Abstract
The accumulation of damaged mitochondria has been proposed as a key factor in aging and the pathogenesis of many common age-related diseases, including Parkinson disease (PD). Recently, in vitro studies of the PD-related proteins Parkin and PINK1 have found that these factors act in a common pathway to promote the selective autophagic degradation of damaged mitochondria (mitophagy). However, whether Parkin and PINK1 promote mitophagy under normal physiological conditions in vivo is unknown. To address this question, we used a proteomic approach in Drosophila to compare the rates of mitochondrial protein turnover in parkin mutants, PINK1 mutants, and control flies. We found that parkin null mutants showed a significant overall slowing of mitochondrial protein turnover, similar to but less severe than the slowing seen in autophagy-deficient Atg7 mutants, consistent with the model that Parkin acts upstream of Atg7 to promote mitophagy. By contrast, the turnover of many mitochondrial respiratory chain (RC) subunits showed greater impairment in parkin than Atg7 mutants, and RC turnover was also selectively impaired in PINK1 mutants. Our findings show that the PINK1–Parkin pathway promotes mitophagy in vivo and, unexpectedly, also promotes selective turnover of mitochondrial RC subunits. Failure to degrade damaged RC proteins could account for the RC deficits seen in many PD patients and may play an important role in PD pathogenesis.
Understanding the mechanisms of mitochondrial quality control is a critical challenge in research on neurodegeneration and aging. The accumulation of damaged mitochondria has been linked to normal aging and multiple age-related disorders, including Alzheimer’s disease, diabetes, and Parkinson disease (PD) (1, 2). Recent research points to two PD-associated proteins as essential mediators of selective autophagic mitochondrial degradation: phosphatase and tensin homolog-induced putative kinase 1 (PINK1), a mitochondrially targeted serine/threonine kinase, and Parkin, a cytosolic E3 ubiquitin ligase. Genetic studies in Drosophila determined that PINK1 acts upstream of Parkin in a common pathway to regulate mitochondrial morphology and integrity (3–8), and led to the hypothesis that this pathway promotes the selective degradation of damaged mitochondria (6, 9). Subsequent experiments, primarily in cultured cells, validated this hypothesis and described the mechanism of action of the pathway (10–12). These studies showed that loss of mitochondrial membrane potential (depolarization) leads to accumulation of PINK1 on the mitochondrial outer membrane, which triggers recruitment of Parkin to the mitochondria. Parkin then ubiquitinates proteins in the outer mitochondrial membrane (13–17), leading to autophagic degradation of the dysfunctional mitochondrion.
Although there is substantial support for the role of the PINK1–Parkin pathway in selective mitochondrial degradation, it is still not clear that this pathway promotes mitochondrial degradation in vivo. PINK1–Parkin-dependent mitophagy has been documented only in toxin-treated cultured cells, usually in the presence of overexpressed Parkin (18). Studies in cultured neurons have yielded mixed results (12, 19, 20), and in vivo experiments have thus far failed to show either Parkin recruitment or mitophagy in neurons with dysfunctional mitochondria (21, 22). In addition, the in vitro findings on PINK1–Parkin-dependent mitophagy represent the cellular response to sudden, catastrophic mitochondrial dysfunction rather than the response to gradual accumulation of damage. Because PINK1–Parkin pathway dysfunction is implicated in aging and slowly progressive disorders, it is necessary to study the mechanisms of mitochondrial turnover under physiological conditions.
To test whether the PINK1–Parkin pathway promotes mitochondrial degradation in vivo, we used a proteomic assay to measure the influence of PINK1 and Parkin on mitochondrial protein turnover in Drosophila. We found that parkin mutant flies had a significantly decreased rate of mitochondrial protein turnover, similar to the decrease produced by general autophagy blockade, supporting the hypothesis that Parkin promotes mitochondrial turnover through autophagy. Results from PINK1 mutants were more complex, suggesting a mitophagy deficit largely masked by compensatory up-regulation of an alternative turnover pathway. In addition, we found evidence that both Parkin and PINK1 are involved in selective turnover of respiratory chain (RC) subunits. Together, our findings indicate that the PINK1–Parkin pathway promotes mitochondrial autophagy in vivo and also mediates selective turnover of mitochondrial RC proteins.
Results
To measure mitochondrial turnover in vivo, we developed a proteomic assay that enabled us to compare the half-lives of mitochondrial proteins in PINK1 and parkin mutants with their half-lives in normal flies. We fed adult flies a stable isotope label [[5,5,5–2H3]-leucine (D3-leucine)] and used mass spectrometry (MS) to monitor the rates at which unlabeled proteins were degraded and replaced by labeled proteins. Data were analyzed with Topograph (23), a recently described software platform that computes protein half-lives based on the distribution of partially and fully labeled peptides. We were thus able to measure the half-lives of many mitochondrial proteins simultaneously, yielding a more detailed picture of mitochondrial turnover than afforded by either radiolabeling or monitoring of individual mitochondrial proteins.
Mitochondrial Proteins Have a Wide Range of Evolutionarily Conserved Half-Lives.
