Abstract
Retinoic acid inducible gene I (RIG-I) senses viral RNAs and triggers innate antiviral responses through induction of type I IFNs and inflammatory cytokines. However, whether RIG-I interacts with host cellular RNA remains undetermined. Here we report that Rig-I interacts with multiple cellular mRNAs, especially Nf-κb1. Rig-I is required for NF-κB activity via regulating Nf-κb1 expression at posttranscriptional levels. It interacts with the multiple binding sites within 3′-UTR of Nf-κb1 mRNA. Further analyses reveal that three distinct tandem motifs enriched in the 3′-UTR fragments can be recognized by Rig-I. The 3′-UTR binding with Rig-I plays a critical role in normal translation of Nf-κb1 by recruiting the ribosomal proteins [ribosomal protein L13 (Rpl13) and Rpl8] and rRNAs (18S and 28S). Down-regulation of Rig-I or Rpl13 significantly reduces Nf-κb1 and 3′-UTR–mediated luciferase expression levels. These findings indicate that Rig-I functions as a positive regulator for NF-κB signaling and is involved in multiple biological processes in addition to host antivirus immunity.
Retinoic acid inducible gene I (RIG-I) has been demonstrated to play a crucial role in initiating innate antiviral immune responses by sensing viral RNAs, recruiting a specific adaptor protein, IFN-β promoter stimulator 1 (IPS-1; also named MAVS, VISA, or Cardif), and activating downstream IFN regulatory factor 3 (IRF3) and NF-κB signaling (1–5). In recent years, much effort has been put into understanding how RIG-I discriminates viral RNAs from host cellular RNAs. A number of studies have identified the different nonself RNA ligands able to be recognized by RIG-I, including short polyinosinic polycytidylic acid (poly I:C), 5′-triphosphate ssRNA, blunt dsRNA and the panhandle structures formed by the 5′ and 3′ ends of negative-sense RNA genomes (1, 6–12). However, ssRNA aptamers have also been shown to activate RIG-I signaling in 5′-triphosphate-RNA-independent form (13). Most recently, the crystal structure analyses of RIG-I in either ligand-free or dsRNA-bound status elucidate the structural basis for the interaction of RIG-I with nonself RNA ligands, and the conformational changes potentially necessary for dsRNA binding and signal transduction (14–17). These studies have greatly expanded our understanding of the mechanism underlying the interaction between RIG-I and its RNA ligands. However, whether RIG-I binds to host cellular RNAs and what the biological function is of such binding remain undetermined.
In fact, RIG-I was originally identified as being induced in the acute promyelocytic leukemia cell line NB4 during all-trans retinoic acid (ATRA)-induced cell differentiation, implying that RIG-I may function as an important mediator in ATRA-induced cell differentiation (18). Structural analysis and in vitro studies demonstrate that RIG-I is a DExD/H-box ATPase/RNA helicase (19). It is well known that members of the DExD/H-box family have diverse functions in regulating gene expression and cellular processes (20, 21). Evolutionally, RIG-I shares high sequence similarities among species from Caenorhabditis elegans to mammals. It is ubiquitously expressed in human and mouse, and induced by ATRA, IFN, LPS, interleukin-1, and RNA viruses in various cell types. RIG-I is involved in the induction of inflammatory factors and chemokines upon inflammatory stimuli (22). We have previously reported that Rig-I–deficient mice developed a colitis-like phenotype due to impaired G protein subunit alpha-i2 (Gαi2) expression and T-cell activation (23), progressive granulocytosis with reduction of interferon consensus sequence binding protein (Icsbp) expression and STAT1 activation (24, 25), and increased susceptibility to infection with Escherichia coli due to impaired LPS-induced phagocytosis of bacteria (26). Therefore, RIG-I seems more likely to be involved in multiple biological processes including recognition of nonself viral RNAs.
Results
Rig-I Interacts with Multiple Cellular mRNAs.
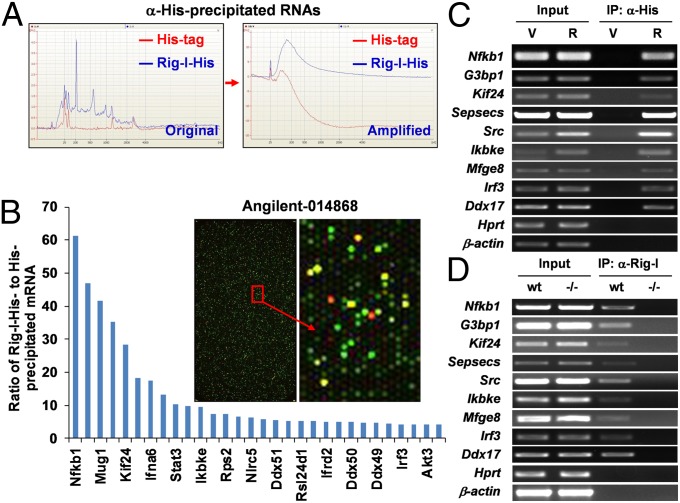
To identify the potential interaction of Rig-I with host cellular mRNA, we performed RNA immunoprecipitation (RIP) by using an anti-His antibody from the total cell lysates of the murine splenic B-cell line (1.B4.B6), which stably expresses His-tag or His-tagged Rig-I. Total RNA extracted from the pooled His-precipitate was amplified in parallel (Fig. 1A), and analyzed by using an Agilent-014868 Whole Mouse Genome Microarray. In comparing RNA abundance between His-tag and His-tagged Rig-I precipitates, we found that the signal intensities of 298 spots are more than quadruple that in control (log ratio >2). Most strikingly, the spot with highest log ratio (5.9) was identified as Nf-κb1/p105 encode large precursor protein p105, which upon proteasome-mediated processing generate the mature Nf-κb1 subunits p50 (Fig. 1B and Table S1). To be more stringent, part of the mRNAs with log ratio >2, including Nf-κb1/p105, Inhibitor of kappaB kinase epsilon (Ikbke), Irf3, Rous sarcoma oncogene (Src), kinesin family member 24 (Kif24), etc., were verified by RT-PCR in the same precipitate (Fig. 1C) and detected in anti–Rig-I precipitate from the cell lysate of WT but not from that of Rig-I−/− murine embryonic fibroblasts (MEFs) (Fig. 1D). Furthermore, the interaction of Rig-I with Nf-κb1 was confirmed in anti–Rig-I precipitate from the in vitro reaction of purified recombinant Rig-I protein with total RNA extracted from WT splenocytes (Fig. S1A). LPS is a potent inducer of the NF-κB–activating signaling cascade (27, 28). To exclude the possible effect of NF-κB activation on such interaction, the same cells as in Fig. 1C were treated with LPS, RIP, and subsequent RT-PCR showed that NF-κB activation had no effect on the interaction of Rig-I with Nf-κb1 mRNA (Fig. S1B). These data suggest that Rig-I interacts with multiple host cellular RNAs, especially Nf-κb1/p105. Other mRNA molecules potentially interacting with Rig-I include Interferon alpha-6 precursor (Ifna6), Interferon gamma receptor 2 (Ifngr2), Interferon alpha B (Ifnab), NLR family, CARD domain containing 5 (Nlrc5), Src, Stat3 and so on. It is known that RNA-binding proteins play an integral and vital role in gene expression via regulating the translation of RNA, and some posttranscriptional events such as RNA splicing and editing (29). Thus, these findings may imply that Rig-I performs its antivirus functions through multilevel mechanisms.
