Abstract
Cytotoxic lymphocytes eliminate virus-infected and cancerous cells by immune recognition and killing through the perforin-granzyme pathway. Traditional killing assays measure average target cell lysis at fixed times and high effector:target ratios. Such assays obscure kinetic details that might reveal novel physiology. We engineered target cells to report on granzyme activity, used very low effector:target ratios to observe potential serial killing, and performed low magnification time-lapse imaging to reveal time-dependent statistics of natural killer (NK) killing at the single-cell level. Most kills occurred during serial killing, and a single NK cell killed up to 10 targets over a 6-h assay. The first kill was slower than subsequent kills, especially on poor targets, or when NK signaling pathways were partially inhibited. Spatial analysis showed that sequential kills were usually adjacent. We propose that NK cells integrate signals from the previous and current target, possibly by simultaneous contact. The resulting burst kinetics and spatial coordination may control the activity of NK cells in tissues.
Decision making by individual leukocytes is crucial to immunity. Removing unhealthy cells from normal tissue requires that immune cells individually interrogate potentially threatening cells, and the outcome of countless single-cell decisions will determine whether a nascent viral focus or cancer will develop into a disease. Cytotoxic lymphocytes, including natural killer (NK) cells, are thought to be involved in this process of immunosurveillance (1), but quantitative relationships between single-cell killing decisions and tissue level immunosurveillance are unclear.
A specific one-to-one immune synapse between an individual cytotoxic cell and its target precedes killing by the perforin/granzyme pathway (2, 3). Sequential attack on successive targets by a single effector, termed serial killing, was suggested in early observations (4–6). However, direct experimental observation of serial killing has been difficult and, as a result, quantitative analysis has been largely absent (7, 8). Most measurements of killing consist of bulk assays, such as 51Cr release (9), or discontinuous methods, such as flow cytometry (10), which obscure single-cell behavior. Instead, we used low magnification imaging to continuously track large numbers of individual NK cells to reveal killing statistics. We also used very low effector-target ratios to facilitate observation of possible serial killing.
Results and Discussion
Cytotoxic T lymphocytes (CTLs) and NK cells deliver cytotoxic granzymes into the cytosol of target cells by using the pore-forming protein perforin to trigger granzyme uptake (11, 12). Granzyme B (GzmB), the most abundantly expressed and well studied granzyme, can kill through both caspase-dependent and caspase-independent pathways. GzmB has a substrate specificity similar to, but not identical with, initiator caspases (13). Using a GzmB-specific substrate sequence (ref. 14; SI Results and Discussion), we developed a genetically encoded reporter of GzmB activity as a specific, sensitive, and rapid single-cell marker of cytotoxic attacks (Fig. 1 A–D). Spontaneous cell death and other granzyme-independent processes did not activate this reporter. GzmB activity, reported by our probe, was a reliable early signal of cytotoxic attack (Movie S1). Its activation was always followed by cell death in our reporter lines as measured by membrane permeabilization, although permeabilization occurred after a long, variable delay and was less suited to kinetic analysis (Fig. S4).
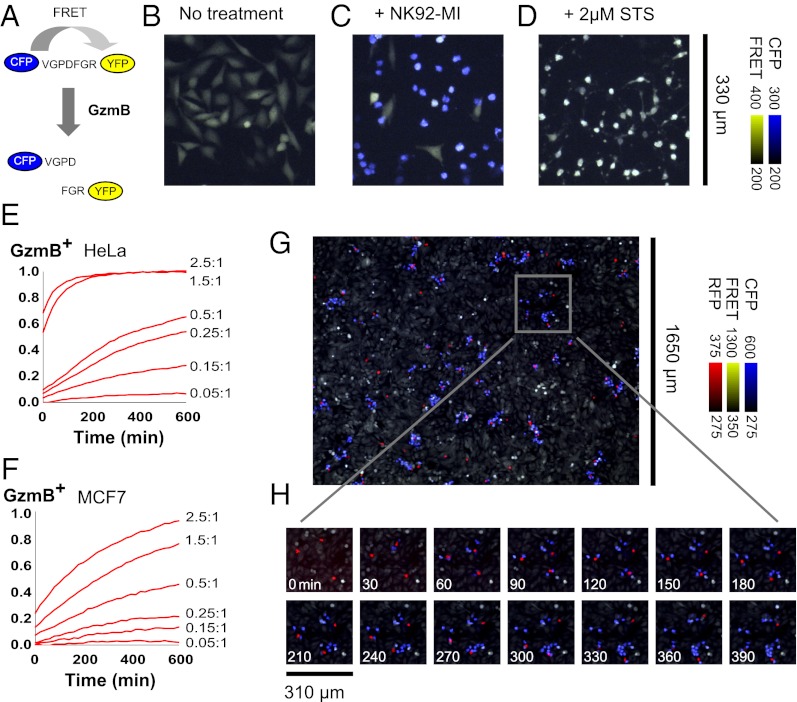
Fig. 1.
