Abstract
DNA binding proteins find their cognate sequences within genomic DNA through recognition of specific chemical and structural features. Here we demonstrate that high-resolution DNase I cleavage profiles can provide detailed information about the shape and chemical modification status of genomic DNA. Analyzing millions of DNA backbone hydrolysis events on naked genomic DNA, we show that the intrinsic rate of cleavage by DNase I closely tracks the width of the minor groove. Integration of these DNase I cleavage data with bisulfite sequencing data for the same cell type’s genome reveals that cleavage directly adjacent to cytosine-phosphate-guanine (CpG) dinucleotides is enhanced at least eightfold by cytosine methylation. This phenomenon we show to be attributable to methylation-induced narrowing of the minor groove. Furthermore, we demonstrate that it enables simultaneous mapping of DNase I hypersensitivity and regional DNA methylation levels using dense in vivo cleavage data. Taken together, our results suggest a general mechanism by which CpG methylation can modulate protein–DNA interaction strength via the remodeling of DNA shape.
Keywords: deoxyribonuclease I, DNA minor groove, functional genomics, protein-DNA recognition, 5-methylcytosine
DNase I is an endonuclease that cleaves the backbone of double-stranded DNA. It approximates the size and nuclear diffusion properties of a typical human transcription factor (TF) (1). The enzyme interacts with DNA via the minor groove (2), where it recognizes approximately six consecutive base pairs (3). In addition to its nearly ubiquitous use in the removal of DNA from cellular extracts, DNase I has been widely used as a structural probe of in vitro and in vivo DNA and chromatin structure (4, 5), and to map regulatory DNA in the human and other genomes (6–9). The interaction of DNase I with specific DNA sequences can be abrogated through steric hindrance by DNA binding proteins, leading to its widespread use as a reagent for studying TF binding (10, 11).
One of the best studied DNA modifications is the methylation of cytosines at position 5 of the pyrimidine ring. This covalent modification, in the context of a CpG dinucleotide, can be found in eukaryotes from plants to humans and is observed on over 70% of CpGs in vertebrate DNA (12, 13). The patterns of methylation can be dynamic (14), can vary between cell lines and in the course of developmental processes, and therefore provide a mechanism for the generation of epigenetic variation at the level of the primary DNA sequence (12). The biological contribution of DNA methylation is both significant and complex. First and foremost, CpG methylation has been linked to transcriptional silencing at promoters of genes on the inactive X chromosome, on imprinted loci and genes rendered inactive in cancers (13). Moreover, gene silencing may be mediated by the recruitment of repressor proteins by methyl CpG binding proteins to promoters (15), or by interference with TF action. Notably, however, some CpG-containing promoters can be both methylated and transcriptionally active (16, 17).
How DNA methylation affects the binding of transcriptional regulators is currently unknown. It has long been speculated that steric occlusion by a bulky methyl group of the cognate recognition sequence of a TF could affect its binding affinity (18). However, this putative mechanism leaves various observations unaccounted for. For instance, some TFs interact with the major groove, yet are not affected by DNA methylation despite the extra methyl group protruding in the major groove. Other TFs interact with the minor groove, yet are affected by DNA methylation. Increased TF occupancy upon DNA methylation within their recognition sites has also been observed (19).
An explanation for these phenomena might be found in the 3D structure of DNA (20). We recently showed that DNA shape plays an important role in protein–DNA recognition (21–24). TFs can form direct and specific contacts with functional groups of the bases in the major groove. This base readout mechanism, however, does not suffice for the minor groove, where instead subtle sequence-dependent variation in DNA shape is read out by charged amino acid side chains via local variation in electrostatic potential (22, 25). Here, we expand this line of thought by analyzing the impact of adding a bulky methyl group in the major groove on the geometry of the minor groove. DNase I is an ideal molecule for asking this question, as it exclusively interacts with DNA via the minor groove.
Under standard conditions, successful molecular recognition of double-stranded DNA by DNase I leaves a permanent record in the form of hydrolysis of the O3′-P bond between the phosphorus and the oxygen attached to the 3′ carbon of the deoxyribose sugar within one of the strands of the recognition sequence (26). We reasoned that massively parallel sequencing could be applied to characterize millions of such events in a single experiment, enabling precise reconstruction of the sequence features that influence this interaction with the genome in vivo. Indeed, as described below, we find that the intrinsic DNase I cleavage rate varies over three orders of magnitude with immediate hexamer context.
Existing cocrystal structures of the DNase I–DNA complex reveal that DNase I docks in the minor groove of DNA (2). It has previously been suggested that sequence dependencies in DNase I cleavage rate might reflect differences in DNA shape, and specifically the configuration of the minor groove (27). Two separate studies have previously used autoradiogram data from DNase I digestions of a small number of end-labeled DNA fragments to quantify the sequence preferences of DNase I (28, 29). Their respective models, however, showed little correlation (29), and consequently the details of the intrinsic specificity of DNase I remained elusive when we began our study.
Since cytosine methylation covalently alters DNA, it may also influence DNase I cleavage rate. Here we show that integrating DNase I cleavage data with bisulfite sequencing data for genomic DNA purified from the same cell type reveals marked (>eightfold) enhancement of DNA backbone cleavage directly adjacent to CpG dinucleotides. Many TFs derive part of their DNA binding specificity from interactions with the minor groove. NMR studies have previously shown that a CpG dinucleotide flanked by A-tracts undergoes a severe narrowing at its center (30). By examining the effect of CpG methylation on DNA geometry for hundreds of sequence contexts, our results provide a specific structural mechanism that may explain how DNA methylation affects regulatory factor binding and gene expression.
Results and Discussion
Data Generation.
