Abstract
Precursor proteins of the solute carrier family and of channel forming Tim components are imported into mitochondria in two main steps. First, they are translocated through the TOM complex in the outer membrane, a process assisted by the Tim9/Tim10 complex. They are passed on to the TIM22 complex, which facilitates their insertion into the inner membrane. In the present study, we have analyzed the function of the Tim9/Tim10 complex in the translocation of substrates across the outer membrane of mitochondria. The purified TOM core complex was reconstituted into lipid vesicles in which purified Tim9/Tim10 complex was entrapped. The precursor of the ADP/ATP carrier (AAC) was found to be translocated across the membrane of such lipid vesicles. Thus, these components are sufficient for translocation of AAC precursor across the outer membrane. Peptide libraries covering various substrate proteins were used to identify segments that are bound by Tim9/Tim10 complex upon translocation through the TOM complex. The patterns of binding sites on the substrate proteins suggest a mechanism by which portions of membrane-spanning segments together with flanking hydrophilic segments are recognized and bound by the Tim9/Tim10 complex as they emerge from the TOM complex into the intermembrane space.
INTRODUCTION
Targeting and translocation of most nuclear-encoded mitochondrial proteins depend on cleavable N-terminal extensions referred to as mitochondrial targeting sequences or presequences (for recent reviews, see Neupert, 1997; Pfanner and Geissler, 2001). However, all proteins of the mitochondrial outer membrane and some proteins of the intermembrane space (IMS) and the inner membrane are devoid of such signals. Among the latter proteins, the family of solute carrier proteins together with some Tim components comprise a subclass of inner membrane proteins that uses a unique pathway for mitochondrial import (Koehler et al., 1999b; Tokatlidis and Schatz, 1999). After their synthesis on cytosolic ribosomes, these precursor proteins are protected against aggregation by interaction with molecular chaperones which also help to direct them to the import receptor Tom70 (Komiya et al., 1997; Young et al., 2003). From the receptor protein, the carrier precursor is transferred to the outer membrane translocation pore. Carrier precursor proteins, like other mitochondrial precursors with internal import signals, cross the TOM complex in a loop structure (Endres et al., 1999; Wiedemann et al., 2001; Curran et al., 2002b; Stan et al., 2003). On their exit from the TOM complex, these precursors interact with complexes of so-called small Tim components. Tim9 and Tim10 are two such proteins. They facilitate the transfer of precursors across the intermembrane space and deliver them to the membrane embedded TIM22 complex (Koehler et al., 1998a; Koehler et al., 1998b; Sirrenberg et al., 1998; Adam et al., 1999; Koehler et al., 1999b; Leuenberger et al., 1999; Tokatlidis and Schatz, 1999; Jensen and Dunn, 2002). The complex in yeast is a heterohexamer which contains three Tim9 and three Tim10 molecules (Luciano et al., 2001; Curran et al., 2002a; Vial et al., 2002). Although highly conserved homologues of Tim9 and Tim10 were found in the genome of many other eukaryotic organisms (Bauer et al., 1999; Kayingo et al., 2000), the Tim9/Tim10 complex has so far been analyzed only in S. cerevisiae. Two other small Tim components, Tim8 and Tim13, also form a heterohexameric complex in the intermembrane space (Koehler et al., 1999a; Kurz et al., 1999; Leuenberger et al., 1999). In contrast to Tim9/Tim10, in yeast they are not essential for viability, and seem to be involved in the import of only a limited number of precursor proteins, such as Tim23 (Davis et al., 2000; Paschen et al., 2000; Curran et al., 2002b; Jensen and Dunn, 2002).
Previous studies have demonstrated the importance of Tim9 and Tim10 for the release of the carrier preprotein from the TOM complex and its insertion into the inner membrane (Luciano et al., 2001; Truscott et al., 2002). It seems that the TOM and Tim9/Tim10 complexes are necessary to translocate the AAC precursor across the outer membrane. However, it is still unclear whether they accomplish this transfer with the help of additional, yet unknown component(s) or whether these two complexes are sufficient for mediating the process. To answer this question we identified and analyzed the Tim9/Tim10 complex from N. crassa. We used a reconstituted system where the TOM core complex was reconstituted into lipid vesicles into which the purified Tim9/Tim10 complex was entrapped. The precursor of ADP/ATP carrier could be translocated across the membrane of these lipid vesicles. Hence, these import components are sufficient for translocation of AAC precursor across the outer membrane.
The ability of the Tim9/Tim10 complex to facilitate membrane translocation of carrier proteins raises the question as to how the Tim9/Tim10 complex interacts with its various precursor substrates. One previous suggestion was that the complex recognizes a specific conserved sequence motif found in each of the three IMS loops of carrier proteins (Sirrenberg et al., 1998; Endres et al., 1999). Alternatively, the Tim9/Tim10 complex could function as a chaperone in the intermembrane space by binding exposed hydrophobic sequences of unfolded precursor proteins. Two recent reports support the latter hypothesis. A recombinant Tim9/Tim10 complex was reported to have a moderate general chaperone activity (Vial et al., 2002). In another study, the Tim9/Tim10 complex was used to screen a library of peptides covering the ADP/ATP carrier; and preferential binding to the transmembrane domains was observed (Curran et al., 2002a). Because all the above-mentioned data were obtained only in the yeast system and the latter study analyzed the binding to only one substrate protein, it is an open question whether these findings can be extrapolated to other substrates and to Tim9/Tim10 complexes from other organisms. To address this question, we have used the native Tim9/Tim10 complex of Neurospora crassa to screen a library of peptides covering various precursor proteins of the mitochondrial inner membrane. Based on our results we propose a working model for how the Tim9/Tim10 complex recognizes its substrates and helps to guide them from the TOM to the TIM22 complex.
MATERIALS AND METHODS
Cloning of N. crassa TIM9 and TIM10
The amino acid sequence of the Saccharomyces cerevisiae Tim10 protein was used as a query to identify an N. crassa homologue in a BLAST search of N. crassa expressed sequence tag databases (http://www.genome.ou.edu/fungal.html). The assembly of three overlapping expressed sequence tags led to a full-length cDNA sequence. The open reading frame (ORF) was amplified in a polymerase chain reaction (PCR) by using a cDNA library from N. crassa as template. A 300-base pair PCR fragment was inserted into the vector pGEM4 (Promega, Madison, WI). The intron-containing gene was amplified in a PCR reaction with genomic DNA as template to generate a tim10 specific Digoxigenin (Roche Applied Science, Indianapolis, IN) labeled probe, and used to screen the N. crassa cosmid library pMOcosX (Fungal Genetics Stock Center, Department of Microbiology, University of Kansas Medical Center, Kansas City, KS). A 3.5-kb fragment containing the tim10 gene of cosmid X25:B10 was sequenced.
N. crassa tim9 gene was found in a BLAST search (http://wwwgenome.wi.mit.edu/annotation/fungi/neurospora/) by using the S. cerevisiae Tim9 protein sequence as a query. The ORF was amplified from a cDNA library in a PCR reaction as described above and inserted into the vector pGEM4. On comparison of the cDNA to the genomic sequence, the existence of two predicted introns was verified.
To create an expression vector for N. crassa a segment containing the tim10 ORF and its upstream 500 base pairs was amplified by PCR and cloned into plasmid pCB1179 (Fungal Genetics Stock Center). A histidinyl tag at the C terminus of the protein was added by using a 3′ primer that contained nine His codons before the stop codon. The downstream 1-kb sequence was amplified by PCR and cloned into the same plasmid, yielding the expression vector pNcTim10His9. Transformation into N. crassa cells and selection of transformants were performed as described previously (Prokisch et al., 2000).
Creation of Sheltered Repeat Induced Point (RIP) Mutant of tim9
The procedure of sheltered RIP mutation was used to inactivate the tim9 gene. Sheltered RIP ensures that nuclei containing nonfunctioning alleles are present in a heterokaryon that also contains nuclei with a wild-type copy of the gene. The rationale and strains used for sheltered RIP are described in detail elsewhere (Harkness et al., 1994). To identify sheltered heterokaryons containing RIPed alleles of the gene, the tim9 region of the genome in these strains was amplified by PCR and the products sequenced directly. One of the strains showing evidence of RIP in the tim9 gene (Tim9RIPedH3-11) was chosen for further analysis.
Biochemical Procedures
Isolation of mitochondria from N. crassa was performed as described previously (Mayer et al., 1993). For production of antibodies against N. crassa Tim9 and Tim10 both proteins were cloned into the pMalCRI plasmid, and expressed in Escherichia coli as fusion proteins with maltose binding protein. Antibodies were raised by injecting the corresponding purified proteins into rabbits. Blotting to polyvinylidene difluoride (PVDF) or nitrocellulose membranes and immunodecoration were according to standard procedures and visualization was by the ECL method (Amersham Biosciences, Piscataway, NJ).
