Abstract
Kluyveromyces lactis zymocin, a heterotrimeric toxin complex, imposes a G1 cell cycle block on Saccharomyces cerevisiae that requires the toxin-target (TOT) function of holo-Elongator, a six-subunit histone acetylase. Here, we demonstrate that Elongator is a phospho-complex. Phosphorylation of its largest subunit Tot1 (Elp1) is supported by Kti11, an Elongator-interactor essential for zymocin action. Tot1 dephosphorylation depends on the Sit4 phosphatase and its associators Sap185 and Sap190. Zymocin-resistant cells lacking or overproducing Elongator-associator Tot4 (Kti12), respectively, abolish or intensify Tot1 phosphorylation. Excess Sit4·Sap190 antagonizes the latter scenario to reinstate zymocin sensitivity in multicopy TOT4 cells, suggesting physical competition between Sit4 and Tot4. Consistently, Sit4 and Tot4 mutually oppose Tot1 de-/phosphorylation, which is dispensable for integrity of holo-Elongator but crucial for the TOT-dependent G1 block by zymocin. Moreover, Sit4, Tot4, and Tot1 cofractionate, Sit4 is nucleocytoplasmically localized, and sit4Δ-nuclei retain Tot4. Together with the findings that sit4Δ and totΔ cells phenocopy protection against zymocin and the ceramide-induced G1 block, Sit4 is functionally linked to Elongator in cell cycle events targetable by antizymotics.
INTRODUCTION
Reversible protein phosphorylation is an important means of regulating cellular processes such as signal transduction, gene expression, or cell cycle progression (Stark, 1998; Kobor and Greenblatt, 2002). In Saccharomyces cerevisiae, execution of the latter requires cyclin-dependent kinase and Sit4, a type 2A protein phosphatase (PP2A) (Cross, 1990; Sutton et al., 1991). Sit4 acts positively for G1 cyclin (CLN1/2) function, which activates cyclin-dependent kinase and G1 exit (Nasmyth and Dirick, 1991; Fernandez-Sarabia et al., 1992). Consistently, sit4ts cells arrest in G1 (Sutton et al., 1991) and SIT4 relates to other cell cycle-relevant genes (CAK1, CLN3, SWI4, and BCK2) (Sutton and Freiman, 1997; Munoz et al., 2003). Once CLN2 is provided from a SIT4-independent promoter, sit4ts cells enter S phase but remain unbudded (Fernandez-Sarabia et al., 1992). So, bud emergence also requires Sit4 and further studies connect Sit4 to protein kinase C, target of rapamycin (TOR), ubiquitination, and the ceramide-induced G1 block, implying that Sit4 is a multifunctional enzyme catalyzing distinctive dephosphorylation events (Di Como and Arndt, 1996; Nickels and Broach, 1996; de la Torre-Ruiz et al., 2002; Singer et al., 2003). The phenotype of sit4 mutants depends on SSD1, a polymorphic gene. Whereas in ssd1-d strains sit4Δ is lethal, SSD1-v alleles allow SIT4 to be deleted, although SSD1-v sit4Δ cells are delayed in G1 (Sutton et al., 1991).
In response to Kluyveromyces lactis zymocin or rapamycin, S. cerevisiae arrests in G1 (Schaffrath and Breunig, 2000; Crespo and Hall, 2002). Rapamycin inhibition of TOR leads to several read-outs that require dissociation of Sit4 from its interactor Tap42 to trigger dephosphorylation of TOR effectors (Di Como and Arndt, 1996; Crespo and Hall, 2002). Unlike rapamycin, zymocin, a three-subunit (αβγ) toxin complex, imposes a G1 block that allows transient macromolecular synthesis (Stark and Boyd, 1986; Butler et al., 1991b). Zymocin docks onto cell wall chitin followed by uptake of its γ-toxin subunit (Butler et al., 1991a; Jablonowski et al., 2001b). The γ-toxin target (TOT) involves Elongator, an RNA polymerase II (pol II) associated histone acetylase (HAT) that facilitates pol II transcription, and several other factors, including PP2A Sit4 (Otero et al., 1999; Frohloff et al., 2001, 2003; Jablonowski et al., 2001a,c; Winkler et al., 2001, 2002; Kim et al., 2002; Fichtner et al., 2002a,b, 2003; Fichtner and Schaffrath, 2002; Mehlgarten and Schaffrath, 2003). As judged from the findings that modulation of pol II carboxy-terminal domain (CTD) phosphorylation alters a cell's response to zymocin, that phospho-CTD stabilizes the Elongator·pol II association, and that removal of an Elongator NLS protects against zymocin, TOT function requires nuclear Elongator·pol II contact (Otero et al., 1999; Jablonowski et al., 2001c; Fichtner et al., 2003).