Live-cell granzyme B reporter enables a single-cell killing assay. (A) Förster resonance energy transfer (FRET) between fluorescent protein variants (37) can report on protease activity. We constructed a reporter for GzmB by using cyan (CFP) and yellow (YFP) fluorescent proteins fused by the peptide linker VGPDFGR. (B–D) HeLa cell stably expressing the GzmB reporter are imaged after 4 h of no treatment (B), treatment with 1:1 NK92-MI cells (C), or treatment with 2 μM staurosporine (EMD Biosciences) (D). An overlay of CFP emission (blue) and FRET emission (yellow) leads to strongly blue cells after cleavage of the GzmB reporter (Fig. S1). The reporter specifically indicates GzmB and not caspase activity (Fig. S2). (E and F) HeLa (E) or MCF7 (F) targets stably expressing the GzmB reporter are incubated at the indicated E:T ratio with NK92-MI, and the fraction of GzmB-positive targets is determined at each time point by using the integrated fluorescence emission of an entire field of view in comparison with standards with zero or complete killing. (G) At an E:T ratio <0.05:1, clusters of GzmB-positive targets cells indicate killing of multiple targets by well-isolated individual effector cells. Low magnification (4×) aided in obtaining measurements from a sufficient number of individual effector cells, labeled with CellTracker Orange (Invitrogen) (red), at this low dilution. The live-cell stain and phototoxicity had minimal effect on killing activity (Fig. S3). (H) Serial killing by individual NK92-MI cells can be tracked in a time lapse.
We imaged killing of adherent HeLa and MCF7 GzmB reporter lines by the IL2-producing NK line NK92-MI (15) at various effector-to-target (E:T) ratios (Fig. 1 E and F and Fig. S5A) while holding the target number constant by plating to confluency. At similar E:T ratios, NK92-MI killed a greater fraction of HeLa than MCF7 targets over a given time interval. However, killing goes to completion for both target lines at high enough E:T ratios or long enough incubation times, and there was no evidence of a resistant fraction or other strong response heterogeneity in bulk killing kinetics (Fig. S5B).
Standard killing assays often use E:T ratios of 10:1 or higher, which are presumably nonphysiological and, furthermore, incompatible with observation of possible serial killing. We used E:T ratios below 0.05:1 to isolate single effector cells. At these low densities on adherent target lines, movement and killing activity of individual effectors was unambiguously tracked over time (Fig. 1 G and H). Low-magnification imaging of multiple fields allowed collection of sufficient statistics (hundreds of cells per experiment). Single-cell analysis of killing is also possible through flow cytometry (10) or microscopy following isolation of preformed effector-target conjugates (16). However, our in situ approach enabled tracking of the entire killing process, repeated over many cycles, for the same effector cell. Our dilution strategy avoided the need for microwells (17).
Individual NK cells often sequentially killed multiple nearby target cells in our assay (Fig. 1H, Movies S2, S3, and S4, and SI Results and Discussion). Over 6 h of imaging, individual kill counts, k, ranged from 0 to 14 targets (Fig. 2 A and B). Our kill counts are higher than reported (4–7), possibly because each NK cell had access to an unrestricted number of targets, and each killing event was clearly visible. We analyzed >100 NK cells participating in serial killing in each experiment.
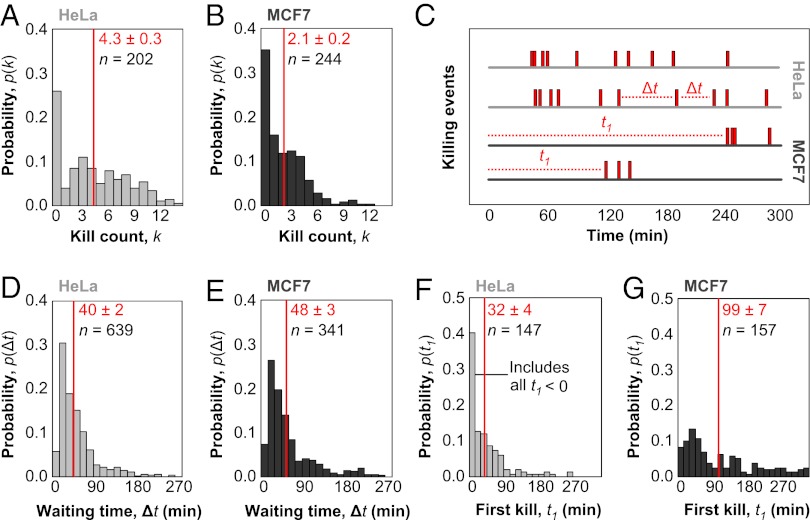
Fig. 2.
Quantitative single-cell measurements of serial killing. (A and B) Histogram of kill count, k, for the total number of targets killed by individual effector cells for HeLa (A) and MCF7 (B) targets after 6 h of coincubation of 500 NK92-MI cells and 2 × 104 HeLa or 3 × 104 MCF7 cells per well of a 96-well plate. For all of the following histograms, mean values and associated SEs are indicated in red, whereas n is the sample size. For A and B, n indicates the total number of NK cells profiled. Each NK cell is assigned a value for k based on the total number of kills over 6 h. The histogram shows the probability that a single NK cell among the n observed will have a particular value of k. (C) Sample killing traces of individual effector cells are shown, with killing events marked along the time axis by red bars. The waiting time, Δt, denotes the interval between successive killing events. The first kill time, t1, denotes the time before the first killing event. (D and E) Histogram of waiting times for HeLa (D) and MCF7 (E) targets. For D and E, n denotes the total number of waiting times recorded when two killing events occur in succession. NK cells with k < 2 do not contribute waiting times, whereas NK cells with k > 2 contribute multiple waiting times. The histogram shows the probability that any individual waiting time has a particular value of Δt. (F and G) Histogram of first kill times for HeLa (F) and MCF7 (G) targets. For F and G, n denotes the total number of first kill times measured, which is equivalent to the number of NK cells with k > 0. The histogram shows the probability that any individual first kill time has a particular value of t1.
