Abstract
The pronounced biological influence of the tumor microenvironment on cancer progression and metastasis has gained increased recognition over the past decade, yet most preclinical antineoplastic drug testing is still reliant on conventional 2D cell culture systems. Although monolayer cultures recapitulate some of the phenotypic traits observed clinically, they are limited in their ability to model the full range of microenvironmental cues, such as ones elicited by 3D cell–cell and cell–extracellular matrix interactions. To address these shortcomings, we established an ex vivo 3D Ewing sarcoma model that closely mimics the morphology, growth kinetics, and protein expression profile of human tumors. We observed that Ewing sarcoma cells cultured in porous 3D electrospun poly(ε-caprolactone) scaffolds not only were more resistant to traditional cytotoxic drugs than were cells in 2D monolayer culture but also exhibited remarkable differences in the expression pattern of the insulin-like growth factor-1 receptor/mammalian target of rapamycin pathway. This 3D model of the bone microenvironment may have broad applicability for mechanistic studies of bone sarcomas and exhibits the potential to augment preclinical evaluation of antineoplastic drug candidates for these malignancies.
Keywords: tissue engineering, tumor model, biological therapy, connective tissue
Despite the primacy of the cancer cell’s dysregulated genotype [e.g., a near universal translocation of the Ewing sarcoma (EWS) breakpoint region 1 gene in EWS cells] as the initial step in malignant transformation, it has become increasingly apparent that the overall tumor phenotype is also dictated by the 3D tumor microenvironment (1–4). Nonetheless, studies of cancer biology and evaluation of antineoplastic drug efficacy remain heavily dependent on conventional 2D cell culture systems despite their limited ability to reflect the 3D tumor architecture, extracellular matrix (ECM), and surrounding cell types that comprise the in vivo tumor milieu.
To overcome some of these constraints, 3D in vitro models such as spheroid and gel systems have been extensively studied and, compared with 2D monolayer culture, appear to better mimic the profound effects that the in vivo 3D environment has upon the human tumor phenotype (5–9). For example, malignant cells cultured in 3D exhibit increased chemoresistance (10, 11) and decreased cell proliferation (12), and assume specific phenotypes inducible only under a 3D context, such as angiogenic capability (13–15). Furthermore, striking differences in signaling pathways targeted by proven and experimental therapies have been observed in 3D tumor models (16–18). Accordingly, heightened awareness of the importance of 3D culture for cancer cells has resulted in the increasing use of spheroid culture systems for cancer research. However, these non–adhesion-mediated systems provide poor control over the tumor architecture and cell–cell interactions; as a result of culture conditions that prohibit cellular attachment onto surrounding surfaces, cells autonomously aggregate and form their own 3D geometry. An emerging strategy to overcome this inherent drawback of spheroid culture is to leverage 3D scaffolding technologies that have been developed for tissue-engineering applications (19), to guide tumor tissue formation in vitro with controlled and tunable architectural complexity.
In the present study, our interdisciplinary team of cancer biologists, tissue engineers, and clinicians sought to develop and validate a drug-testing tool that would elicit in vivo-like drug sensitivity and potentially advance mechanistic studies of disease biology through precise control of the in vitro tumor architecture. Focusing specifically on bone sarcomas, we established a 3D model of EWS, the second most common pediatric bone malignancy, by culturing TC-71 human EWS cells within electrospun polymeric scaffolds fabricated from poly(ε-caprolactone) (PCL), a biologically inert synthetic polymer previously investigated for use in bone and other tissue-engineering applications (20–23). These scaffolds have several favorable features, including high porosity, a large surface area-to-volume ratio for cellular attachment, tunable fiber diameter, low cost, ease of fabrication and handling, and high reproducibility (24, 25). With these 3D PCL scaffolds, we observed that ex vivo EWS traits were very similar to those in vivo with respect to morphology, growth kinetics, and protein expression, and were remarkably different from their monolayer counterparts throughout the insulin-like growth factor-1 receptor (IGF-1R)/mammalian target of rapamycin (mTOR)-related signaling cascade. IGF-1R and mTOR, in particular, are potential therapeutic targets for EWS under active investigation in human clinical trials, as their combined abrogation promoted striking antineoplastic responses in a subset of patients with chemotherapy-resistant EWS.
Results and Discussion
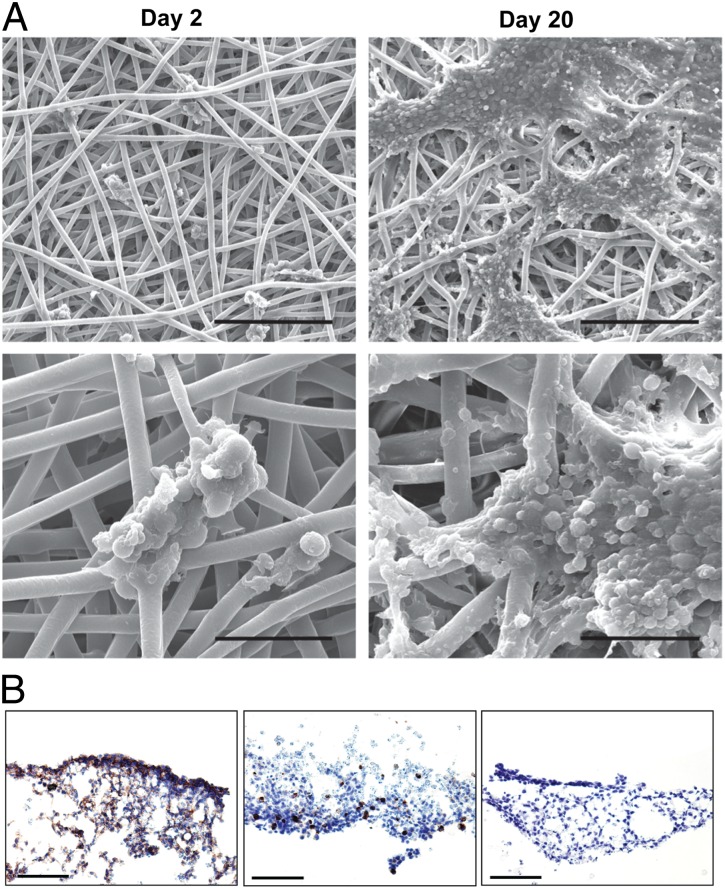
We engineered an ex vivo EWS model by culturing TC-71 cells in a porous 3D electrospun PCL scaffold. At 2 d in culture, TC-71 cells adhered in small, sparse clusters to individual PCL fibers and were located primarily on the top surface of the scaffold. With increased duration of culture (20 d), cells were found throughout the scaffold, with the majority located within the upper one-fifth of the scaffold (Fig. S1). Representative scaffolds collected after short- and long-term culture were imaged using scanning electron microscopy (Fig. 1A). Careful review of the resulting images by an experienced sarcoma pathologist identified the small round-cell morphology characteristic of human EWS tumors. Additionally, we used a well-validated panel of immunohistochemical markers routinely used for diagnostic purposes in patients (CD99+, keratin−, and smooth muscle actin−), which confirmed that our established in vitro 3D tumor model preserves a well-differentiated EWS-like phenotype (Fig. 1B).
