Abstract
The crystal structure of the open conformation of a bacterial voltage-gated sodium channel pore from Magnetococcus sp. (NaVMs) has provided the basis for a molecular dynamics study defining the channel’s full ion translocation pathway and conductance process, selectivity, electrophysiological characteristics, and ion-binding sites. Microsecond molecular dynamics simulations permitted a complete time-course characterization of the protein in a membrane system, capturing the plethora of conductance events and revealing a complex mixture of single and multi-ion phenomena with decoupled rapid bidirectional water transport. The simulations suggest specific localization sites for the sodium ions, which correspond with experimentally determined electron density found in the selectivity filter of the crystal structure. These studies have also allowed us to identify the ion conductance mechanism and its relation to water movement for the NavMs channel pore and to make realistic predictions of its conductance properties. The calculated single-channel conductance and selectivity ratio correspond closely with the electrophysiology measurements of the NavMs channel expressed in HEK 293 cells. The ion translocation process seen in this voltage-gated sodium channel is clearly different from that exhibited by members of the closely related family of voltage-gated potassium channels and also differs considerably from existing proposals for the conductance process in sodium channels. These studies simulate sodium channel conductance based on an experimentally determined structure of a sodium channel pore that has a completely open transmembrane pathway and activation gate.
Voltage-gated cation channels are proteins that produce electrical signals in neurons and other excitable cells to regulate muscle contraction, gene expression, and release of hormones and neurotransmitters among other functions. In response to a change in transmembrane electrical potential, the channels open pores through which ions move passively across the membrane. The large family of cation channels includes those selective for sodium, potassium, or calcium. The opening and closing of these ion-specific channels is carefully choreographed to produce the electrical signals required by the nervous system for rapid signal transduction (1).
Voltage-gated sodium channels have been causally linked with a wide range of neurological and cardiovascular diseases and hence are important pharmaceutical drug-development targets (2, 3). Eukaryotic voltage-gated sodium channels are large, single-chain polypeptides, consisting of 24 transmembrane (TM) helices that form four homologous repeats, each contributing both a voltage sensor and a pore domain; the latter are arranged to form a central Na+-selective transmembrane pathway. Bacterial voltage-gated sodium channels are far simpler, consisting of four polypeptide chains, each of which is composed of six TM segments, with segments TM1–TM4 forming the voltage sensors and TM5–TM6 forming the pore domains. The TM5–TM6 segments associate as a homotetramer to form a similar central Na+-selective pore. The sodium channel from Bacillus halodurans (NaChBac), the first bacterial sodium channel to be identified, has been well-characterized with respect to its conductance properties (4). Since then a large number of other members of the prokaryotic voltage-gated sodium channel family have been identified and shown to be close sequence and structural homologs (5, 6).
Several crystal structures of bacterial sodium channels have been published recently (7–10); in these, the proteins are present in different functional states. The first structure had an activated voltage sensor and a closed pore (7), and others were in potentially inactivated states (8, 9), but none of these had open activation gates or a complete TM hydrophilic pathway that would enable the flow of ions between the cytoplasmic and extracellular surfaces. However, the recent determination of the crystal structure of an open conformation (10) of a pore-only construct of a sodium channel now enables examination and simulation of the full process of ion passage across a sodium channel structure based on an experimentally determined structure.
Results
Effects of Simulation Procedures on the Channel Structure.
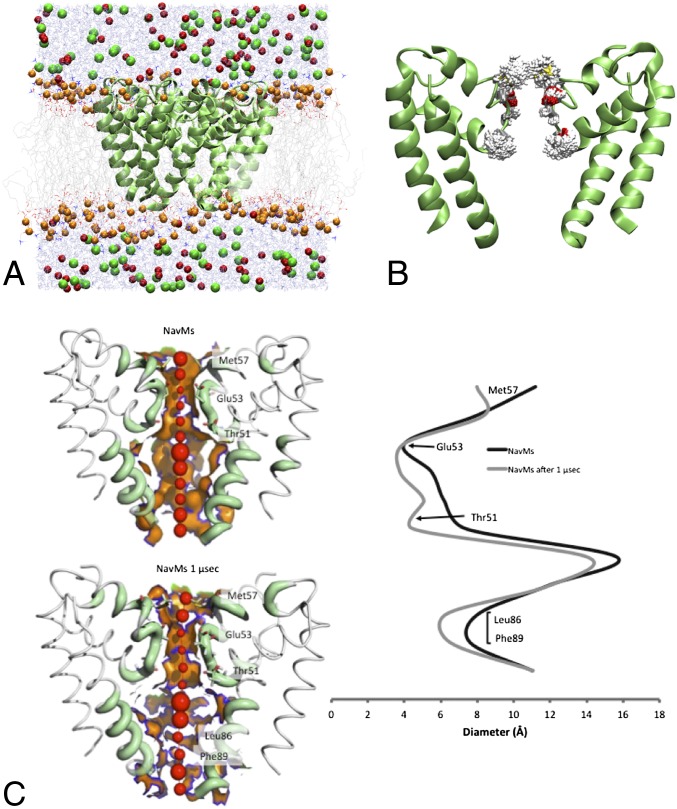
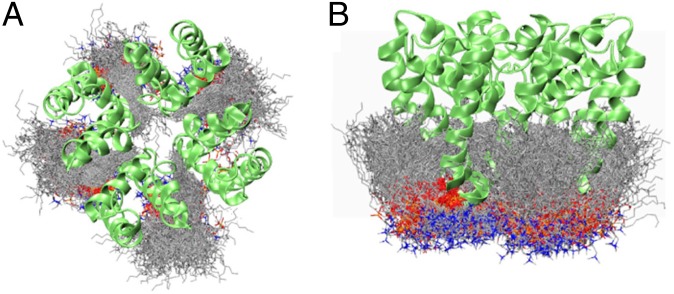
The structure of the bacterial NavMs channel pore in its open conformation was inserted into a model preformed palmitoyloleoyl phosphatidylcholine (POPC) lipid bilayer (Fig. 1A) and subjected to molecular dynamics (MD) simulations. The starting pore structure did not contain any water or ions; these were initially placed in the bath surrounding the bilayer and rapidly migrated into and through the pore during the simulations. Because in the absence of harmonic restraints the pore rapidly closes at the cytoplasmic gate (observed for both OPLS and CHARMM simulations), harmonic restraints (1 kcal/mol/Å2) were applied to the α-carbon atoms of the TM helices during the simulations to retain the open configuration in the absence of the voltage-sensing domain. Over a typical 250-ns simulation period, while the positions of the backbone atoms of the protein, including the residues in the selectivity filter (SF), remain close to their positions in the crystal structure, the SF side chains broadly sample the rotameric space available (Fig. 1B). At the end of the simulation, the protein conformation and the size and shape of the central pore have changed only slightly (Fig. 1C).