To quantify the sequence sensitivity spectrum of DNase I cleavage, we digested purified, deproteinated DNA from human fibroblast (IMR90) cells to an average size of ∼300 bp using DNase I (SI Methods). DNA fragment ends were resolved to the nearest 3′-strand cleavage through end repair (SI Methods) and sequencing adapter ligation. We then obtained 15 million single-end 36 bp Illumina sequence reads and mapped these to the human genome sequence, discarding any reads that did not map to unique genomic positions. This provided us with a large sample of individual, nucleotide-resolution cleavage events across the genome (SRA accession SRX247626).
Modeling Intrinsic DNase I Specificity Reveals Strong Sequence Preferences.
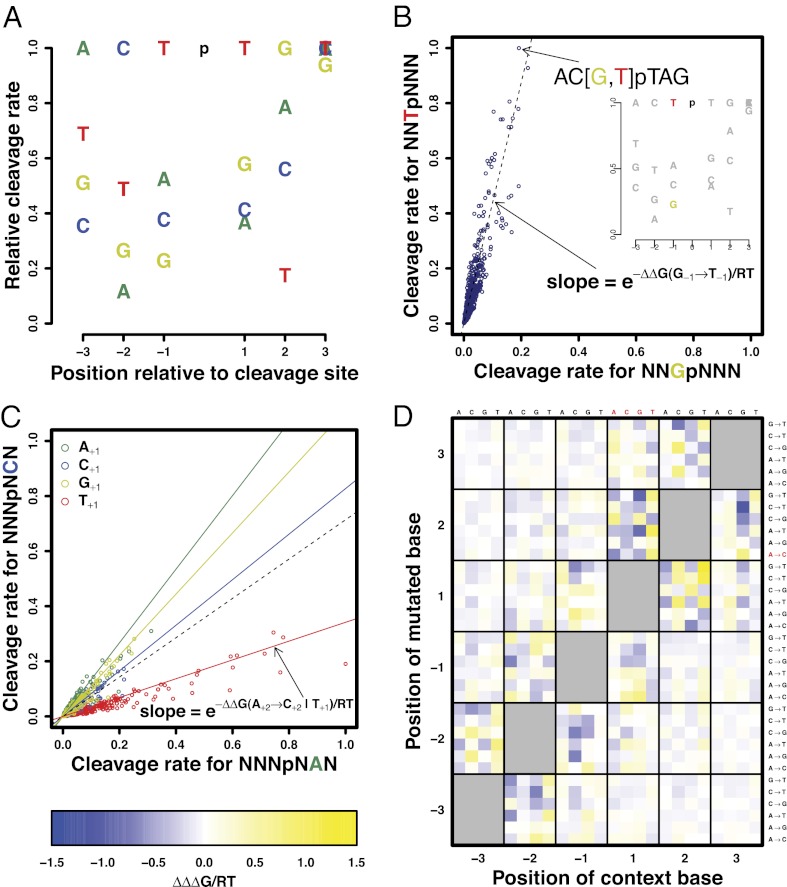
We next developed a model that quantifies the relative rate of cleavage by DNase I in terms of local DNA sequence context. We first asked over what spatial range of nucleotide positions this rate depends on base pair identity. As only relative rates are meaningful, we normalized by the most cleavable base at each nucleotide position (Table S1). Far enough from the cleaved bond, these relative rates are expected to become equal to unity. Indeed, a plot of information content versus nucleotide position (Fig. S1) shows that the dependence on base identity is largely limited to a window from 3 nt upstream (position −3) to 3 nt downstream (position +3) of the cleaved bond (Fig. 1A). This finding is consistent with crystallographic data on the protein–DNA interface of the DNase I–DNA complex (26). To the extent that the sequence sensitivity of DNase I cleavage is dominated by variation in its equilibrium DNA binding affinity (26), these relative cleavage rates are given by exp(−ΔΔG/RT), where ΔΔG represents the difference in binding free energy with the optimal DNA sequence context (see SI Methods for details).
Fig. 1.
Deep sequencing reveals striking positional dependencies between nucleotide positions within the DNase I recognition site. (A) Position-specific relative cleavage rate parameters as derived from DNase I digestion of human genomic DNA (normokaryotypic IMR90 fibroblasts) under the assumption of independence between nucleotides. Dependence on local sequence context is largely limited to a hexamer centered at the cleaved backbone bond. (B) Comparison between cleavage rates for pairs of hexamers that are related by a single-nucleotide substitution. The slope of the dashed line corresponds to the position-specific cleavage rate in panel A, and is directly related to the “unconditional” ΔΔG, the change in binding free energy associated with the point mutation. The fold change in cleavage rate due to a mutation from G to T at position −1 is largely independent of the base identity of the five neighboring nucleotides. (C) Breakdown of the independence assumption (dashed line). The effect on cleavage rate of a point mutation from A to C at position +2 is highly dependent on the base identity at the “modulating” position +1. Using a “conditional” ΔΔG for each possible base at position +1 (colored lines) provides a far more accurate description. (D) The strength of the positional dependencies can be quantified in terms of a new quantity “ΔΔΔG,” defined as the difference between the conditional and unconditional ΔΔG. The values in the highlighted row and columns correspond to the ratio in slope between each of the colored solid lines and the dashed line in C. Far away from the diagonal, ΔΔΔG becomes numerically small (white in heat map), indicating an increasing degree of independence.
The richness and depth of our dataset enabled us to estimate the relative cleavage rate for each of the 4,096 possible hexamer contexts. To this end, we divided the number of observed mappable cleavage events by the number of mappable genomic positions for each hexamer, and normalized by the highest such ratio (Table 1, Dataset S1). Unexpectedly, we found this rate to vary with local hexamer context over almost three orders of magnitude (Fig. S2A). We also found the cleavage to exhibit strong strand specificity (Fig. S2B). To assess reproducibility, we randomly partitioned the mappable genomic positions into training and test sets of equal size. The rates inferred from each set (Fig. S3A) are highly correlated (R2 = 0.99), indicating high reproducibility.
Table 1.