For cross-linking experiments of the purified Tim9/Tim10 complex, glutaraldehyde was added for the indicated time periods at 25°C. Excess cross-linker was quenched by the addition of glycine, pH 8.8, to 100 mM, and the reactions were kept for 15 min at 25°C. Aliquots were removed before and after addition of the cross-linking reagent. Cross-linking of Tim23 precursor was performed by adding 500 μM m-maleimidobenzoyl-N-hydroxysuccinimide ester (MBS) during incubation of the precursor with deenergized mitochondria. After 30 min at 15°C, excess cross-linker was quenched as described above, and immunoprecipitation was performed as described previously (Rapaport et al., 1997). Coimmunoprecipitation experiments were performed by dissolving mitochondria (75 μg) to a final concentration of 2.5 mg/ml in a buffer containing 0.5% digitonin, 0.05% bovine serum albumin, 100 mM NaCl, 1 mM phenylmethylsulfonyl fluoride (PMSF), 25 μM zinc acetate, 50 mM NaPi, pH 8.0, for 1 h at 4°C. The solubilized mitochondria were centrifuged (20 min; 90,700 × g), and the supernatant was incubated for 1 h with antibodies prebound to protein A-Sepharose beads. The protein A-Sepharose beads were then washed twice with digitonin-containing buffer, and bound proteins were eluted with sample buffer and subjected to SDS-PAGE. The gels were blotted and immunodecorated with antibodies against various mitochondrial proteins.
Size-exclusion chromatography of mitochondrial proteins was performed by solubilizing 1 mg of mitochondria in 200 μl of buffer containing 0.5% β-dodecyl maltoside, 20 mM HEPES, 50 mM NaCl, 2.5 mM MgCl2, 1 mM EDTA, 10% glycerol, pH 7.4. Unsolubilized material was pelleted, and the supernatant was loaded on a Superose-6 size-exclusion column (Amersham Biosciences). Proteins were eluted with the same buffer containing 0.05% β-dodecyl maltoside, and fractions of 500 μl were collected.
Blue native gel electrophoresis (BNGE) was performed using a 6-16% linear polyacrylamide gradient (Schägger et al., 1994). Mitochondria were solubilized in a buffer containing 0.5% digitonin, 50 mM NaCl, 1 mM Tris-(2-carboxyethyl)phopshine (TCEP), 1 mM PMSF, 25 μM zinc acetate, 50 mM NaPi, 10% glycerol, pH 8.0, for 1 h at 4°C. The solubilized mitochondria were centrifuged (20 min; 90,700 × g), and the supernatant was analyzed by BNGE.
Import of Preproteins into Isolated Mitochondria
Radiolabeled precursor proteins were synthesized in rabbit reticulocyte lysate in the presence of [35S]methionine (Amersham Biosciences) after in vitro transcription by SP6 polymerase from pGEM4 vectors containing the gene of interest. Protein import in yeast mitochondria was performed in import buffer [0.25% BSA (wt/vol), 0.6 M sorbitol, 50 mM HEPES-KOH, 80 mM KCl, 10 mM Mg acetate, 2 mM KH2PO4, 2 mM ATP, 5 mM NADH, pH 7.2]. Protease treatment of mitochondria was performed by incubation with proteinase K (PK) for 15 min on ice, followed by addition of 1 mM PMSF. Import was analyzed by SDS-PAGE, autoradiography, and densitometry.
Purification of Tim9/Tim10 Complex
Isolated mitochondria (30-50 g of proteins) were solubilized in 3-5 liters of buffer A containing 300 mM NaCl, 20 mM imidazole, 10% glycerol, 1 mM PMSF, 1 mM TCEP, 1% Triton X-100, 50 mM MOPS-NaOH, pH 8.0. After 1 h incubation, a clarifying spin was performed to remove unsolubilized mitochondria (50 min; 15,900 × g). The solubilized material was loaded at a flow rate of 5.5 ml/min onto a Superflow Ni-NTA agarose column (QIAGEN, Valenica, CA). The column was then washed with six column volumes of buffer A containing 40 mM imidazole, and 25 μM zinc acetate, followed by another 14 volumes of buffer A with 10 mM NaCl. The column was eluted with buffer containing 20 mM Tris acetate, 300 mM imidazole, 10% glycerol, 1 mM TCEP, 25 μM zinc acetate, pH 7.5. The fractions containing the Tim9/Tim10 complex were pooled and loaded onto a ResourceQ ion-exchanger column. The flow-through fractions of this column contained Tim9/Tim10 complex, whereas the contaminants left after the Ni-NTA column were bound to the ion-exchange column and eluted with higher salt concentrations. The fractions containing the Tim9/Tim10 complex were dialyzed against a buffer containing 20 mM Tris acetate, 25 μM zinc acetate, pH 7.5, and concentrated using 10-kDa cut-off Centricones (PALL, Boston, MA).
Reconstitution of the TOM and Tim9/Tim10 Complex into Liposomes
Lipid vesicles were prepared from E. coli lipids by sonication. Lipids were purchased in chloroform/methanol solution (Avanti Polar Lipids, Birmingham, AL). The solvents were evaporated under a stream of nitrogen, and residual organic solvents were removed under high vacuum for 5 h. The lipids were resuspended in water by vortex mixing, and the resulting lipid suspension was sonicated on ice under nitrogen atmosphere for 10 min. The lipid concentration of the solution was determined by phosphorus analysis to be ca. 10 mg/ml.
Proteoliposomes were prepared from these lipid vesicles. TOM core complex was isolated as described previously (Ahting et al., 1999). Liposomes (3.2 mg/ml) were solubilized with β-dodecyl maltoside [0.2% (wt/vol)] at room temperature for 30 min. Then, TOM core complex (400 μg/ml) was added and incubated further for 30 min under gentle stirring. In some cases, the reconstitution mix also contained the purified Tim9/Tim10 complex (100-200 μg/ml) or lactate dehydrogenase (40 μg/ml). The detergent was then adsorbed onto SM2 Bio-Beads (Bio-Rad, Hercules, CA) at room temperature for 2 h. To remove nonreconstituted proteins, proteoliposomes were finally centrifuged through a 0-18% sucrose gradient at 215,000 × g at 2°C. Proteoliposomes were harvested at a sucrose concentration of ∼15%.
Isolation of ScAAC2
Samples enriched in AAC2 were prepared according to published procedure with some modification (Klingenberg et al., 1995). Total membrane fraction from yeast wild-type cells was collected by differential centrifugation after cell disruption by glass beads. Lysate cleaned from glass beads and unbroken cells was centrifuged at 40,000 × g for 60 min at 4°C. The pelleted membranes were resuspended at a concentration of 10 mg/ml in buffer containing 6% Triton X-100, 100 mM Na2SO4, 50 mM Tris-HCl, 1 mM EDTA, proteases inhibitors mix (Complete; Roche Applied Science), pH 7.4. The solubilized membranes were loaded on a 50-ml hydroxyapatite column. The flow-through of this column was collected and was dialyzed extensively against buffer containing 100 mM sucrose, 10 mM MOPS, 80 mM KCl, 5 mM MgCl2, 0.1% Triton X-100, pH 7.2, and concentrated on a C30-Centricon.
Screening of Peptide Scans with Tim9/Tim10 Complex
The Tim9/Tim10 complex was purified as described above. Cellulose-bound peptide scans were prepared by automated spot synthesis by Jerini AG (Berlin, Germany) (Frank, 1992; Kramer and Schneider-Mergener, 1998). Multiple 13mer peptides with a 10-amino acid residue overlap were synthesized according to the sequence of the indicated proteins and were attached to the cellulose membrane via their C terminus. The membranes were incubated with 150 nM of the isolated Tim9/Tim10 complex in binding buffer as described previously (Brix et al., 1999; Stan et al., 2003). After washing, the bound proteins were transferred to a PVDF membrane, followed by detection with antibodies against either Tim9 or Tim10. Binding was quantified by scanning laser densitometry. The mean of at least three independent experiments for each peptide was used.
RESULTS
tim9 and tim10 Genes of N. crassa
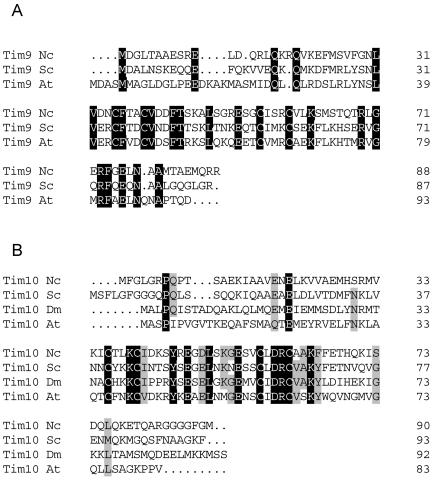
The amino acid sequences of N. crassa Tim9 and Tim10 were determined based on a BLAST search by using the sequences of yeast Tim9 and Tim10 as queries. Primers were constructed according to the sequences obtained from the database, and the cDNAs were amplified using a N. crassa cDNA library as template (see details in MATERIALS AND METHODS). Based on comparison of the Tim9 cDNA to the corresponding region on the chromosome two introns were identified (GenBank accession no. AY141127). The Tim10 gene is interrupted by one intron (GenBank accession no. AF343077). Both proteins contain the conserved twin CX3C motif characteristic for all predicted small Tim proteins (Figure 1) (Bauer et al., 1999). N. crassa Tim9 comprises 88-amino acid residues and has 40% identity to yeast Tim9 (Figure 1A), whereas the Tim10 gene encodes a protein of 90-amino acid residues that is 44% identical to the yeast Tim10 (Figure 1B). Our searches in various databases failed to identify a N. crassa homologue of Tim12, an essential protein in yeast that facilitates the transfer of carrier proteins from Tim9/Tim10 to the TIM22 complex (Koehler et al., 1998a; Sirrenberg et al., 1998). In fact, Tim12 has not been found in any organism other than S. cerevisiae (Bauer et al., 1999; Lister et al., 2002).
Figure 1.