Fig. 1.
MD Simulations of NavMs. (A) System used for simulation: NavMs (in green ribbons) embedded in a POPC lipid bilayer (in gray). Sodium ions are red, chloride ions green, water molecules blue, and lipid head groups orange. (B) Effects of simulations on the protein structure: Overlay of the conformational space sampled by NavMs during 250 ns of a 1-μs simulation. The backbone Cα atoms of the transmembrane helices were weakly restrained to retain the open conformation of the channel, whereas the SF was not restrained. (C) (Left) The accessibility of the internal cavity of the initial (crystal) and final (simulation) structures, Upper and Lower, respectively. (Right) Plots showing the internal dimensions of the crystal and final simulation structures.
Ion Permeation, Selectivity, and Conductance.
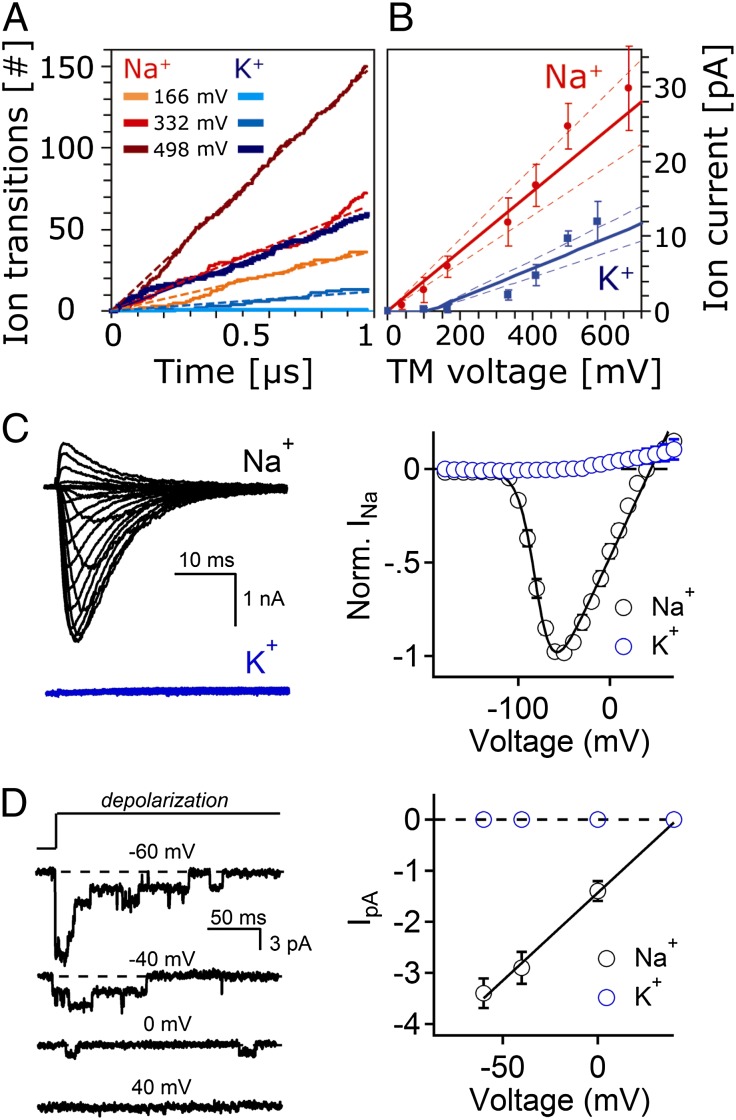
MD simulations were used to directly calculate the rate of ion permeation through the NavMs channel (Fig. 2A). The simulations were conducted in the presence of 0.5-M solutions of NaCl or KCl. A temperature of 37° C was used in the simulations, not only for biological relevance, but also to speed up ion kinetics and to enable the capture of more transition events on the 1- to 2-μs simulation timescale. To drive ions through the channel (from the extracellular to the intracellular compartments), different electric fields between 4 and 75 mV/nm were applied (Table S1), corresponding to transmembrane potentials of 39–665 mV. Fig. 2A shows the cumulative translocations of Na+ and K+ through the pore at selected voltages. These indicate that the channel is capable of conducting both Na+ and K+, with a preference for Na+, as would be expected for a sodium channel. The selectivity ratios (Table 1) were calculated from the current–voltage (I-V) curves for Na+ and K+ (Fig. 2B). The I-V curve for Na+ is linear; however, the K+ curve exhibits an ∼70-mV lag, resulting in a two-phase curve. As a result, two different selectivity ratios are calculated for the high and low voltage ranges. At high voltages there is a modest (about three- to sixfold) preference of Na+ over K+, whereas at lower applied voltages the ratios are in the 11–18:1 range (the latter calculations are more poorly converged due to few K+ conductance events observed under these conditions, but represent a more physiologically relevant regime). The I-V curve (Fig. 2B) was used to obtain the predicted conductance value of γNa = 40 ± 7 pS.
Fig. 2.