Hexamer-based model of relative DNase I cleavage rate
| Hexamer | Observed cuts | Genomic position | Ratio | Scaled ratio |
| ACTpTAG | 90,964 | 1,092,889 | 0.08323 | 1.00000 |
| ACTpTGT | 99,223 | 1,284,748 | 0.07723 | 0.92790 |
| ACTpTGG | 91,281 | 1,360,831 | 0.06708 | 0.80590 |
| ACTpTAA | 119,341 | 1,840,040 | 0.06486 | 0.77924 |
| TCTpTAG | 85,512 | 1,335,788 | 0.06402 | 0.76912 |
| CGGpTTT | 10 | 201,805 | 0.00005 | 0.00060 |
| CGCpGCG | 3 | 81,371 | 0.00004 | 0.00044 |
| GACpGCG | 0 | 49,356 | 0.00000 | 0.00000 |
For each subclass of phosphates, as defined by the sequence of a hexamer window centered at each phosphate, the total number of cleavage events and the total number of mappable genomic positions were determined. Ratios of these counts were then taken and scaled to a maximum of unity for the most cleavable hexamer.
We also performed a direct comparison both with the trimer-based hexamer model for relative cleavage rate defined by Brukner et al. (29) and the weight-matrix for preferred hexamer contexts defined by Herrera and Chaires (28). Reassuringly, we find rather good agreement between our hexamer-level cleavage rate table and the model of Herrera and Chaires (R2 = 0.54). Interestingly, Brukner et al. (29) reported that their model showed little correlation with that of Herrera and Chaires (28). Our own analysis confirms this (R2 = 0.01). We also observe low correlation between Brukner’s trimer-based predictions and our model (R2 = 0.01). A plausible explanation is that the model of Brukner et al. (29) alone assumes reverse-complement symmetry, whereas we find our tables to be strongly strand-specific (R2 = 0.33 when comparing forward hexamers with their reverse complement).
Dissecting Dependencies Between Nucleotide Positions.
Analysis of position-specific cleavage rates for each recognized hexamer revealed significant dependencies between nucleotide positions (Fig. S3B). In some cases, single-nucleotide variations in the hexamer sequence behaved independently. For example, in Fig. 1B, a single parameter (the slope of the dashed line) suffices to summarize all 45 different point mutations of type NNT|NNN → NNG|NNN (with the mutated base in bold and the site of cleavage indicated). However, most sensitivity to sequence variation was highly interdependent. Fig. 1C shows that for the substitution A+2→C+2 a single slope (dashed line) does not suffice to summarize its effect on cleavage rate. Rather, there are two distinct diagonals, with different slopes. The points on the lower diagonal can be perfectly demarcated by the occurrence of a T at the modulating position +1 (Fig. 1C). As expected, the strength of the dependency between the mutated and modulating position—quantified here as the difference, “ΔΔΔG,” between the conditional and unconditional ΔΔG values—tends to be largest when these positions are adjacent (Fig. 1D, Fig. S4 A and B). Still, dependencies of high statistical significance can be detected throughout the binding site (Fig. S5), underscoring the power of the massive sequence sampling approach.
Minor Groove Width Profile Is Predictive of DNase I Cleavage Rate.
The positional dependencies identified above hinted at the importance of 3D DNA structure. Indeed DNase I is known to interact with the minor groove of DNA (2, 26). We therefore asked whether a quantitative relationship exists between minor groove width (MGW) and cleavage rate. To this end, we used a high-throughput (HT) approach that can predict MGW at the center of any pentanucleotide to predict MGW across all six nucleotide positions for each of the 46 possible hexamers. This model was derived from a database of Monte Carlo (MC) simulations for a large number of free DNA sequences (see SI Methods for details). Since the hexamers occur as part of longer double-stranded DNA sequences, we accounted for the influence of flanking sequence by averaging over all possible ways of adding a dinucleotide flank on each side. We used a base-pair–centric coordinate system in which the MGW at position +1 is measured between the phosphate group connecting the +2 and +3 nucleosides on the forward strand and that connecting the −2 and −1 position on the reverse strand, etc.
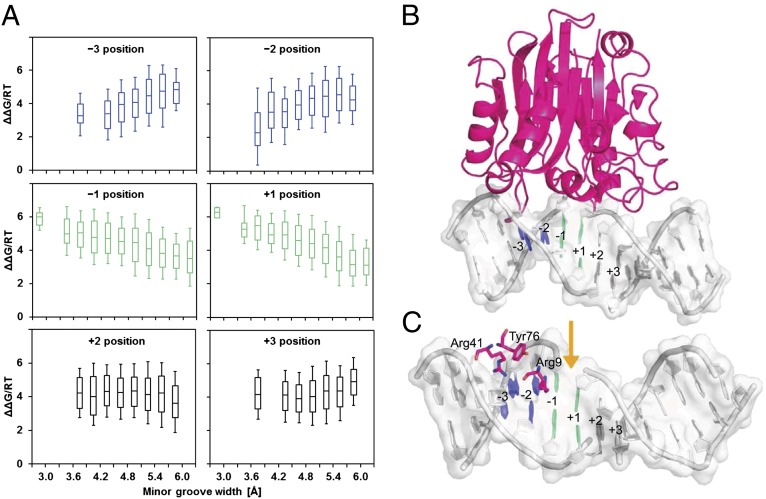
To assess to what extent the variation in DNA shape might explain the observed variation in DNase I cleavage rate, we first plotted the negative of the logarithm of the relative DNase I cleavage rate as a function of MGW at each base pair position. We interpret this negative logarithm as a binding free energy difference ΔΔG between a given sequence and the optimal sequence for DNase I cleavage. This analysis revealed a clear partitioning of the hexamer into three parts (Fig. 2A): at positions −3 and −2 a narrow minor groove is highly significantly associated with higher cleavage rate (with the t values measuring the regression coefficient in units of its SE equal to +19.8 and +15.1, respectively); at positions −1 and +1 this relationship is reversed but still highly significant (t values −15.6 and −26.3); at positions +2 and +3 a less strong association is observed (t values −6.0 and +6.4). The spatial profile of correlation between MGW and DNase I cleavage rate is consistent with features of a crystal structure of a complex of DNase I with a nicked DNA octamer duplex (31) (Fig. 2B). In that structure, an arginine, Arg41, from DNase I can be seen to interact with the minor groove near the −3 position, while a second arginine, Arg9, contacts the minor groove between the −2 and −1 positions (Fig. 2C). The narrower the minor groove is in the 5′ region of the hexamer at the −3 and −2 positions, the higher the cleavage rate is.