We measured the fractions of active NK cells with specific numbers of kills, k, for each target line over 6 h (Fig. 2 A and B). These kill count distributions, p(k), for HeLa and MCF7 cells were not well fit by Poisson or geometric distributions (Fig. S6), even accounting for a possibly inactive fraction. Thus, serial killing is not a uniform kinetic process described by a single rate constant and the degree of serial killing is not determined by a single stochastic event. There was a peak at zero kills, which presumably reflects inactive cells, but the majority of effectors participated in killing, with p(k > 0)HeLa = 0.74 and p(k > 0)MCF7 = 0.65 for HeLa and MCF7 targets, respectively. The mean kill counts for only active effectors indicate the extent of serial killing as 〈k, k > 0〉HeLa = 5.9 and 〈k, k > 0〉MCF7 = 3.3 for HeLa and MCF7 targets, respectively. Thus, a larger fraction of NK cells attacked HeLa targets than MCF7 targets, and of the NK cells that attacked, the number of kills per NK cell was greater for HeLa targets than MCF7 targets. These two metrics provide absolute descriptors of target susceptibility at the limit of a single effector cell and, therefore, allow direct comparison across target lines or conditions. In contrast, bulk lysis-release metrics are influenced by cell lysis properties (18), dependent on the exact E:T ratio, and ambiguous in their interpretation (19).
The waiting time, Δt, between successive killing events by individual NK cells is defined in Fig. 2C. The distribution of waiting times for HeLa and MCF7 targets (Fig. 2 D and E) were fit by an exponential decay, excluding the short rise time (Fig. S7A). Thus, a single rate constant may dominate killing of subsequent targets, although targets still exhibit more long waiting times events than predicted by simple kinetic models (Fig. S7B). Interestingly, the HeLa and MCF7 waiting time distributions were not statistically different from each other (Fig. S7C). Accordingly, we found that the average waiting times were similar for HeLa and MCF7 targets (Fig. 2 D and E). The slightly longer average waiting time for MCF7 targets results from a small number of killing events with very long waiting times. Although MCF7 cells are significantly worse targets than HeLa cells based on bulk killing kinetics (Fig. 1 E and F) and average number of kills (Fig. 2 A and B), the waiting time between kills does not significantly vary between them.
The first kill time is also defined in Fig. 2C and plotted in Fig. 2 F and G. The first kill time, t1, is the time between the start of the experiment and the first killing event by a particular NK cell. Because the zero time point is poorly defined in the experiment (SI Results and Discussion and Fig. S8), we instead compared the relative difference between 〈t1〉 for HeLa and MCF7 targets for samples from the same experimental trial. We found that the difference 〈t1〉MCF7 − 〈t1〉HeLa had a value of 67 min in one experiment (Fig. 2 F and G) and 69 min in a second independent experiment (Fig. 3D). The difference means that MCF7 is a much poorer target than HeLa for only the first kill. Killing of subsequent MCF7 and HeLa targets occurs with nearly identical kinetics based on 〈Δt〉.
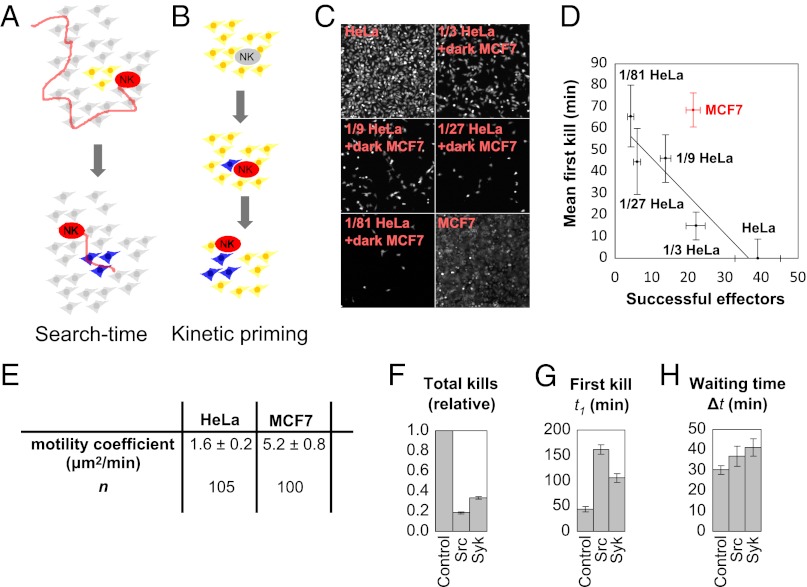
Fig. 3.
Bursting kinetics in serial killing. (A) In a search-time model, the long t1 accounts for the time to find a cluster of easily killed targets, followed by rapid subsequent kills with a short 〈Δt〉. (B) In a kinetic-priming model, the long t1 accounts for poor target recognition of the first target, followed by a change in effector state that leads to rapid subsequent kills with a short 〈Δt〉. (C) A mixture of fluorescent, well-killed HeLa targets on a background of nonfluorescent MCF7 targets mimics the scenario from the search-time model. (D) The changes in first kill time and number of successful effectors as a function of HeLa target dilution estimates the contribution of search time to the killing kinetics (see Results and Discussion). We set 〈t1〉 = 0 for 100% HeLa targets and report the relative 〈t1〉 for the other cases. (E) The average motility coefficient for effector cells on MCF7 targets is larger than on HeLa targets. (F) Killing is partially repressed at intermediate concentrations of the Src inhibitor PP2 (700 ng/mL; Sigma) and the Syk inhibitor BAY 61–3606 (500 ng/mL; Santa Cruz Biotechnology) after 6 h of incubation of 500 NK92-MI cells and 2 × 104 HeLa or 3 × 104 MCF7 cells per well of a 96-well plate. Samples sizes of 406, 143, and 218 total kills were measured for the control, Src-inhibitor, and Syk-inhibitor samples, respectively. (G) Average first kill times 〈t1〉 for untreated and inhibitor treated samples. Sample sizes of 104, 96, and 116 first kill times were measured for the control, Src-inhibitor, and Syk-inhibitor samples, respectively. (H) Average waiting times, 〈Δt〉, for treated and inhibitor treated samples. Sample sizes of 334, 100, and 157 waiting times were measured for the control, Src-inhibitor, and Syk-inhibitor samples, respectively. Waiting times are less sensitive than first kill times to partial Src and Syk inhibition, even when total killing is greatly reduced. All error bars in this figure reflect SEs.