Fig. 1.
Multiple cellular RNAs are immunoprecipitated with Rig-I. (A) Quantitative or qualitative evaluation for the total RNA extracted from the pooled anti-His precipitates from total cell lysates of 1.B4.B6 cell line stably expressing His-tag or His-tagged Rig-I. The graphs shown are of RNA abundance against molecular size before or after two-round amplification. (B) Equal amounts of labeled cRNA samples were analyzed by using Agilent 44k Whole Mouse Genome Microarray (Agilent; 014868). Some of the positive mRNAs were shown in the histogram. The indicated mRNAs were further detected by RT-PCR in either anti-His precipitates from the cell lysates of the 1.B4.B6 cell line stably expressing His-tagged Rig-I (C) or anti–Rig-I precipitates from the cell lysates of WT but not from that of Rig-I−/− MEFs (D). X-box binding protein 1 (Xbp1), hypoxanthine-guanine phosphoribosyltransferase (Hprt), and β-actin are shown as negative controls.
Rig-I Interacts with Nf-κb1 3′-UTR mRNA and Positively Regulates Nf-κb1 Expression.
Further characterization of the interaction of Rig-I with Nf-κb1 mRNA revealed that Rig-I is able to bind to its 3′ untranslated region (3′-UTR), as demonstrated by the following three experiments. First, we showed that only 3′ but not 5′ fragments were precipitated with Rig-I antibody from the reaction of recombinant Rig-I with in vitro transcribed p105 mRNA fragments and detected by RT-PCR using p105-specific primers located within the overlapping region between two fragments (Fig. 2A). Second, GST pull-down assay showed that the same fragment but labeled with biotin-UTP with or without 5′ 7-methylguanosine cap (5′m7G cap) was further detected in the GST–Rig-I precipitates by using IRDye800CW-conjugated streptavidin (Fig. S2A). Third, cotransfection of the mouse embryo fibroblast cell line NIH 3T3 with His-tagged Rig-I expression vector and reporter construct with p105 3′-UTR inserted downstream of the luciferase coding region in the pGL3 luciferase reporter vector (Luc-p105 3′-UTR) showed that anti-His antibody was able to precipitate Luc-p105 3′-UTR and endogenous p105 (internal control) transcripts, as demonstrated by RT-PCR using luciferase and p105-specific primer pairs, respectively (Fig. 2B). These data suggest that Rig-I is able to interact with 3′-UTR of p105 mRNA. To illustrate the biological significance of such interaction, we transfected pGL3 [with Simian virus 40 (SV40) poly(A) signal] or Luc-p105 3′-UTR vector into MEFs, and found that pGL3-derived luciferase activity showed no significant difference between WT and Rig-I−/− MEFs. However, Luc-p105 3′-UTR–derived luciferase activity was significantly decreased in Rig-I−/− cells compared with that in WT cells (Fig. 2C). By cotransfection of Rig-I−/− MEFs with Rig-I–expressing vector and pGL3 or Luc-p105 3′-UTR vector, we found that Luc-p105 3′-UTR–derived luciferase activity was fully rescued in a dose-dependent manner compared with pGL3 control (Fig. 2D). A similar response of p105 3′-UTR to Rig-I in luciferase activity was also observed in NIH 3T3 cells (Fig. S2B). These data suggest that Rig-I is required for Nf-κb1 expression through binding to 3′-UTR.
Fig. 2.
Rig-I interacts with Nf-κb1 3′-UTR mRNA and positively regulates Nf-κb1 expression. (A) Nf-κb1/p105 mRNA fragments as indicated in IgG or anti–Rig-I precipitates from the reactions of Rig-I protein with 5′ or 3′ mRNA fragments are detected by RT-PCR using the primers located within overlapping regions between two fragments. (B) RT-PCR for p105 and Luc in anti-His precipitates from the lysates of NIH 3T3 cells cotransfected with His-tagged Rig-I expression vector and Luc-p105 3′-UTR construct in which p105 3′-UTR is inserted downstream of the luciferase coding region. (C) Luc activity in MEFs transfected with pGL3 control or Luc-p105 3′-UTR construct is shown as mean ± SD (n = 3). (D) Luc activity in Rig-I−/− MEFs cotransfected with Rig-I expression vector and either pGL3 control or Luc-p105 3′-UTR vector is expressed as a percentage relative to that with Rig-I empty vector (mean ± SE, n = 3). (E) RT-PCR for p105 and Luc in anti-His precipitates from the lysates of NIH 3T3 cells cotransfected with His-tagged Rig-I expression vector and Luc reporter constructs with insertion of p105 3′-UTR fragments as indicated. (F) MEFs were transfected with pGL3 control or Luc-p105 3′-UTR constructs harboring the indicated p105 3′-UTR fragments. The dependence of Luc activity expression on Rig-I is expressed as percentage of 1 minus the ratio of Luc activity in Rig-I−/− MEFs to that in WT cells (mean ± SE, n = 3). (G) Immunoprecipitation of Luc mRNA transcribed from Luc-p105 3′-UTR construct and p105 mRNA with Myc-tagged, intact Rig-I, or individual domains (CD, HD, and RD) from the lysates of cotransfected NIH 3T3 cells. (H) HD or RD expression vector and the same reporter constructs as in E were used in the same experiment as in G. *P < 0.05; **P < 0.01. One of three independent experiments is shown for C, D, and F.