Fig. 2.
MGW is predictive of DNase I cleavage rate. (A) ΔΔG derived from the negative logarithm of cleavage rate as a function of MGW at the six positions of all 4,096 unique hexamers. MGW of this region was predicted for naked binding sites based on a pentamer-based HT shape prediction approach (SI Methods). HT predictions for all possible 16 dinucleotide flanks were averaged and values of MGW that fall within intervals of 0.3 Å assigned to groups of sequences for which cleavage rates are shown as box plots. (B) DNase I–DNA complex based on crystal structure (Protein Data Bank ID code 2DNJ). Base pairs at positions −3 and −2, where DNase I cleavage anticorrelates with MGW, are highlighted in blue. Base pairs at positions −1 and +1, where DNase I cleavage correlates positively with MGW, are highlighted in green. Regions where no correlation could be detected are shown in gray. The color code of the base pairs in the crystal structure is equivalent to the one used for the box plots. (C) DNase I–minor groove contacts within a distance of 5 Å from any base atom are shown for the same crystal structure. Arg41 and Arg9 bind upstream of the cleavage site, where MGW anticorrelates with DNase I cleavage (blue base pairs). This anticorrelation likely arises from the attraction between the positively charged arginine residues and the locally enhanced negative electrostatic potential. The cleavage site (indicated by the orange arrow), by contrast, is located in a region where MGW correlates positively with DNase I cleavage (green base pairs).
The relationship between MGW and DNase I cleavage rate indicates a recognition mechanism similar to the recently described binding of arginine residues to narrow regions of the minor groove (23). Such minor groove shape readout is based on the enhancement of negative electrostatic potential in narrow groove regions, which in turn allows for a stronger interaction with positively charged arginine residues (22). The increase in DNase I cleavage rate with narrowing of the minor groove is likely to be based on the attraction of the two arginine side chains through such locally enhanced negative electrostatic potential. The opposite sign of the correlation between MGW and cleavage rate at the −1 and +1 positions (Fig. 2A) also makes structural sense. Earlier reports have shown that the phosphodiester backbone at purine–pyrimidine (RpY) dinucleotides, which intrinsically widen the minor groove, are cleaved by DNase I at higher rates (32, 33). Having a widened minor groove where the backbone is cleaved would thus seem to be beneficial (34).
CpG Methylation Greatly Enhances Adjacent DNase I Cleavage.
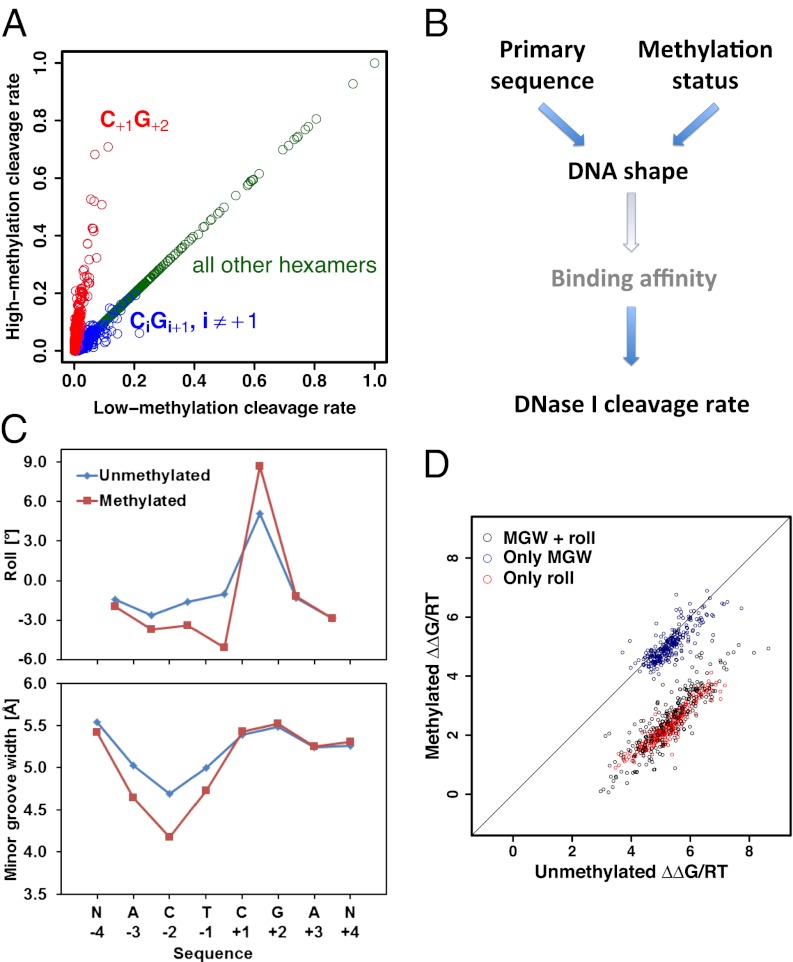
The results above indicate that molecular recognition of DNA by DNase I is subject to significant dependencies between nucleotides, consistent with readout of specific features of DNA shape (29). Since DNA methylation has the potential to alter the structural properties of DNA (35), we sought to analyze the influence of methylation on protein–DNA binding. To this end, we used whole-genome shotgun bisulfite sequencing data obtained from IMR90 cells (36) to define two subsets of phosphate positions, with hexamer contexts containing only hypermethylated or only hypomethylated CpG dinucleotides (SI Methods). Direct comparison of cleavage rates between both sets revealed a striking dependency on methylation status for a subset of the hexamers (Fig. 3A). A systematic search for DNA sequence features that could explain this dependency (Fig. S6) revealed that it is almost completely explained by the occurrence of a CpG dinucleotide immediately downstream of the cleaved bond (Fig. 3A). Upon methylation of the two cytosines within the C+1G+2 base pair step, the rate of cleavage by DNase I is enhanced ∼eightfold (red points in Fig. 3A), and for the most cleavable CpG-containing hexamer (ACT|CGA) increases from ∼7% to ∼68% of the maximum. Our findings are consistent with, but greatly extend, an earlier observation that methylation of the central cytosine in the sequence GCGC renders the 5′ phosphate more susceptible to cleavage by DNase I (37, 38).