We first performed general characterization of fly protein half-lives. To reduce sample complexity and enrich for neural tissue, we performed all experiments on homogenates from adult heads. We fed 1-d-old adult flies from three different genetic backgrounds yeast paste fully labeled with D3-leucine (Fig. 1A). At each labeling time point, we prepared postnuclear protein homogenates from heads and subjected the homogenates to trypsin digestion, MS, and Topograph analysis. The 524 proteins identified from MS that met our quality control standards (Materials and Methods) showed an extensive range of half-lives, with mitochondrial proteins generally longer-lived than nonmitochondrial proteins (Fig. S1 and Dataset S1). The broad range of mitochondrial protein half-lives is consistent with the fact that, although autophagy degrades mitochondria as units, mitochondrial protein turnover also occurs through mitochondrial proteases (24) and the ubiquitin–proteasome system (25, 26). Protein half-life values were highly reproducible, with low sensitivity to genetic background (Fig. S2 A and B). Protein half-life was also evolutionarily conserved: half-lives of proteins from fly heads correlated strongly with half-lives of homologous proteins in mouse brain (Fig. S2 C and D) [mouse data from the work by Price et al. (27)].
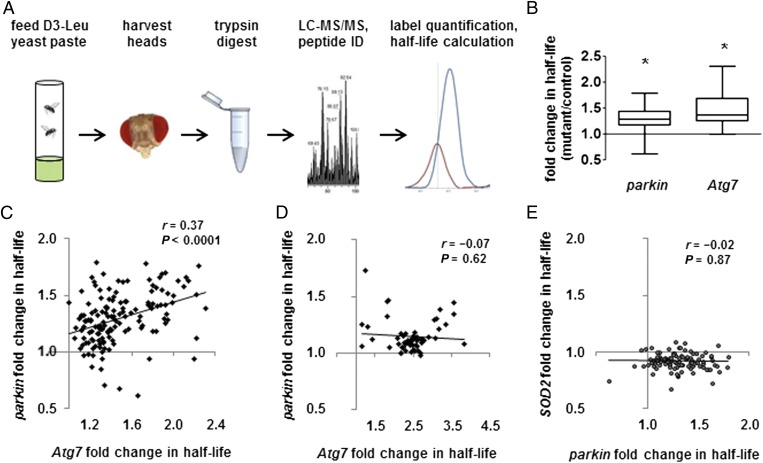
Fig. 1.
Parkin promotes mitophagy in vivo. (A) Experimental workflow. (B) parkin and Atg7 mutations prolong mitochondrial protein half-life. Box-and-whisker plots of fold change in half-life show median, quartiles, and extreme values. parkin mean and SD = 1.30 ± 0.22; Atg7 = 1.47 ± 0.30. *P < 0.005, mutant vs. control, by nested ANOVA. (C and D) The effects of parkin and Atg7 mutations on half-life correlate significantly for (C) individual mitochondrial proteins but not for (D) proteins from other organellar targets of autophagy (ribosomes, endoplasmic reticulum, and peroxisomes). n = 147 mitochondrial proteins; n = 58 other organellar proteins. All correlations reported as Pearson r. (E) The effects of parkin mutation on mitochondrial protein half-life do not correlate significantly with the effects of SOD2 deficiency (n = 103).
Parkin Is Required for Mitophagy in Vivo.
We next compared the half-lives of mitochondrial proteins in parkin null flies and controls. We hypothesized that mitochondrial protein half-lives would be prolonged in parkin null mutants because of impaired mitophagy. We also measured half-lives in autophagy-deficient Atg7 null mutants as a positive control. Because Atg5/Atg7-dependent autophagy acts downstream from Parkin (10, 28) and also mediates nonselective forms of mitochondrial degradation (29), we hypothesized that Atg7 null mutants would have a deficit in mitochondrial protein turnover similar to, but more severe than, the deficit in parkin null mutants. To test these hypotheses, we compared the half-lives of 156 mitochondrial proteins from parkin mutants and 170 mitochondrial proteins from Atg7 mutants with their half-lives in control flies. All proteins that met quality control standards in each dataset were included, and all conclusions remained valid when analyses were restricted to proteins that were in both datasets (n = 147). For each protein, we divided mutant half-life by control half-life to compute fold change in half-life.
Consistent with our hypothesis, mitochondrial proteins in parkin and Atg7 mutants had significantly prolonged half-lives compared with controls (Fig. 1B and Dataset S2). Also as hypothesized, lack of Atg7 had a stronger effect on mitochondrial protein turnover than lack of Parkin (Fig. 1B). Furthermore, we found a significant positive correlation between the effects of parkin and Atg7 mutations on the half-lives of the 147 mitochondrial proteins that were in both datasets (Fig. 1C). This relationship was specific to mitochondrial proteins; there was no such correlation between the effects of parkin and Atg7 mutations on the half-lives of proteins from other organellar targets of autophagy (Fig. 1D). Moreover, the parkin–Atg7 correlation did not simply reflect general mitochondrial dysfunction; the effect of mutation in another mitochondrial quality control gene, superoxide dismutase 2 (SOD2), did not correlate with the effect of mutation in either parkin or Atg7 (Fig. 1E and Fig. S3A). Together, these findings support the model that Parkin promotes mitophagy in vivo.