Deduced amino acid sequences of N. crassa Tim9 and Tim10. Protein sequence alignments of N. crassa Tim9 (A) and Tim10 (B) with homologues from other organisms are presented. Identical residues occurring in all organisms are indicated in black. Similar residues are indicated by gray background. Nc, Neurospora crassa; Sc, S. cerevisiae; At, Arabidopsis thaliana; Dm, Drosophila melanogaster.
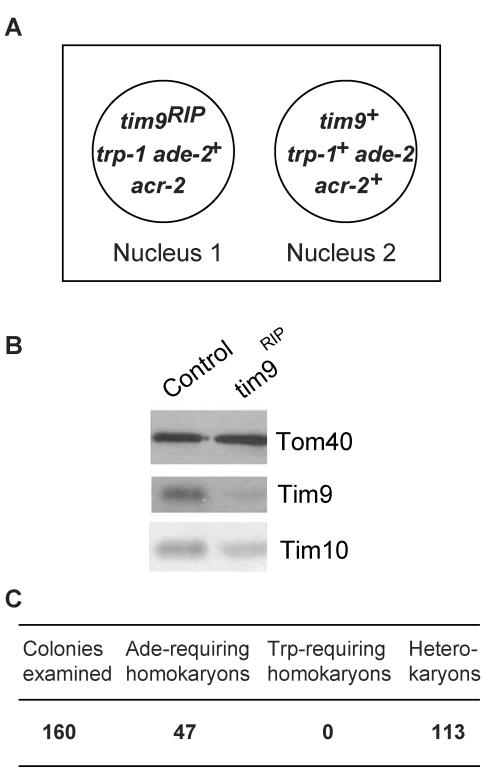
Because Tim9 was shown to be the product of an essential gene in yeast (Koehler et al., 1998b; Adam et al., 1999), we used the procedure of sheltered RIP to generate tim9 mutants in N. crassa. The product of the procedure was a heterokaryotic strain in which the tim9 gene in one nucleus was inactivated by RIP, whereas the other nucleus retained a wild-type copy of the gene (Figure 2A). Growth of the heterokaryon in the presence of acriflavine and tryptophan resulted in the RIPed nucleus becoming numerically superior because resistance to the drug is supplied by this nucleus and the addition of tryptophan supplies its nutritional requirements. Under these conditions the cells showed a slow growth rate compared with acriflavine resistant controls (our unpublished data). The reduction in growth rate was correlated with a decreased level of Tim9 in mitochondria (Figure 2B). The level of Tim10 was also slightly reduced in these cultures.
Figure 2.
tim9 gene is essential in N. crassa. (A) Sheltered heterokaryon containing the tim9RIP mutant. The box depicts the heterokaryon produced during the sheltered RIP cross described in MATERIALS AND METHODS. Circles represent the genetically distinct nuclei that make up the heterokaryon. Nucleus 1 contains only the nonfunctional RIPed version of tim9, whereas nucleus 2 contains a wild-type version of the gene. Only genes relevant to the growth and manipulation of the sheltered heterokaryon are shown. (B) The level of Tim9 is reduced in cultures where the nucleus containing the RIPed gene is increased in number. The sheltered heterokaryon (A) and an acriflavine resistant control strain (Host III) were grown in the presence of acriflavine and tryptophan and mitochondria were isolated. Mitochondrial proteins were separated by SDS-PAGE and blotted to nitrocellulose. The blot was decorated with antibodies against the indicated proteins. (C) Scoring of single colonies from the heterokaryotic strain described in A (ade, adenine; trp, tryptophan).
To determine whether tim9 is an essential gene in N. crassa, conidiaspores produced by the heterokaryon were streaked onto medium containing all the nutritional requirements of both nuclei in the strain (Figure 2A). Testing of nutritional requirements of individual colonies produced from these conidia revealed the tim9RIP nucleus to be inviable (Figure 2C). To confirm that the effects of RIP were specific to the tim9 gene, the sheltered heterokaryon was transformed with a bleomycin resistance plasmid containing a wild-type copy of tim9. Viable tryptophan-requiring homokaryotic strains were recovered (our unpublished data). In conclusion, tim9 is an essential gene in N. crassa.
Tim10 was not a desirable candidate for RIP mutagenesis because of the size of the duplication required to act as a RIP substrate and the proximity of tim10 to a potential tRNA synthetase gene.
Tim9 and Tim10 Form a Heterooligomer in the Intermembrane Space of Mitochondria
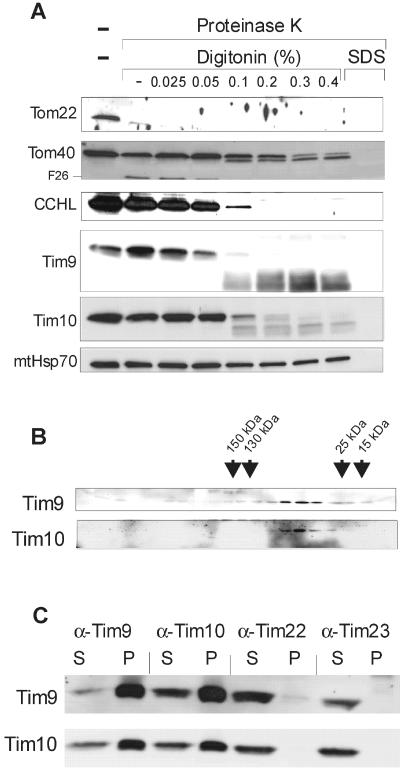
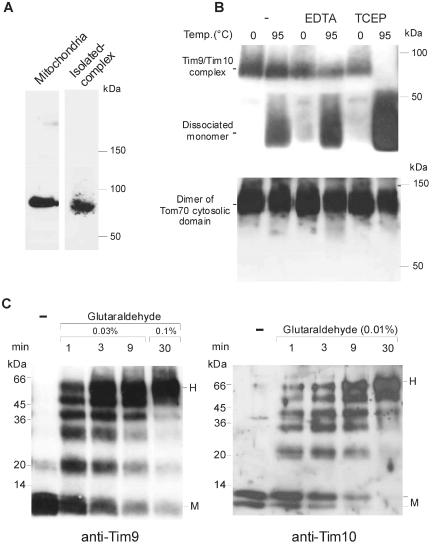
To determine the intramitochondrial localization of Tim9 and Tim10 in N. crassa, isolated mitochondria were fractionated by treating them with increasing amounts of the mild detergent digitonin. Digitonin preferentially opens the outer membrane, and permeabilizes the inner membrane only at higher concentrations (Hartl et al., 1986). Fractionation was performed in the presence of proteinase K to assay for the accessibility of the various proteins. Tim9 and Tim10 are located in the intermembrane space as shown by their accessibility to protease only upon opening of the outer membrane (Figure 3A). Degradation of both proteins occurred at the same digitonin concentrations as that of the IMS protein CCHL and of the N and C termini of Tom40, which are also localized in the IMS (Künkele et al., 1998). A proteinase K-resistant core structure could be observed in both Tim9 and Tim10, suggesting a tight folding of this domain (Figure 3A).
Figure 3.
Characterization of the Tim9/Tim10 complex. (A) Localization of Tim9 and Tim10 in mitochondria. Mitochondria were subfractionated in the presence of proteinase K (250 μg/ml) by the addition of increasing concentrations of digitonin [0-0.4% (wt/vol)]. Solubilization with SDS (0.1%) as control is also shown. Proteins in the samples were precipitated with trichloroacetic acid and analyzed by SDS-PAGE and immunodecoration with the indicated antibodies. The typical N-terminal 26-kDa fragment of Tom40 is indicated (F26). (B) Analysis of Tim9 and Tim10 by size-exclusion chromatography. Mitochondria were solubilized in a buffer containing 0.5% β-dodecyl maltoside and subjected to size-exclusion chromatography. Tim9 and Tim10 in fractions were detected by immunodecoration. The peaks of elution of various marker proteins with the indicated molecular masses are marked by arrows. (C) Analysis of Tim9/Tim10 complex by coimmunoprecipitation. Mitochondria (75 μg/lane) were lysed in a buffer containing 1% digitonin and added to protein A-coupled Sepharose beads containing prebound antibodies against the indicated Tim components. After incubation, the beads were pelleted and proteins in the supernatant (S) and the pellets (P) were subjected to SDS-PAGE, blotting, and immunodecoration with the antibodies indicated at the left side.
We next determined the oligomeric states of Tim9 and Tim10. Mitochondria were solubilized in buffer containing 0.5% β-dodecyl maltoside and subjected to gel filtration. Both proteins were found in a complex of molecular mass of ca. 60 kDa (Figure 3B). To demonstrate that the two proteins are indeed components of the same complex, mitochondria were solubilized and the ability of antibodies against one protein to coprecipitate its partner protein was tested. Antibodies against Tim9 efficiently precipitated both Tim9 and Tim10. Likewise, antibodies against Tim10 immunoprecipitated Tim9 and Tim10 (Figure 3C). Tim9 and Tim10 were not precipitated by antibodies against mitochondrial inner membrane proteins such as Tim23 or Tim44 (our unpublished data). Antibodies against Tim22 precipitated a negligible amount of Tim9 and Tim10 under these conditions (Figure 3C). It seems that the majority of these proteins is not attached to the TIM22 complex. Hence, N. crassa Tim9 and Tim10 interact tightly and form a heteromeric complex in the IMS.