To further define the binding region of Rig-I within p105 3′-UTR, different fragments derived from p105 3′-UTR as illustrated in Fig. S2C were inserted downstream of the luciferase coding region in pGL3, respectively. An in vitro binding assay by using recombinant Rig-I and in vitro transcribed mRNA fragments containing luciferase and p105 3′-UTR sequences as indicated in Fig. S2D revealed that all but fragment d were positive in the precipitates, suggesting that Rig-I interacts with at least fragments a and c. The presence of full-length p105 3′-UTR mRNA (a–d, positive control) and the individual fragments a, b, c but not d (negative control) in the precipitates with Rig-I was further confirmed by using biotin-UTP–labeled fragments (Fig. S2E). We further fused fragments a and b with fragment d [containing poly(A)] and inserted them into the same position in pGL3, respectively. Cotransfection and RIP as for Fig. 2B showed that all but fragment d of 3′-UTR, as well as endogenous p105 were precipitated with Rig-I (Fig. 2E). These findings indicate that there are at least three Rig-I binding sites in the region of p105 3′-UTR mRNA. The question is whether such bindings are functional in regulating luciferase activity. To address this question, we transfected WT or Rig-I−/− MEFs with pGL3 or Luc-p105 3′-UTR constructs harboring different 3′-UTR fragments as shown in Fig. 2F. The ratio of luciferase activity in Rig-I−/− over that in WT MEFs showed that Luc-p105 3′-UTR constructs with the insertion of an intact 3′-UTR or individual fragments (a, b, and c) produced substantially less luciferase activity in Rig-I−/− MEFs than in WT cells, meaning more dependence on Rig-I in regulating luciferase activity. However, pGL3 control vector or pGL3 with insertion of fragment d displayed no difference in luciferase activity between two genotypes (Fig. 2F). These data suggest that binding of Rig-I to the specific 3′-UTR sequences of p105 mRNA plays an indispensable role in protein translation regulation.
The existence of multiple binding sites for Rig-I in three sequential mRNA fragments within p105 3′-UTR suggests that different fragments may be recognized by different domains of Rig-I because the three fragments share no sequence similarities. With this in mind, NIH 3T3 cells were cotransfected with Luc-p105 3′-UTR and the vectors encoding Myc-tagged, intact, or truncated Rig-I, respectively. RIP revealed that luciferase and endogenous p105 were precipitated with the ATPase/helicase domain (HD) and the C-terminal regulatory domain (RD) but not the N-terminal caspase recruitment domain (CD) of Rig-I (Fig. 2G), indicating the interaction of Rig-I with p105 3′-UTR mRNA is mediated through both HD and RD domains. In this case, the same assay was performed by cotransfecting HD or RD domain-expressing vector with the reporter constructs harboring different fragments of p105 3′-UTR. Surprisingly, mRNA fragments a, b, and c of p105 3′-UTR were precipitated with both HD and RD domains albeit with no conserved motif between the two (Fig. 2H), suggesting that such interactions is more likely dependent on the primary sequence-based secondary structures.
Relevant to these findings, we asked whether p105 3′-UTR mRNA fragments that bind with Rig-I are capable of activating Rig-I signaling as viral dsRNA and RNAs with 5′-triphosphate ends are. To this end, WT or Rig-I−/− MEFs were transfected with pGL3 or Luc-p105 3′-UTR constructs harboring p105 3′-UTR fragments. It was found that IFN-β secretion in culture supernatants were unchanged in the presence of p105 3′-UTR fragments, whereas normal response to LPS was observed compared with basal or pGL3-transfected cells (Fig. S2F). Furthermore, neither WT nor Rig-I−/− MEFs, which were directly transfected with p105 3′-UTR mRNA fragments, showed IFN-β responses to p105 3′-UTR mRNA fragments, whereas vesicular stomatitis virus infection indeed induced IFN-β production in WT but not Rig-I−/− MEFs (Fig. S2G). These data suggest that Rig-I binding with cellular mRNA is functionally different from sensing nonself viral RNA. It is more likely that Rig-I plays a role in regulating protein translation via binding to cellular mRNA rather than inducing type I IFN production.
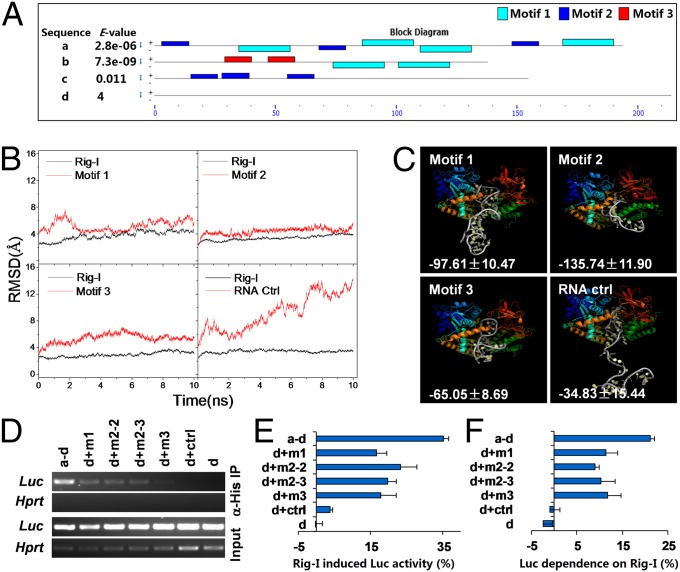
Rig-I Can Recognize Three Distinct Tandem Motifs Enriched in Nf-κb1 3′-UTR mRNA.