Sodium and potassium ion permeation parameters. (A) Cumulative ion translocation over 1 μs at selected voltages. The calculations used 0.5 M concentrations of NaCl and KCl. All curves show linear behavior for the ion permeation events. (B) Calculated I-V curves for Na+ (red) and K+ (blue) used for calculation of single-channel conductances. There is no voltage lag for Na+, allowing for a simple linear fit, but K+ has a 70-mV voltage lag and is fitted to the expression I = aΔV/[1 − exp(−bΔV)]; ΔV = V − V0 (41). Dashed lines indicate the 60% confidence interval, and error bars are from block averaging over three blocks. (C) Sample whole-cell current traces from NavMs-transfected HEK 293T cells elicited by depolarizing pulses from a −180-mV holding potential. Currents were recorded from the same cell in the presence of either 150 mM NaCl (black) or KCl (blue). The resulting current–voltage relationship (Right) was normalized to the peak of Na+ current for each cell (n = 9). The Nernst equation reversal potential for Na+ in these conditions is 41 mV, which is close to the averaged measured values of 43 ± 1 in the extracellular sodium condition. The expected reversal potential for K+ under these conditions was 128 mV. (D, left) Sample single-channel events recorded from inside-out patches where sodium is the charge carrier. Channel-opening events were triggered by variable depolarizations from −180 mV. The resulting event amplitudes are plotted (right) as a function of voltage, and the conductance was estimated by fitting the data to a linear relationship (n = 4).
Table 1.
Selectivity calculated as a function of applied voltage
| VTM* (mV) | ETM* (mV/nm) | INa† (pA) | IK† (pA) | Selectivity (INa/IK) |
| 498 | 55.9 | 24.7 | 9.7 | 2.5 |
| 410 | 46.0 | 16.9 | 4.8 | 3.5 |
| 332 | 37.3 | 11.9 | 2.1 | 5.6 |
| 166 | 18.6 | 5.9 | 0.33 | 18 |
| 100 | 11.3 | 2.8 | 0.25 | 11 |
Applied voltage and electric field.
Calculated Na+ and K+ currents.
To compare NavMs ion permeability estimated from the MD simulations to that measured in cells, patch-clamped HEK 293 cells were transfected with the full-length NavMs channel. As inferred from the large current (3,592 ± 98 pA/pF in whole-cell patch clamp), NavMs expression was robust. The voltage-dependent properties of activation (Act. V1/2), inactivation (Inact. V1/2), and the inactivation kinetics (τ Inact. V1/2) of NavMs were compared with the bacterial sodium channel NaChBac (4) and the human neuronal sodium channel (hNav1.1) (Fig. S1 A–C and Table S2). The NavMs voltage dependence of activation and inactivation is >50 mV more hypopolarized than NaChBac and hNaV1.1, indicating that the NavMs channel requires more energy in the form of a hypolarized membrane potential to stay closed. NavMs enters the inactivation state 12× more slowly than hNaV1.1 but 7× faster than NaChBac. Similar biophysical properties were recently reported for the NavAb channel (8), but use dependence of slow inactivation did not occur. NavMs is selective for sodium over potassium ions (permeability ratio Na+/K+ >24); no voltage-dependent inward current was observed when K+ was substituted for Na+ in the extracellular saline solution (Fig. 2C). To rule out anomalous Na+ blockade of the potassium conductance, additional experiments used a very low concentration (5 mM) of either sodium or potassium in the intracellular saline solution (Fig. S1D). Again, no inward potassium currents were observed. Single-channel events were then recorded with a patch pipette filled with either sodium or potassium. High-resistance seals (10–16 GΩ) were achieved and voltage-dependent single-channel events were observed only when the pipette contained Na+ (γNa = 33 pS) (Fig. 2D). Because the simulation was done under high Na+ conditions (500 mM) (to view as many events as possible), currents were measured experimentally in response to increasing extracellular Na+ ions to estimate the concentration at which conductance saturates (Fig. S1E). An exponential fit of these results indicates that the concentration limit is 445 mM. Because the simulations and experiments were conducted at different temperatures, an additional simulation was done at 22 °C (corresponding to the experimental temperature) to estimate the temperature effect on the single-channel conductance. The calculated fluxes differed (Table S1, lines 6 and 7) by a factor of 0.84 (i.e., 31 vs. 37); hence it is expected that the calculated single-channel conductance at the experimental temperature would be 34 ± 6 pS, which is very close to the measured value. We conclude that the NavMs channel is selective for sodium over potassium ions, as was seen in the simulation, and that it has a single-channel conductance of ˜33 pS, as indicated by both experiment and simulation.
Identification of Ion-Binding Sites.