Fig. 3.
Observation and analysis of the effect of methylation on DNase I cleavage rate. (A) The rate of cleavage depends strongly on the DNA methylation status. We used a positional map of DNA methylation in IMR90 (36) to delineate subsets of genomic positions with low/high degrees of CpG methylation, respectively. Comparison between the hexamer cleavage rates derived from these respective subsets shows an ∼eightfold increase in cleavage rate for hexamers with a methylated CpG immediately downstream of the cleaved phosphate (red points). (B) Interplay between DNA sequence and methylation status, DNA geometry, and DNase I cleavage suggested by our analysis. (C) Roll and MGW of methylated and unmethylated versions of the same hexamer based on the average of MC predictions for three different flanking sequences (see SI Methods for details). Methylation leads to an increase in the positive roll angle at the CpG dinucleotide and a narrowing of the MGW at position −2 by roughly 0.5 Å. (D) The effect of methylation on DNase I cleavage can be predicted in silico by training a model to predict the cleavage rates of unmethylated DNA sequences of type NNNCGN using information on DNA MGW and roll angle along these same unmethylated sequences. An increase in cleavage rate (i.e., data points shifting downward) is predicted when MGWs and roll angles for the methylated versions of the sequences are supplied as input to the model.
CpG Methylation Narrows the Minor Groove at Adjacent Positions.
So far, we have described two independent observations regarding DNase I as it acts on naked DNA. First, its cleavage rate depends on the primary sequence context via the width of the minor groove (Fig. 2A). Second, this rate increases by a multiplicative factor when the cytosines in the CpG base pair step immediately 3′ of the cleaved phosphate are methylated (Fig. 3A). We wondered if a direct relationship exists between methylation and MGW, as that would have the potential to unify both observed phenomena (Fig. 3B). Specifically, we asked whether methylation intrinsically leads to a narrowing of the minor groove, which in turn would explain the observed increase in cleavage rate upon methylation. To test this hypothesis, we extended the MC algorithm (see SI Methods for details) so that it could also predict the shape of free DNA molecules containing 5-methylcytosine bases. We first applied it to the most cleavable hexamer, ACTCGA. Strikingly, we observed that CpG methylation leads to an increased roll angle at the CpG step and a narrowing of the minor groove (Fig. 3C). Roll is the angle between two adjacent base pair planes describing the opening of a dinucleotide to either the minor or major groove. The narrowing of the minor groove is most pronounced (∼0.5 Å) at position −2, which is exactly where according to Fig. 2A we expect it to have the biggest positive impact on cleavage rate (Figs. S7–S9).
Modulation of DNA Shape Explains the Methylation Sensitivity of DNase I.
Encouraged by the above result, we reasoned that if DNase I cleavage rate indeed only depends on primary sequence and methylation status to the extent that the latter modulates DNA shape, we should be able to test this explicitly. To this end, we performed multiple linear regression of ΔΔG on a set of structural parameters that together quantify the local shape of the double-stranded DNA around the site of imminent cleavage. As predictor variables we used both the MGW at each base pair position within the hexamer and the roll angle associated with each base pair step, derived from MC simulations of 256 sequences in their unmethylated and methylated forms. This combined model explains a third (adjusted R2 = 0.34) of the variance in ΔΔG across all 256 unmethylated sequences of type NNNCGN. The most statistically significant regression coefficients are those for the roll between positions +1 and +2 and that for the MGW at position −2. This again is consistent with what is known about the structure of the DNase I–DNA complex. The predictive power of the model is diminished when only MGW or only roll parameters are used (adjusted R2 = 0.08 and 0.22, respectively).
Having thus constructed a model capable of predicting cleavage rate from DNA shape for unmethylated sequences, we used it to analyze the functional consequences of the DNA shape changes caused by cytosine methylation. Specifically, we predicted the value of DNase I cleavage ΔΔG for the methylated and unmethylated versions of each NNNCGN hexamer from its shape parameters alone (as predicted by the MC method), using the coefficients from a model trained on unmethylated shape and cleavage data for the remaining 255 hexamers. The results are shown as red points in Fig. 3D, and are characterized by a striking shift in ΔΔG upon methylation, which is largely independent of the identity of the hexamer. Here again, including the roll angles as predictors in the model is crucial for capturing the full effect of methylation (compare with black and blue points in Fig. 3D). A plausible explanation is that roll angles between adjacent base pairs are more directly related to changes in the chemical structure of the nucleobases than is the MGW. We note that an additive change in ΔΔG upon CpG methylation is consistent with the multiplicative change for the relative cleavage rate itself seen in Fig. 3A. The average shift in binding free energy predicted by the full model (ΔΔG/RT = 3.3) is equivalent to a ∼25-fold increase in cleavage rate. This is of the same order of magnitude but smaller than the eightfold seen in Fig. 3A. The latter, however, should be taken as a lower bound, as our classification of CpG dinucleotides in the genome in terms of their methylation status was necessarily imperfect given the ∼15× coverage level of the bisulfite sequencing data used.