Fig. 1.
Morphological characterization of 3D EWS model. (A) Scanning electron micrographs of TC-71 cells seeded in electrospun 3D PCL scaffolds at low magnification (Upper; scale bar, 200 μm) and high magnification (Lower; scale bar, 50 μm). By the 20th day in culture, cells formed sheet-like clusters and exhibited small, round-cell morphology, bearing close morphological resemblance to human EWS tumors. (B) Immunohistochemical validation of the EWS phenotype of cells cultured within the 3D PCL scaffold for 20 d using the clinical diagnostic markers CD99 (Left), keratin (Center), and smooth muscle actin (Right). (Scale bar, 100 μm.)
Paralleling a rapid increase in regulatory approval of biologically targeted drugs and “omics”-based technologies that have made tumor profiling commonplace, some of the most promising experimental agents being tested in the treatment of EWS in early-phase clinical trials exert their effect by antagonizing IGF-1R, mTOR, or other proteins in their shared signaling cascades (26, 27). Although a thorough discussion of the roles of IGF-1R and mTOR in EWS carcinogenesis is beyond the scope of this article, two decades of research indicate that their expression is critical, as the pathognomonic EWS breakpoint region 1–erythroblast transformation specific (EWSR1–ETS) translocation alone is insufficient to promote and maintain EWS oncogenesis, growth, and invasion (28–31).
Despite a wealth of preclinical information and two recent clinical trials demonstrating marked tumor regression in a subset of patients with EWS treated with combined antagonists of the IGF-1R/mTOR pathway, we noted a surprising lackluster cellular response to IGF-1R antagonists in conventional 2D monolayer culture (32, 33). In seeking to elucidate biomarkers of response and mechanisms of de novo and acquired resistance, we hypothesized that this difference in preclinical and clinical response may result from inherent changes in the IGF-1R/mTOR signaling cascade when tumor cells are removed from their native microenvironment, which we sought to recapitulate using our 3D PCL scaffold while maintaining control over experimental parameters.
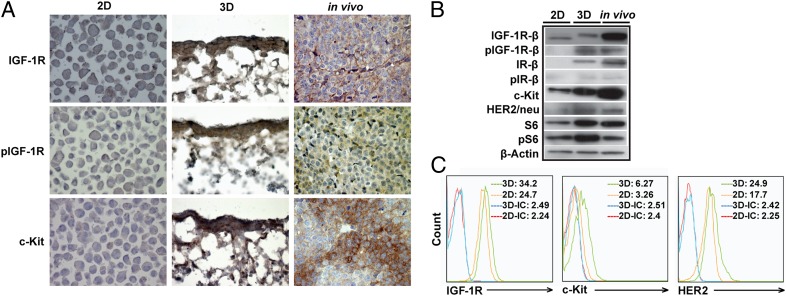
Striking differences were observed in the expression of proteins associated with the IGF-1R/mTOR signaling pathway using immunohistochemistry, Western blotting, and flow cytometry. First, expression of both IGF-1R and, in particular, phosphorylated IGF-1R (pIGF-1R), was significantly higher in cells that were cultured within the 3D PCL scaffolds and grown as EWS xenografts than cells in 2D monolayer culture, suggesting more in vivo-like, vigorous constitutive activation of IGF-1R signaling (Fig. 2 A and B). Additionally, we examined the expression of proteins associated with putative mechanisms of resistance to IGF-1R–targeted therapy, including c-kit and human epidermal growth factor receptor 2 (HER2/neu), and found higher expression of both proteins in the TC-71 xenografts as well as cells that were cultured within our 3D PCL scaffolds, compared with cells in 2D monolayer culture (Fig. 2 A and B). We confirmed this observation using flow cytometry, as indicated by the mean fluorescence levels for c-kit (3.26 versus 6.27) and HER2/neu (17.7 versus 24.9) in cells in 2D monolayer culture and the 3D PCL scaffolds, respectively (Fig. 2C). Taken together, these observations suggest that cells cultured in the 3D PCL scaffold may serve as a reliable ex vivo surrogate for xenografts, and potentially human EWS tumors, for evaluating the IGF-1R/mTOR pathway under both physiological and perturbed states.
Fig. 2.
Comparison of IGF-1R/mTOR signaling in TC-71 cells cultured in 2D monolayer culture to 80% confluency, in 3D PCL scaffold for 10 d or in vivo as subcutaneous (s.c.) tumors for 12 d. (A) Representative IGF-1R, pIGF-1R, and c-kit immunohistochemical staining of TC-71 cells cultured in 2D monolayer culture, in 3D PCL scaffold, and as xenograft tumors (in vivo). Weak expression of IGF-1R and pIGF-1R and lack of expression of c-kit in the 2D TC-71 cells was observed. In contrast, the expression of IGF-1R, pIGF-1R, and c-kit is much more pronounced when the same cells were grown in 3D. Importantly, the levels of expression of these proteins in the 3D culture conditions show more consistency with their in vivo levels of expression when TC-71 cells were implanted in nude mice (original magnification, 400×). (B) Comparison of expression of IGF-1R/mTOR pathway-related proteins in TC-71 cells cultured in 2D monolayer culture, in 3D PCL scaffold, and as xenograft tumors. Expression of pIGF-1R was strikingly higher in the 3D scaffolds than in 2D culture, suggesting constitutive activation of IGF-1R signaling (similar to that observed in vivo). Higher c-kit and HER2/neu expression in the 3D scaffolds than in 2D culture was also observed. (C) Flow-cytometric analysis of IGF-1R, c-kit, and HER2/neu expression in TC-71 cells cultured within 2D monolayer culture or in 3D PCL scaffold.
