M. (1999). Requirement of Drosophila I(2)gl function for survival of the germline cells and organization of the follicle cells in a columnar epithelium during oogenesis. Int. J. Dev. Biol. 43, 207-217. [PubMed] [Google Scholar]
- de Nadal, E., Calero, F., Ramos, J., and Arino, J. (1999). Biochemical and genetic analyses of the role of yeast casein kinase 2 in salt tolerance. J. Bacteriol. 181, 6456-6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drees, B.L., et al. (2001). A. protein interaction map for cell polarity development. J. Cell Biol. 154, 549-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunbar, L.A., and Caplan, M.J. (2001). Ion pumps in polarized cells: sorting and regulation of the Na+, K+- and H+, K+-ATPases. J. Biol. Chem. 276, 29617-19620. [DOI] [PubMed] [Google Scholar]
- Engelberg-Kulka, H., and Glaser, G. (1999). Addiction modules and programmed cell death and antideath in bacterial cultures. Annu. Rev. Microbiol. 53, 43-70. [DOI] [PubMed] [Google Scholar]
- Fröhlich, K.U., and Madeo, F. (2000). Apoptosis in yeast - a monocellular organism exhibits altruistic behaviour. FEBS Lett. 473, 6-9. [DOI] [PubMed] [Google Scholar]
- Fröhlich, K.U., and Madeo, F. (2001). Apoptosis in yeast: a new model for aging research. Exp. Gerontol. 37, 27-31. [DOI] [PubMed] [Google Scholar]
- Gateff, E. (1978). Malignant neoplasms of genetic origin in Drosophila melanogaster. Science 200, 1448-1459. [DOI] [PubMed] [Google Scholar]
- Haro, R., Garciadeblas, B., and Rodriguéz-Navarro, A. (1991). A novel P-type ATPase from yeast involved in sodium transport. FEBS Lett. 291, 189-191. [DOI] [PubMed] [Google Scholar]
- Huh, G.H., Damsz, B., Matsumoto, T.K., Reddy, M.P., Rus, A.M., Ibeas, J.I., Narasimhan, M.L., Bressan, R.A., and Hasegawa, P.M. (2002). Salt causes ion disequilibrium-induced programmed cell death in yeast and plants. Plant J. 29, 649-659. [DOI] [PubMed] [Google Scholar]
- Humbert, P., Russell, S., and Richardson, H. (2003). Dlg, Scribble and Lgl in cell polarity, cell proliferation and cancer. Bioessays 25, 542-553. [DOI] [PubMed] [Google Scholar]
- Joza, N., Kroemer, G., and Penninger, J.M. (2002). Genetic analysis of the mammalian cell death machinery. Trends Genet. 18, 142-149. [DOI] [PubMed] [Google Scholar]
- Katsuhara, M. (1997). Apoptosis like cell death in barley roots under salt stress. Plant Cell Physiol. 38, 1091-1093. [Google Scholar]
- Kerr, J.F. (2002). History of the events leading to the formulation of the apoptosis concept. Toxicology 181-182, 471-474. [DOI] [PubMed] [Google Scholar]
- Larsson, K., Böhl, F., Sjöström, I., Akhtar, N., Strand, D., Mechler, B., Grabowski, R., and Adler, L. (1998). The Saccharomyces cerevisiae SOP1 and SOP2 genes, which act in cation homeostasis, can be functionally substituted by the Drosophila lethal(2)giant larvae tumor suppressor gene. J. Biol. Chem. 273, 33610-33618. [DOI] [PubMed] [Google Scholar]
- Larsson, K., Eriksson, P., Ansell, R., and Adler, L. (1993). A gene encoding sn-glycerol 3-phosphate dehydrogenase (NAD+) complements an osmosensitive mutant of Saccharomyces cerevisiae. Mol. Microbiol. 10, 1101-1111. [DOI] [PubMed] [Google Scholar]
- Latterich, M., Fröhlich, K.U., and Schekman, R. (1995). Membrane fusion and the cell cycle: Cdc48p participates in the fusion of ER membranes. Cell 82, 885-893. [DOI] [PubMed] [Google Scholar]
- Laun, P., Pichova, A., Madeo, F., Fuchs, J., Ellinger, A., Kohlwein, S., Dawes, I., Fröhlich, K.U., and Breitenbach, M. (2001). Aged mother cells of Saccharomyces cerevisiae show markers of oxidative stress and apoptosis. Mol. Microbiol. 39, 1166-1173. [PubMed] [Google Scholar]