Predicting Genomic DNA Methylation Status from in Vivo DNase I Profiles.
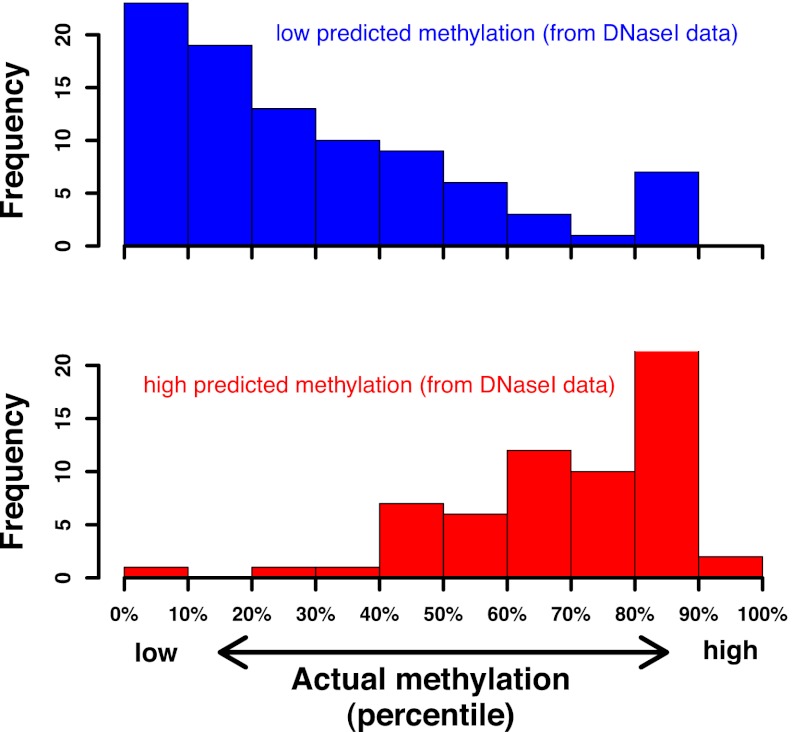
The striking enhancement of DNase I cleavage adjacent to methylated CpG dinucleotides motivated us to ask whether DNA methylation status could be inferred directly from in vivo DNase I profiles. To this end, we mapped 200 million in vivo DNase I cleavages from IMR90 fibroblasts (SI Methods; GEO accession GSM723024). We inferred regional CpG methylation within 2.5 kb genomic windows attaining a threshold read depth of >400 CpG-adjacent cleavages (corresponding to a sample error of at most 5%). For each window, we computed the expected number of CpG-adjacent cuts, as a function of the DNA sequence, the total number of cuts, and our hexamer-based, methylation-blind model of DNase I specificity. The observed–expected ratio for each window then served as a predictor of its methylation status. Fig. 4 shows results of these computations for 455 nonoverlapping windows, together accounting for 1.13 Mbp of genomic DNA. A given window was predicted to be hypermethylated whenever the number of CpG-adjacent cleavages was at least 120% of the expected value (SI Methods), and hypomethylated when that number was below 50% of expected. To validate our predictions, we inspected the actual methylation level in the IMR90 cells (36). We found that this level was below the median in 80% of the windows predicted to be hypomethylated and above the median in 84% of those predicted to be hypermethylated. These results demonstrate that regional DNA methylation status can indeed be inferred from the in vivo DNase I profile with reasonable accuracy as long as the local coverage is dense enough.
Fig. 4.
Genomic methylation status can be predicted from dense in vivo DNase I footprints. Starting from in vivo DNase I footprinting data for the IMR90 cell line, a set of nonoverlapping windows (2,500 bp long) containing at least 400 cleavage events with hexamer context NNNpCGN was identified. Next, for each window, the observed number of cleavages upstream of CpG dinucleotides was compared with the expected number. This allowed us to infer the methylation status of the corresponding DNA. To validate our predictions, we ranked all windows by their actual degree of methylation as measured by Lister et al. (36). Shown is the distribution of ranks for the subset of windows predicted to be hypomethylated (red) and hypermethylated (blue), respectively.
Conclusion
In this work, we have discovered the existence of a strong DNA-shape–driven sequence dependence of DNase I cleavage, which can be leveraged to map DNA shape at single-nucleotide resolution on a genomic scale. DNase I emerges from our study as a highly sensitive probe of the geometry of the minor groove. The latter serves as an important recognition site within the DNA-binding interface of many regulatory proteins. It has been previously noted that DNA binding proteins such as DNase I may be regarded as structural probes of DNA conformation and flexibility (33). Moreover, although the interaction between DNA and its binding proteins is known to result in conformational changes in both molecules (31, 39, 40), the present work shows that intrinsic DNA shape is an important recognition signal. The depth of the HT data used in this study has allowed us to study this phenomenon at an unprecedented level of resolution and quantification.
The intrinsic DNase I cleavage rate changes greatly with each single-nucleotide shift in position along the genome, and it self-averages over genomic windows as small as a few dozen nucleotides. Taking into account our hexamer model, therefore, does not significantly affect the detection sensitivity of regional DNase I hypersensitivity, nor does the intrinsic sequence-dependent variation in cleavage rate seem to result in false-positive DNase I footprints (41). However, the unprecedented accuracy of our model for intrinsic DNase I cleavage specificity has the potential to enhance the interpretation of high-resolution DNase I cleavage patterns (11, 42).
Additionally, we show that DNase I is so exquisitely sensitive to the changes in DNA shape caused by CpG methylation that in vivo DNA methylation patterns can be inferred directly from high-density in vivo DNase I cleavage profiles. Our demonstration that it is possible to harness the methylation sensitivity of DNase I to infer cell-type–specific DNA methylation status provides a unique and complementary tool for analysis of domain-level methylation patterns in conjunction with chromatin state changes during development (43) or disease (44).