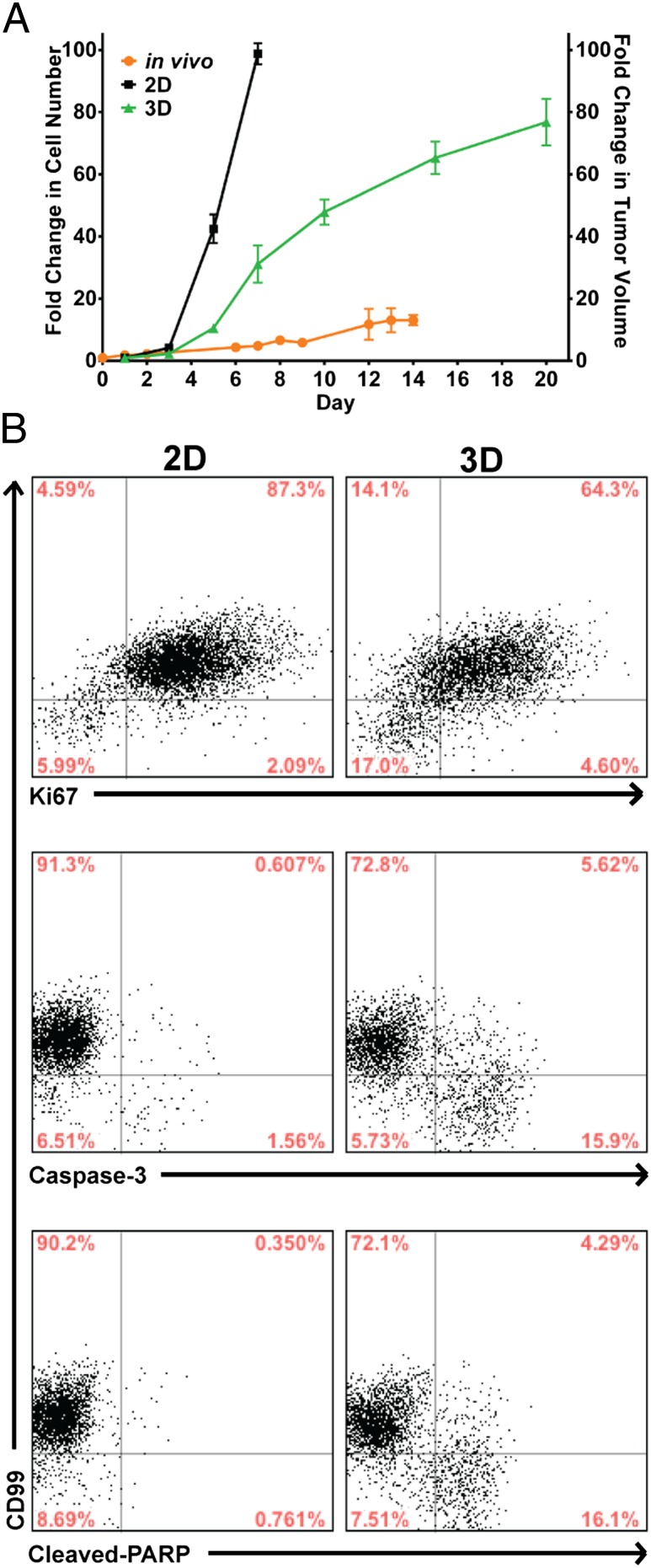
To demonstrate that our 3D EWS model exhibits tumor growth characteristics necessary to serve as a reliable platform for testing promising drug candidates, particularly those that exert their antineoplastic effect on actively dividing cells, we compared the rate of proliferation of TC-71 cells cultured in 3D PCL scaffolds with that of cells in 2D monolayer culture and human EWS xenografts (Fig. 3A) using DNA content as an index of proliferation. In contrast to 2D monolayer culture, the proliferation rate of TC-71 cells within the 3D PCL scaffolds was substantially slower and more closely aligned with that of cells grown in vivo as xenografts, which serves as the current preclinical gold standard for assessing drug efficacy. As determined by flow cytometry, this finding can be attributed to both decreased proliferation and increased apoptosis as evidenced by altered intracellular expression of Ki-67, caspase-3, and cleaved poly(ADP-ribose) polymerase (PARP) protein in TC-71 cells cultured within the 3D PCL scaffolds (Fig. 3B).
Fig. 3.
Growth profile of TC-71 EWS cells in 2D, 3D, and in vivo. (A) EWS growth in 2D monolayer culture, in 3D PCL scaffold, or in vivo as xenografts. TC-71 cells proliferated at a substantially slower rate in the 3D scaffolds than in 2D culture (left axis, fold change in cell number), with the former bearing closer similarity to the in vivo tumor growth rate (right axis, fold change in tumor volume). Data represent the mean fold change in cell number (2D and 3D) or tumor volume (in vivo) ± SEM (n = 4 for 2D culture and in vivo, n = 6 for 3D scaffolds). (B) Levels of Ki-67, caspase-3, and cleaved-PARP expression in TC-71 cells in 3D PCL scaffolds (cultured for 10 d) and 2D monolayer culture as measured using flow cytometry. The majority of the cells in the 3D PCL scaffolds were still proliferative even at 10 d in culture, with a small percentage of apoptotic cells present.
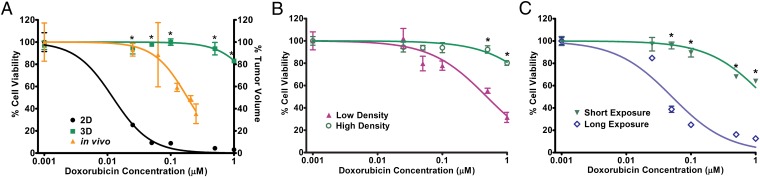
We further characterized the sensitivity of the 3D EWS model to doxorubicin, a prototypical cytotoxic chemotherapeutic agent with proven clinical value in EWS patients. Comparable with increased doxorubicin resistance observed in TC-71 xenografts that grew in CB17/severe combined immunodeficient (SCID) mice [half-maximal inhibitory concentration (IC50), 0.184 µM], TC-71 cells cultured in 3D PCL scaffolds demonstrated greater resistance than cells in 2D monolayer culture (IC50, 2.738 and 0.012 µM, respectively) (Fig. 4A). We expected this result given the slower proliferation rate associated with growth in the 3D PCL scaffolds and doxorubicin’s known mechanism of cytotoxicity. However, this may also be attributed to restoration of structurally complex in vivo-like cell–cell contact in the 3D PCL scaffolds similar to previously reported observations of drug and radiation resistance in tumor spheroids (34).