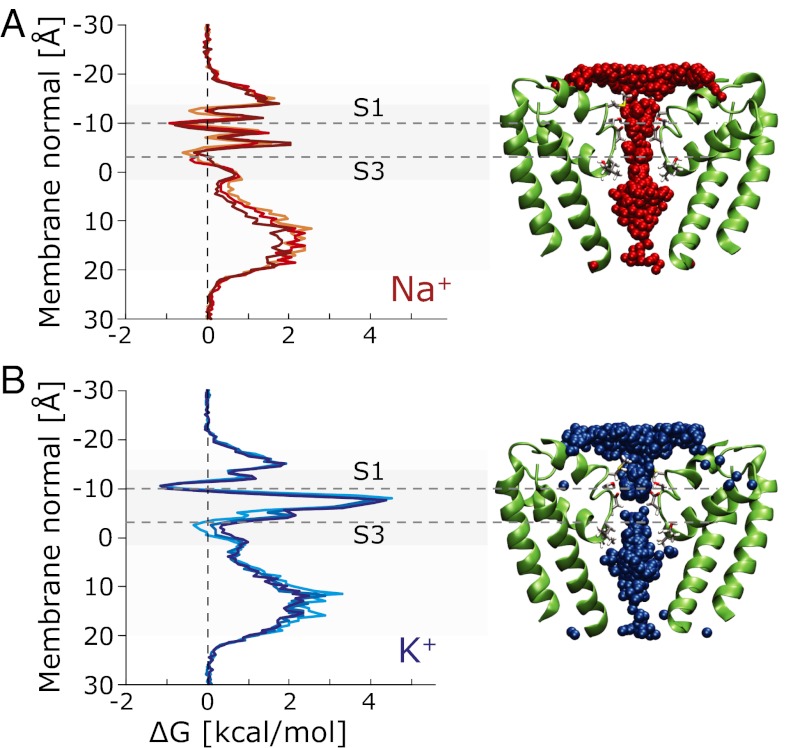
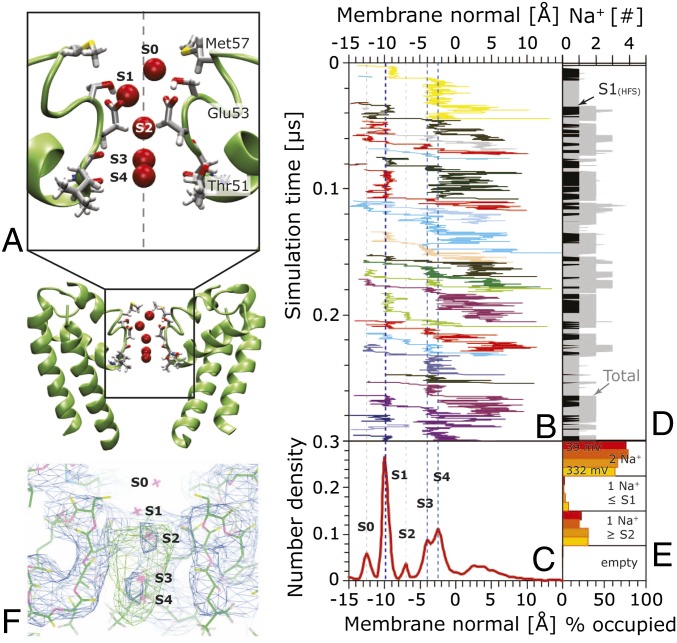
Ion-binding sites were identified from the free-energy profiles developed from cumulative positions in the 1 μs simulations (Fig. 3). Permeating Na+ can apparently reside at five distinct binding sites in the region of the SF, designated S0–S4 (Fig. 4). Of these, S1 (called SiteHFS in ref. 7) is the dominant site (based on number density calculations, Fig. 4E); at S1, the ion is directly coordinated with the carboxylate group of one of the Glu53 residues in the SF. As a consequence, the S1 site is asymmetrically disposed along the pore direction (Fig. 4A and Fig. S2A), whereas the sites farther down (S2–S4) are aligned on the axis of the SF. The simulations also suggest the presence of a minor external site, S0, at the vestibule of the SF. This short-lived (τ < 5 ns) site is also asymmetric and loosely coordinates Na+ with the Ser54 and Met57 side chains. As suggested previously (7, 10, 11), Na+ is coordinated at S1 by the Glu53 carboxylate and Ser54 hydroxyl groups and two water molecules while forming hydrogen bonds to two other Glu53 side chains. In marked contrast to K+ in potassium channels, the sodium ions in this channel are hydrated to some extent at all sites in the NavMs SF. Ion coordination numbers, defined as the number of oxygen atoms within the first solvation shell (3.1 Å for Na+, 3.4 Å for K+), are shown in Fig. S2B. Both Na+ and K+ are hexa-coordinated throughout (mostly by water molecules); only at S1 (and to a minor extent at S3) do pore residues replace water molecules in the coordination.
Fig. 3.
Ion permeation profiles for sodium and potassium. (A) Free-energy profile (Left) constructed from the total Na+ positions (Right) sampled during a 1 μs simulation (4,000 frames spaced 0.25 ns apart), showing the free-energy barriers and minima (corresponding to the sites S0–S4). (B) Free-energy profile (Left) constructed from the K+ positions (Right) sampled during a 1 μs simulation The high free-energy barrier for potassium is clearly visible as a population gap in the SF between S1 and S3. Because many conductance events require more than one ion, these are multi-ion free-energy profiles.
Fig. 4.
Ion-binding sites in the selectivity filter. (A) Five Na+-binding sites, labeled S0–S4, were identified in the equilibrium simulations. These sites were identical for all simulations, irrespective of applied voltage. S0 is at the vestibule of the SF, contacting Ser54 and Met57 side chains; S1 (corresponding to the site designated “HFS” in ref. 7) is the only site within the SF that is off the symmetry axis of the channel due to strong contact of the ion with one Glu53; S2 is a minor site just beyond the contact distance of the Glu side chains; S3 (corresponding to site “CEN” in ref. 7) and S4 (corresponding to site “IN” in ref. 7) are at the height of Leu52 and Thr51 backbone carbonyls, respectively. S0, S2, S3, and S4 are stabilized by full water-hydration shells. S1 has a partial water hydration shell. (B) Representative positions of individual permeating Na+ (varying colors) along the membrane normal (x axis) versus simulation time (first 300 ns, 332 mV). (C) Multi-ion number density histogram over the entire simulation showing the five ion-binding sites. Ions enter the SF either directly or loiter at the external site S0 before being stopped at site S1. The ions then move to the lower end of the SF at S3 and S4. Some ions also call transiently at the intermediate site S2, but S1 and S2 are never populated at the same time due to their proximity. Once leaving S3/S4, the ions extensively explore the cavity and can rebind to S3/S4 before ultimately leaving through the open gate. (D) Total number of ions in the SF versus time. There are typically one to two ions in the SF at any given time. (E) Average occupancy of the SF throughout the simulations. Most commonly (∼70%), there are two ions, one at S0–S1 and another at S2–S4. At 0.5 M NaCl, the channel is never observed to be empty. (F) Overlay of the calculated ion positions and the experimental (crystallographic) electron density map. The calculated atom positions (S0–S4) are indicated as red Xs. The 2Fo-Fc electron density map (no ions were included in the map calculation, contoured at 1.5σ) is colored in blue, and the Fo-Fc omit map (contoured at 3σ) is colored in green. The electron density matches the S2 and S3/S4 sites seen in the simulations.