Isolation of the Tim9/Tim10 Complex
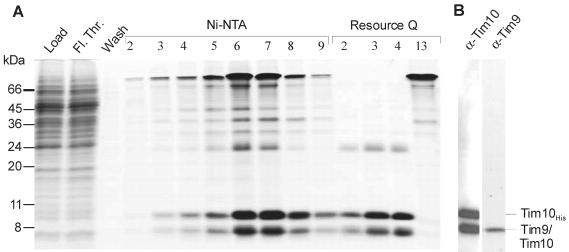
Are TOM core and Tim9/Tim10 complexes sufficient to translocate the AAC carrier across the outer membrane? The best way to answer this question is to use purified components in a reconstituted system. Therefore, a N. crassa strain expressing a nonahistidinyl-tagged version of Tim10 in addition to the wild-type protein was constructed. This strain was similar to the parental wild-type strain in its growth behavior (our unpublished data). Mitochondria were isolated and used to purify the Tim9/Tim10 complex (see MATERIALS AND METHODS). Fractions eluted from a Ni-NTA column were pooled and purified further by ion-exchange chromatography (Figure 4A). Identification of the bands of the purified complex by mass spectroscopy (our unpublished data) and immunodecoration (Figure 4B) demonstrate the presence of Tim9 and wild-type Tim10 in a complex with his-tagged Tim10. No such complex was observed, when solubilization was performed after denaturing the proteins by the addition of urea (our unpublished data). Because the wild-type and his-tagged versions of Tim10 were copurified the complex contains at least two molecules of Tim10, most probably three (Curran et al., 2002a; Vial et al., 2002).
Figure 4.
Purification of the Tim9/Tim10 complex. (A) The Tim9/Tim10 complex was isolated from mitochondria of a Neurospora strain carrying a Tim10 protein with a histidinyl tag. The mitochondria were lysed with digitonin and subjected to Ni-NTA chromatography. Fractions 5-8 from the Ni-NTA column were combined and loaded onto a Q-Sepharose ion-exchange column. Column fractions were analyzed by SDS-PAGE, and proteins were stained with Coomassie Blue. Tim 9, Tim10, and Tim10His are indicated. (B) The proteins in fraction 3 from the ion-exchange column were analyzed by SDS-PAGE and immunodecoration with antibodies against Tim9 or Tim10. Fl. Thr., flow-through.
The purified complex was compared with the complex set free from mitochondria by lysis with digitonin. Both complexes displayed an apparent molecular weight of 70-80 kDa when analyzed by BNGE (Figure 5A). Earlier reports suggest that Tim10 binds Zn2+ (Sirrenberg et al., 1998). On the other hand, recent studies failed to detect Zn2+ in the Tim9/Tim10 complex or in the homologous Tim8/Tim13 complex. It was proposed that the cysteine residues are engaged in disulfide bonds (Curran et al., 2002a,b). To determine whether Zn2+ is essential for the oligomeric state of the purified N. crassa Tim9/Tim10 complex we tested the ability of the complex to refold under various conditions (Curran et al., 2002a,b). At low temperature, the complex was stable in the presence of the metal chelator EDTA or the reducing agent TCEP (Figure 5B). When the complex was heated to 95°C in the presence or absence of EDTA and allowed to cool on ice, refolding to its native structure was observed and only minor amounts of monomer were dissociated from the complex. In contrast, the presence of TCEP during the procedure led to complete dissociation of the complex to the monomers form (Figure 5B, top). Similar results were obtained using antibodies against Tim10 (our unpublished data), suggesting that the 70-80-kDa complex contains both proteins. As a control, we analyzed the dimerization of the cytosolic domain of Tom70, which contains three cysteine residues. The dimer could reform in the presence of either reducing agent or metal chelator (Figure 5B, bottom). Thus, the effect of the reducing agent on the stability of the Tim9/Tim10 complex is not a general feature of every cysteine-containing protein. These results suggest that a complex purified from mitochondria isolated under non-reducing conditions does not require Zn2+ for its oligomeric structure because the cysteine residues are forming disulfide bonds.
Figure 5.
N. crassa Tim9 and Tim10 form a heterohexamer. (A) Analysis of the Tim9/Tim10 complex by BNGE. Mitochondria solubilized in 0.5% digitonin and the purified Tim9/Tim10 complex were analyzed by BNGE, followed by immunodecoration with antibodies against Tim9. (B) The oligomeric structure of the purified Tim9/Tim10 complex depends on oxidative conditions. The purified Tim9/Tim10 complex (top) and the cytosolic domain of Tom70 (bottom) were incubated for 10 min at either 0 or 95°C in the absence or presence of EDTA (2 mM) or TCEP (1 mM). The samples were then kept on ice for a further 10 min. Samples were then analyzed by BNGE followed by immunodecoration with antibodies against either Tim10 (top) or Tom70 (bottom). (C) Tim9 and Tim10 form a hexamer. The purified Tim9/Tim10 complex was incubated with the indicated concentrations of glutaraldehyde for various time periods. Cross-linked products were analyzed by SDS-PAGE, followed by immunodecoration with antibodies against either Tim9 (left) or Tim10 (right). The position of the monomer (M) and hexamer (H) are indicated at the right side.
The oligomeric state of the purified Tim9/Tim10 complex was further studied by chemical cross-linking. Purified complex was treated with the cross-linking reagent glutaraldehyde, and cross-linking products were analyzed by SDS-PAGE and immunostaining with antibodies against Tim9 (Figure 5C, left) or Tim10 (Figure 5C, right). In the absence of cross-linker both Tim9 and Tim10 were found to be a monomer. Bands corresponding to oligomeric species (dimer to hexamer) were observed when the complex was treated with the cross-linker (Figure 5C). Because the higher oligomeric forms can include the authentic and/or his-tagged version of Tim10, decoration with antibody against Tim10 revealed several bands representing the dimeric and trimeric forms. Such heterogenicity is not observed for the higher oligomeric forms probably because the gel system cannot resolve these differences in the higher molecular weight region. The presence of metal chelators (10 mM EDTA, or 10 mM EGTA and 10 mM o-phenanthroline) in the cross-linking reactions did not influence hexamer formation (our unpublished data). Thus, the formation of a heterohexamer by Tim9 and Tim10 is not dependent on metal ions.
The Purified Tim9/Tim10 Complex Can Bind a Substrate Protein
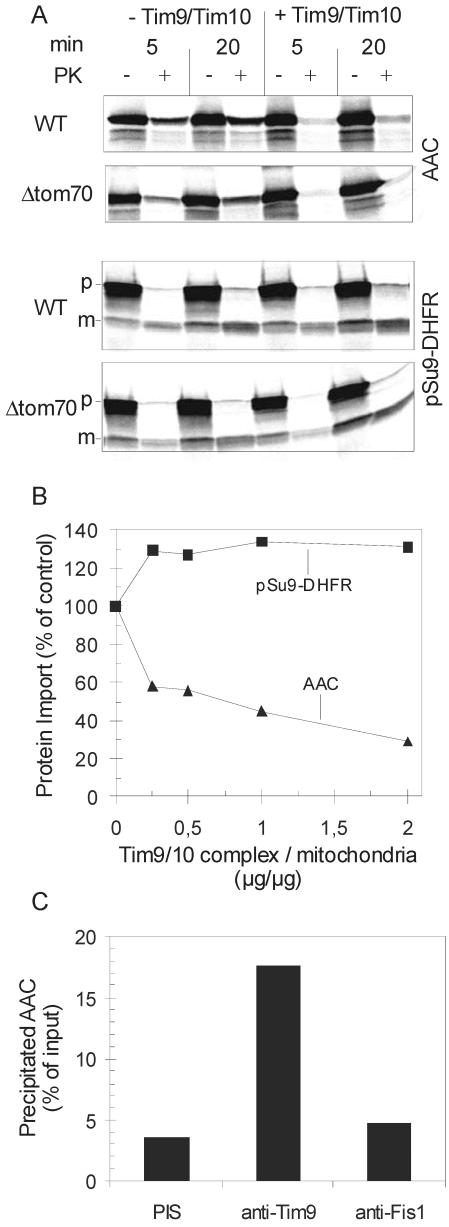
Functionality of the purified Tim9/Tim10 complex was verified by measuring its ability to interact with a physiological substrate, the precursor of AAC. Radiolabeled precursors of AAC, and of the matrix destined precursor pSu9-DHFR as control, were added to isolated mitochondria in the presence or absence of competing amounts of the purified complex. Addition of the Tim9/Tim10 complex almost completely inhibited the import of AAC, whereas the import of the matrix-destined protein was unaffected or even slightly increased (Figure 6, A and B). Because chaperones are known to be required to keep preproteins in an import-competent conformation, we believe that the slight increase in the import of pSu9-DHFR in the presence of the Tim9/Tim10 complex is due to the general chaperone character of this complex (Vial et al., 2002). In contrast, the binding of the Tim9/Tim10 complex to the AAC precursor probably occurs with higher affinity and most likely hinders the required interaction with the TOM complex. The mitochondrial outer membrane remained intact under these conditions because IMS proteins such as d-lactate dehydrogenase or Tim13 were not degraded by the added protease (our unpublished data). This inhibition was not due to a possible interaction of Tim9/Tim10 with Tom70, because import into mitochondria from a strain lacking the receptor Tom70 was also reduced to a similar degree. In the absence of Tom70 AAC is imported via a less efficient bypass pathway (Figure 6A) (Hines et al., 1990).
Figure 6.