Fig. 4.
Response of TC-71 EWS cells to doxorubicin. (A) Percentage of viable TC-71 cells in 2D monolayer culture, in 3D PCL scaffold, and in vivo as xenografts after doxorubicin exposure. Cells in 3D scaffolds (IC50, 2.738 µM) and in vivo (IC50, 0.184 µM) demonstrated greater drug resistance than did cells in 2D culture (IC50, 0.0122 µM). P < 0.05 for 3D versus 2D; the asterisk (*) indicates P < 0.05 at each respective concentration. (B) Percentage of viable TC-71 cells cultured in 3D PCL scaffolds at low and high densities (representing little and extensive cell–cell contact, respectively) after doxorubicin exposure. High-density culture resulted in greater doxorubicin resistance (IC50, 4.225 µM) than did low-density culture (IC50, 0.485 µM). P < 0.05 for low versus high density; the asterisk (*) indicates P < 0.05 at each respective concentration. (C) Percentage of viable TC-71 cells in 3D PCL scaffolds after short (3 d) or long (16 d) doxorubicin exposure. Cells exposed to doxorubicin for a longer duration (IC50, 0.051 µM) exhibited lower drug resistance than did those exposed to it for a shorter duration (IC50, 1.397 µM). P < 0.05 for short versus long duration; the asterisk (*) indicates P < 0.05 at each respective concentration. Data represent the mean percentage viability (2D and 3D; left axis) and tumor volume (in vivo; right axis) ± SEM normalized against untreated controls (n = 4 for 2D culture and in vivo, n = 6 for 3D scaffolds).
To elucidate the potential role of cell–cell contact in doxorubicin resistance in our 3D model, we investigated the effect of cell density as a surrogate measure of cell–cell contact in the scaffolds. As shown in Fig. 4B, densely seeded cell cultures were associated with 8.7-fold greater chemoresistance (IC50, 4.225 µM) than were their more sparsely growing counterparts (IC50, 0.485 µM), suggesting a negative correlation between cell–cell contact and drug sensitivity. However, the exact signaling mechanisms implicated in drug resistance are still unclear, and current studies are looking at whether differential sensitivity is a direct consequence of enhanced cell–cell contact alone or interplay among other factors (e.g., intracellular changes, paracrine signaling, modifications in the supporting matrix). The well-recognized phenomenon of chemoresistance previously observed in another spatially complex tumor model (tumor spheroids) results from poor drug penetration into the innermost cell layers, but we found nothing to suggest that this occurred in the 3D PCL scaffolds (35, 36). Indeed, as drug transport and interaction with the scaffold may have an impact on the efficacy of the drug, we conducted an additional study to characterize the potential interaction between doxorubicin and the 3D PCL scaffold (Fig. S2). Although significantly more doxorubicin was found to adsorb onto the PCL scaffolds compared with tissue culture plastic controls, Fig. 4A shows that, within the concentration range of doxorubicin after adsorption onto the PCL scaffold, cytotoxicity in 2D is still greater than 90%, indicating that the increased resistance observed in 3D is not due to decreased availability of the drug after adsorption onto the scaffold.
Given the lower proliferative index of cells in the 3D PCL scaffolds than in 2D monolayer culture that better mimics human tumor growth, this model may be particularly appropriate for investigating the long-term impact of drug exposure on cancer cells, which is a challenging endeavor with 2D culture systems, given that confluency limits the duration of culture. Fig. 4C shows that prolonged exposure to doxorubicin ultimately elicited significant cell death despite negligible short-term antineoplastic effects of the drug (IC50, 1.397 and 0.051 µM for short and long doxorubicin exposure, respectively). Hence, in addition to its greater fidelity to the in vivo EWS tumor phenotype, our 3D EWS model may be an exceptionally useful tool for conducting long-term studies necessary for determining the often subtle and delayed antineoplastic effects exerted by biologically targeted therapy. Notably, as the vast majority of cytotoxic and biologically targeted therapies exert their antineoplastic effects well within the long doxorubicin exposure period investigated in this study, we did not extend this time frame beyond 16 d.