When a sodium ion translocates from site S1, it frequently stops at site S2, which is ∼3 Å below S1. S2 is only weakly populated and short lived (<0.1–5 ns). There the sodium ion is fully hydrated by water molecules hydrogen-bonded to the carboxylate groups of Glu53 and the backbone carbonyls of Leu52. Similarly, an entire hydration shell surrounds the ions in the subsequent closely spaced positions S3 and S4 [called SiteCEN and SiteIN, respectively, in the NavAb closed structure (7)]. These positions are unlikely to be occupied at the same time due to the overlapping spreads of their distributions (Fig. 4E). These sites are located at the lower end of the SF; the surrounding water molecules are hydrogen-bonded to the carbonyl oxygens of Leu52 (at S3) and Thr51 (at S4). After detaching from S4, the ions explore the cavity for a significant time (∼1–30 ns) during which they can also rebind to S3/S4. Ultimately, they leave through the open gate.
Interestingly, the crystal structure electron density map (10) and the MD-defined sites correlate very well to sites S2 and S3/S4 (Fig. 4F); however, no density was seen corresponding to S1. This may be because S1 is the only degenerate off-center site, due to the four equivalent Glu residues, and steric constraints mean only one of these can be bound at any one time, which would result in low occupancy at each of the individual sites. In the crystal structure, the density seen was not conclusively assigned as being sodium ions due to the resolution of the data, but it was noted that the sites were likely to be Na+ (rather than water molecules) due to their scattering density being of the correct magnitude for sodium ions, because the Rfactor decreased on placement of sodiums at these positions, and because of the requirement of the crystallization conditions for sodium salts (10). In contrast, no ordered density (either for ions or water) was found in the SF of the closed NavAb X-ray structure (7). The colocalization of the experimental electron density in the open state with the computed sodium sites strengthens the cases for both the identity of the experimental density and the simulation results.
Potassium ion conductance through the channel is markedly different from that of sodium ions (Fig. S3). Site S2 does not appear to be occupied, and sites S3 and S4 are more diffuse. Potassium ions appear to halt at S1, with a large barrier preventing further translocation.
Ion Flux.
Although single Na+ passage through NavMs is observed in the simulations (Fig. S2 and Movies S1–S5), there is typically more than one ion in the channel at any given time (Fig. S4A), with an overall time-average number (Ntotal) of ∼1.8 Na+, which is independent of the applied voltage (Table S2). The channel is never observed to be empty in 0.5 M NaCl (Fig. S3E). Two Na+ occupy the pore for ∼70% of the time, with one at site S0/S1 and another at site S2/S3/S4. For the other half of the time, there is only one ion either at S0/S1 (3%) or at S2/S3/S4 (26%) (Fig. 4D). In contrast, passage of K+ requires two ions at S1 (Fig. S4B and Table S3); this appears to be the rate-limiting step, explaining the lower conductance of K+. Whereas the occupancy distribution of K+ is very different from Na+, Ntotal for K+ is also ∼1.8.
The dwell time (Δt) of Na+ in the channel (Table S4) depends greatly on the applied voltage, as expected. At 166 mV, its dwell time in the channel is 40 ns. The mean time between consecutive permeation events (Δt′ = Nconducted/simulation time) is 24 ns at 166 mV, which is shorter than the dwell time. The ratio Δt/Δt′ = 1.7 is constant for all voltages (except at the lower voltages that are poorly converged). This ratio is equal to Ntotal when there are no instances where an ion binds the SF but does not transit the pore. This is essentially the case for Na+, but not for K+, where Δt/Δt′ is voltage-dependent (Tables S3 and S4); this means that there are numerous unsuccessful K+ transit attempts and that K+ conduction is slower than Na+ conduction through NavMs channels. This can be compared with K+ conductance through potassium channels, where a constant Δt/Δt′ = 2.4 was reported (12); hence, on average more potassium ions are in the potassium channel SF alone than there are sodium ions in the entire NavMs channel at any given time.
Water Transport.
Although no water molecules were placed inside the channel initially, during the simulation water molecules sampled all parts of the pore (Fig. S2D). The water conductance rate is calculated to be ∼3 × 109 s−1 and is equal in both directions (Fig. S2C), resulting in a zero net water transport through the channel, except at the largest applied voltage, where ionic drag results in a slight net transport of water molecules in the direction of sodium flux. Only water molecules that traversed the entire length of the channel were considered in this analysis. The zero net transport is a result of the balance of two large and opposing water fluxes of equal magnitude. Interestingly, the magnitude of the bidirectional water flux is not correlated with the ion flux rates, the latter of which are strongly dependent on the applied voltage (Fig. 2A). Furthermore, the magnitude of the flux is similar in the simulations with sodium and potassium ions, indicating that water flux is independent of ionic flux. This is in contrast to potassium channels, where water and ion transport are strongly coupled (13, 14).
Occupancy of the Fenestrations.
Of particular interest was the nature of the contents of the channel fenestrations during the simulations. The fenestrations, which are openings in the sides of the TM regions of the protein that have been proposed to be entry portals for hydrophobic molecules such as ligands and drugs (7, 10), were initially empty. During the simulations these sites rapidly filled with one or more phospholipid fatty acid chains from the bilayer lipids (Fig. 5A). Only lipids from the cytoplasmic bilayer leaflet (the one in which the central cavity is located) entered the fenestrations (Fig. 5B and Fig. S5). Lipid tails enter and exit the fenestrations many times over the course of a simulation, and there is a frequent exchange of these lipids with bulk lipids, suggesting that, although there is access, there is no strong binding site for the lipid tails in the fenestration. The fatty acid chains that breached the fenestrations were found to enter deeply into the channel cavity. In several instances, both fatty acid chains of a single lipid entered a fenestration. However, no blocking of the channel conductance pathway by the lipid chains was observed over the duration of the simulations.
Fig. 5.
Occupancy of the fenestrations. (A) The fenestrations that were initially empty rapidly filled with the lipid fatty acid chains. The conformational space sampled by lipids (tails in gray, head groups in red) viewed from the cytoplasmic surface shows that they entered the cavity over the course of the simulation. Each fenestration is occupied by at least one lipid tail for 90% of the time. Nevertheless, there is frequent exchange of lipids in the fenestrations with the bulk bilayer. (B) Image in A rotated 90° showing that only lipids from the cytoplasmic bilayer leaflet enter the fenestrations.