To identify the specific sequence and structural features in these fragments for Rig-I binding, the individual binding fragments of p105 3′-UTR mRNA were used as input material for Meta-MEME software to search the potential motifs enriched in these binding fragments. Such a motif finding process enabled us to identify three enriched distinct motifs among three binding fragments. We named them motif 1 ([AG]A[TG]G[AT][AG][AC]T[GC][GC]AGT[AG][AGT]C[AC][ACG]C[GA][CG]), motif 2 (G[CG]TGT[CG]C[CT]TTC), and motif 3 (GCTCAG[CG]TGCA), respectively. Interestingly, these motifs were found to be distributed in double or triple tandem, and none of them was identified in fragment d, which was unable to bind Rig-I through inspection by using MAST software with default parameters (Fig. 3A). This finding stimulated us to perform the extensive bioinformatics analyses on these potential motifs. First, 3D RNA structure prediction and folding analysis using iFoldRNA software (30) revealed that three dual-tandem motifs (27–45 bp) were able to efficiently fold and they formed more double helix structure than control RNA (45 bp, 70% vs. 50% of population) (Fig. S3A). Then, three tandem motifs and control RNA were docked with RIG-I using Discovery Studio 2.5, based on the structural information of RIG-I–RNA complex extracted from Brookhaven Protein DataBank (PDB code: 2YKG) (17). The constructed complexes were used for molecular dynamics simulation using the AMBER11 simulation package (31, 32). It was found that rmsds of RNA motifs were between 2 Å and 5 Å for the complexes of RIG-I and three tandem motifs; whereas control RNA in the complex, showed much higher rmsd value (∼13 Å) (Fig. 3B). Finally,the structural simulation displayed well-matched binding interfaces between RIG-I and tandem RNA motifs but not control RNA. The binding-free energy was calculated using the Molecular Mechanics/Poisson-Boltzmann Surface Area method. It was shown that much lower binding-free energy was needed for forming the complexes between RIG-I and the dual-tandem motifs in comparison with the complex between RIG-I and control RNA, suggesting that the complexes with three motifs are more stable than those with RNA control (Fig. 3C). All these data pointed to the possibility that these tandem RNA motifs might mediate the binding of RIG-I with p105 3′-UTR mRNA. To address this possibility, we inserted double- or triple-tandem motifs (46–66 bp) or control sequence (60 bp) between luciferase coding region and fragment d in Luc-p105 3′-UTR construct. Under the same experimental settings as in the previous experiment (Fig. 2E), the luciferase mRNA, derived from the Luc-p105 3′-UTR constructs only with intact 3′-UTR, motifs 1, 2, and 3, but not with control sequence or d alone, was detected in His–Rig-I precipitates (Fig. 3D). More importantly, the luciferase activity expressed by Luc-p105 3′-UTR constructs with the insertions of motifs 1, 2, and 3 was efficiently induced by Rig-I overexpression. However, the reporter constructs with control sequence inserted or d alone had no response to Rig-I (Fig. 3E). Moreover, the same experiment as for Fig. 2F was performed using the constructs harboring these motifs. The results similar to that shown in Fig. 2F indicated that Luc-p105 3′-UTR constructs with these tandem motifs were dependent on the presence of Rig-I in the expression of luciferase activity compared with that in control sequence inserted or fragment d alone (Fig. 3F). To further confirm the binding capacity of these tandem motifs with Rig-I, the motif search was performed by using MAST software with default parameters in those with experimental evidence for Rig-I binding (Fig. 1 C and D). The data show that most mRNAs except for Sep (O-phosphoserine) tRNA:Sec (selenocysteine) tRNA synthase (Sepsecs) harbor two to seven conserved motifs arrayed in homo- or heterotandem (Fig. S3B), whereas none or only one motif is identified in those experimentally demonstrated not to interact with Rig-I (Fig. S3C). These findings support the Rig-I interactions with the cellular mRNAs and further strengthen the role of these motifs in tandem in mediating Rig-I binding. Taken together, we may conclude that these tandem motifs in p105 3′-UTR mRNA are responsible for Rig-I binding to p105 3′-UTR mRNA. Such bindings are functionally required for protein translation regulation.
Fig. 3.
Structural features of p105 3′-UTR mRNA for Rig-I binding. (A) The distribution of the three enriched motifs in p105 3′-UTR mRNA fragments. (B) The C5′ and Cα atom rmsds for dual-tandem motifs or control RNA and Rig-I protein are shown. (C) The binding model between Rig-I and dual-tandem motifs or control RNA. The binding-free energy data are labeled in each panel. (D) The reporter constructs harboring the indicated tandem motifs or control sequence were cotransfected with His-tagged Rig-I expression vector into NIH 3T3 cells. Anti-His antibody-precipitated Luc mRNA was detected by RT-PCR. (E) The same experimental settings as in D but Luc activity was measured and expressed as a percentage relative to that of Rig-I empty vector. (F) The same experiment as shown in Fig. 2F. One of two independent experiments is shown as mean ± SE (n = 3) for E and F, respectively.
Rig-I Regulates Nf-κb1 Translation via Interacting with Ribosomal Components.