A slow initial kill, followed by rapid subsequent kills describes bursting kinetics, in which events occur in temporally correlated clusters. Against MCF7 targets, within the imaging time constraints (SI Results and Discussion), most NK cells likely undergo only one burst of killing, such that an analysis of first killing times and subsequent waiting times is sensitive to bursting (Fig. 2C, Lower). Against HeLa targets, the rate of first kills is high enough that bursting is less significant (Fig. 2C, Upper). Further analysis of the waiting time distribution is consistent with bursting kinetics in both HeLa and MCF7 targets (SI Results and Discussion and Fig. S9).
Bursting kinetics can arise for several reasons (Fig. 3 A and B). We ruled out the first search-time model with a series of experiments. In the search-time model, the time to find the first target is long because only a small fraction of targets are killable (Fig. 3A). That first target, however, could be surrounded by a cluster of other killable targets, because of inheritable (sister cells) or noninheritable (neighbor–neighbor interactions) reasons. Such heterogeneous death responses have been reported for death ligand-induced apoptosis (20). To assess the search-time model, we examined killing in a mixture containing sparse, well-killed targets surrounded by poorly killed targets. Because the waiting times for HeLa and MCF7 targets are similar, the search-time model hypothesizes that HeLa targets and sparse, well-killed MCF7 targets are kinetically equivalent. We therefore plated different densities of the GzmB reporter HeLa line on a confluent background of unlabeled MCF7 cells (Fig. 3C) to construct a search-time scenario. We measured the relationship between 〈t1〉 and the number of successful effectors for the different HeLa densities (Fig. 3D) as well as MCF7. The data for MCF7 appears as an outlier and does not support the search-time model (SI Results and Discussion).
To further evaluate the search-time model, we also measured the motility coefficients of effector cells on HeLa and MCF7 targets to test whether slower motility contributes to a longer search time. We found that the motility coefficient of effectors was larger on MCF7 targets than on HeLa targets (Fig. 3E and Fig. S10), which would decrease the search time for MCF7, rendering the search-time model even more unlikely. Measured motility coefficients were also within the range of previously reported values for lymphocytes (21). Thus, the dilution results suggest that the search time for the first target is insufficient to explain the slow first kill time in MCF7 and associated bursting kinetics (SI Results and Discussion). As an additional test, we also found that exchanging conditioned media did not affect the kinetics (Fig. S11).
We propose kinetic priming as an alternative mechanism for bursting kinetics (Fig. 3B), where the kinetics of particular NK–target interactions depends on the recent history of killing events. In the absence of recent killing, the next kill is slow, and in the presence of recent killing, the next kill is fast. One possible mechanism for kinetic priming would be that gene expression driven by the first killing event increases the potency of the effector cell. Consistent with this possibility, NK receptor signaling is known to rapidly alter transcription factor activities (22). We found that treatment with cycloheximide had no effect on serial killing kinetics in our 6-h assay (Fig. S12), so new gene expression is not a factor in kinetic priming.
We compared the sensitivity of first and subsequent kills to small-molecule kinase inhibitors that inhibit NK signaling pathways. Killing was completely abolished by high doses of Src and Syk kinase inhibitors (Fig. S13 A–C), as expected since both families of kinases are involved in signaling downstream of NK receptors (23, 24). We found intermediate doses that partially blocked killing and verified that drug activities were stable for the course of an experiment (Fig. S13). We then examined the kinetics of partially inhibited killing. The fraction of NK cells that were active in killing decreased, but the active cells still exhibited serial killing (Fig. 3 F–H and Fig. S14). Moderate Src or Syk inhibition had little effect on time between kills, 〈Δt〉, but strongly increased the time to first kills, 〈t1〉, for HeLa targets (Fig. 3 G and H). Partial drug inhibition thus made killing kinetics for a good target (HeLa) similar to that of a poor target (MCF7) in the absence of drugs. Thus, some signaling effect from the first kill overcomes weak signaling in subsequent kills, whether the weak signaling is due to partial drug inhibition or a less-susceptible population. Short-term memory of a previous killing event, giving rise to kinetic priming, could explain this observation.
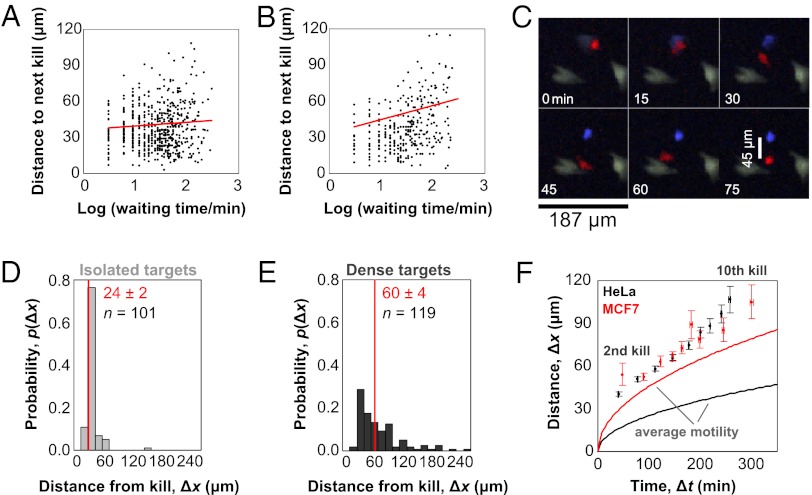
We next analyzed the spatial distribution of kills, because physical coordination between immune synapses might provide a possible memory mechanism for kinetic priming. Although waiting times between killing events spanned two orders of magnitude, the distance between subsequent targets was constant at approximately one target diameter or 40 microns (Fig. 4 A and B). The distance suggests that subsequent targets are almost always directly adjacent to the prior target, and it is possible that an NK cell simultaneously contacts both targets. Simultaneous contact with multiple targets has been reported for cytotoxic T lymphocytes (16, 25).