Purified Tim9/Tim10 complex can bind AAC precursor. (A) Purified Tim9/Tim10 complex added to an import reaction containing intact mitochondria inhibits the import of AAC precursor. Mitochondria from wild-type and from a Tom70 null strain were either incubated with purified Tim9/Tim10 complex or mock treated for 2 min at 0°C (+/- Tim9/Tim10). They were then added to import buffer containing radiolabeled AAC, and as a control, pSu9-DHFR. After incubation for 5 or 20 min, mitochondria were reisolated, resuspended in SEM buffer, and divided into two halves. Both halves were kept on ice without (-PK) or with addition of proteinase K (+PK). Imported proteins were analyzed by SDS-PAGE. p, precursor; m, mature form. (B) The indicated amounts of purified Tim9/Tim10 complex were incubated for 10 min at 0°C with radiolabeled AAC and as a control, pSu9-DHFR. These mixtures were then added to import buffer containing 40 μg of wild-type mitochondria. After incubation for 10 min at 25°C proteinase K was added. Imported proteins were analyzed by SDS-PAGE. The amount of import in the absence of added Tim9/Tim10 complex was taken as 100%. (C) Purified Tim9/Tim10 complex was incubated with a mixture of porin and AAC for 30 min at 4°C. The mixture was then split into three aliquots that were added to protein A-coupled Sepharose beads containing prebound antibodies from preimmune serum or antibodies against either Tim9 or Fis1. After incubation for 3 h at 4°C, the beads were pelleted and proteins in the pellets were subjected to SDS-PAGE, blotting and immunodecoration with antibodies against AAC. The amount of added AAC was taken as 100%.
Further evidence for a direct interaction between Tim9/Tim10 complex and its substrate was obtained by immunoprecipitation. On incubation of the complex with a mixture of porin and AAC2 isolated from yeast cells, antibodies against Tim9 could precipitate significant amounts of the AAC2 protein but not of porin (Figure 6C; our unpublished data). Specific precipitation was also observed when radiolabeled AAC synthesized in vitro was used as substrate (our unpublished data). Only background levels of precipitation were observed when either the complex was omitted from the reaction mixture or when preimmune serum or antibody against an unrelated protein, Fis1 were used (Figure 6C; our unpublished data). Together, we conclude that the purified Tim9/Tim10 complex can specifically recognize its physiological substrate even when it is not presented in the context of the import pathway.
The TOM Core Complex and the Tim9/Tim10 Complex Are Sufficient for Import of ADP/ATP Carrier across the Outer Membrane of Mitochondria
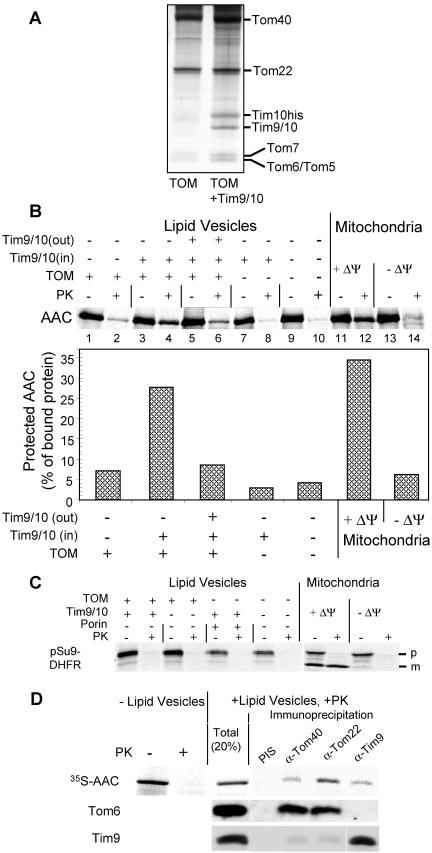
To study whether receptor proteins or yet unknown components in the internal compartments of mitochondria are required for the translocation of AAC precursor across the outer membrane, we established a reconstituted system. Proteoliposomes were formed from a detergent-containing solution of purified TOM core complex and E. coli lipids by removal of the detergent through addition of polystyrene beads. To verify the presence of the TOM complex in the lipid vesicles, lactate dehydrogenase was enclosed in the vesicles. The accessibility of the enzyme to externally added substrates NADH and pyruvate was measured (Diekert et al., 2001). The production of lactate confirmed the presence of TOM pores in the proteoliposomes (our unpublished data). Figure 7A shows the protein composition of lipid vesicles containing either only the TOM core complex or in addition enclosed Tim9/Tim10 complex.
Figure 7.
Translocation of the AAC precursor into proteoliposomes containing the TOM core complex and Tim9/Tim10 complex. (A) The proteins of proteoliposomes containing either the TOM core complex (TOM) only or, in addition, Tim9/Tim10 complex (TOM+Tim9/10) were analyzed by SDS-PAGE and silver staining. (B) Import of AAC into proteoliposomes containing TOM core complex and TIM9/10 complex. Radiolabeled AAC precursor was incubated with proteoliposomes (50 μg of lipids) in the absence (lanes 1-4 and 7-10) or presence (lanes 5 and 6) of externally added Tim9/Tim10 complex. The following proteoliposomes were used: lanes 1 and 2, liposomes containing TOM core complex; lanes 3-6, liposomes containing TOM complex and enclosed Tim9/Tim10 complex; lanes 7 and 8, liposomes containing enclosed Tim9/Tim10 complex; and lanes 9 and 10, liposomes without reconstituted proteins. Import of AAC precursor into intact mitochondria in the absence (+ΔΨ, lanes 11 and 12) or presence (-ΔΨ, lanes 13 and 14) of valinomycin is shown for comparison. After import, vesicles and mitochondria were reisolated by centrifugation, resuspended in buffer and halved. One-half was left untreated (-PK), whereas the other was treated with 50 μg/ml proteinase K (+PK). The samples were precipitated with trichloroacetic acid and subjected to SDS-PAGE and autoradiography (top). The proportions of bound AAC precursor that were resistant to proteinase K treatment were determined (bottom). (C) Import of pSu9-DHFR into proteoliposomes containing TOM core complex and TIM9/10 complex. Radiolabeled pSu9-DHFR precursor was incubated with proteoliposomes or with mitochondria as described above. The precursor and the mature forms of pSu9-DHFR are indicated (p and m, respectively). (D) AAC precursor interacts simultaneously with TOM and Tim9/Tim10 complexes. Radiolabeled AAC precursor was incubated for 30 min at 25°C with proteoliposomes containing the TOM core and the Tim9/Tim10 complexes. After the import reaction, proteoliposomes were treated with proteinase K and reisolated. Vesicles were solubilized in digitonin (1%), and immunoprecipitation was performed as described in the legend to Figure 3C. Precipitated proteins were analyzed by SDS-PAGE, blotting, and autoradiography (35S-AAC) and immunodecoration with the antibodies indicated at the left side.
The AAC precursor was incubated with the TOM complex containing proteoliposomes. Translocation of radiolabeled AAC precursor across the membrane of the liposome was followed by analyzing the precursor molecules that became resistant to externally added proteinase K. Only minor amounts of the bound AAC molecules were protected when the TOM core or Tim9/Tim10 complex were reconstituted separately into the vesicles (Figure 7B, lanes 2 and 8, respectively). In contrast, when the Tim9/Tim10 complex was enclosed in vesicles containing the TOM core complex, the amount of protected material was increased to levels similar to those observed with intact mitochondria (Figure 7B, lanes 4 and 12). The observation that proteoliposomes can provide better protection than deenergized mitochondria (Figure 7B, lanes 4 and 14) could be explained by the significantly higher amounts of TOM and Tim9/Tim10 complexes in reconstituted system. The large amount of AAC seen to be associated with the vesicles before PK treatment is not unexpected because AAC has many hydrophobic segments (Figure 7B).
The specificity of the translocase-mediated transport of AAC is demonstrated by the following control experiments: 1) the import can be competed out by addition of external Tim9/Tim10 complex (Figure 7B, lane 6); 2) imported AAC was completely degraded by proteinase K upon solubilization of the liposomes by Triton X-100 (our unpublished data); 3) matrix-destined precursor (pSu9-DHFR) and a peroxisomal transporter (Ant1) were completely degraded by proteinase K under all experimental conditions (Figure 7C; our unpublished data); and 4) when the pore-forming protein, porin, was present in the membrane of the Tim9/Tim10-containing vesicles instead of the TOM complex, the small molecules pyruvate and NADH could cross the membrane, but only background levels of translocation of AAC precursor were observed (our unpublished data). We conclude that the TOM core and Tim9/Tim10 complexes are sufficient to facilitate transfer of the ADP/ATP carrier across the outer membrane of mitochondria.
Tim23 was found in our study to be another substrate of the N. crassa Tim9/Tim10 complex (see below). However, significant amounts of protease-protected radiolabeled Tim23 were not observed when it was added to the above proteoliposomes (our unpublished data). As the other tiny Tims (Tim8/Tim13) were suggested to be involved in the import of Tim23 (Koehler et al., 1999a; Paschen et al., 2000), we propose that the absence of these latter components from our reconstituted system could explain why AAC was translocated in our system, whereas Tim23 was not.
We next analyzed the environment of the AAC precursor in its protease-protected site. Antibodies against Tom components and against Tim9 could precipitate AAC precursor at this site (Figure 7D). The TOM and Tim9/Tim10 complexes, however, did not form a combined complex under these conditions. Antibodies against Tom components could coprecipitate Tom6 but not Tim9. Likewise, Tim9 was precipitated only by antibodies against Tim9 (Figure 7D). Thus, those molecules of AAC that are translocating across the outer membrane interact with Tim9/Tim10 complex while still being engaged by the TOM complex. These results are in line with previous observations that a soluble AAC intermediate bound only to Tim9/Tim10 complex could not be isolated.