Parkin Mediates Selective Turnover of Mitochondrial Respiratory Chain Proteins.
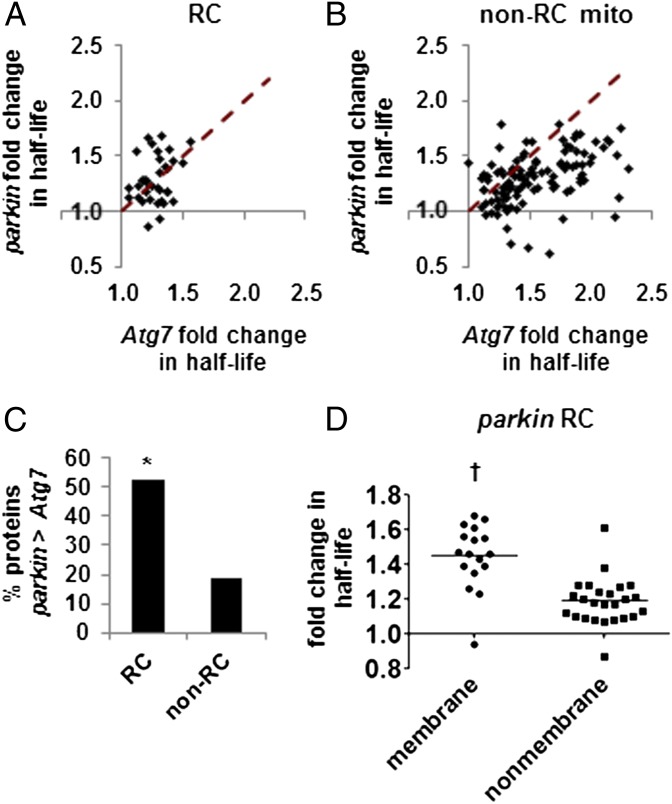
The turnover of most mitochondrial proteins showed greater dependence on Atg7 than Parkin, which was expected if Atg7 acts downstream from Parkin and also mediates nonselective mitochondrial turnover. However, the turnover of 40 mitochondrial proteins showed greater dependence on Parkin than Atg7 (Dataset S2 has a full list). Such proteins potentially represent targets of a Parkin-dependent turnover process that is independent of Atg7. Among these 40 proteins, RC components were strikingly overrepresented, which was confirmed by χ2 analysis (10/40 predicted, 19/40 observed, P = 0.003). The effect of parkin mutation on turnover exceeded the effect of Atg7 for fully 53% of all RC proteins detected, compared with only 19% of all other mitochondrial proteins (Fig. 2 A–C). The RC proteins with greater turnover dependence on Parkin represented all five respiratory complexes and included the single mitochondrially encoded RC protein present in both datasets, indicating that inefficient import of nuclear-encoded RC proteins into dysfunctional mitochondria is an unlikely explanation for the selective effects of parkin on RC proteins. A disproportionately large number of membrane-bound RC subunits (11 of 14 total membrane-bound subunits vs. 8 of 22 subunits not directly anchored to a membrane) was included among the 19 RC subunits that showed greater dependence on Parkin than Atg7. Other mitochondrial membrane proteins were not overrepresented, suggesting that Parkin’s greater effects on membrane-bound proteins are specific to the RC. In addition, Parkin had a greater mean effect on turnover of membrane-bound RC subunits than on turnover of nonmembrane subunits (Fig. 2D). Our findings thus suggest that, in addition to its role in mitophagy, Parkin has a selective effect on the turnover of RC proteins.
Fig. 2.
Parkin has a selective effect on turnover of RC proteins. (A and B) Plots comparing the influence of parkin and Atg7 on the half-lives of RC proteins (A) and all other mitochondrial proteins (B). Dashed lines indicate equal effect from both mutations; the half-lives of proteins above the dashed line are more greatly influenced by parkin mutation than Atg7 mutation. (C) The percentage of RC and non-RC proteins with half-lives that are more greatly affected by parkin than Atg7 mutation (n = 36 RC proteins; n = 111 non-RC). The RC is significantly enriched in proteins with greater parkin than Atg7 effect on half-life. *P = 0.003 by χ2 test. (D) Mutation in parkin has a larger effect on the half-lives of membrane-bound RC subunits than on those of nonmembrane RC subunits (mean fold change = 1.45 ± 0.19 vs. 1.19 ± 0.14). Horizontal lines indicate the median. †P = 4.8 × 10−6 by Student t test.
PINK1 Is Required for Selective RC Turnover.