Fig. 4.
Spatiotemporal coordination of serial killing. (A and B) Scatterplots of distance and time between subsequent HeLa (A) or MCF7 (B) targets. Trendlines (red) of the log-transformed data indicate relatively uniform distances to the next target over two order of magnitude of waiting times. (C) The distance between the first observed kill (blue) and the effector (red) is measured at the end of a time course. (D and E) Five hundred fluorescent HeLa cells stably expressing the GzmB reporter are plated in the well of a 96-well plate either in isolation (D) or on a confluent background of nonfluorescent HeLa cells (E). Distances between the centers of effectors and their first fluorescent targets are recorded at the end of a 6-h assay. Effectors do not detach after killing isolated targets but readily release killed targets in the presence of dense adjacent targets. (F) The average distance and time interval between subsequent targets and the first target are plotted for HeLa (black dots) and MCF7 (red dots) to illustrate the propagation of serial killing activity. In comparison, the average cell motilities are plotted for HeLa (black line) and MCF7 (red line). Although average motilities differ on the two targets, killing activity propagates in a similar, accelerated manner.
We also recorded distances between effectors and their first target at the end of the assay (Fig. 4C). When HeLa targets are plated at very low densities, NK cells remain attached to their first targets and did not dissociate over 6 h (Fig. 4D and Movie S5). Persistent interaction and signaling between an effector and target has been observed (25). At high target densities, NK cells readily detached from old targets and moved to new targets (Fig. 4E and Movie S6). These data suggest that the interaction with an old target remains until interaction with a new target triggers detachment. Thus, a simple hypothetical scenario for kinetic priming is that an NK kills the first target and remains attached until encountering a second adjacent target, resulting in continuous signaling after the first kill that persists until formation of a new synapse. If the state of sustained signaling is broken, it would take a longer time to reinitiate killing, explaining the long waiting time events in the distribution of Fig. 2E and the long distance events in the scatterplot of Fig. 4B for MCF7 targets. In a three-dimensional solid tissue environment in vivo, however, the old effector-target conjugate would usually be in contact with a new potential target, so persistence at an isolated old target is unlikely to be observed.
Spatial analysis reveals one final consequence of bursting kinetics. We plotted the “motility” of killing activity by calculating the distance and time of subsequent targets from the first target (Fig. 4F) in comparison with the average motility of NK cells on HeLa and MCF7 targets. Although NK cells displayed different motilities on HeLa and MCF7 targets, during the course of serial killing, killing proceeded equally fast, not just in time, but also across distances for both target types. Thus, the kinetics of NK recycling between targets determines both serial killing and NK cell migration.
In summary, single cell analysis revealed that serial killing by an NK model showed bursting kinetics, with a long delay before the first kill followed by short times between subsequent kills. The boost provided by the first kill was able to overcome partial inhibition of signaling pathways, or the effects of a less attractive target. Spatial analysis suggested a possible mechanism underlying kinetic priming, and the decision to kill may often occur in the context of a ternary complex, consisting of the effector, old target, and new target. In this proposed scenario, the old target helps signal, and the new target accelerates detachment from the old target. Our simplified experimental model enabled quantitative characterization of higher-level regulatory principles (26) that can have broader implications in the complex environment of tissues. For example, in a tumor, individual tumor cells can have diverse phenotypes, and context-dependent signaling may impact cancer-immune responses through bystander immune killing (27, 28) or other mechanisms.
The behavior of individual CTL or NK cells infiltrating solid tumors has been imaged in mouse models (29). Despite the differences between the in vivo and in vitro models and their accompanying cell types, some results are similar. CTLs were found to form stable contacts with tumor cells, in contrast to NK cells, which formed transient contacts (29). Direct observation of killing events is difficult in vivo, but in vitro work suggests that CTLs may be able to simultaneously bind and attack multiple targets, with each synapse operating independently (25). Thus, the spatial coordination of serial killing observed for CTLs in vitro is consistent with the in vivo observation of stable CTL-target contacts. Our in vitro work shows that NK cells release old targets when attacking new targets (Fig. 4E), and the release is consistent with the in vivo observation of transient NK-target contacts.
Intravital imaging studies of single lymphocytes have tended to focus on motility and binding properties, because direct observations of single-cell tumor killing events in vivo is technically challenging. As a surrogate, Mempel et al. (30) imaged individual CTL attacks of antigen-pulsed B cells in mouse lymph nodes, staining B cells by lysis-reporter dyes before injection. For direct observation of CTL killing in solid tumors, Breart et al. (31) used a genetically encoded caspase-3 FRET reporter. In both cases, however, the total number of observed killing events was limited to tens of events, restricting the statistical analysis of single-cell killing kinetics.