Finally, and perhaps most importantly, our in-depth study of DNase I allowed us to uncover a structural mechanism that plausibly answers the long-standing question of how cytosine methylation modulates protein–DNA interaction. Our data strongly suggest that DNA methylation generally acts to narrow the minor groove. DNase I activity is sensitive to this change in DNA conformation, and this explains our observation of greatly enhanced cleavage adjacent to methylated CpG base pair steps. However, we believe that our insight could apply much more widely across many families of nucleotide-binding proteins. Narrowing of the MGW may thus be the general mechanism by which the addition or removal of methyl groups in the major groove influences gene expression. An intriguing possibility is that nucleosome positioning might be influenced by methylation (45, 46). Recently, an observed correlation between these two variables was interpreted as influence of nucleosome positioning on methylation patterning (45). However, electrostatic interactions between arginines and the minor groove occur in the nucleosome (22, 47), and the minor groove narrowing associated with cytosine methylation could enhance these. The methylation patterns might therefore also be a partial determinant of nucleosome position, with methylated CpG dinucleotides giving rise to stronger electrostatic interactions with histones, increasing the stability of nucleosomes.
Methods
Please see SI Methods for additional details on cell culture and DNA extraction; DNase I treatment of purified DNA; and digital DNase I mapping in IMR90 cells. There is also information on library construction and sequencing; single-nucleotide model; hexamer model, outlier removal, dependency between mutated and modulating positions, statistical significance of nucleotide dependencies, high-throughput (HT) prediction of minor groove width (MGW), Monte Carlo (MC) prediction of DNA structures for unmetylated and methylated DNA, dependence of DNase I cleavage rate on DNA methylation status, and inferring methylation status from in vivo DNase I footprints.
Supplementary Material
Acknowledgments
The authors wish to thank other members of the Bussemaker, Rohs, and Stamatoyannopoulos labs for valuable discussions. This research was funded by National Institutes of Health Grants R01HG003008 and U54CA121852 (to H.J.B.), U54HG004592 (to J.S.), USC start-up funds and Grant IRG-58-007-51 from the American Cancer Society (to R.R.). H.J.B. is a John Simon Guggenheim Fellow, and R.R. is an Alfred P. Sloan Research Fellow.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1216822110/-/DCSupplemental.
References
- 1.Oliveri M, et al. DNase I behaves as a transcription factor which modulates Fas expression in human cells. Eur J Immunol. 2004;34(1):273–279. doi: 10.1002/eji.200223817. [DOI] [PubMed] [Google Scholar]
- 2.Suck D, Lahm A, Oefner C. Structure refined to 2A of a nicked DNA octanucleotide complex with DNase I. Nature. 1988;332(6163):464–468. doi: 10.1038/332464a0. [DOI] [PubMed] [Google Scholar]
- 3.Weston SA, Lahm A, Suck D. X-ray structure of the DNase I-d(GGTATACC)2 complex at 2.3 A resolution. J Mol Biol. 1992;226(4):1237–1256. doi: 10.1016/0022-2836(92)91064-v. [DOI] [PubMed] [Google Scholar]
- 4.Hogan ME, Roberson MW, Austin RH. DNA flexibility variation may dominate DNase I cleavage. Proc Natl Acad Sci USA. 1989;86(23):9273–9277. doi: 10.1073/pnas.86.23.9273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heddi B, Abi-Ghanem J, Lavigne M, Hartmann B. Sequence-dependent DNA flexibility mediates DNase I cleavage. J Mol Biol. 2010;395(1):123–133. doi: 10.1016/j.jmb.2009.10.023. [DOI] [PubMed] [Google Scholar]
- 6.Dorschner MO, et al. High-throughput localization of functional elements by quantitative chromatin profiling. Nat Methods. 2004;1(3):219–225. doi: 10.1038/nmeth721. [DOI] [PubMed] [Google Scholar]
- 7.Sabo PJ, et al. Discovery of functional noncoding elements by digital analysis of chromatin structure. Proc Natl Acad Sci USA. 2004;101(48):16837–16842. doi: 10.1073/pnas.0407387101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.John S, et al. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat Genet. 2011;43(3):264–268. doi: 10.1038/ng.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sabo PJ, et al. Genome-scale mapping of DNase I sensitivity in vivo using tiling DNA microarrays. Nat Methods. 2006;3(7):511–518. doi: 10.1038/nmeth890. [DOI] [PubMed] [Google Scholar]
- 10.Galas DJ, Schmitz A. DNAse footprinting: A simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978;5(9):3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hesselberth JR, et al. Global mapping of protein-DNA interactions in vivo by digital genomic footprinting. Nat Methods. 2009;6(4):283–289. doi: 10.1038/nmeth.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pennings S, Allan J, Davey CS. DNA methylation, nucleosome formation and positioning. Brief Funct Genomics Proteomics. 2005;3(4):351–361. doi: 10.1093/bfgp/3.4.351. [DOI] [PubMed] [Google Scholar]
- 13.Suzuki MM, Bird A. DNA methylation landscapes: Provocative insights from epigenomics. Nat Rev Genet. 2008;9(6):465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 14.Kangaspeska S, et al. Transient cyclical methylation of promoter DNA. Nature. 2008;452(7183):112–115. doi: 10.1038/nature06640. [DOI] [PubMed] [Google Scholar]