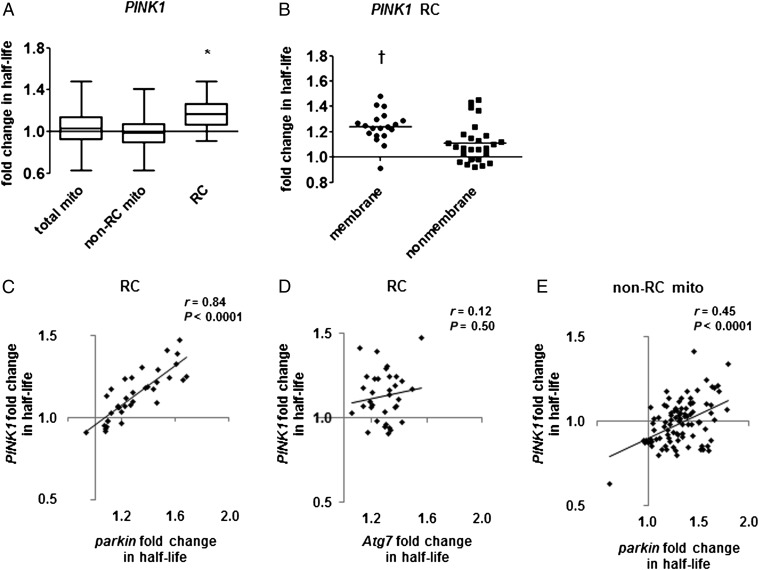
Previous work has shown that PINK1 is required for Parkin-mediated mitophagy after treatment of cultured cells with mitochondrial depolarizing agents (11, 12), and genetic studies in Drosophila show that PINK1 null mutants have similar phenotypes to parkin null mutants (4, 5). We therefore anticipated that PINK1 mutants would have mitochondrial protein turnover defects similar to those seen in parkin mutants. However, although there was a trend toward increased half-lives of mitochondrial proteins in PINK1 mutants, this difference did not reach significance (Fig. 3A). Additional analysis revealed that the RC was the only mitochondrial protein subgroup with a significantly increased mean half-life in PINK1 mutants (Fig. 3A). Like parkin mutants, PINK1 mutants showed greater mean fold change in half-life for membrane-bound than for nonmembrane RC subunits (Fig. 3B). The effect of PINK1 mutation on RC turnover correlated strongly with the effect of parkin mutation on the same proteins (Fig. 3C), but neither the PINK1 nor the parkin effect on RC correlated with the effect of Atg7 (Fig. 3D and Fig. S3B). Together, our findings suggest that the PINK1–Parkin pathway promotes selective turnover of RC proteins in a manner independent of conventional autophagy.
Fig. 3.
PINK1 null mutants have a selective impairment of RC protein turnover. (A) PINK1 null mutation prolongs the mean half-life of RC proteins but not other mitochondrial proteins. Box-and-whisker plots of fold change in half-life show median, quartiles, and extreme values. Mean fold change: total mito = 1.04 ± 0.16 (n = 147 proteins; P = 0.082 by nested ANOVA), non-RC mito = 0.99 ± 0.13 (n = 102), RC = 1.17 ± 0.16 (n = 45). *P < 0.0005, mutant vs. control, by nested ANOVA. (B) Mutation in PINK1 has a larger effect on membrane-bound than on nonmembrane RC subunits (mean fold change = 1.24 ± 0.13 vs. 1.11 ± 0.16). Horizontal lines indicate the median. †P < 0.005 by t test. (C and D) The effects of PINK1 mutation on RC protein half-lives strongly correlate with (C) the effects of mutation in parkin but not (D) the effects of mutation in Atg7 (n = 36 for parkin; n = 34 for Atg7). (E) The effects of PINK1 mutation on the half-lives of non-RC mitochondrial proteins correlate significantly with the effects of parkin mutation (n = 94). The regression line has a negative y intercept, suggesting a uniform shift to faster mitochondrial protein turnover in PINK1.
Compensatory Turnover Masks a Mitophagy Deficit in PINK1 Mutants.
The apparent lack of impairment in general mitochondrial protein turnover in PINK1 mutants is surprising given that PINK1 is essential for Parkin-mediated mitophagy in vitro (12). However, studies in Drosophila indicate that Parkin retains some activity in the absence of PINK1 (4, 5, 13). This residual Parkin activity may be sufficient to support basal mitophagy, which seems to occur at a low rate; the longest-lived mitochondrial proteins had half-lives in excess of 30 d in normal flies (Dataset S1). PINK1 may thus be required for mitophagy only under conditions of extreme mitochondrial stress, such as acute ischemia (30) or toxin treatment of cultured cells. Alternatively, because increased autophagy has been described in PINK1-deficient cells and animals (31–33), PINK1 mutants could have a deficit in mitophagy that is masked by compensatory increase in another form of protein turnover. Although these possibilities are not mutually exclusive, our data seem to support the idea of compensation in PINK1 mutants. Although PINK1 mutants show no mean change in turnover of non-RC mitochondrial proteins, the effect of PINK1 mutation on those proteins correlates well with the effect seen in parkin mutants (Fig. 3E). This correlation seems to reflect a uniform increase in mitochondrial protein turnover in PINK1 mutants relative to parkin mutants, such that the proteins with the smallest turnover impairment in parkin mutants actually have slightly increased turnover rates in PINK1 mutants relative to control. Our findings thus suggest that compensatory degradation masks a mitophagy defect in PINK1 mutants.