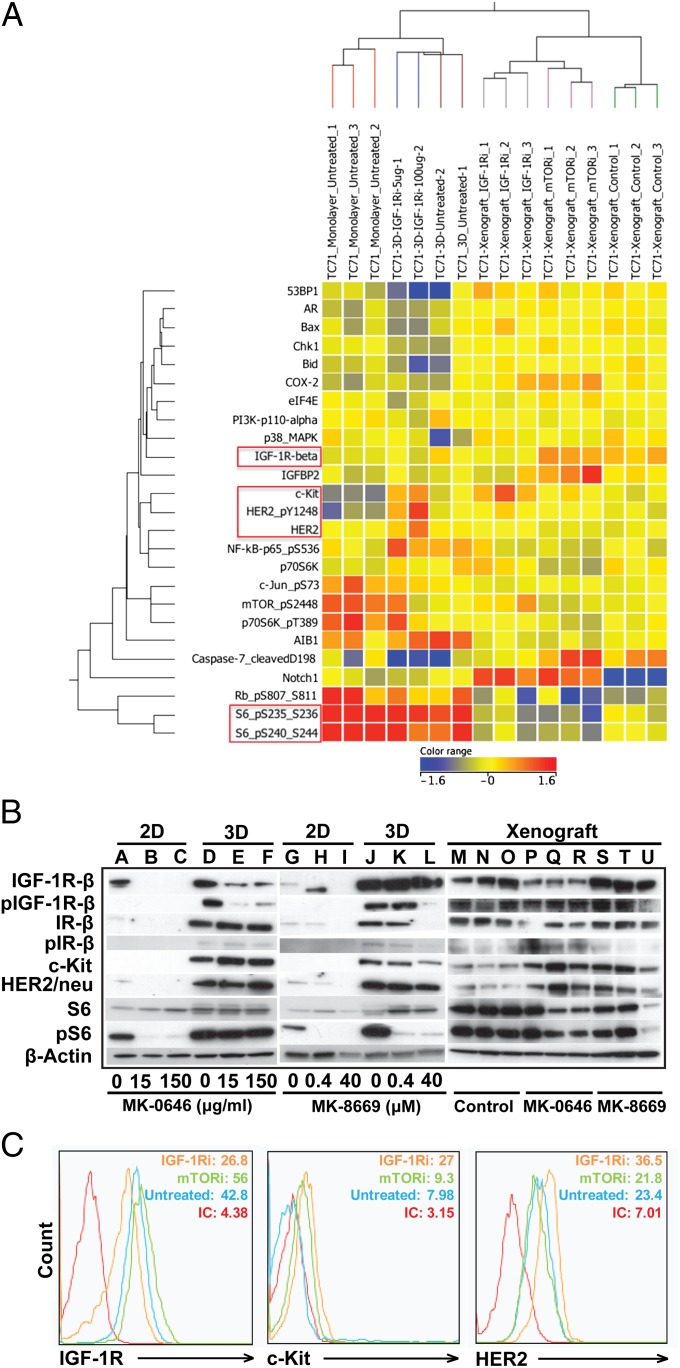
As we observed striking differences in the IGF-1R/mTOR pathway signaling pattern in EWS cells in our 3D PCL scaffold and 2D monolayer culture, we next sought to investigate whether we could elicit more in vivo-like drug sensitivity to inhibitors of IGF-1R and mTOR. We treated TC-71 cells grown under the three conditions (2D monolayer, 3D PCL scaffold, and as xenografts) with MK-0646, a humanized IgG1 monoclonal antibody against IGF-1R. We observed an up-regulation of HER2/neu and c-kit expression in the 3D PCL scaffolds, which is in concordance with the expression pattern in xenografts (Fig. 5 A–C). Additionally, in agreement with published data implicating the insulin receptor (IR) as a major contributor of resistance to IGF-1R–targeted therapy (via formation of hybrid IGF-1R/IR-α receptors) (37), our data demonstrated that IGF-1R inhibition led to constitutive phosphorylated IR-β protein activation in TC-71 cells cultured in our 3D PCL scaffold and in xenograft tumors but not in 2D monolayer culture (Fig. 5B). Furthermore, treatment with the small-molecule mTOR inhibitor MK-8669 (ridaforolimus) had no effect on IGF-1R, c-kit, or HER2/neu expression despite suppressed phosphorylated S6, suggesting that our 3D model is able to mimic the expected in vivo pharmacodynamic response of mTOR inhibition. Overall, these results offer a unique perspective on IGF-1R/mTOR signaling in a biomimetic 3D preclinical model of EWS.
Fig. 5.
Response of TC-71 EWS cells to IGF-1R and mTOR inhibition. (A) Reverse-phase protein array (RPPA) analysis of selected proteins in the IGF-1R/mTOR pathway (red, increased signal; blue, decreased signal). Protein lysates were harvested from TC-71 cells in 3D PCL scaffolds, in 2D monolayer culture, or in vivo. RPPA findings were validated using (B) Western blot and (C) flow-cytometric analysis. Similar alterations in the expression of IGF-1R, c-kit, and HER2/neu were observed in cells in the 3D PCL scaffolds and in vivo xenografts.
Conclusion
We developed an in vitro human EWS model that exhibits morphological and biochemical features of in vivo tumors in stark contrast with conventional 2D models that poorly represent in vivo EWS tumor biology. The remarkable similarity between the engineered EWS tumor model and in vivo xenograft EWS tumors suggests that tumor cells cultured within the 3D PCL scaffold may represent a better model than 2D culture systems for mechanistic studies of standard chemotherapies and/or biologically targeted therapies for EWS under preclinical investigation.
Accelerated development of antineoplastic drugs is critically dependent on preclinical models that simulate in vivo tumor growth and intracellular signaling as accurately as possible. Two-dimensional culture systems that poorly recapitulate the in vivo 3D tumor microenvironment and, hence, in vivo signaling cascades may possess limited utility in this regard. Dysregulated signaling cascades propagated in 2D culture may lead to erroneous scientific conclusions that complicate our understanding of cancer biology and mask resistance mechanisms that would otherwise be elucidated under 3D culture conditions.
With superior fidelity to the in vivo phenotype, our 3D model of EWS provides the unique opportunity to examine the biology of this cancer in a preclinical in vivo-like system that is still amenable to standard experimental techniques. Furthermore, the enhanced constitutive activation of IGF-1R in this 3D system may facilitate the identification of critical nodes in the IGF-1R signaling network that may be exploited therapeutically. Moreover, because our 3D model reproduces several putative mechanisms of drug resistance known to be found clinically (e.g., increased reliance on alternative receptor tyrosine kinases such as c-kit and IGF-1R/IR-α hybrid receptors that activate the mTOR pathway independent of IGF-1R), it may serve as a reliable platform for testing combined antagonists of the IGF-1R/mTOR signaling cascade. Last, given the in vivo-like sensitivity of EWS cells to doxorubicin in our 3D model, this model may also facilitate the pursuit of combinational therapeutic strategies involving both IGF-1R blockade and standard cytotoxic chemotherapy.
Although our understanding of the profound influence of 3D culture conditions on EWS growth and survival has advanced with the use of the engineered PCL scaffolds, many questions remain to be answered. For example, as the effect of oxygen, nutrient, and drug concentration gradients on the tumor phenotype and drug efficacy are important considerations in 3D tumor models, further studies are warranted to elucidate the potential impact of these parameters. Additionally, given the role of tumor-associated cells in influencing tumor behavior, a logical progression of the work reported herein is to recapitulate the in vivo cellular cross talk and address whether the coculture of EWS cells with other tumor-associated cells (such as stromal and endothelial cells) within our 3D PCL scaffold affects the activation status of the IGF-1R signaling cascade. Furthermore, is the in vivo-like signaling cascade observed in 3D induced in part by altered adhesion of tumor cells to the scaffold surface via focal adhesion kinases or attributable to changes in the expression of cell–cell adhesion molecules? Will the use of flow perfusion culture conditions to mimic the mechanical aspect of the bone microenvironment affect EWS cell differentiation and signaling? The answers to these questions and others must ultimately be derived from the use of a biologically relevant 3D environment.