Discussion
Voltage-gated ion channels exhibit different ion selectivities and functional properties based on the nature of their SFs. In homo-tetrameric potassium channels, each of the four monomers contributes to the binding of a dehydrated K+ through their main chain carbonyls (15). In contrast, the SFs of bacterial sodium channels are produced by side chains that have the consensus sequence TLESWS (Fig. S6), with the principal binding site in the extended SF expected to be produced by one glutamate side chain contributed by each of the monomers. The carbonyl oxygen atoms of a threonine residue on the intracellular side of the filter were also hypothesized to interact through hydrogen bonds with water molecules from the hydration shell of a Na+ (7). The present MD study using the open pore structure provides evidence for new binding sites within the extended SF and for the dynamics of occupation of these sites during the process of ion translocation. Furthermore, although no ions or water molecules were visible in the SF of the crystal structure of the closed conformation of the NavAb channel (7), the crystal structure of the open conformation of the NavMs pore (10) exhibited electron density at multiple sites in the center of the SF. The correspondence between the positions of the MD-calculated sites described here and the experimental electron density supports its identification as arising from sodium ions.
The long time simulations in this study demonstrated that the mechanism of conductance through the NavMs channel pore differs greatly from the mechanism used by potassium channels. In NavMs, the sodium ions are always hydrated to some extent and can even be accommodated next to each other in the SF, a situation very different from potassium channels where dehydrated potassium ions go through the SF in a single-file manner. Although single Na+ transport is observed in NavMs, heterogeneous multiple ion occupancy of the channel is a frequent occurrence, with an average of 1.8 Na+ in the channel at any time. Although single ion/multisite models had previously been suggested based on unidirectional flux measurements of Na channels (16–19), the multi-ion occupancies observed in the present simulations agree with both the experimentally observed nonindependence of ion movement (18) and the proposals based on recent Brownian dynamics simulations of simplified sodium channel models (20).
The present MD simulations also clearly demonstrate that water flux through the channel is not correlated with ion flux and that there is no systematic net water transport. This, too, is in contrast to potassium channels, where the rate of ionic flux limits the maximum water flux through the channel (12, 21). The key difference is that the SF of the sodium channel is much wider than that of the potassium channel. NavMs can accommodate at least four water molecules simultaneously at any position along the filter, even at the most narrow regions, whereas potassium channels constrain ionic and water flux to a single-file arrangement in the filter. The average dwell time of a water molecule in the sodium channel is two orders of magnitude lower than that of a sodium ion, confirming that ionic conductance and water flow are not coupled as they are in potassium channels. This differs from previous molecular dynamics simulations based on the closed NavAb channel, where exchange of water molecules between the bulk and cavity through the SF did not dislodge sodium ions resting in sites S1 and S3 (22).
The origin of sodium channel selectivity was previously investigated by MD (22) and computational potential of mean force (PMF) studies (11, 23) using the closed NavAb crystal structure (7); those studies suggested that differing water structures at S1 were responsible for selectivity. The use of unbiased MD simulations (as performed here) has the advantage that the free-energy profiles derive directly from the calculated multi-ion occupancy of the channel (i.e., they are multi-ion PMFs). In this case, convergence was excellent, as was seen by the negligible dependence on the applied voltage. The main barrier, between site S1 and S3, is 2 kcal/mol for Na+, resulting in a value for the ΔΔGbarrier of 3.5 kcal/mol.
Because of the use of an open channel structure and the long time simulations (which permitted observation of complete ion translocation events), it was possible to quantitatively determine conductance parameters and make direct comparisons of experimental and calculated electrophysiological values for a bacterial sodium channel. Previous experimental determinations of bacterial sodium channel selectivities for NaChBac and homologs have reported Na:K ratios of between 5:1 and 171:1 (7, 24). In this study, the experimental selectivity ratio of NavMs was shown to be between these values (24:1). The calculated selectivity ratios derived from the MD studies of NavMs (∼11–18:1 for V < 170 mV) were a close match to this experimental value. Single-channel conductance data are also available for several of the bacterial sodium channel homologs: NaChBac has reported conductances ranging widely from ∼12 (4) to ∼120 pS (25), depending on the method used; the estimated conductance of the NavSp homolog is ∼30 pS (24) and for NavAb is 37 pS (7). In this study, the experimental NavMs conductance for sodium was estimated to be ∼33 pS, very close to the computed conductance at this temperature of ∼34 pS. Hence, these correlations are a strong indication that the simulations reported here not only have provided information on the dynamics and mechanism of ion translocation through a sodium channel, but also could accurately reproduce quantitative experimental values for the channel.
Finally, our simulations showed that lipid tails can spontaneously enter the channel fenestrations, but have no specific strong binding sites. Hence they provide dynamic evidence to support the proposals (7, 10) that small hydrophobic molecules such as drugs could enter through the fenestrations and reach their binding sites deep inside the pore by this mechanism.
In summary, long time simulations based on the crystal structure of the open form of a bacterial sodium channel pore have been compared with experimental electrophysiological measurements on the NavMs channel and have provided significant insight into the processes, dynamics, and structural features associated with ion conduction and ion selectivity in sodium channels.
Methods
Molecular Dynamics Simulations.
The symmetric tetramer model of the open conformation of the NavMs pore from Magnetococcus sp. (strain MC-1) (10), derived from the crystal structure [Protein Data Bank (PDB) code 4F4L], was used for the protein structure. The model included residues 8–94 (numbering based on the NavMs pore in the PDB file), encompassing the TM5 and TM6 helices and the intervening P1, P2, and SF regions (Fig. S6). No crystallographic waters were included in the structure used for the MD simulations. Ionizable residues were set to their charge configuration at neutral pH in aqueous solution, except Glu53, which was simulated in two different protonation states (Table S1) (one where all were charged and one where only two were protonated). The protein was inserted into preformed atomic detail POPC lipid bilayers (composed of 204 lipid molecules) in the presence of 0.1–0.5 M salt solutions and equilibrated for 20 ns. There was a mismatch of 12 in the number of lipids present in the two leaflets (due to the shape of the protein). Before production runs, each system was equilibrated for 100 ns in the NPT (constant number, pressure, and temperature) ensemble, with strong harmonic restraints placed on the protein backbone.