To further understand the biological significance of Rig-I binding to Nf-κb1 3′-UTR mRNA, we first checked whether such interaction affects the stabilization of Nf-κb1 mRNA. Unexpectedly, neither semi-Q RT-PCR (Fig. S4A) nor Northern blot (Fig. S4B) for Nf-κb1/p105 transcript in splenocytes upon LPS stimulation showed significant differences in p105 as well as p65 mRNA levels between WT and Rig-I−/− cells. Under the same conditions, p105 mRNA level remained unchanged in the 1.B4.B6 cell line expressing Rig-I siRNA (Fig. S4C). Furthermore, the primary splenocytes or 1.B4.B6 cells stably expressing Rig-I were treated with 10 μg/mL actinomycin D (ActD), followed by LPS stimulation. Semi-Q RT-PCR revealed that Nf-κb1 mRNA level remained stable either in the absence of Rig-I (Fig. S4D) or upon exogenous Rig-I expression (Fig. S4E) compared with their controls. Then, we analyzed the protein levels of p50 and its precursor p105 in primary splenocytes by Western blotting. Surprisingly, both p105 and p50 proteins were significantly reduced in Rig-I−/− cells with or without LPS stimulation compared with that in WT cells, whereas the protein level of p65 in the same extracts was comparable (Fig. 4A). Similar results were observed in Rig-I–silenced cells (Fig. S5A). Interestingly, exogenous Rig-I expression in the same cell line induced an enhanced response of p105/p50 expression to LPS stimulation (Fig. 4B). These findings thus indicate that Rig-I is crucial for normal expression of p105/p50 protein. To address whether Rig-I is involved in a p105/p50 degradation process, both WT and Rig-I−/− splenocytes were treated with 10 μg/mL cycloheximide (CHX), followed by LPS stimulation. Western blot analysis revealed that both p105 and p50 declined at a comparable rate between two genotypes, albeit at relatively lower levels of p105/p50 in Rig-I−/− splenocytes (Fig. S5B). We further tested Nf-κb1/p50 nuclear translocation or activation upon LPS stimulation. We found that compared to WT cells, p105 and p50 were significantly reduced in total cell lysates and cytosolic fractions of Rig-I−/− cells with or without LPS stimulation. Strikingly, Nf-κb1/p50 in nuclear fractions of Rig-I−/− cells was undetectable even after LPS stimulation (Fig. 4C). A similar result was also observed with immunofluorescence staining for p105/p50 (Fig. 4D). Consistent with these findings, NF-κB activity was found significantly decreased and poorly responded to LPS stimulation in Rig-I−/− MEFs (Fig. 4E). It could be fully rescued by Rig-I in a dose-dependent manner (Fig. 4F). Similar changes in the protein levels of p105/p50 and NF-κB activity were further observed in MEFs with LPS or poly (I:C) treatment at different time points (Fig. S6). Based on these findings, we believe that Rig-I is crucial for NF-κB activity via regulating Nf-κb1 expression at posttranscriptional stage, especially protein translation.
Fig. 4.
Rig-I is a positive regulator for NF-κB activity. Western blots for the indicated proteins in splenocytes (A) and the 1.B4.B6 cell line (B) stably expressing Rig-I. (C) Immunoblots of Nf-κb1/p105/p50 in total cell lysate, cytoplasmic and nuclear fractions from splenocytes. (D) Confocal images for Nf-κb1/p105/p50 (green) distribution in splenic B lymohocytes with an original magnification of 400×. (E) Luc activity in MEFs transfected with Nf-κb-Luc reporter construct followed by LPS treatment. (F) Induction of NF-κB activity in RIG-I−/− MEFs cotransfected with Rig-I expression vector and Nf-κb1-Luc reporter construct is expressed as a ratio of Luc activity with Rig-I expression to that of empty vector. One of three independent experiments is shown as mean ± SD (n = 3, **P < 0.01) in E and F, respectively.
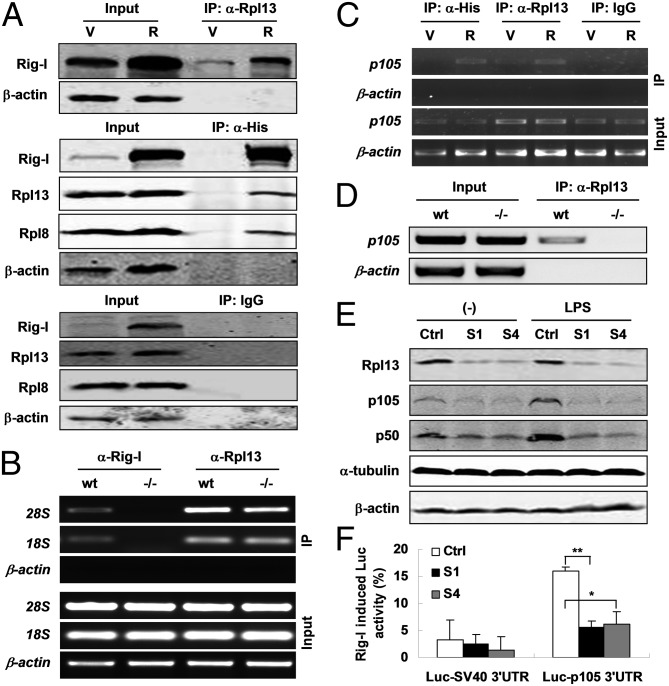
However, the question that remains to be addressed is how Rig-I regulates p105 translation via binding to its 3′-UTR mRNA. One of the possible answers could be that Rig-I recruits translational machinery proteins while binding to p105 3′-UTR to facilitate protein translation. To this end, a relative and absolute quantification mass spectrometry (iTRAQ-MS) assay was performed with the pooled immunoprecipitates with anti-His antibody from the cell lysates of 1.B4.B6 cells stably expressing His-tagged Rig-I. By comparing with the same cells but expressing His alone, we were able to identify 28 proteins (total ion score confidence interval > 80%) potentially interacting with Rig-I (Table S2). Among them, special attention was paid to a ribosomal protein that is a component of the large 60S subunit of ribosome, Rpl13 (60S ribosomal protein L13). Following this clue, we carried out RNA or protein immunoprecipitation assay by using antibodies against Rpl13, His-tag, and Rig-I, respectively. Total cell lysates of 1.B4.B6 cells stably expressing His-tag (shown as V in fig. 5A) or His-tagged Rig-I (shown as R in fig. 5A) were subjected to immunoprecipitation with a monoclonal antibody to Rpl13. As expected, trace amounts of endogenous Rig-I or overexpressed His-tagged Rig-I can be detected in the precipitates. On the contrary, endogenous Rpl13, as well as Rpl8, another component of the 60S subunit can be coimmunoprecipitated from the same lysates with His-tagged Rig-I by using an anti-His antibody. As negative controls, no detectable Rig-I, Rpl13, Rpl8, or β-actin were found in the IgG precipitates from the same lysates (Fig. 5A). Given iTRAQ-MS results, these data