Materials and Methods
Scaffold Preparation.
Nonwoven electrospun PCL scaffolds were electrospun using the electrospinning apparatus as previously described (25). In brief, the setup consists of a syringe pump, power supply, and a grounded, square copper plate. A 30-mL syringe was filled with PCL (inherent viscosity range, 1.0–1.3; DURECT Corporation) dissolved in a 5:1 (vol/vol) chloroform/methanol solution to 18 wt%, and fitted with a 16-gauge blunt needle. A copper ring was placed in-between the needle and the copper plate to stabilize the electric field. The positive lead from the power supply was split and connected to both the needle and the copper ring. Fibers were collected on a glass plate placed in front of the copper plate during the electrospinning process. PCL microfiber mats (microfiber diameter, 11.6 ± 1.7 µm) were electrospun to a thickness of 1 ± 0.1 mm, and individual 8-mm–diameter PCL discs were punched out using a biopsy dermal punch. Scanning electron microscopy was used to assess each disk’s fiber morphology and diameter. Before cell seeding, electrospun scaffolds were sterilized in ethylene oxide (AN73; Andersen Products) for 12 h and aerated to remove residual fumes. Scaffolds were then prewetted in an ethanol gradient [100–30% (vol/vol)], rinsed three times with PBS, and soaked in culture medium for 2 d.
Cell Culture and Seeding.
The human EWS cell line, TC-71, generously provided by John Trent (University of Texas M. D. Anderson Cancer Center, Houston, TX), was used in all experiments. Cells were maintained in RPMI medium 1640 (Mediatech) containing 10% (vol/vol) FBS (Gemini Bio-Products) and antibiotics (100 IU/mL penicillin and 100 μg/mL streptomycin; Mediatech) in a humidified incubator at 37 °C and in a 5% CO2 atmosphere. The medium was replaced every 2 d. For monolayer cell culture, 50,000 TC-71 cells were seeded onto 12-well plates containing 3 mL of medium per well. Two milliliters of the medium was replaced daily. For 3D cell culture, the prewetted scaffolds were press-fitted into individual cylindrical cassettes designed to confine the cell suspension and placed in 12-well plates for seeding. A total of 250,000 TC-71 cells in 200 μL of medium was then seeded onto each prewetted scaffold. The cells were allowed to adhere to the scaffolds for 4–5 h before medium was gently added to immerse the cassettes completely. Seeded scaffolds were removed from the cassettes at 24 h and placed in 12-well plates containing 3 mL of medium, which was replaced daily.
Scanning Electron Microscopy.
Three-dimensional constructs were fixed in 2.5% (vol/vol) glutaraldehyde for at least 30 min and then dehydrated in an ethanol gradient [70–100% (vol/vol)]. Constructs were then air-dried overnight, sputter-coated with gold, and imaged using an FEI Quanta 400 emission scanning electron microscope (FEI Company).
Cell Proliferation Assay.
DNA content was used as an indirect measure of cell number in both 2D monolayer and 3D PCL scaffold. The DNA content of cells was quantified using a Quant-iT PicoGreen dsDNA assay kit (Life Technologies) as described in SI Materials and Methods.
Immunohistochemical Analysis.
Constructs were fixed in 10% (vol/vol) neutral buffered formalin and then immersed in 70% (vol/vol) ethanol before being embedded in HistoPrep freezing medium (Fisher Scientific). The 7-µm–thick frozen sections of the constructs were cut using a CM1850 UV cryostat (Leica Biosystems) and mounted onto Superfrost Plus microslides (VWR). Slides were allowed to thaw at room temperature after cryosectioning, baked overnight at 60 °C, and analyzed histologically (SI Materials and Methods).
Flow Cytometry.
Cells in monolayer culture or 3D PCL scaffolds were harvested using an enzyme-free cell dissociation buffer (Life Technologies), washed in PBS, and processed as described in SI Materials and Methods.
Doxorubicin Studies.
The dose–response for doxorubicin (Pfizer) was evaluated in 2D and 3D culture at concentrations of 0, 0.025, 0.05, 0.1, 0.5, and 1 µM. Cells were cultured as 2D monolayers for 1 d or in 3D PCL scaffolds for 16 d before a 3-d exposure to doxorubicin. In assessing the effect of cell density on doxorubicin sensitivity, cells were cultured for 3 or 16 d, which represented low and high density, respectively, before being exposed to doxorubicin for 3 d. The effect of drug exposure duration was also investigated, where cells were cultured for 16 d before being exposed to doxorubicin over a short (3 d) or long (16 d) culture duration. Doxorubicin-containing medium was replaced every 2 d during the drug exposure period. Percentage cell viability was calculated by normalizing the average number of cells at each doxorubicin concentration against the average number of cells in the untreated controls. The percentage tumor volume was calculated by normalizing the average tumor volume at each doxorubicin concentration against the average tumor volume in the untreated controls.
IGF-1R and mTOR Studies.
Cells in monolayer were either exposed to MK-0646 (humanized antibody against IGF-1R; Merck) at concentrations of 0, 15, and 150 µg/mL for 3 d, or to MK-8669 (small-molecule inhibitor of mTOR; Merck) at concentrations of 0, 0.4, and 40 µM for 1 d. Three-dimensional constructs were cultured for 10 d, before being exposed to the same MK-0646 and MK-8669 concentrations for 7 subsequent days. Medium change was carried out every 2 d.
Protein Isolation and Quantification for Western Blot and Reverse-Phase Protein Array Analysis.
Lysis buffer [1% Triton X-100 (vol/vol), 50 mM Hepes, pH 7.4, 150 mM NaCl, 1.5 mM MgCl2, 1 mM EGTA, 100 mM NaF, 10 mM Na pyrophosphate, 1 mM Na3VO4, 10% (vol/vol) glycerol] containing a freshly added protease inhibitor mixture and phosphatase inhibitors (Roche Applied Science) was used to lyse frozen EWS tumors via homogenization (excised tumors), and TC-71 monolayer and 3D constructs via cold incubation. The protein concentration for each scaffold was measured using a Micro BCA protein assay kit (Thermo Fisher Scientific). Lysates were then stored at −80 °C until analyzed.