The use of planar adherent cells and low magnification, in conjunction with a rapid and specific granzyme reporter, allowed our in vitro approach to provide detailed mechanistic analysis of single-cell killing kinetics, complementary to the in vivo observations. Aspects of our in vitro approach may be of value for in vivo imaging. For example, the cited intravital works used short windows of continuous imaging ranging from 35 to 60 min (30, 31). It is possible that many targets attacked by CTLs did not lyse or sufficiently activate caspases in that narrow timeframe. Furthermore, cells in a tumor environment may also have different rates or capacities for lysis or caspase-dependent death. A granzyme reporter, as used in our work, is a more rapid and direct readout of cytotoxic attack and could aid in more efficient detection of individual killing events in vivo.
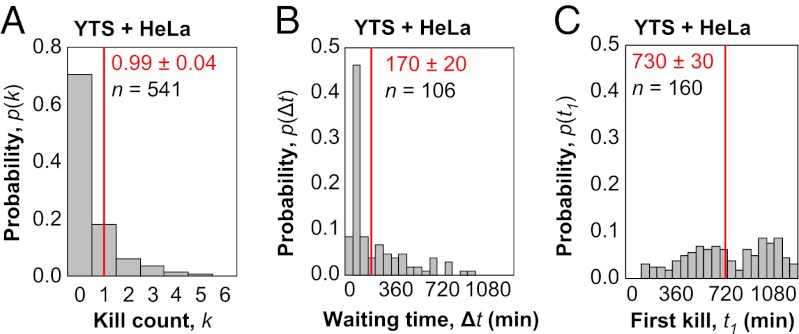
The main advantage of the method we present, however, is for detailed analysis of single-cell killing dynamics in vitro. NK cells from different cell lines, growth conditions, and histories are expected to have diverse responses to potential target cells, and our approach can measure a range of responses. To demonstrate the method's utility, we imaged single-cell killing by the YTS NK line against HeLa targets (Movie S7 and Fig. S15). Killing of HeLa targets by YTS effectors was much poorer than killing by NK92-MI effectors, and less than one-third of YTS cells participated in killing over 18 h of imaging. Of the YTS cells that did kill, approximately 40% participated in serial killing of more than one target, with the degree of serial killing reaching up to five consecutive kills, lower than NK92-MI achieved over a shorter 6-h window (Fig. 5A). The waiting time histogram for serial killing by YTS cells strongly peaked at 60 min (Fig. 5B). The first killing time, however, was relatively uniform across the entire 18 h of imaging (Fig. 5C). Thus, slow first kill times, and greatly accelerated subsequent kills, is even more apparent in YTS cells than NK92-MI cells. Because the frequency and absolute rate of killing events made collection of sufficient statistics slow for YTS cells, we did not provide the same detailed analysis as for NK92-MI cells. However, the main observation of bursting kinetics is still observed in YTS cells, although on a different timescale. In addition, although we used low resolution imaging for collection of serial killing statistics, higher resolution imaging with the GzmB reporter is possible as well (Fig. S16). Thus, our method distinguishes diverse outcomes between various effector-target combinations at the level of individual killing events, and future work can explore how microscopic mechanisms contribute to these specific quantitative outcomes.
Fig. 5.
Bursting kinetics in serial killing by the YTS NK line on a slower timescale. (A) Histogram of kill count, k, for the total number of targets killed by individual effector cells over 18 h of coincubation of 1,000 YTS cells and 2 × 104 HeLa cells per well of a 96-well plate. For all following histograms, mean values and associated SEs are indicated in red, whereas n indicates the total number of YTS effectors profiled. (B) Histogram of waiting times for HeLa targets attacked by YTS effectors. The sample size n denotes the total number of waiting times recorded when two killing events occur in succession. (C) Histogram of first kill times for HeLa targets attacked by YTS effectors. The sample size n denotes the total number of first kill times measured, which is equivalent to the number of NK cells with k > 0. When serial killing occurs, the waiting time between subsequent kills is typically much faster than the time needed for the first kill.
Materials and Methods
Cell Lines and Plasmids.
NK92-MI cells (ATCC; CRL-2408) were propagated according to the provider’s instructions, YTS cells (gift of J. Strominger, Harvard University, Cambridge, MA) in RPMI with 10% (vol/vol) FBS, HeLa cells in DMEM with 10% (vol/vol) FBS, and MCF7 cells in RPMI with 10% (vol/vol) FBS. Coculture imaging experiments were performed in RPMI with 10% (vol/vol) FBS. Media components for all experiments were obtained from Cellgro (RPMI, 10-040 and DMEM, 10-014) and Invitrogen (FBS, 26140-079). Standard techniques were used to construct the CFP-VGDPFGR-YFP GzmB reporter in the pLNCX2 (Clontech) retroviral backbone, followed by infection of HeLa and MCF7 cells and clonal selection to generate the HeLa-GzmB and MCF7-GzmB lines. CFP and YFP genes were PCR amplified from the effector caspase reporter protein (EC-RP) plasmid (ref. 32; gift of P. Sorger, Harvard Medical School, Cambridge, MA). The calcium indicator R-GECO1 from CMV-R-GECO1 (ref. 33; Addgene 32444) was transferred to the lentiviral backbone, after excision of the existing GFP-RIF1 insert, from pHR’CMVGFP_hRIF1(406-2446)IREShygro (ref. 34; Addgene 23135). Infection of HeLa targets stably expressing the GzmB reporter and flow sorting of positive clones generates a cell line with simultaneous red calcium and GzmB reporters. The calcium indicator GCaMP3 (ref. 35; Addgene 22692) was transferred to the lentiviral backbone, after excision of the existing GFP insert, from pLenti-CMV-GFP (ref. 36; Addgene 17447). Infection of HeLa targets and selection by G418 resulted in cells stably expressing the green calcium reporter.
Microscopy.