- 15.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16(1):6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 16.Eckhardt F, et al. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat Genet. 2006;38(12):1378–1385. doi: 10.1038/ng1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weber M, et al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39(4):457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- 18.Rozenberg JM, et al. All and only CpG containing sequences are enriched in promoters abundantly bound by RNA polymerase II in multiple tissues. BMC Genomics. 2008;9:67. doi: 10.1186/1471-2164-9-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rishi V, et al. CpG methylation of half-CRE sequences creates C/EBPalpha binding sites that activate some tissue-specific genes. Proc Natl Acad Sci USA. 2010;107(47):20311–20316. doi: 10.1073/pnas.1008688107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rohs R, West SM, Liu P, Honig B. Nuance in the double-helix and its role in protein-DNA recognition. Curr Opin Struct Biol. 2009;19(2):171–177. doi: 10.1016/j.sbi.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Joshi R, et al. Functional specificity of a Hox protein mediated by the recognition of minor groove structure. Cell. 2007;131(3):530–543. doi: 10.1016/j.cell.2007.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rohs R, et al. The role of DNA shape in protein-DNA recognition. Nature. 2009;461(7268):1248–1253. doi: 10.1038/nature08473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rohs R, et al. Origins of specificity in protein-DNA recognition. Annu Rev Biochem. 2010;79:233–269. doi: 10.1146/annurev-biochem-060408-091030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Slattery M, et al. Cofactor binding evokes latent differences in DNA binding specificity between Hox proteins. Cell. 2011;147(6):1270–1282. doi: 10.1016/j.cell.2011.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kitayner M, et al. Diversity in DNA recognition by p53 revealed by crystal structures with Hoogsteen base pairs. Nat Struct Mol Biol. 2010;17(4):423–429. doi: 10.1038/nsmb.1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suck D. DNA recognition by DNase I. J Mol Recognit. 1994;7(2):65–70. doi: 10.1002/jmr.300070203. [DOI] [PubMed] [Google Scholar]
- 27.Drew HR, Travers AA. DNA structural variations in the E. coli tyrT promoter. Cell. 1984;37(2):491–502. doi: 10.1016/0092-8674(84)90379-9. [DOI] [PubMed] [Google Scholar]
- 28.Herrera JE, Chaires JB. Characterization of preferred deoxyribonuclease I cleavage sites. J Mol Biol. 1994;236(2):405–411. doi: 10.1006/jmbi.1994.1152. [DOI] [PubMed] [Google Scholar]
- 29.Brukner I, Sánchez R, Suck D, Pongor S. Sequence-dependent bending propensity of DNA as revealed by DNase I: Parameters for trinucleotides. EMBO J. 1995;14(8):1812–1818. doi: 10.1002/j.1460-2075.1995.tb07169.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marcourt L, Cordier C, Couesnon T, Dodin G. Impact of C5-cytosine methylation on the solution structure of d(GAAAACGTTTTC)2. An NMR and molecular modelling investigation. Euro J Biochem. 1999;265(3):1032–1042. doi: 10.1046/j.1432-1327.1999.00819.x. [DOI] [PubMed] [Google Scholar]
- 31.Lahm A, Suck D. DNase I-induced DNA conformation. 2 A structure of a DNase I-octamer complex. J Mol Biol. 1991;222(3):645–667. doi: 10.1016/0022-2836(91)90502-w. [DOI] [PubMed] [Google Scholar]
- 32.Lomonossoff GP, Butler PJ, Klug A. Sequence-dependent variation in the conformation of DNA. J Mol Biol. 1981;149(4):745–760. doi: 10.1016/0022-2836(81)90356-9. [DOI] [PubMed] [Google Scholar]
- 33.Brukner I, Jurukovski V, Savic A. Sequence-dependent structural variations of DNA revealed by DNase I. Nucleic Acids Res. 1990;18(4):891–894. doi: 10.1093/nar/18.4.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bishop EP, et al. A map of minor groove shape and electrostatic potential from hydroxyl radical cleavage patterns of DNA. ACS Chem Biol. 2011;6(12):1314–1320. doi: 10.1021/cb200155t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adams RL. DNA methylation. The effect of minor bases on DNA-protein interactions. Biochem J. 1990;265(2):309–320. doi: 10.1042/bj2650309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lister R, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462(7271):315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fox KR. The effect of HhaI methylation on DNA local structure. Biochem J. 1986;234(1):213–216. doi: 10.1042/bj2340213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kochanek S, Renz D, Doerfler W. Differences in the accessibility of methylated and unmethylated DNA to DNase I. Nucleic Acids Res. 1993;21(25):5843–5845. doi: 10.1093/nar/21.25.5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.N’soukpoé-Kossi CN, Diamantoglou S, Tajmir-Riahi HA. DNase I - DNA interaction alters DNA and protein conformations. Biochem Cell Biol. 2008;86(3):244–250. doi: 10.1139/o08-039. [DOI] [PubMed] [Google Scholar]
- 40.Meijsing SH, et al. DNA binding site sequence directs glucocorticoid receptor structure and activity. Science. 2009;324(5925):407–410. doi: 10.1126/science.1164265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Neph S, et al. An expansive human regulatory lexicon encoded in transcription factor footprints. Nature. 2012;489(7414):83–90. doi: 10.1038/nature11212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang X, et al. Using RNase sequence specificity to refine the identification of RNA-protein binding regions. BMC Genomics. 2008;9(Suppl 1):S17. doi: 10.1186/1471-2164-9-S1-S17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lister R, et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471(7336):68–73. doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hansen KD, et al. Increased methylation variation in epigenetic domains across cancer types. Nat Genet. 2011;43(8):768–775. doi: 10.1038/ng.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chodavarapu RK, et al. Relationship between nucleosome positioning and DNA methylation. Nature. 2010;466(7304):388–392. doi: 10.1038/nature09147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kelly TK, et al. Genome-wide mapping of nucleosome positioning and DNA methylation within individual DNA molecules. Genome Res. 2012;22(12):2497–2506. doi: 10.1101/gr.143008.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.West SM, Rohs R, Mann RS, Honig B. Electrostatic interactions between arginines and the minor groove in the nucleosome. J Biomol Struct Dyn. 2010;27(6):861–866. doi: 10.1080/07391102.2010.10508587. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.