Sequences in Mitochondrial Precursor Proteins That Are Recognized by the Tim9/Tim10 Complex
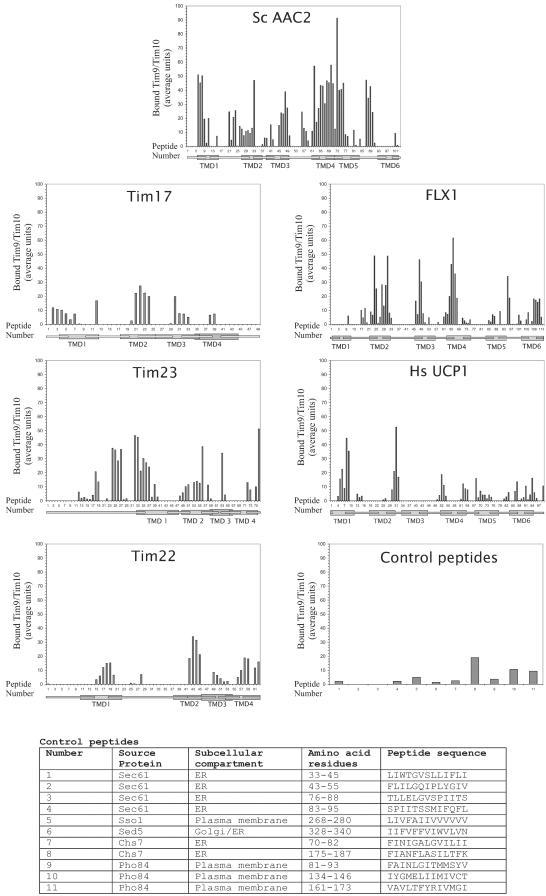
What are the sequences in mitochondrial precursor proteins that are recognized by the Tim9/Tim10 complex? Peptide libraries consisting of 13mers with an overlap of 10 residues were scanned. The following proteins were analyzed: S.c. AAC2, N.c. Tim17, N.c. Tim22, N.c. Tim23, N.c. FLX1 (a homologue of the yeast FLX1, a carrier-type FAD-transporter), and human uncoupler protein 1 (UCP1). We chose to investigate the yeast AAC rather than the N. crassa protein to allow comparison with the results of the peptide scan performed with yeast Tim9/Tim10 complex (Curran et al., 2002a). Control peptides corresponding to transmembrane domains of various nonmitochondrial proteins were spotted on the same membrane (origin and sequences of control peptides are given in Figure 8). The purified Tim9/Tim10 complex was incubated with the peptide-bearing membrane. Bound complex was transferred to a PVDF membrane that was then immunodecorated with antibodies against Tim10.
Figure 8.
Substrate recognition by the Tim9/Tim10 complex. A peptide library on a cellulose membrane covering the indicated proteins and control peptides was incubated with purified Tim9/Tim10 complex. Bound proteins were blotted to PVDF membranes and decorated with antibodies against Tim9. Binding was quantified by scanning densitometry of three independent experiments. The intensity of the strongest spot from each experiment was set to 100. Below each peptide it is indicated whether its sequence covers a predicted TMD, a predicted loop, or its sequence stretches over both types of structure.
All peptides were on one membrane allowing us to directly compare the various proteins. The highest affinity among the proteins analyzed was observed toward the AAC2 protein (Figure 8). Binding occurred mainly to peptides containing residues from both the transmembrane domains (TMD) and their flanking regions. The strongest interaction was with peptides covering parts of the fourth TMD, followed by TMDs 1, 3, and 5. The recently published recognition pattern of AAC2 by the yeast Tim9/Tim10 complex (Curran et al., 2002a) allows direct comparison with the pattern observed here with the N. crassa complex. Both complexes have high affinity to peptides representing TMDs 2, 3, and 4 and did not bind peptides covering TMD 6. The binding pattern to exposed regions differs in the two complexes. Whereas the complex from N. crassa binds with relatively high affinity to the region before TMD 1 and to the loops between TMD1-TMD2, TMD4-TMD5, and TMD5-TMD6, no such binding was observed with the Tim9/Tim10 complex from yeast. Thus, although the recognition pattern of the complexes from both organisms exhibit overall similarity, significant deviations also exist.
Binding to peptides representing the N. crassa Tim17, Tim22, FLX1, and to the human homologue of UCP1 was generally lower than to AAC. The highest affinities were usually seen with TMDs or their immediate flanking regions (Figure 8). Binding to Tim22 was relatively weak and occurred mainly with TMDs rather than intermembrane space loops. In the case of Flx1, the strongest binding was to peptides in the central TMDs 2, 3, and 4. The binding to the human Ucp1 was less restricted to TMDs, although the highest affinity was toward putative TMDs 1 and 2.
Hydrophobicity, however, is not a critical parameter for recognition by Tim9/Tim10 because weak binding was observed to hydrophobic nonmitochondrial control peptides (Figure 8). Furthermore, among the 20 peptides displaying the highest affinity in both AAC (Figure 9) and Flx1 (our unpublished data) 19 peptides contained at least one charged residue within their sequence. The Tim9/Tim10 complex did not bind with the highest affinity to peptides in the core of the putative TMDs but rather to peptides covering the interface between the membrane spanning segment and the soluble loops (Figure 9). Interestingly, the strongest binding among peptides derived from AAC2 was toward a segment covering a part of an intermembrane space loop and the beginning of the fifth transmembrane segment. A peptide covering exactly the same region of the phosphate carrier was found to be among several internal segments that interact with Tom receptors and was the only peptide to interact with purified Tim22 (Brix et al., 1999; Kovermann et al., 2002). Amino acid sequences related to this stretch are conserved in a number of other proteins from the carrier family (Nelson et al., 1998).
Figure 9.
Binding of the Tim9/Tim10 complex to a peptide library covering AAC2. The sequences of the 103 peptides covering the sequence of AAC2 are presented together with the level of binding to each peptide. The sequences of the six putative transmembrane domains are underlined and bold. Binding levels higher than 30% of the maximal value are indicated by bigger digits.
Tim23 Is a Substrate of Tim9/Tim10 Complex in N. crassa
The pattern of binding of the N. crassa Tim9/Tim10 complex to peptides representing Tim23 from N. crassa differed significantly from that observed with yeast Tim9/Tim10 complex and yeast Tim23 (Curran et al., 2002a). Whereas the yeast Tim9/Tim10 complex was reported not to bind yeast Tim23, significant binding to two segments of the N. crassa protein was observed with Tim9/Tim10 from Neurospora (Figure 8). One segment lies in the putative intermembrane space domain (peptides 24-27, residues 70-91), whereas the other covers the interface between the intermembrane space domain and the first predicted transmembrane segment (peptides 33-37, residues 97-121) (Figure 8). Both segments are highly conserved among Tim23 proteins from various organisms.
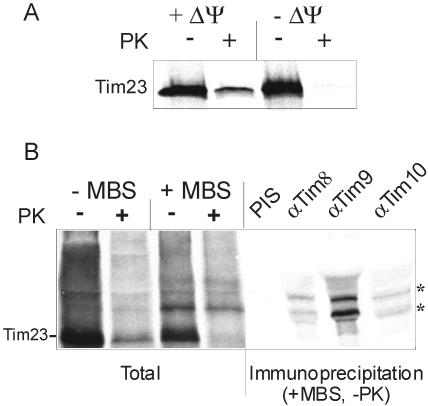
To demonstrate a direct interaction of a precursor of Tim23 with the Tim9/Tim10 complex we used a chemical cross-linking approach. On incubation of radiolabeled precursor of Tim23 with deenergized mitochondria, an import intermediate that is partly in the intermembrane space but still accessible to external protease is formed (Figure 10A). This intermediate is equivalent to the well defined stage III intermediate of the AAC precursor. (Pfanner and Neupert, 1987). Under these conditions, the addition of the bifunctional cross-linking reagent MBS resulted in the formation of specific cross-linking adducts. These adducts could be immunoprecipitated with antibodies against Tim9, Tim10, and Tim8 (Figure 10B). Interestingly, the bound precursor was accessible to externally added proteases, but the Tim23-Tim9 cross-linking adduct was protected. These results suggest that those precursor molecules that are associated with the Tim9/Tim10 complex are pulled through the TOM complex deeper into the intermembrane space. Thus, on its route into the inner membrane the precursor of N. crassa Tim23 interacts with both the Tim9/Tim10 and the Tim8/13 complexes.
Figure 10.
The precursor of Tim23 interacts with the Tim9/Tim10 complex in the intermembrane space. (A) Tim23 is translocated across the outer membrane in a ΔΨ-dependent manner. Radiolabeled Tim23 precursor was incubated with either energized (+ΔΨ) or deenergized mitochondria (-ΔΨ) for 15 min at 25°C. The samples were then halved. One-half was left untreated (-PK), whereas the other was treated with proteinase K (+PK). (B) Tim23 on its insertion pathway is in the vicinity of Tim9, Tim10, and Tim8. Radiolabeled Tim23 precursor was incubated with deenergized mitochondria in the presence or absence of the chemical cross-linker MBS. Then reactions were made 100 mM glycine (pH 8.0), and one-half of the sample was treated with PK. The mitochondria of all four samples were pelleted and subjected to SDS-PAGE. Another sample treated with MBS but not with PK (+MBS, -PK) was subjected to immunoprecipitation with antibodies against the indicated Tim proteins or with preimmune serum to Tim9 (PIS). The immunoprecipitates were solubilized in sample buffer and analyzed by SDS-PAGE and autoradiography. Cross-linked adducts of Tim23 and Tim proteins are labeled by asterisks.