further suggest a physical interaction of Rig-I with Rpl13 and Rpl8, both belonging to the 60S subunit of ribosome and potentially interacting with each other (www.compbio.dundee.ac.uk/www-pips/). Relevant to this finding, the question of whether Rig-I interacts with the small 40S subunit of ribosome was raised. The answer was positive because both 28S (60S subunit) and 18S (40S subunit) rRNAs were able to be immunoprecipitated from the cell lysates of WT or Rig-I−/− MEFs using an anti-Rpl13 antibody but only from that of WT MEFs when using a monoclonal antibody to Rig-I (Fig. 5B), implying that Rig-I interacts with not only the large but also small ribosomal subunits that participate in p105 translation. It has been shown that Rig-I can bind with p105 mRNA. Does Rpl13 also bind with p105 mRNA, at least via Rig-I protein? To address this question, we performed an additional RNA immunoprecipitation assay by using IgG (negative control) or antibodies against Rpl13 and His-tag (positive control) proteins. It was found that p105 mRNA was precipitated with endogenous Rpl13 from the cell lysates with or without overexpression of Rig-I. However, the amount of precipitated p105 mRNA appeared related to Rig-I dose (Fig. 5C), suggesting that Rpl13 interacts with p105 mRNA through Rig-I protein. To make the point clearer, a similar experiment was performed in WT and Rig-I−/− MEFs. It was shown that p105 mRNA was detected in the precipitates with Rpl13 only from the cell lysates of WT MEFs (Fig. 5D). To evaluate the effect of Rpl13 on p105 expression, the RNA silencing experiment in NIH 3T3 cells was performed using two siRNA oligos against Rpl13. As shown in Fig. 5E, p105/p50 protein levels were significantly decreased and the responses to LPS stimulation were abolished with down-regulation of Rpl13. Furthermore, p105 3′-UTR–mediated luciferase activity was efficiently induced by cotransfected Rig-I–expressing vector compared with empty vector, but the induction was significantly impaired upon Rpl13 silencing (Fig. 5F). These data indicate that Rpl13 is required for p105 translation through the interaction between p105 3′-UTR and Rig-I to which Rpl13 binds.
Fig. 5.
Rig-I regulates Nf-κb1 translation via interacting with ribosomal components. (A) Immunoblots for the indicated proteins in IgG, anti-His, and anti-Rpl13 precipitates from the lysates of 1.B4.B6 cell line stably transfected with empty vector (V) or His-tagged Rig-I expression vector (R). (B) RT-PCR for 28S, 18S rRNAs in anti-Rig-I, or anti-Rpl13 precipitates from the lysates of WT or Rig-I−/− MEFs. (C) p105 mRNA by RT-PCR in the precipitates under the same experimental settings as in A. (D) RT-PCR for p105 mRNA in anti-Rpl13 precipitates as in B. (E) Western blots for the indicated proteins in NIH 3T3 cells stably expressing Rpl13 siRNA (S1 and S4) and control siRNA (Ctrl). (F) Luc activity in the same cell lines as in E but cotransfected with Rig-I expression and Luc-reporter constructs with p105 or SV40 3′-UTR is expressed as a percentage relative to that of Rig-I empty vector. One of three independent experiments is shown as mean ± SD (n = 3, *P < 0.05; **P < 0.01).
Discussion
Previous studies reveal that RIG-I selectively interacts with certain RNA molecules with specific structural features. Given the results of crystal structure analyses on RIG-I in either ligand-free or dsRNA-bound status, it seems more likely that primary sequence-dependent tertiary configuration of RNA ligands could be required for RIG-I recognition. In these circumstances, it could be hard to exclude the possibility that RIG-I recognizes or interacts with the cellular RNAs with certain structural features. Our data uniquely indicate that Rig-I could regulate Nf-κb1 translation through interacting with cellular Nf-κb1 mRNA. To elucidate the structural basis in p105 3′-UTR that contributes to Rig-I binding, the extensive bioinformatics analyses on p105 3′-UTR fragments that interact with Rig-I are performed. Interestingly, we find that: (i) three distinct motifs appear enriched in Rig-I binding fragments (a, b, and c) but not in fragment d, which has no interaction with Rig-I. The three motifs spanning 11–21 bp are arrayed as clusters composed of three to seven motifs in homo- or heterotandem among three Rig-I binding fragments. (ii) Three-dimensional RNA structure prediction and folding analyses reveal that three motifs in homotandem with two of each (48, 24, and 29 bp in length, respectively) can efficiently fold and form more double helix structure than control RNA (44 bp). (iii) Molecular dynamics simulations display that well-matched binding interfaces between RIG-I and dual-tandem motifs, and more stable RIG-I complexes with three dual-tandem motifs but not with RNA control can be formed. (iv) The binding-free energy needed to form such complexes is much lower than that in controls. (v) Three motifs in homo dual or triple tandem (24–51 bp) indeed mediate the binding of RIG-I with p105 3′-UTR. (vi) Finally, the response or dependence of luciferase activity to, or on RIG-I, suggest the functional importance of these RIG-I binding motifs in regulating Nf-κb1/p105 expression. Taken together, we have demonstrated that RIG-I interacts with 3′-UTR of Nf-κb1/p105 mRNA through three distinct conserved motifs in double helix structure, implying that primary sequence-dependent tertiary configuration of RNA ligands is more crucial for Rig-I binding.