Discussion
We have provided evidence that the PINK1–Parkin pathway promotes mitophagy in vivo and described an additional role for this pathway in the selective nonmitophagic turnover of RC proteins. Loss of PINK1 and/or Parkin activity has previously been reported to cause RC deficits, particularly in complex I (34–36), and complex I dysfunction has repeatedly been implicated in the pathogenesis of PD (37). Our findings suggest that selective impairment of RC protein turnover may explain the RC deficits seen in both familial and sporadic PD patients. Impaired turnover could, for example, lead to the accumulation of misfolded RC proteins, previously noted in PINK1 and parkin mutant flies (38). Complex I, the largest and most intricate of the RC complexes, may be the most vulnerable to dysfunction under such conditions.
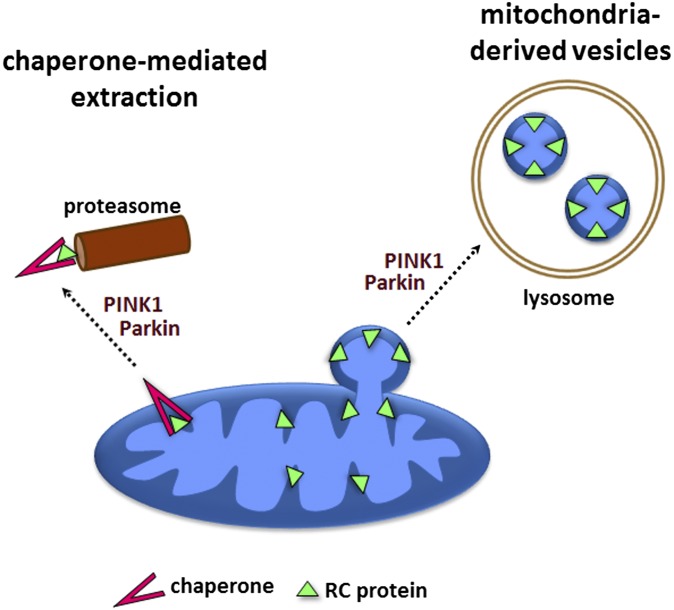
The mechanism by which PINK1 and Parkin promote selective RC turnover requires additional elucidation. Although there are several possible explanations for our findings, existing literature suggests two particularly plausible models (Fig. 4). One model involves chaperone-mediated extraction of mitochondrial proteins, which is described by Margineantu et al. (39). Another possible model involves mitochondria-derived vesicles, which have been shown to transport selected mitochondrial cargo to the lysosome in an autophagy-independent manner (40). Such vesicles have been shown to transport a membrane-bound complex IV subunit and contain inner mitochondrial membrane (41), thus offering a potential explanation for the greater effect of PINK1 and parkin mutations on membrane-bound RC components. Additional studies will be needed to determine both the mechanism of RC turnover and the contribution of RC turnover defects to the pathogenesis of PD.
Fig. 4.
Possible mechanisms of selective RC turnover. (1) Chaperone-mediated extraction and proteasomal degradation. (2) Transport to the lysosome through mitochondria-derived vesicles.
Materials and Methods
Drosophila Strains and Culture.
Fly stocks were maintained on standard cornmeal molasses food at 25 °C. The park25, parkrvA, PINK1B9, PINK1rv, Atg7d4, Atg7d77, SOD2n283, and SOD2wk alleles have been previously described (3, 5, 42–44). Other strains and alleles were obtained from the Bloomington Stock Center. Detailed fly genotypes are listed in SI Materials and Methods and Table S1.
In Vivo Stable Isotope Labeling of Flies.
D3-leucine (99 atom percent deuterium) was obtained from Isotec/Sigma-Aldrich. Synthetic complete medium without leucine was supplemented with glucose and 60 mg/L D3-leucine. A strain of Saccharomyces cerevisiae auxotrophic for leucine (BB14-3A; Brewer Laboratory, University of Washington) (45) was grown to saturation at 30°, and then spun down, flash-frozen in liquid nitrogen, lyophilized, and stored at −80 °C until needed. Groups of 10–50 male flies were selected on the day of eclosion and housed in perforated plastic flasks, where they received plain yeast paste for 24 h. They were then provided with D3-leucine–labeled yeast paste, which was replaced every 2–3 d, and they were maintained in humidified containers at 25 °C. After 120 or 240 h of labeling (the shortest time points that allowed adequate labeling of mitochondrial proteins), flies were flash-frozen in liquid nitrogen. Three biological replicates (50–115 heads each) were obtained for each genotype and time point.
Sample Preparation.
Frozen flies were vortexed to remove heads, and the isolated heads were homogenized in 0.1% RapiGest solution in 50 mM ammonium bicarbonate (Waters Corporation) using a Wheaton 0.2-mL micro tissue grinder (Wheaton). Homogenates were centrifuged at 4 °C at 1,600 × g for 10 min and then 6,000 × g for 10 min to remove debris and nuclei. The supernatants were then boiled for 7 min and incubated with DTT (final concentration = 5 mM) at 60 °C for 30 min. Iodoacetamide was added to a final concentration of 15 mM, and the samples were incubated at room temperature in the dark for 30 min. Trypsin (Promega) was added at a ratio of 1 μg trypsin per 50 μg protein and incubated for 1 h at 37 °C with shaking. RapiGest was hydrolyzed by adding HCl to a final concentration of 200 mM, followed by incubation at 37 °C with shaking for 45 min. The samples were then centrifuged for 10 min at 4 °C at 20,000 × g, and the supernatant was collected.