DISCUSSION
In the present study, we have identified the Tim9/Tim10 complex in N. crassa and studied its role in the translocation of carrier precursor proteins across the mitochondrial outer membrane. The complex was purified and was found to be a heterohexamer composed of three Tim9 and three Tim10 molecules. These structural characteristics are shared by the yeast complex, suggesting that the oligomeric structure has an important functional role. Tim9 and Tim10 contain a potential zinc-binding motif that is also present in the other small Tim components Tim12, Tim8, and Tim13. Recent reports, however, raised the possibility that the cysteine residues within this domain are engaged in disulfide bridges rather than coordinating zinc (Curran et al., 2002a,b). Our observations suggest that both the reduced and oxidized forms of Tim9/Tim10 complex are of physiological relevance. The structure of the Tim9/Tim10 complex in vivo might be subject to oxidation-reduction reactions. In the reducing environment of the mitochondria, the zinc-bound form may be the dominant one, whereas upon solubilization of mitochondria in the presence of atmospheric oxygen, the equilibrium is shifted to the disulfide bridged form (Lutz et al., 2003). The conserved heat shock protein Hsp33 is a known example of a redox regulated molecular chaperone (Graf and Jakob, 2002). The redox sensor in Hsp33 is a cysteine center that coordinates zinc under reducing, inactivating conditions and that forms two intramolecular disulfide bonds under oxidizing, activating conditions.
Our ability to purify both native TOM core and Tim9/Tim10 complexes enabled us to address a number of questions related to the role of the Tim9/Tim10 and its mechanism of action. In a reconstituted system containing only the TOM core complex, the AAC precursor was transferred to the internal side of the vesicles membrane. Apparently, import by the TOM core complex follows the “bypass” route observed in the absence of import receptors (Pfaller et al., 1989). Parts of the AAC precursor molecule were still exposed to the cytosol. The inclusion of the Tim9/Tim10 complex into the interior of the vesicles led to a “pulling” of the AAC precursor further across the membrane to a site where it was not exposed to the external milieu. Precursor molecules at this site interacted with both the TOM and the Tim9/Tim10 complexes. It thus seems that neither additional proteins in the mitochondrial inner membrane or intermembrane space nor structural elements such as import contact sites are required for this translocation step. The interaction of the small Tims with the incoming precursor prevented its retrograde sliding out of the translocation machinery and mediated a vectorial movement across the membrane. We propose that the TOM core complex and Tim9/Tim10 form the minimal machinery for translocation of the ADP/ATP carrier across the mitochondrial outer membrane.
How does the Tim9/Tim10 complex recognize its various substrate proteins? Initially, it was proposed that charged amino acid residues in the zinc-binding motif of Tim10 and Tim12 could interact with specific charged residues in the extramembrane loops of carrier protein, the so-called carrier signature motif (Sirrenberg et al., 1998). However, some of the substrates of the Tim9/Tim10 complex lack sequence similarity to carrier proteins. This makes the possibility of sequence-specific recognition unlikely. Indeed, no binding to peptides corresponding to the carrier signature was observed. The Tim9/Tim10 complex was suggested to interact preferentially with peptides covering the predicted membrane spanning domains of the AAC (Curran et al., 2002a). Based on our results with a variety of Tim9/Tim10 substrates, we propose a modified concept for the recognition of precursor proteins by the Tim9/Tim10 complex. We observed highest affinity of the complex toward peptides containing residues from both the membrane-spanning domains and the loops between them. Preferential binding to such regions may trap segments that emerge at the IMS side of the TOM complex, whereas most of the precursor is still present within the TOM complex. In such a way, the membrane-spanning segments could be transferred from the TOM to the Tim9/Tim10 complex without the hydrophobic stretches being exposed to the IMS. Such a mechanism of recognition and binding is in agreement with recent findings showing that the Tim9/Tim10 complex is required for precursor release from the TOM complex (Truscott et al., 2002).
Our results shed new light on the import pathway of the Tim23 precursor. It is widely accepted that Tim23 is inserted into the inner membrane via the TIM22 complex (Kerscher et al., 1997; Káldi et al., 1998; Leuenberger et al., 1999). However, experiments with yeast were not conclusive regarding the involvement of Tim9/Tim10 complex in the import of Tim23. Two groups reported that the Tim23 precursor could be cross-linked mainly to Tim8 and Tim13 with little, if any, adduct to Tim9 and Tim10 (Leuenberger et al., 1999; Paschen et al., 2000). A third group reported a major interaction of Tim8/Tim13 complex with Tim23, but observed a weak interaction of the Tim9/Tim10 complex with the COOH-terminal hydrophobic domain of Tim23. They suggested that the Tim9/Tim10 complex plays an important role in the import of Tim23 (Davis et al., 2000; Jensen and Dunn, 2002). Our results with peptide scans and cross-linking in N. crassa point to a recognition of Tim23 by the native Tim9/Tim10 complex. Whether the interactions of the Tim23 precursor with the Tim9/Tim10 complex occur concomitantly with its interaction with the Tim8/Tim13 complex, or at a later stage of its import pathway is unknown. Recent studies using peptide scans with yeast proteins suggested that peptides representing Tim23 were bound to the Tim8/Tim13 complex but not the Tim9/Tim10 complex (Curran et al., 2002a,b). A possible explanation of these differences between Neurospora and yeast could be the absence of Tim12 in N. crassa. Conceivably, Tim12 in yeast might be involved in the transfer of the Tim23 precursor to the TIM22 complex. In N. crassa, a similar function may be fulfilled by the Tim9/Tim10 complex.
Acknowledgments
We thank H. Germeroth, U. Staudinger, and M. Malesic for excellent technical assistance; K. Hell for critically reading the manuscript; D. Mokranjac for helpful discussions; T. Stan for providing the cytosolic domain of Tom70; B. Müller and L. Eichacker for performing mass spectroscopy analysis; and C. Koehler and K. Tokatlidis for providing yeast strains. This work was supported by Sonderforschungsbereich 594 and grant RA 1028/1-1 of the Deutsche Forschungsgemeinschaft, a grant of the Bundesministerium für Bildung und Forschung (MITOP), the Fonds der Chemischen Industrie (to W.N.) and by a grant from the Canadian Institutes of Health Research (to F.E.N.).
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03-05-0272. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-05-0272.