Imaging was performed in an enclosed incubator at 37 °C and 5% CO2 on a Nikon Ti microscope with a motorized stage, Plan Fluor 10×/0.3 N.A. or Plan Apo 4×/0.2 N.A. objective, Hamamatsu ORCA-AG CCD camera, Intensilight light source, the NIS Elements software, and 96-well plates. Images were collected with 2 × 2 pixel binning. FRET imaging was performed with a dual CFP/YFP dichroic (Chroma 89002) and standard excitation and emission filters. Labeled effector cells were imaged with an RFP filter set (Chroma). Before imaging, effector cells were stained in 1 μM CellTracker Orange CMRA (Invitrogen) for 15 min in serum-free media at 37 C, followed by 1 h of recovery in complete media. NK cells in 50 μL of media were added to wells of a 96-well plate containing adherent target cells and 50 μL of media. X, Y, and Z positions were then set for automated time-lapse recording, typically for approximately 40 positions in one experiment, as limited by the number of positions that may be scanned within the acquisition rate. Time-lapse movies of NK92-MI cells were imaged every 3 min for a total of 6 h, whereas time-lapse movies of YTS cells were imaged every 10 min for a total of 18 h.
Data Analysis.
Image processing was performed with the aid of NIS Elements (Nikon) and MetaMorph (Molecular Devices). Spatial tracking was performed by using the tracking module in NIS Elements with manual correction. Location and timing of target killing events were identified manually. Custom Python scripts were used to extract motility coefficients and killing statistics.
Supplementary Material
Acknowledgments
We thank J. Tallaric, Y. Feng, and J. Lieberman for discussions; J. Paulsson for critical reading of the manuscript; and members of the T.J.M. group for technical assistance. The Nikon Imaging Center at Harvard Medical School provided assistance with microscopy. This work is supported by a collaborative arrangement with Novartis Institutes for Biomedical Research.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1221312110/-/DCSupplemental.
References
- 1.Dunn GP, Bruce AT, Ikeda H, Old LJ, Schreiber RD. Cancer immunoediting: From immunosurveillance to tumor escape. Nat Immunol. 2002;3(11):991–998. doi: 10.1038/ni1102-991. [DOI] [PubMed] [Google Scholar]
- 2.Grakoui A, et al. The immunological synapse: A molecular machine controlling T cell activation. Science. 1999;285(5425):221–227. doi: 10.1126/science.285.5425.221. [DOI] [PubMed] [Google Scholar]
- 3.Davis DM, et al. The human natural killer cell immune synapse. Proc Natl Acad Sci USA. 1999;96(26):15062–15067. doi: 10.1073/pnas.96.26.15062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zagury D, Bernard J, Thierness N, Feldman M, Berke G. Isolation and characterization of individual functionally reactive cytotoxic T lymphocytes: Conjugation, killing and recycling at the single cell level. Eur J Immunol. 1975;5(12):818–822. [Google Scholar]
- 5.Sanderson CJ. The mechanism of T cell mediated cytotoxicity. II. Morphological studies of cell death by time-lapse microcinematography. Proc R Soc Lond B Biol Sci. 1976;192(1107):241–255. doi: 10.1098/rspb.1976.0011. [DOI] [PubMed] [Google Scholar]
- 6.Rothstein TL, Mage M, Jones G, McHugh LL. Cytotoxic T lymphocyte sequential killing of immobilized allogeneic tumor target cells measured by time-lapse microcinematography. J Immunol. 1978;121(5):1652–1656. [PubMed] [Google Scholar]
- 7.Bhat R, Watzl C. Serial killing of tumor cells by human natural killer cells—enhancement by therapeutic antibodies. PLoS ONE. 2007;2(3):e326. doi: 10.1371/journal.pone.0000326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Isaaz S, Baetz K, Olsen K, Podack E, Griffiths GM. Serial killing by cytotoxic T lymphocytes: T cell receptor triggers degranulation, re-filling of the lytic granules and secretion of lytic proteins via a non-granule pathway. Eur J Immunol. 1995;25(4):1071–1079. doi: 10.1002/eji.1830250432. [DOI] [PubMed] [Google Scholar]
- 9.Brunner KT, Mauel J, Cerottini JC, Chapius B. Quantitative assay of the lytic action of immune lymphoid cells on 51Cr-labelled allogenic target cells in vitro; Inhibition by isoantibody and by drugs. Immunol. 1968;14(2):181–196. [PMC free article] [PubMed] [Google Scholar]
- 10.Goldberg JE, Sherwood SW, Clayberger C. A novel method for measuring CTL and NK cell-mediated cytotoxicity using annexin V and two-color flow cytometry. J Immunol Methods. 1999;224(1-2):1–9. doi: 10.1016/s0022-1759(98)00038-6. [DOI] [PubMed] [Google Scholar]
- 11.Chowdhury D, Lieberman J. Death by a thousand cuts: Granzyme pathways of programmed cell death. Annu Rev Immunol. 2008;26:389–420. doi: 10.1146/annurev.immunol.26.021607.090404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thiery J, et al. Perforin pores in the endosomal membrane trigger the release of endocytosed granzyme B into the cytosol of target cells. Nat Immunol. 2011;12(8):770–777. doi: 10.1038/ni.2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thornberry NA, et al. A combinatorial approach defines specificities of members of the caspase family and granzyme B. Functional relationships established for key mediators of apoptosis. J Biol Chem. 1997;272(29):17907–17911. doi: 10.1074/jbc.272.29.17907. [DOI] [PubMed] [Google Scholar]
- 14.Packard BZ, Telford WG, Komoriya A, Henkart PA. Granzyme B activity in target cells detects attack by cytotoxic lymphocytes. J Immunol. 2007;179(6):3812–3820. doi: 10.4049/jimmunol.179.6.3812. [DOI] [PubMed] [Google Scholar]