Molecular dynamics simulations were performed using the Anton machine at the Pittsburgh Supercomputing Center with CHARMM27 protein (26) and CHARMM36 lipid force fields (27) and TIP3P water models (28). Ion parameters were taken from ref. 29. Initial configurations for the Anton runs were created using DESMOND software (30). Additional simulations were performed with GROMACS 4.5 (31) and hippo beta (www.biowerkzeug.com) software with the OPLS all-atom protein force field (32) and TIP3P water model. OPLS united-atom lipid parameters were used (33). For all simulations, electrostatic interactions were computed using particle mesh Ewald electrostatic treatment. A cutoff of 10 Å was used for van der Waals interactions. Bonds involving hydrogen atoms were restrained using LINCS (34). The integration time step was 2 fs, and neighbor lists were updated every five steps. Water, lipids, and the protein were each coupled separately to a heat bath with a time constant of τT = 0.1 ps using weak temperature coupling (35). Atmospheric pressure of 1 bar was maintained using weak semi-isotropic pressure coupling with compressibility κz = κxy = 4.6 × 10−5 bar–1 and time constant τP = 1 ps (36). Homogeneous external electric fields Ez were applied in the z-direction (membrane normal), the corresponding transmembrane voltage was calculated via V = Ez × Lz with Lz equal to the time average of the box dimension in the z-direction (89.1 Å) (37), and the electrostatic potential (Fig. S7) was verified using Poisson’s equation. The components and characteristics of each of the simulations undertaken and the results from each are listed in Table S1.
Structures were displayed using VMD software (38), electron density maps were displayed in COOT (39), and calculations of the accessible area in the pore were done using Pore Walker software (40).
Electrophysiology.
HEK 293T cells were transiently transfected with full-length NavMs, seeded onto glass coverslips, and placed in a perfusion chamber for experiments where extracellular conditions could be altered. For whole-cell experiments, the extracellular solution contained (in mM) NaCl or KCl (150), CaCl2 (1.8), MgCl2 (1), and Hepes (10) (pH 7.4). The intracellular (pipette) solution contained (in mM) CsF (110), NaCl (30), Hepes (10), EGTA (5), MgCl2 (2), KCl (1) (pH 7.5) except for those in experiments in Fig. S1D where either NaCl or KCl (5 mM) was used with CsF (130 mM). Data were analyzed by Igor Pro-6.00 (Wavemetrics). Residual leak (> −100 pA) and capacitance were subtracted using a standard P/4 protocol. Current–voltage relationships were fit with (V − Vrev)/{1 + exp[(V − V1/2)/k], where Vrev is the extrapolated reversal potential. Experimental conditions were the same for inside-out patches except the pipette and bath saline were switched. The equation for the exponential fits used in Fig. S1A was: f(x) = B + Aexp[(1/τ)x], where τ is the half-time constant or half concentration of sodium, and x is the time or concentration of sodium. The PNa/PK permeablity ratios where PNa and PK are the sodium and potassium permeabilities, respectively, were calculated by the equation ΔErev = (RT/zF)ln PNa [ion]/PK [ion], where R is the universal gas constant (8.314 J⋅K−1/mol−1), T is the temperature (K = 295°), F is Faraday’s constant (96,485 C⋅mol−1), and z = 1e−.
Supplementary Material
Acknowledgments
P.G.D., M.D., and D.E.C. acknowledge Betsy Navarro and Nat Blair for their advice and technical assistance. This work was supported by grants from the UK Biotechnology and Biological Science Research Council (to B.A.W.) and by a European Union Marie Curie Fellowship (to M.B.U.). Anton computer time was provided by the National Resource for Biomedical Supercomputing and the Pittsburgh Supercomputing Center (Grant PSCA10059P).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1214667110/-/DCSupplemental.
References
- 1.Hille B. Local anesthetics: Hydrophilic and hydrophobic pathways for the drug-receptor reaction. J Gen Physiol. 1977;69(4):497–515. doi: 10.1085/jgp.69.4.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Catterall WA. Sodium channel mutations and epilepsy. In: Noebels JL, Avoli M, Rogawski MA, Olsen RW, Delgado-Escueta AV, editors. Chapter 52. Jasper’s Basic Mechanisms of the Epilepsies. Bethesda, MD: National Center for Biotechnology Information; 2012. [PubMed] [Google Scholar]
- 3.Nieto FR, et al. Tetrodotoxin (TTX) as a therapeutic agent for pain. Mar Drugs. 2012;10(2):281–305. doi: 10.3390/md10020281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ren D, et al. A prokaryotic voltage-gated sodium channel. Science. 2001;294(5550):2372–2375. doi: 10.1126/science.1065635. [DOI] [PubMed] [Google Scholar]
- 5.Koishi R, et al. A superfamily of voltage-gated sodium channels in bacteria. J Biol Chem. 2004;279(10):9532–9538. doi: 10.1074/jbc.M313100200. [DOI] [PubMed] [Google Scholar]
- 6.Irie K, et al. Comparative study of the gating motif and C-type inactivation in prokaryotic voltage-gated sodium channels. J Biol Chem. 2010;285(6):3685–3694. doi: 10.1074/jbc.M109.057455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Payandeh J, Scheuer T, Zheng N, Catterall WA. The crystal structure of a voltage-gated sodium channel. Nature. 2011;475(7356):353–358. doi: 10.1038/nature10238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Payandeh J, Gamal El-Din TM, Scheuer T, Zheng N, Catterall WA. Crystal structure of a voltage-gated sodium channel in two potentially inactivated states. Nature. 2012;486(7401):135–139. doi: 10.1038/nature11077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang X, et al. Crystal structure of an orthologue of the NaChBac voltage-gated sodium channel. Nature. 2012;486(7401):130–134. doi: 10.1038/nature11054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCusker EC, et al. Structure of a bacterial voltage-gated sodium channel pore reveals mechanisms of opening and closing. Nat Commun. 2012;3:1102. doi: 10.1038/ncomms2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corry B, Thomas M. Mechanism of ion permeation and selectivity in a voltage gated sodium channel. J Am Chem Soc. 2012;134(3):1840–1846. doi: 10.1021/ja210020h. [DOI] [PubMed] [Google Scholar]