In eukaryotic cells, protein synthesis is a complex process that consists of four major steps: initiation, elongation, termination, and ribosome recycling. All of the steps involved in protein synthesis can be the targets for regulation (33). Because Rig-I is shown to positively regulate Nf-κb1/p105 translation through direct binding to its 3′-UTR mRNA, we focus on the potential effect of this binding on protein translation. Indeed, 3′-UTRs of mRNAs play crucial roles in posttranscriptional regulation of gene expression by modulating mRNA localization, stability, and translation (34, 35). Apart from having binding sites for miRNAs, 3′-UTRs harbor certain motifs that interact with specific RNA-binding proteins (36). Translational control mechanisms often involve interactions between the 3′- or 5′-UTRs of mRNA and RNA-binding proteins. One such mechanism requires circularization of the mRNA molecule involving protein–protein interactions between the mRNA termini (37).
As one of 3′-UTR binding proteins, Rig-I is possible to interact with other proteins while binding to 3′-UTR, especially the proteins involved in protein translation. As expected, iTRAQ-MS analysis on the precipitate with anti-His antibody from the total cell lysate of the splenic B-cell line stably expressing His-tagged Rig-I, and subsequent immunoprecipitation and Western blotting, reveal that Rig-I is able to interact with ribosomal protein L13 (Rpl13), as well as ribosomal protein L8 (Rpl8), both are 60S subunit components. RPL13 belongs to the L13E family of ribosomal proteins. It has been shown that ribosomal proteins may have extraribosomal functions apart from ribosome and protein biosynthesis (38). Rpl8 belongs to the L2P family of ribosomal proteins. It is located in the cytoplasm. In rat, the protein associates with the 5.8S rRNA, very likely participates in the binding of aminoacyl-tRNA, and is a constituent of the elongation factor 2-binding site at the ribosomal subunit interface (http://omim.org/entry/604177). Moreover, Rpl8 is predicted to potentially interact with initiation factor eIF5B (www.compbio.dundee.ac.uk/www-pips/), a ribosome-dependent GTPase that mediates displacement of initiation factors from the 40S ribosomal subunit in 48S initiation complexes and joining of 40S and 60S subunits (39). Thus, we may propose that RIG-I participates in the processes of ribosome assembly, mRNA circularization, or ribosome recycling. In support of this notion, 18S and 28S rRNAs, the components of 40S and 60S ribosomal subunits, are shown to interact with Rig-I. Further evidence emerges from Rpl13 silencing experiments. The data show that Nf-κb1/p105 protein levels significantly decrease with the effective down-regulation of Rpl13. Consistent with this finding, p105 3′-UTR–mediated luciferase activity is reduced in NIH 3T3 cells stably expressing Rpl13 siRNA, once again, demonstrating a crucial role of the 3′-UTR in regulating Nf-κb1/p105 translation.
In conclusion, our data demonstrate that Rig-I functions as a positive regulator for NF-κB signaling. The regulatory effect of Rig-I is dependent on the interaction with the multiple sites within 3′-UTR of Nf-κb1 mRNA through its ATPase/helicase domains and C-terminal regulatory domain. The double helix structure formed with the tandem motifs enriched within p105 3′-UTR is crucial for Rig-I binding and protein translation. In addition, Rig-I is proposed to be involved in ribosome assembly, mRNA circularization, or ribosome recycling by recruiting ribosomal components while binding to p105 3′-UTR mRNA. Given the important roles of Rig-I in host antivirus responses, regulating NF-κB activity, and protein translation, as well as the complex interaction network, which has emerged in this study, further intensive studies on this “puzzling” molecule are needed in future. It is believed that such effort will be greatly beneficial for better understanding of biological processes and drug development research.
Materials and Methods
All of the experiments were processed according to the standard protocols. The detailed materials and methods are provided as SI Materials and Methods. Primers used in this study are listed in Table S3.
Supplementary Material
Acknowledgments
We thank Dr. Amy. L. Kenter for the B-cell line; Dr. Qiang-su Guo and Jia-qi Xiao for technical assistance; and Profs. Klaus Rajewsky, Xue-tao Cao, and Hua Gu for critically reading the manuscript and for informative discussion. This work was partially supported by grants from the Chinese National Science Fund for Distinguished Young Scholars (39925023), the National Natural Science Foundation of China (30530390 and 31000408), the Ministry of Science and Technology of China (2006BAI23B02,2011BAI15B02), grants from the Science and Technology Commission of Shanghai municipality (10DZ2251500, 10JC1410300, and 11DZ2292400), and the E-Institutes of Shanghai Municipal Education Commission (E03003).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1304432110/-/DCSupplemental.