- 15.Tam YK, et al. Characterization of genetically altered, interleukin 2-independent natural killer cell lines suitable for adoptive cellular immunotherapy. Hum Gene Ther. 1999;10(8):1359–1373. doi: 10.1089/10430349950018030. [DOI] [PubMed] [Google Scholar]
- 16.Zagury D, Bernard J, Jeannesson P, Thiernesse N, Cerottini JC. Studies on the mechanism of T cell-mediated lysis at the single effector cell level. I. Kinetic analysis of lethal hits and target cell lysis in multicellular conjugates. J Immunol. 1979;123(4):1604–1609. [PubMed] [Google Scholar]
- 17.Varadarajan N, et al. A high-throughput single-cell analysis of human CD8+ T cell functions reveals discordance for cytokine secretion and cytolysis. J Clin Invest. 2011;121(11):4322–4331. doi: 10.1172/JCI58653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Macken CA, Perelson AS. A multistage model for the action of cytotoxic T lymphocytes in multicellular conjugates. J Immunol. 1984;132(4):1614–1624. [PubMed] [Google Scholar]
- 19.Pollock RE, Zimmerman SO, Fuchshuber P, Lotzová E. Lytic units reconsidered: Pitfalls in calculation and usage. J Clin Lab Anal. 1990;4(4):274–282. doi: 10.1002/jcla.1860040408. [DOI] [PubMed] [Google Scholar]
- 20.Spencer SL, Gaudet S, Albeck JG, Burke JM, Sorger PK. Non-genetic origins of cell-to-cell variability in TRAIL-induced apoptosis. Nature. 2009;459(7245):428–432. doi: 10.1038/nature08012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cahalan MD, Parker I. Choreography of cell motility and interaction dynamics imaged by two-photon microscopy in lymphoid organs. Annu Rev Immunol. 2008;26:585–626. doi: 10.1146/annurev.immunol.24.021605.090620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pandey R, DeStephan CM, Madge LA, May MJ, Orange JS. NKp30 ligation induces rapid activation of the canonical NF-kappaB pathway in NK cells. J Immunol. 2007;179(11):7385–7396. doi: 10.4049/jimmunol.179.11.7385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lanier LL. Up on the tightrope: Natural killer cell activation and inhibition. Nat Immunol. 2008;9(5):495–502. doi: 10.1038/ni1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Watzl C, Long EO. 2010. Signal transduction during activation and inhibition of natural killer cells. Curr Prot Immunol 90:11.9B.1–11.9B.17.
- 25.Wiedemann A, Depoil D, Faroudi M, Valitutti S. Cytotoxic T lymphocytes kill multiple targets simultaneously via spatiotemporal uncoupling of lytic and stimulatory synapses. Proc Natl Acad Sci USA. 2006;103(29):10985–10990. doi: 10.1073/pnas.0600651103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Benoist C, Germain RN, Mathis D. A plaidoyer for ‘systems immunology’. Immunol Rev. 2006;210:229–234. doi: 10.1111/j.0105-2896.2006.00374.x. [DOI] [PubMed] [Google Scholar]
- 27.Burrows SR, Fernan A, Argaet V, Suhrbier A. Bystander apoptosis induced by CD8+ cytotoxic T cell (CTL) clones: Implications for CTL lytic mechanisms. Int Immunol. 1993;5(9):1049–1058. doi: 10.1093/intimm/5.9.1049. [DOI] [PubMed] [Google Scholar]
- 28.Ando K, et al. Perforin, Fas/Fas ligand, and TNF-α pathways as specific and bystander killing mechanisms of hepatitis C virus-specific human CTL. J Immunol. 1997;158(11):5283–5291. [PubMed] [Google Scholar]
- 29.Deguine J, Breart B, Lemaître F, Di Santo JP, Bousso P. Intravital imaging reveals distinct dynamics for natural killer and CD8(+) T cells during tumor regression. Immunity. 2010;33(4):632–644. doi: 10.1016/j.immuni.2010.09.016. [DOI] [PubMed] [Google Scholar]
- 30.Mempel TR, et al. Regulatory T cells reversibly suppress cytotoxic T cell function independent of effector differentiation. Immunity. 2006;25(1):129–141. doi: 10.1016/j.immuni.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 31.Breart B, Lemaître F, Celli S, Bousso P. Two-photon imaging of intratumoral CD8+ T cell cytotoxic activity during adoptive T cell therapy in mice. J Clin Invest. 2008;118(4):1390–1397. doi: 10.1172/JCI34388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Albeck JG, et al. Quantitative analysis of pathways controlling extrinsic apoptosis in single cells. Mol Cell. 2008;30(1):11–25. doi: 10.1016/j.molcel.2008.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao Y, et al. An expanded palette of genetically encoded Ca2+ indicators. Science. 2011;333(6051):1888–1891. doi: 10.1126/science.1208592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu L, Blackburn EH. Human Rif1 protein binds aberrant telomeres and aligns along anaphase midzone microtubules. J Cell Biol. 2004;167(5):819–830. doi: 10.1083/jcb.200408181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tian L, et al. Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nat Methods. 2009;6(12):875–881. doi: 10.1038/nmeth.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Campeau E, et al. A versatile viral system for expression and depletion of proteins in mammalian cells. PLoS ONE. 2009;4(8):e6539. doi: 10.1371/journal.pone.0006529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heim R, Tsien RY. Engineering green fluorescent protein for improved brightness, longer wavelengths and fluorescence resonance energy transfer. Curr Biol. 1996;6(2):178–182. doi: 10.1016/s0960-9822(02)00450-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.