- 12.Jensen MØ, et al. Principles of conduction and hydrophobic gating in K+ channels. Proc Natl Acad Sci USA. 2010;107(13):5833–5838. doi: 10.1073/pnas.0911691107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ando H, Kuno M, Shimizu H, Muramatsu I, Oiki S. Coupled K+-water flux through the HERG potassium channel measured by an osmotic pulse method. J Gen Physiol. 2005;126(5):529–538. doi: 10.1085/jgp.200509377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alcayaga C, Cecchi X, Alvarez O, Latorre R. Streaming potential measurements in Ca2+-activated K+ channels from skeletal and smooth muscle. Coupling of ion and water fluxes. Biophys J. 1989;55(2):367–371. doi: 10.1016/S0006-3495(89)82814-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doyle DA, et al. The structure of the potassium channel: Molecular basis of K+ conduction and selectivity. Science. 1998;280(5360):69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 16.Begenisich T, Busath D. Sodium flux ratio in voltage-clamped squid giant axons. J Gen Physiol. 1981;77(5):489–502. doi: 10.1085/jgp.77.5.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rakowski RF, Gadsby DC, De Weer P. Single ion occupancy and steady-state gating of Na channels in squid giant axon. J Gen Physiol. 2002;119(3):235–249. doi: 10.1085/jgp.20028500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Begenisich TB, Cahalan MD. Sodium channel permeation in squid axons. II: Non-independence and current-voltage relations. J Physiol. 1980;307:243–257. doi: 10.1113/jphysiol.1980.sp013433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hille B. The permeability of the sodium channel to metal cations in myelinated nerve. J Gen Physiol. 1972;59(6):637–658. doi: 10.1085/jgp.59.6.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vora T, Corry B, Chung S-H. Brownian dynamics study of flux ratios in sodium channels. Eur Biophys J. 2008;38(1):45–52. doi: 10.1007/s00249-008-0353-5. [DOI] [PubMed] [Google Scholar]
- 21.Saparov SM, Pohl P. Beyond the diffusion limit: Water flow through the empty bacterial potassium channel. Proc Natl Acad Sci USA. 2004;101(14):4805–4809. doi: 10.1073/pnas.0308309101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carnevale V, Treptow W, Klein ML. Sodium ion binding sites and hydration in the lumen of a bacterial ion channel from molecular dynamics simulations. J Phys Chem Lett. 2011;2(19):2504–2508. [Google Scholar]
- 23.Furini S, Domene C. On conduction in a bacterial sodium channel. PLOS Comput Biol. 2012;8(4):e1002476. doi: 10.1371/journal.pcbi.1002476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shaya D, et al. Voltage-gated sodium channel (NaV) protein dissection creates a set of functional pore-only proteins. Proc Natl Acad Sci USA. 2011;108(30):12313–12318. doi: 10.1073/pnas.1106811108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Studer A, Demarche S, Langenegger D, Tiefenauer L. Integration and recording of a reconstituted voltage-gated sodium channel in planar lipid bilayers. Biosens Bioelectron. 2011;26(5):1924–1928. doi: 10.1016/j.bios.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 26.MacKerell AD, et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102(18):3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 27.Klauda JB, et al. Update of the CHARMM all-atom additive force field for lipids: Validation on six lipid types. J Phys Chem B. 2010;114(23):7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79(2):926–935. [Google Scholar]
- 29.Joung IS, Cheatham TE., III Determination of alkali and halide monovalent ion parameters for use in explicitly solvated biomolecular simulations. J Phys Chem B. 2008;112(30):9020–9041. doi: 10.1021/jp8001614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bowers KJ, et al. 2006. Scalable algorithms for molecular dynamics simulations on commodity clusters. Proceedings of the ACM/IEEE Conference on Supercomputing (Institute of Electrical and Electronics Engineers, Tampa)
- 31.Berendsen HJC, van der Spoel D, van Drunen R. GROMACS: A message-passing parallel molecular dynamics implementation. Comput Phys Commun. 1995;95:43–56. [Google Scholar]
- 32.Jorgensen WL, Maxwell DS, Tirado-Rives J. Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J Am Chem Soc. 1996;118(45):11225–11236. [Google Scholar]
- 33.Ulmschneider JP, Ulmschneider MB. United atom lipid parameters for combination with the optimized potentials for liquid simulations all-atom force field. J Chem Theory Comput. 2009;5(7):1803–1813. doi: 10.1021/ct900086b. [DOI] [PubMed] [Google Scholar]
- 34.Hess B, Bekker H, Berendsen HJC, Fraaije JGEM. LINCS: A linear constraint solver for molecular simulations. J Comput Chem. 1997;18(12):1463–1472. [Google Scholar]
- 35.Bussi G, Donadio D, Parrinello M. Canonical sampling through velocity rescaling. J Chem Phys. 2007;126(1):014101. doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 36.Berendsen HJC, Postma JPM, Vangunsteren WF, Dinola A, Haak JR. Molecular dynamics with coupling to an external bath. J Chem Phys. 1984;81(8):3684–3690. [Google Scholar]
- 37.Roux B. The membrane potential and its representation by a constant electric field in computer simulations. Biophys J. 2008;95(9):4205–4216. doi: 10.1529/biophysj.108.136499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J Mol Graph. 1996;14(1):33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 39.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 40.Pellegrini-Calace M, Maiwald T, Thornton JM. Pore-Walker: A novel tool for the identification and characterization of transmembrane protein channels from their three-dimensional structure. PLOS Comput Biol. 2009;5(7):1–16. doi: 10.1371/journal.pcbi.1000440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goychuk I, Hänggi P. Ion channel gating: A first-passage time analysis of the Kramers type. Proc Natl Acad Sci USA. 2002;99(6):3552–3556. doi: 10.1073/pnas.052015699. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.