- Lehman, K., Rossi, G., Adamo, J.E., and Brennwald, P. (1999). Yeast homologues of tomosyn and lethal giant larvae function in exocytosis and are associated with the plasma membrane SNARE, Sec9. J. Cell Biol. 146, 125-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeo, F., Fröhlich, E., and Fröhlich, K.U. (1997). A yeast mutant showing diagnostic markers of early and late apoptosis. J. Cell Biol. 139, 729-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeo, F., Fröhlich, E., Ligr, M., Grey, M., Sigrist, S.J., Wolf, D.H., and Fröhlich, K.U. (1999). Oxygen stress: a regulator of apoptosis in yeast. J. Cell Biol. 145, 757-767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeo, F., et al. (2002). A caspase-related protease regulates apoptosis in yeast. Mol. Cell 9, 911-917. [DOI] [PubMed] [Google Scholar]
- Matsuyama, S., Nouraini, S., and Reed, J.C. (1999). Yeast as a tool for apoptosis research. Curr. Opin. Microbiol. 2, 618-623. [DOI] [PubMed] [Google Scholar]
- Ohshiro, T., Yagami, T., Zhang, C., and Matsuzaki, F. (2000). Role of cortical tumour-suppressor proteins in asymmetric division of Drosophila neuroblast. Nature 408, 593-596. [DOI] [PubMed] [Google Scholar]
- Peng, C.Y., Manning, L., Albertson, R., and Doe, C.Q. (2000). The tumour-suppressor genes lgl and dlg regulate basal protein targeting in Drosophila neuroblasts. Nature 408, 596-600. [DOI] [PubMed] [Google Scholar]
- Qi, H., Li, T.-K., Kuo, D., Nur-El-Kamal, A., and Li, L.F. (2003). Inactivation of Cdc13p triggers MEC1-dependent apoptotic signals in yeast. J. Biol. Chem. 278, 15136-15141. [DOI] [PubMed] [Google Scholar]
- Serrano, R., et al. (1999). A glimpse of the mechanisms of ion homeostasis during salt stress. J. Exp. Bot. 50, 1023-1036. [Google Scholar]
- Severin, F.F., and Hyman, A.A. (2002). Pheromone induces programmed cell death in S. cerevisiae. Curr. Biol. 12, R233-R235. [DOI] [PubMed] [Google Scholar]
- Uetz, P., et al. (2000). A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403, 623-627. [DOI] [PubMed] [Google Scholar]
- Uren, A.G., and Vaux, D.L. (1996). Molecular and clinical aspects of apoptosis. Pharmacol. Ther. 72, 37-50. [DOI] [PubMed] [Google Scholar]
- Wadskog, I., and Adler, L. (2003). Ion homeostasis in Saccharomyces cerevisiae under NaCl stress. In: S. Hohmann and P. Mager, eds. Topics in current genetics: yeast stress responses (vol. 1). Berlin, Springer-Verlag, 201-239. [Google Scholar]
- Wadskog, I. (2003). Functional studies of the Sro yeast homologues of the Drosophila lethal(2) giant larvae tumor suppressor. [Ph.D. thesis]. Göteborg, Sweden: Götenborg University.
- Vaux, D.L., and Korsmeyer, S.J. (1999). Cell death in development. Cell 96, 245-254. [DOI] [PubMed] [Google Scholar]
- Weaver, V.M., Lelievre, S., Lakins, J.N., Chrenek, M.A., Jones, J.C., Giancotti, F., Werb, Z., and Bissell, M.J. (2002). beta4 Integrin-dependent formation of polarized three-dimensional architecture confers resistance to apoptosis in normal and malignant mammary epithelium. Cancer Cell 2, 205-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb, S., Harrison, D.J., and Wyllie, A.H. (1997). Apoptosis: an overview of the process and its relevance in disease. Adv. Pharmacol. 41, 1-34. [DOI] [PubMed] [Google Scholar]
- Wieland, J., Nietsche, A.M., Strayle, J., Steiner, H., and Rudolph, H.K. (1995). The PMR2 gene cluster encodes functionally distinct isoforms of a putative Na+ pump in the yeast plasma membrane. EMBO J. 14, 3870-3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wodarz, A. (2000). Tumor suppressors: linking cell polarity and growth control. Curr. Biol. 10, R624-R626. [DOI] [PubMed] [Google Scholar]
- Wyllie, A.H., et al. (1999). Apoptosis and carcinogenesis. Br. J. Cancer 80, 34-37. [PubMed] [Google Scholar]
- Wyllie, A.H., Kerr, J.F., and Currie, A.R. (1980). Cell death: the significance of apoptosis. Int. Rev. Cytol. 68, 251-306. [DOI] [PubMed] [Google Scholar]