MS.
MS was performed on an LTQ-Orbitrap instrument (Thermo Fisher) with NanoACQUITY UPLC system (Waters Corporation). Two analytical replicates were obtained for each biological replicate. SI Materials and Methods has additional details.
Protein Half-Life Calculations.
Protein half-lives were calculated using the Topograph software platform, version 1.0 (23). A protein’s half-life was computed based on data from all peptides detected. Each protein was represented by at least 15 total values of percent newly synthesized per genotype and at least two peptides. Peptides that could be the product of more than one gene were excluded from analysis. Data points from all biological replicates were pooled for half-life calculations. SI Materials and Methods has a full description of Topograph settings.
We did not analyze the few proteins with a control half-life ≥1,000 h, which accumulated too little tracer within the study period to ensure reliable quantification. We also excluded proteins with excessive variability of percent newly synthesized values, defined as follows. We divided the 95% confidence interval for each half-life (generated by Topograph) by the half-life value itself, creating a measure analogous to coefficient of variation. Proteins with a 95% confidence interval/half-life ratio ≥0.3 were excluded from analysis.
Half-life means for mitochondrial proteins were compared across genotypes using one-way nested ANOVA (biological replicates nested under genotypes). For significance testing, a separate half-life was computed for each biological replicate.
Protein Abundance Measurement.
Topograph half-life computations assume that protein synthesis rate equals degradation rate, and therefore that protein abundance remains constant throughout the experiment. Although constant protein abundance is important for the accurate calculation of half-life, the accurate calculation of fold change in half-life between genotypes requires only that any change in protein abundance is the same in both genotypes. To address this issue, we assessed protein abundance change over the study period in both mutants and controls. SI Materials and Methods has details of calculation and statistical analysis, and Dataset S3 shows statistics on individual proteins. There was no significant difference between mutant and control in abundance pattern over time for any mitochondrial protein in the SOD2 dataset. In the PINK1 dataset, 0.7% of mitochondrial proteins (1 of 145 proteins for which significance could be tested; mitochondrial sublocalization unknown) showed different abundance patterns over time in mutant and control. In the parkin dataset, 3.4% of mitochondrial proteins (5 of 149) showed different abundance patterns. Four of these proteins were matrix proteins, and one was a non-RC protein of the mitochondrial inner membrane. In the Atg7 dataset, 3.0% of mitochondrial proteins (5 of 166) showed different abundance patterns (2 mitochondrial proteins with mitochondrial sublocalization unknown, 1 inner membrane mitochondrial protein, and 2 RC mitochondrial proteins). Excluding the proteins with differential abundance change from our analyses had no significant effect on mean fold change in half-life for mitochondrial proteins in any dataset (Table S2). Similarly, the reported correlations of mutation effect (e.g., parkin vs. Atg7) were not significantly altered by exclusion of proteins with differential abundance change (Table S3). Our findings thus cannot be explained in terms of abundance change.
Protein Classification and Annotation.
Drosophila protein localization and orthology to mouse proteins were determined from a variety of resources, including gene and protein information databases (FlyBase, MitoDrome, National Center for Biotechnology Information, and UniProt), protein localization prediction algorithms (WoLF PSORT, MitoProt, Predotar, SignalP, NucPred, and PTS1 Predictor), BLAST, and primary literature. Proteins were classified as exclusively mitochondrial (M), nonmitochondrial (N), or mitochondrial with significant secondary localization (M/N). The inclusion of M/N proteins did not significantly alter mean fold change in half-life in any dataset. Also, M/N proteins showed the same relationship between parkin and Atg7 effects as M proteins (no difference in r values by Fisher r-to-z test, P = 0.77). M/N proteins were therefore included in all mitochondrial protein analyses.
Comparison with Mouse Half-Lives.
Mouse brain half-life data were obtained from Price et al. (27). Rate constant values (K0) for mouse brain proteins were converted to half-lives and compared with half-lives of orthologous proteins in the fly head data from Dataset S1.
Supplementary Material
Acknowledgments
We thank E. Hsieh and M. Bereman for advice on data analysis and J. K. Chung, A. Duttaroy, and T. Neufeld for fly stocks. We thank the Brewer Laboratory for assistance with yeast culture and G. Findlay for advice on isotope labeling. We also thank the following people for feedback on the manuscript: D. Gottschling, A. Hughes, C. Link, A. Whitworth, and members of the L.J.P. laboratory. This work was supported by National Institutes of Health Grants P30AG013280 (to M.J.M.), R01DK069386 (to M.J.M.), 5R01GM086394 (to L.J.P.), 3R01GM086394-01S1 (to L.J.P.), and P41GM103551 (to the Yeast Resource Center). This work was also supported by the University of Washington's Proteomics Resource (UWPR95794).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1221132110/-/DCSupplemental.