References
- Adam, A., Endres, M., Sirrenberg, C., Lottspeich, F., Neupert, W., and Brunner, M. (1999). Tim9, a new component of the TIM22.54 translocase in mitochondria. EMBO J. 18, 313-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahting, U., Thun, C., Hegerl, R., Typke, D., Nargang, F.E., Neupert, W., and Nussberger, S. (1999). The TOM core complex: the general protein import pore of the outer membrane of mitochondria. J. Cell Biol. 147, 959-968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer, M.F., Rothbauer, U., Muhlenbein, N., Smith, R.J., Gerbitz, K., Neupert, W., Brunner, M., and Hofmann, S. (1999). The mitochondrial TIM22 preprotein translocase is highly conserved throughout the eukaryotic kingdom. FEBS Lett. 464, 41-47. [DOI] [PubMed] [Google Scholar]
- Brix, J., Rudiger, S., Bukau, B., Schneider-Mergener, J., and Pfanner, N. (1999). Distribution of binding sequences for the mitochondrial import receptors Tom20, Tom22, and Tom70 in a presequence-carrying preprotein and a noncleavable preprotein. J. Biol. Chem. 274, 16522-16530. [DOI] [PubMed] [Google Scholar]
- Curran, S.P., Leuenberger, D., Oppliger, W., and Koehler, C.M. (2002a). The Tim9p-Tim10p complex binds to the transmembrane domains of the ADP/ATP carrier. EMBO J. 21, 942-953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran, S.P., Leuenberger, D., Schmidt, E., and Koehler, C.M. (2002b). The role of the Tim8p-Tim13p complex in a conserved import pathway for mitochondrial polytopic inner membrane proteins. J. Cell Biol. 158, 1017-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, A.J., Sepuri, N.B., Holder, J., Johnson, A.E., and Jensen, R.E. (2000). Two intermembrane space TIM complexes interact with different domains of Tim23p during its import into mitochondria. J. Cell Biol. 150, 1271-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diekert, K., de Kroon, A.I.P.M., Ahting, U., Niggemeyer, B., Neupert, W., de Kruijff, B., and Lill, R. (2001). Apocytochrome c requires the TOM complex for translocation across the mitochondrial outer membrane. EMBO J. 20, 5626-5635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endres, M., Neupert, W., and Brunner, M. (1999). Transport of the ADP/ATP carrier of mitochondria from the TOM complex to the TIM22.54 complex. EMBO J. 18, 3214-3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank, R. (1992). Spot synthesis: an easy technique for the positionally addressable, parallel chemical synthesis on a membrane support. Tetrahedron 48, 9217-9232. [Google Scholar]
- Graf, P.C., and Jakob, U. (2002). Redox-regulated molecular chaperones. Cell. Mol. Life Sci. 59, 1624-1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harkness, T.A., Metzenberg, R.L., Schneider, H., Lill, R., Neupert, W., and Nargang, F.E. (1994). Inactivation of the Neurospora crassa gene encoding the mitochondrial protein import receptor MOM19 by the technique of “sheltered RIP”. Genetics 136, 107-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl, F.U., Schmidt, B., Wachter, E., Weiss, H., and Neupert, W. (1986). Transport into mitochondria and intramitochondrial sorting of the Fe/S protein of ubiquinol-cytochrome c reductase. Cell 47, 939-951. [DOI] [PubMed] [Google Scholar]
- Hines, V., Brandt, A., Griffiths, G., Horstmann, H., Brütsch, H., and Schatz, G. (1990). Protein import into yeast mitochondria is accelerated by the outer membrane protein MAS70. EMBO J. 9, 3191-3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen, R.E., and Dunn, C.D. (2002). Protein import into and across the mitochondrial inner membrane: role of the TIM23 and TIM22 translocons. Biochim. Biophys. Acta 1592, 25-34. [DOI] [PubMed] [Google Scholar]
- Káldi, K., Bauer, M.F., Sirrenberg, C., Neupert, W., and Brunner, M. (1998). Biogenesis of Tim23 and Tim17, integral components of the TIM machinery for matrix targeted preproteins. EMBO J. 17, 1569-1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayingo, G., Potier, S., Hohmann, S., and Prior, B.A. (2000). Isolation and characterization of the TIM10 homologue from the yeast Pichia sorbitophila: a putative component of the mitochondrial protein import system. Yeast 16, 589-596. [DOI] [PubMed] [Google Scholar]
- Kerscher, O., Holder, J., Srinivasan, M., Leung, R.S., and Jensen, R.E. (1997). The Tim54p-Tim22p complex mediates insertion of proteins into the mitochondrial inner membrane. J. Cell Biol. 139, 1663-1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingenberg, M., Winkler, E., and Huang, S. (1995). ADP/ATP carrier and uncoupling protein. Methods Enzymol. 260, 369-389. [DOI] [PubMed] [Google Scholar]
- Koehler, C.M., Jarosch, E., Tokatlidis, K., Schmid, K., Schweyen, R.J., and Schatz, G. (1998a). Import of mitochondrial carriers mediated by essential proteins of the intermembrane space. Science 279, 369-373. [DOI] [PubMed] [Google Scholar]
- Koehler, C.M., Leuenberger, D., Merchant, S., Renold, A., Junne, T., and Schatz, G. (1999a). Human deafness dystonia syndrome is a mitochondrial disease. Proc. Natl. Acad. Sci. USA 96, 2141-2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler, C.M., Merchant, S., Oppliger, W., Schmid, K., Jarosch, E., Dolfini, L., Junne, T., Schatz, G., and Tokatlidis, K. (1998b). Tim9p, an essential partner subunit of Tim10p for the import of mitochondrial carrier proteins. EMBO J. 17, 6477-6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler, C.M., Merchant, S., and Schatz, G. (1999b). How membrane proteins travel across the mitochondrial intermembrane space. Trends Biochem. Sci. 24, 428-432. [DOI] [PubMed] [Google Scholar]
- Komiya, T., Rospert, S., Schatz, G., and Mihara, K. (1997). Binding of mitochondrial precursor proteins to the cytoplasmic domains of the import receptors Tom70 and Tom20 is determined by cytoplasmic chaperones. EMBO J. 16, 4267-4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovermann, P., Truscott, K.N., Guiard, B., Rehling, P., Sepuri, N.B., Müller, H., Jensen, R.E., Wagner, R., and Pfanner, N. (2002). Tim22, the essential core of the mitochondrial protein insertion complex, forms a voltage-activated and signal-gated channel. Mol. Cell 9, 363-373. [DOI] [PubMed] [Google Scholar]
- Kramer, A., and Schneider-Mergener, J. (1998). Synthesis and screening of peptide libraries on continuous cellulose membrane supports. Methods Mol. Biol. 87, 25-39. [DOI] [PubMed] [Google Scholar]
- Künkele, K.-P., Juin, P., Pompa, C., Nargang, F.E., Henry, J.-P., Neupert, W., Lill, R., and Thieffry, M. (1998). The isolated complex of the translocase of the outer membrane of mitochondria. J. Biol. Chem. 273, 31032-31039. [DOI] [PubMed] [Google Scholar]
- Kurz, M., Martin, H., Rassow, J., Pfanner, N., and Ryan, M.T. (1999). Biogenesis of Tim proteins of the mitochondrial carrier import pathway: differential targeting mechanisms and crossing over with the main import pathway. Mol. Biol. Cell 10, 2461-2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leuenberger, D., Bally, N.A., Schatz, G., and Koehler, C.M. (1999). Different import pathways through the mitochondrial intermembrane space for inner membrane proteins. EMBO J. 18, 4816-4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister, R., Mowday, B., Whelan, J., and Millar, A.H. (2002). Zinc-dependent intermembrane space proteins stimulate import of carrier proteins into plant mitochondria. Plant J. 30, 555-566. [DOI] [PubMed] [Google Scholar]
- Luciano, P., Vial, S., Vergnolle, M.A., Dyall, S.D., Robinson, D.R., and Tokatlidis, K. (2001). Functional reconstitution of the import of the yeast ADP/ATP carrier mediated by the TIM10 complex. EMBO J. 20, 4099-4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz, T., Neupert, W., and Herrmann, J.M. (2003). Import of small Tim proteins into the mitochondrial intermembrane space. EMBO J. 22, 4400-4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer, A., Lill, R., and Neupert, W. (1993). Translocation and insertion of precursor proteins into isolated outer membranes of mitochondria. J. Cell Biol. 121, 1233-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, D.R., Felix, C.M., and Swanson, J.M. (1998). Highly conserved chargepair networks in the mitochondrial carrier family. J. Mol. Biol. 277, 285-308. [DOI] [PubMed] [Google Scholar]
- Neupert, W. (1997). Protein import into mitochondria. Annu. Rev. Biochem. 66, 863-917. [DOI] [PubMed] [Google Scholar]
- Paschen, S.A., Rothbauer, U., Kaldi, K., Bauer, M.F., Neupert, W., and Brunner, M. (2000). The role of the TIM8-13 complex in the import of Tim23 into mitochondria. EMBO J. 19, 6392-6400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaller, R., Pfanner, N., and Neupert, W. (1989). Mitochondrial protein import. Bypass of proteinaceous surface receptors can occur with low specificity and efficiency. J. Biol. Chem. 264, 34-39. [PubMed] [Google Scholar]
- Pfanner, N., and Geissler, A. (2001). Versatility of the mitochondrial protein import machinery. Nat. Rev. Mol. Cell. Biol. 2, 339-349. [DOI] [PubMed] [Google Scholar]
- Pfanner, N., and Neupert, W. (1987). Distinct steps in the import of the ADP/ATP carrier into mitochondria. J. Biol. Chem. 262, 7528-7536. [PubMed] [Google Scholar]
- Prokisch, H., Neupert, W., and Westermann, B. (2000). Role of MMM1 in maintaining mitochondrial morphology in Neurospora crassa. Mol. Biol. Cell 11, 2961-2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapaport, D., Neupert, W., and Lill, R. (1997). Mitochondrial protein import. Tom40 plays a major role in targeting and translocation of preproteins by forming a specific binding site for the presequence. J. Biol. Chem. 272, 18725-18731. [DOI] [PubMed] [Google Scholar]
- Schägger, H., Cramer, W.A., and von Jagow, G. (1994). Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal. Biochem. 217, 220-230. [DOI] [PubMed] [Google Scholar]
- Sirrenberg, C., Endres, M., Fölsch, H., Stuart, R.A., Neupert, W., and Brunner, M. (1998). Zinc finger-like proteins Tim10/Mrs11p and Tim12/Mrs5p mediating import of carrier proteins into mitochondria. Nature 391, 912-915. [DOI] [PubMed] [Google Scholar]
- Stan, T., Brix, J., Schneider-Mergener, J., Pfanner, N., Neupert, W., and Rapaport, D. (2003). Mitochondrial protein import: recognition of internal import signals of BCS1 by the TOM complex. Mol. Cell. Biol. 23, 2239-2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokatlidis, K., and Schatz, G. (1999). Biogenesis of mitochondrial inner membrane proteins. J. Biol. Chem. 274, 35285-35288. [DOI] [PubMed] [Google Scholar]
- Truscott, K.N., Wiedemann, N., Rehling, P., Müller, H., Meisinger, C., Pfanner, N., and Guiard, B. (2002). Mitochondrial import of the ADP/ATP carrier: the essential TIM complex of the intermembrane space is required for precursor release from the TOM complex. Mol. Cell. Biol. 22, 7780-7789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vial, S., Lu, H., Allen, S., Savory, P., Thornton, D., Sheehan, J., and Tokatlidis, K. (2002). Assembly of Tim9 and Tim10 into functional chaperone. J. Biol. Chem. 277, 36100-36108. [DOI] [PubMed] [Google Scholar]
- Wiedemann, N., Pfanner, N., and Ryan, M.T. (2001). The three modules of ADP/ATP carrier cooperate in receptor recruitment and translocation into mitochondria. EMBO J. 20, 951-960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young, J.C., Hoogenraad, N.J., and Hartl, F.-U. (2003). Molecular chaperones Hsp90 and Hsp70 deliver preproteins to the mitochondrial import receptor Tom70. Cell 112, 41-50. [DOI] [PubMed] [Google Scholar]