Western Blotting.
Proteins were resolved in SDS-polyacrylamide gel electrophoresis and transferred to PVDF membranes. The membranes were blocked using 5% (wt/vol) milk and hybridized with different primary antibodies: phosphorylated IGF-1Rβ, IGF-1Rβ, HER2, anti-phosphorylated HER2, phosphorylated IRβ, IRβ, S6 ribosomal protein, phosphorylated S6 ribosomal protein, c-kit, and β-actin. All primary antibodies were from Cell Signaling Technology except for phosphorylated HER2 (Millipore) and c-kit (Epitomics). Signals were captured using horseradish peroxidase-conjugated secondary anti-rabbit IgG and anti-mouse IgG antibodies (Cell Signaling Technology) and visualized using SuperSignal West Dura chemiluminescent substrate (Thermo Fisher Scientific). The level of immunoreactive protein was measured using chemiluminescent Hyperfilm ECL (GE Healthcare) and quantified using an ImageQuant TL computing densitometer (GE Healthcare).
Reverse-Phase Protein Array and Bioinformatic Analysis.
Reverse-phase protein array experiments were performed as described previously (38). Analysis of 3D constructs, 2D monolayer, and xenograft TC-71 tumor samples was performed simultaneously using the same array. Lysates were normalized to 1 µg/µL and boiled in a solution containing SDS (90%) and β-mercaptoethanol (10%). Supernatants were manually diluted in fivefold serial dilution with lysis buffer and processed as described in SI Materials and Methods.
In Vivo TC-71 Xenograft Tumor Growth.
Male nonobese diabetic/SCID−/− and CB17/SCID−/− mice (The Jackson Laboratory) were used to generate s.c. (106 cells injected per animal) TC-71 xenografts. All mice were maintained under barrier conditions, and experiments were conducted using protocols and conditions approved by the University of Texas M. D. Anderson Cancer Center Institutional Animal Care and Use Committee as outlined in SI Materials and Methods.
Supplementary Material
Acknowledgments
This research is supported in part by University of Texas M. D. Anderson Cancer Center Support Grant CA016672 and National Institutes of Health Grants R01 AR057083 and R01 CA151533. E.L.S.F. acknowledges funding from the National University of Singapore–Overseas Graduate Scholarship. The contents of this paper are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1221403110/-/DCSupplemental.
References
- 1.Bissell MJ, Radisky D. Putting tumours in context. Nat Rev Cancer. 2001;1(1):46–54. doi: 10.1038/35094059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jacks T, Weinberg RA. Taking the study of cancer cell survival to a new dimension. Cell. 2002;111(7):923–925. doi: 10.1016/s0092-8674(02)01229-1. [DOI] [PubMed] [Google Scholar]
- 3.Weaver VM, et al. Reversion of the malignant phenotype of human breast cells in three-dimensional culture and in vivo by integrin blocking antibodies. J Cell Biol. 1997;137(1):231–245. doi: 10.1083/jcb.137.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fischbach C, et al. Engineering tumors with 3D scaffolds. Nat Methods. 2007;4(10):855–860. doi: 10.1038/nmeth1085. [DOI] [PubMed] [Google Scholar]
- 5.Kunz-Schughart LA. Multicellular tumor spheroids: Intermediates between monolayer culture and in vivo tumor. Cell Biol Int. 1999;23(3):157–161. doi: 10.1006/cbir.1999.0384. [DOI] [PubMed] [Google Scholar]
- 6.Friedrich J, Ebner R, Kunz-Schughart LA. Experimental anti-tumor therapy in 3-D: Spheroids—old hat or new challenge? Int J Radiat Biol. 2007;83(11–12):849–871. doi: 10.1080/09553000701727531. [DOI] [PubMed] [Google Scholar]
- 7.Weigelt B, Bissell MJ. Unraveling the microenvironmental influences on the normal mammary gland and breast cancer. Semin Cancer Biol. 2008;18(5):311–321. doi: 10.1016/j.semcancer.2008.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lawlor ER, Scheel C, Irving J, Sorensen PH. Anchorage-independent multi-cellular spheroids as an in vitro model of growth signaling in Ewing tumors. Oncogene. 2002;21(2):307–318. doi: 10.1038/sj.onc.1205053. [DOI] [PubMed] [Google Scholar]
- 9.Kang HG, et al. E-cadherin cell-cell adhesion in ewing tumor cells mediates suppression of anoikis through activation of the ErbB4 tyrosine kinase. Cancer Res. 2007;67(7):3094–3105. doi: 10.1158/0008-5472.CAN-06-3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.David L, et al. Hyaluronan hydrogel: An appropriate three-dimensional model for evaluation of anticancer drug sensitivity. Acta Biomater. 2008;4(2):256–263. doi: 10.1016/j.actbio.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 11.Chitcholtan K, Sykes PH, Evans JJ. The resistance of intracellular mediators to doxorubicin and cisplatin are distinct in 3D and 2D endometrial cancer. J Transl Med. 2012;10:38. doi: 10.1186/1479-5876-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bates RC, Edwards NS, Yates JD. Spheroids and cell survival. Crit Rev Oncol Hematol. 2000;36(2–3):61–74. doi: 10.1016/s1040-8428(00)00077-9. [DOI] [PubMed] [Google Scholar]