References
- 1.Yoneyama M, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5(7):730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 2.Kawai T, et al. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6(10):981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 3.Meylan E, et al. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437(7062):1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 4.Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122(5):669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 5.Xu LG, et al. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol Cell. 2005;19(6):727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 6.Hornung V, et al. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314(5801):994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 7.Pichlmair A, et al. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 2006;314(5801):997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 8.Schlee M, et al. Recognition of 5′ triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity. 2009;31(1):25–34. doi: 10.1016/j.immuni.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schmidt A, et al. 5′-triphosphate RNA requires base-paired structures to activate antiviral signaling via RIG-I. Proc Natl Acad Sci USA. 2009;106(29):12067–12072. doi: 10.1073/pnas.0900971106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Habjan M, et al. Processing of genome 5′ termini as a strategy of negative-strand RNA viruses to avoid RIG-I-dependent interferon induction. PLoS ONE. 2008;3(4):e2032. doi: 10.1371/journal.pone.0002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y, et al. Structural and functional insights into 5′-ppp RNA pattern recognition by the innate immune receptor RIG-I. Nat Struct Mol Biol. 2010;17(7):781–787. doi: 10.1038/nsmb.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu C, et al. The structural basis of 5′ triphosphate double-stranded RNA recognition by RIG-I C-terminal domain. Structure. 2010;18(8):1032–1043. doi: 10.1016/j.str.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hwang SY, et al. 5′-Triphosphate-RNA-independent activation of RIG-I via RNA aptamer with enhanced antiviral activity. Nucleic Acids Res. 2012;40(6):2724–2733. doi: 10.1093/nar/gkr1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Civril F, et al. The RIG-I ATPase domain structure reveals insights into ATP-dependent antiviral signalling. EMBO Rep. 2011;12(11):1127–1134. doi: 10.1038/embor.2011.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang F, et al. Structural basis of RNA recognition and activation by innate immune receptor RIG-I. Nature. 2011;479(7373):423–427. doi: 10.1038/nature10537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kowalinski E, et al. Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell. 2011;147(2):423–435. doi: 10.1016/j.cell.2011.09.039. [DOI] [PubMed] [Google Scholar]
- 17.Luo D, et al. Structural insights into RNA recognition by RIG-I. Cell. 2011;147(2):409–422. doi: 10.1016/j.cell.2011.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu TX, et al. Gene expression networks underlying retinoic acid-induced differentiation of acute promyelocytic leukemia cells. Blood. 2000;96(4):1496–1504. [PubMed] [Google Scholar]
- 19.Takahasi K, et al. Nonself RNA-sensing mechanism of RIG-I helicase and activation of antiviral immune responses. Mol Cell. 2008;29(4):428–440. doi: 10.1016/j.molcel.2007.11.028. [DOI] [PubMed] [Google Scholar]
- 20.de la Cruz J, Kressler D, Linder P. Unwinding RNA in Saccharomyces cerevisiae: DEAD-box proteins and related families. Trends Biochem Sci. 1999;24(5):192–198. doi: 10.1016/s0968-0004(99)01376-6. [DOI] [PubMed] [Google Scholar]
- 21.Rocak S, Emery B, Tanner NK, Linder P. Characterization of the ATPase and unwinding activities of the yeast DEAD-box protein Has1p and the analysis of the roles of the conserved motifs. Nucleic Acids Res. 2005;33(3):999–1009. doi: 10.1093/nar/gki244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu F, Gu J. Retinoic acid inducible gene-I, more than a virus sensor. Protein Cell. 2011;2(5):351–357. doi: 10.1007/s13238-011-1045-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y, et al. Rig-I-/- mice develop colitis associated with downregulation of G alpha i2. Cell Res. 2007;17(10):858–868. doi: 10.1038/cr.2007.81. [DOI] [PubMed] [Google Scholar]
- 24.Zhang NN, et al. RIG-I plays a critical role in negatively regulating granulocytic proliferation. Proc Natl Acad Sci USA. 2008;105(30):10553–10558. doi: 10.1073/pnas.0804895105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang LJ, et al. RA-inducible gene-I induction augments STAT1 activation to inhibit leukemia cell proliferation. Proc Natl Acad Sci USA. 2011;108(5):1897–1902. doi: 10.1073/pnas.1019059108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kong L, et al. An essential role for RIG-I in toll-like receptor-stimulated phagocytosis. Cell Host Microbe. 2009;6(2):150–161. doi: 10.1016/j.chom.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 27.Siebenlist U, Franzoso G, Brown K. Structure, regulation and function of NF-kappa B. Annu Rev Cell Biol. 1994;10:405–455. doi: 10.1146/annurev.cb.10.110194.002201. [DOI] [PubMed] [Google Scholar]
- 28.May MJ, Ghosh S. Rel/NF-kappa B and I kappa B proteins: An overview. Semin Cancer Biol. 1997;8(2):63–73. doi: 10.1006/scbi.1997.0057. [DOI] [PubMed] [Google Scholar]
- 29.Glisovic T, Bachorik JL, Yong J, Dreyfuss G. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 2008;582(14):1977–1986. doi: 10.1016/j.febslet.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sharma S, Ding F, Dokholyan NV. iFoldRNA: Three-dimensional RNA structure prediction and folding. Bioinformatics. 2008;24(17):1951–1952. doi: 10.1093/bioinformatics/btn328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Duan Y, et al. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J Comput Chem. 2003;24(16):1999–2012. doi: 10.1002/jcc.10349. [DOI] [PubMed] [Google Scholar]
- 32. Kollman PA, et al. (1997) The Development/Application of a ‘Minimalist’ Organic/Biochemical Molecular Mechanic Force Field using a Combination of ab Initio Calculations and Experimental Data. In Computer Simulation of Biomolecular Systems, eds. van Gunsteren WF, Weiner PK, Wilkinson AJ (Kluwer Academic Publishers, Dordrecht, the Netherlands), pp 83–96.
- 33.Gal-Ben-Ari S, et al. Consolidation and translation regulation. Learn Mem. 2012;19(9):410–422. doi: 10.1101/lm.026849.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuersten S, Goodwin EB. The power of the 3′ UTR: Translational control and development. Nat Rev Genet. 2003;4(8):626–637. doi: 10.1038/nrg1125. [DOI] [PubMed] [Google Scholar]
- 35.Mazumder B, Seshadri V, Fox PL. Translational control by the 3′-UTR: The ends specify the means. Trends Biochem Sci. 2003;28(2):91–98. doi: 10.1016/S0968-0004(03)00002-1. [DOI] [PubMed] [Google Scholar]
- 36.Wilkie GS, Dickson KS, Gray NK. Regulation of mRNA translation by 5′- and 3′-UTR-binding factors. Trends Biochem Sci. 2003;28(4):182–188. doi: 10.1016/S0968-0004(03)00051-3. [DOI] [PubMed] [Google Scholar]
- 37.Mazumder B, Seshadri V, Imataka H, Sonenberg N, Fox PL. Translational silencing of ceruloplasmin requires the essential elements of mRNA circularization: Poly(A) tail, poly(A)-binding protein, and eukaryotic translation initiation factor 4G. Mol Cell Biol. 2001;21(19):6440–6449. doi: 10.1128/MCB.21.19.6440-6449.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen FW, Ioannou YA. Ribosomal proteins in cell proliferation and apoptosis. Int Rev Immunol. 1999;18(5-6):429–448. doi: 10.3109/08830189909088492. [DOI] [PubMed] [Google Scholar]
- 39.Unbehaun A, et al. Position of eukaryotic initiation factor eIF5B on the 80S ribosome mapped by directed hydroxyl radical probing. EMBO J. 2007;26(13):3109–3123. doi: 10.1038/sj.emboj.7601751. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.