See Commentary on page 6252.
References
- 1.Karbowski M, Neutzner A. Neurodegeneration as a consequence of failed mitochondrial maintenance. Acta Neuropathol. 2012;123(2):157–171. doi: 10.1007/s00401-011-0921-0. [DOI] [PubMed] [Google Scholar]
- 2.Horan MP, Pichaud N, Ballard JW. Review: Quantifying mitochondrial dysfunction in complex diseases of aging. J Gerontol A Biol Sci Med Sci. 2012;67(10):1022–1035. doi: 10.1093/gerona/glr263. [DOI] [PubMed] [Google Scholar]
- 3.Greene JC, et al. Mitochondrial pathology and apoptotic muscle degeneration in Drosophila parkin mutants. Proc Natl Acad Sci USA. 2003;100(7):4078–4083. doi: 10.1073/pnas.0737556100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clark IE, et al. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature. 2006;441(7097):1162–1166. doi: 10.1038/nature04779. [DOI] [PubMed] [Google Scholar]
- 5.Park J, et al. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature. 2006;441(7097):1157–1161. doi: 10.1038/nature04788. [DOI] [PubMed] [Google Scholar]
- 6.Poole AC, et al. The PINK1/Parkin pathway regulates mitochondrial morphology. Proc Natl Acad Sci USA. 2008;105(5):1638–1643. doi: 10.1073/pnas.0709336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deng H, Dodson MW, Huang H, Guo M. The Parkinson’s disease genes pink1 and parkin promote mitochondrial fission and/or inhibit fusion in Drosophila. Proc Natl Acad Sci USA. 2008;105(38):14503–14508. doi: 10.1073/pnas.0803998105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang Y, et al. Pink1 regulates mitochondrial dynamics through interaction with the fission/fusion machinery. Proc Natl Acad Sci USA. 2008;105(19):7070–7075. doi: 10.1073/pnas.0711845105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Whitworth AJ, et al. Increased glutathione S-transferase activity rescues dopaminergic neuron loss in a Drosophila model of Parkinson’s disease. Proc Natl Acad Sci USA. 2005;102(22):8024–8029. doi: 10.1073/pnas.0501078102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183(5):795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Narendra DP, et al. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8(1):e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vives-Bauza C, et al. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc Natl Acad Sci USA. 2010;107(1):378–383. doi: 10.1073/pnas.0911187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poole AC, Thomas RE, Yu S, Vincow ES, Pallanck L. The mitochondrial fusion-promoting factor mitofusin is a substrate of the PINK1/parkin pathway. PLoS One. 2010;5(4):e10054. doi: 10.1371/journal.pone.0010054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ziviani E, Tao RN, Whitworth AJ. Drosophila parkin requires PINK1 for mitochondrial translocation and ubiquitinates mitofusin. Proc Natl Acad Sci USA. 2010;107(11):5018–5023. doi: 10.1073/pnas.0913485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang X, et al. PINK1 and Parkin target Miro for phosphorylation and degradation to arrest mitochondrial motility. Cell. 2011;147(4):893–906. doi: 10.1016/j.cell.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chan NC, et al. Broad activation of the ubiquitin-proteasome system by Parkin is critical for mitophagy. Hum Mol Genet. 2011;20(9):1726–1737. doi: 10.1093/hmg/ddr048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geisler S, et al. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010;12(2):119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 18.Jin SM, Youle RJ. PINK1- and Parkin-mediated mitophagy at a glance. J Cell Sci. 2012;125(Pt 4):795–799. doi: 10.1242/jcs.093849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Laar VS, et al. Bioenergetics of neurons inhibit the translocation response of Parkin following rapid mitochondrial depolarization. Hum Mol Genet. 2011;20(5):927–940. doi: 10.1093/hmg/ddq531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cai Q, Zakaria HM, Simone A, Sheng ZH. Spatial parkin translocation and degradation of damaged mitochondria via mitophagy in live cortical neurons. Curr Biol. 2012;22(6):545–552. doi: 10.1016/j.cub.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sterky FH, Lee S, Wibom R, Olson L, Larsson NG. Impaired mitochondrial transport and Parkin-independent degeneration of respiratory chain-deficient dopamine neurons in vivo. Proc Natl Acad Sci USA. 2011;108(31):12937–12942. doi: 10.1073/pnas.1103295108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee S, et al. Mitofusin 2 is necessary for striatal axonal projections of midbrain dopamine neurons. Hum Mol Genet. 2012;21(22):4827–4835. doi: 10.1093/hmg/dds352. [DOI] [PubMed] [Google Scholar]