- 13.Fischbach C, et al. Cancer cell angiogenic capability is regulated by 3D culture and integrin engagement. Proc Natl Acad Sci USA. 2009;106(2):399–404. doi: 10.1073/pnas.0808932106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Verbridge SS, Chandler EM, Fischbach C. Tissue-engineered three-dimensional tumor models to study tumor angiogenesis. Tissue Eng Part A. 2010;16(7):2147–2152. doi: 10.1089/ten.tea.2009.0668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu H, Radisky DC, Wang F, Bissell MJ. Polarity and proliferation are controlled by distinct signaling pathways downstream of PI3-kinase in breast epithelial tumor cells. J Cell Biol. 2004;164(4):603–612. doi: 10.1083/jcb.200306090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kenny PA, et al. The morphologies of breast cancer cell lines in three-dimensional assays correlate with their profiles of gene expression. Mol Oncol. 2007;1(1):84–96. doi: 10.1016/j.molonc.2007.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weigelt B, Lo AT, Park CC, Gray JW, Bissell MJ. HER2 signaling pathway activation and response of breast cancer cells to HER2-targeting agents is dependent strongly on the 3D microenvironment. Breast Cancer Res Treat. 2010;122(1):35–43. doi: 10.1007/s10549-009-0502-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muranen T, et al. Inhibition of PI3K/mTOR leads to adaptive resistance in matrix-attached cancer cells. Cancer Cell. 2012;21(2):227–239. doi: 10.1016/j.ccr.2011.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee SJ, Atala A. Scaffold technologies for controlling cell behavior in tissue engineering. Biomed Mater. 2013;8(1):010201. doi: 10.1088/1748-6041/8/1/010201. [DOI] [PubMed] [Google Scholar]
- 20.Thibault RA, Scott Baggett L, Mikos AG, Kasper FK. Osteogenic differentiation of mesenchymal stem cells on pregenerated extracellular matrix scaffolds in the absence of osteogenic cell culture supplements. Tissue Eng Part A. 2010;16(2):431–440. doi: 10.1089/ten.tea.2009.0583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liao J, Guo X, Nelson D, Kasper FK, Mikos AG. Modulation of osteogenic properties of biodegradable polymer/extracellular matrix scaffolds generated with a flow perfusion bioreactor. Acta Biomater. 2010;6(7):2386–2393. doi: 10.1016/j.actbio.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mountziaris PM, Tzouanas SN, Mikos AG. Dose effect of tumor necrosis factor-alpha on in vitro osteogenic differentiation of mesenchymal stem cells on biodegradable polymeric microfiber scaffolds. Biomaterials. 2010;31(7):1666–1675. doi: 10.1016/j.biomaterials.2009.11.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang YZ, Su B, Venugopal J, Ramakrishna S, Lim CT. Biomimetic and bioactive nanofibrous scaffolds from electrospun composite nanofibers. Int J Nanomedicine. 2007;2(4):623–638. [PMC free article] [PubMed] [Google Scholar]
- 24.Cipitria A, Skelton A, Dargaville TR, Dalton PD, Hutmacher DW. Design, fabrication and characterization of PCL electrospun scaffolds-a review. J Mater Chem. 2011;21(26):9419–9453. [Google Scholar]
- 25.Pham QP, Sharma U, Mikos AG. Electrospun poly(epsilon-caprolactone) microfiber and multilayer nanofiber/microfiber scaffolds: Characterization of scaffolds and measurement of cellular infiltration. Biomacromolecules. 2006;7(10):2796–2805. doi: 10.1021/bm060680j. [DOI] [PubMed] [Google Scholar]
- 26.Olmos D, et al. Targeting the insulin-like growth factor 1 receptor in Ewing's sarcoma: Reality and expectations. Sarcoma. 2011;2011:402508. doi: 10.1155/2011/402508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ludwig JA, Lamhamedi-Cherradi S-E, Lee H-Y, Naing A, Benjamin R. Dual targeting of the insulin-like growth factor and collateral pathways in cancer: Combating drug resistance. Cancers. 2011;3(3):3029–3054. doi: 10.3390/cancers3033029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scotlandi K, et al. Insulin-like growth factor I receptor-mediated circuit in Ewing’s sarcoma/peripheral neuroectodermal tumor: A possible therapeutic target. Cancer Res. 1996;56(20):4570–4574. [PubMed] [Google Scholar]
- 29.Scotlandi K, et al. Blockage of insulin-like growth factor-I receptor inhibits the growth of Ewing’s sarcoma in athymic mice. Cancer Res. 1998;58(18):4127–4131. [PubMed] [Google Scholar]
- 30.Scotlandi K, et al. Effectiveness of insulin-like growth factor I receptor antisense strategy against Ewing’s sarcoma cells. Cancer Gene Ther. 2002;9(3):296–307. doi: 10.1038/sj.cgt.7700442. [DOI] [PubMed] [Google Scholar]
- 31.Toretsky JA, Kalebic T, Blakesley V, LeRoith D, Helman LJ. The insulin-like growth factor-I receptor is required for EWS/FLI-1 transformation of fibroblasts. J Biol Chem. 1997;272(49):30822–30827. doi: 10.1074/jbc.272.49.30822. [DOI] [PubMed] [Google Scholar]
- 32.Naing A, et al. Insulin growth factor-receptor (IGF-1R) antibody cixutumumab combined with the mTOR inhibitor temsirolimus in patients with refractory Ewing’s sarcoma family tumors. Clin Cancer Res. 2012;18(9):2625–2631. doi: 10.1158/1078-0432.CCR-12-0061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schwartz GK, et al. Cixutumumab and temsirolimus for patients with bone and soft-tissue sarcoma: A multicentre, open-label, phase 2 trial. Lancet Oncol. 2013 doi: 10.1016/S1470-2045(13)70049-4. 10.1016/s1470-2045(13)70049-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Olive PL, Durand RE. Drug and radiation resistance in spheroids: Cell contact and kinetics. Cancer Metastasis Rev. 1994;13(2):121–138. doi: 10.1007/BF00689632. [DOI] [PubMed] [Google Scholar]
- 35.Burdett E, Kasper FK, Mikos AG, Ludwig JA. Engineering tumors: A tissue engineering perspective in cancer biology. Tissue Eng Part B Rev. 2010;16(3):351–359. doi: 10.1089/ten.TEB.2009.0676. [DOI] [PubMed] [Google Scholar]
- 36.Sutherland RM, Eddy HA, Bareham B, Reich K, Vanantwerp D. Resistance to adriamycin in multicellular spheroids. Int J Radiat Oncol Biol Phys. 1979;5(8):1225–1230. doi: 10.1016/0360-3016(79)90643-6. [DOI] [PubMed] [Google Scholar]
- 37.Garofalo C, et al. Efficacy of and resistance to anti-IGF-1R therapies in Ewing’s sarcoma is dependent on insulin receptor signaling. Oncogene. 2011;30(24):2730–2740. doi: 10.1038/onc.2010.640. [DOI] [PubMed] [Google Scholar]
- 38.Hennessy BT, et al. A technical assessment of the utility of reverse phase protein arrays for the study of the functional proteome in non-microdissected human breast cancers. Clin Proteomics. 2010;6(4):129–151. doi: 10.1007/s12014-010-9055-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.