- 23.Hsieh EJ, et al. Topograph, a software platform for precursor enrichment corrected global protein turnover measurements. Mol Cell Proteomics. 2012;11(11):1468–1474. doi: 10.1074/mcp.O112.017699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koppen M, Langer T. Protein degradation within mitochondria: Versatile activities of AAA proteases and other peptidases. Crit Rev Biochem Mol Biol. 2007;42(3):221–242. doi: 10.1080/10409230701380452. [DOI] [PubMed] [Google Scholar]
- 25.Neutzner A, Youle RJ, Karbowski M. Outer mitochondrial membrane protein degradation by the proteasome. Novartis Found Symp. 2007;287(2007):4–14. [PubMed] [Google Scholar]
- 26.Clarke KJ, et al. A role for ubiquitinylation and the cytosolic proteasome in turnover of mitochondrial uncoupling protein 1 (UCP1) Biochim Biophys Acta. 2012;1817(10):1759–1767. doi: 10.1016/j.bbabio.2012.03.035. [DOI] [PubMed] [Google Scholar]
- 27.Price JC, Guan S, Burlingame A, Prusiner SB, Ghaemmaghami S. Analysis of proteome dynamics in the mouse brain. Proc Natl Acad Sci USA. 2010;107(32):14508–14513. doi: 10.1073/pnas.1006551107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsuda N, et al. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J Cell Biol. 2010;189(2):211–221. doi: 10.1083/jcb.200910140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mizushima N. Autophagy: Process and function. Genes Dev. 2007;21(22):2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 30.Huang C, et al. Preconditioning involves selective mitophagy mediated by Parkin and p62/SQSTM1. PLoS One. 2011;6(6):e20975. doi: 10.1371/journal.pone.0020975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dagda RK, et al. Loss of PINK1 function promotes mitophagy through effects on oxidative stress and mitochondrial fission. J Biol Chem. 2009;284(20):13843–13855. doi: 10.1074/jbc.M808515200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu S, Lu B. Reduction of protein translation and activation of autophagy protect against PINK1 pathogenesis in Drosophila melanogaster. PLoS Genet. 2010;6(12):e1001237. doi: 10.1371/journal.pgen.1001237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qi Z, et al. Loss of PINK1 function decreases PP2A activity and promotes autophagy in dopaminergic cells and a murine model. Neurochem Int. 2011;59(5):572–581. doi: 10.1016/j.neuint.2011.03.023. [DOI] [PubMed] [Google Scholar]
- 34.Morais VA, et al. Parkinson’s disease mutations in PINK1 result in decreased Complex I activity and deficient synaptic function. EMBO Mol Med. 2009;1(2):99–111. doi: 10.1002/emmm.200900006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mortiboys H, et al. Mitochondrial function and morphology are impaired in parkin-mutant fibroblasts. Ann Neurol. 2008;64(5):555–565. doi: 10.1002/ana.21492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Amo T, et al. Mitochondrial membrane potential decrease caused by loss of PINK1 is not due to proton leak, but to respiratory chain defects. Neurobiol Dis. 2011;41(1):111–118. doi: 10.1016/j.nbd.2010.08.027. [DOI] [PubMed] [Google Scholar]
- 37.Zhu J, Chu CT. Mitochondrial dysfunction in Parkinson’s disease. J Alzheimers Dis. 2010;20(Suppl 2):S325–S334. doi: 10.3233/JAD-2010-100363. [DOI] [PubMed] [Google Scholar]
- 38.Pimenta de Castro I, et al. Genetic analysis of mitochondrial protein misfolding in Drosophila melanogaster. Cell Death Differ. 2012;19(8):1308–1316. doi: 10.1038/cdd.2012.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Margineantu DH, Emerson CB, Diaz D, Hockenbery DM. Hsp90 inhibition decreases mitochondrial protein turnover. PLoS One. 2007;2(10):e1066. doi: 10.1371/journal.pone.0001066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Soubannier V, et al. A vesicular transport pathway shuttles cargo from mitochondria to lysosomes. Curr Biol. 2012;22(2):135–141. doi: 10.1016/j.cub.2011.11.057. [DOI] [PubMed] [Google Scholar]
- 41.Neuspiel M, et al. Cargo-selected transport from the mitochondria to peroxisomes is mediated by vesicular carriers. Curr Biol. 2008;18(2):102–108. doi: 10.1016/j.cub.2007.12.038. [DOI] [PubMed] [Google Scholar]
- 42.Juhász G, Erdi B, Sass M, Neufeld TP. Atg7-dependent autophagy promotes neuronal health, stress tolerance, and longevity but is dispensable for metamorphosis in Drosophila. Genes Dev. 2007;21(23):3061–3066. doi: 10.1101/gad.1600707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Duttaroy A, Paul A, Kundu M, Belton A. A Sod2 null mutation confers severely reduced adult life span in Drosophila. Genetics. 2003;165(4):2295–2299. doi: 10.1093/genetics/165.4.2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Paul A, et al. Reduced mitochondrial SOD displays mortality characteristics reminiscent of natural aging. Mech Ageing Dev. 2007;128(11–12):706–716. doi: 10.1016/j.mad.2007.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McCune HJ, et al. The temporal program of chromosome replication: Genomewide replication in clb5Delta Saccharomyces cerevisiae. Genetics. 2008;180(4):1833–1847. doi: 10.1534/genetics.108.094359. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.