sit4Δ cells phenocopy Elongator mutants, suggesting a link between Sit4 and TOT (Jablonowski et al. 2001a). Although both TOT function and the TOR pathway require SIT4, the finding that tap42ts cells are zymocin sensitive but rapamycin resistant implies there is little or no TOT-TOR cross talk (Jablonowski et al., 2001a). Apart from Tap42, Sap155, Sap185, and Sap190 associate with Sit4 as interdependent activators (Luke et al., 1996). Multicopy SAP155 confers zymocin resistance that is abrogated by excess Sap185/190, suggesting high Sap155 titrates binding of Sap185/190 to Sit4 (Jablonowski et al., 2001a). Consistently, sap185Δsap190Δ cells are zymocin resistant, implying that Sit4·Sap185/190 is crucial for zymocin action. Dephosphorylation of Elongator subunit Tot1 (Elp1) is shown here to be suppressed in zymocin-resistant sit4Δ cells. Similarly, zymocin resistance of sap185Δsap190Δ cells is accompanied by high phospho-Tot1 levels and strongly suggests that Tot1 dephosphorylation is Sit4·Sap185/190 dependent and required for TOT proficiency. Zymocin resistance due to removal of Elongator-associator Tot4 (Kti12) abolishes Tot1 phosphorylation, whereas excess Tot4 (and zymocin protection) is antagonized by elevated Sit4·Sap190. Our study reveals an opposing link between Tot4 and Sit4 on Elongator phosphomodification that controls the TOT-dependent G1 cell cycle block by zymocin.
MATERIALS AND METHODS
Yeast Strains, Media, K. lactis Zymocin Methods, and DNA Constructs
Yeast strains used and constructed throughout this study are listed in Table 1. Yeast cells were grown in routine yeast extract, peptone, and dextrose (YPD) or galactose (YPG) rich media (Sherman, 1991). Synthetic complete (SC) medium was prepared as described by Sherman (1991) with either glucose or galactose as carbon source. Testing the effect of C2 ceramide (Calbiochem, San Diego, CA) involved addition (100-200 μM) to YPD plates. Zymocin sensitivity tests of S. cerevisiae used the colony interaction killer eclipse assay essentially as described by Kishida et al. (1996) together with K. lactis killer strains (AWJ137) and nonkiller strains (NK40) (Table 1). Analysis of gene dosage effects on zymocin sensitivity used yeast transformation (Gietz et al., 1992) with 2 μ yeast shuttle vectors YEplac181 (LEU2) carrying TOT4/KTI12 (pJHW27) and YEp24 (URA3) harboring SAP4 (CB2925), SAP155 (CB2643), SAP185 (CB2819), and SAP190 (CB2606) (Butler et al., 1994; Luke et al., 1996; Jablonowski et al., 2001a). If required for marker convenience, the SAP185 and SAP190 genes were moved into YEplac112 (2 μ TRP1) (Gietz and Sugino, 1988) or into pRS423 (2 μ HIS3) (Sikorski and Hieter, 1989) by using cloning strategies described by Luke et al. (1996). Testing multicopy SIT4 involved cloning a SIT4 PCR product essentially as described previously by Posas et al. (1991) into YEplac181 (2 μ LEU2) (Gietz and Sugino, 1988) to yield YEpSIT4. To combine cells maintaining SIT4 and TOT4 in multicopy, the TOT4 gene was removed from pJHW27 and cloned into YEplac112 (2 μ TRP1) by directional EcoRI/HindIII cloning (Butler et al., 1994).
Table 1.
Yeast strains used in this study
| Strain | Description | Source |
|---|---|---|
| K. lactis | ||
| AWJ137 | α leu2 trp1 [k1+ k2+] (killer) | Frohloff et al. (2001) |
| NK40 | α ade1 ade2 leu2 [k1° k2+] (nonkiller) | Frohloff et al. (2001) |
| S. cerevisiae | ||
| JA100 | MATα ura3-52 leu2-3-112 his4 trp1-1 can-1r | Joaquin Ariño |
| EDN75 | As JA100, but ppz1::KAN | Joaquin Ariño |
| JA103 | As JA100, but ppz2::TRP1 | Joaquin Ariño |
| EDN76 | As JA100, but ppz1::KAN ppz2::TRP1 | Joaquin Ariño |
| W303-1A | MATa ade2-1 his3-11, 15 leu2-3, −112 trp1-1 ura3-1 can1-100 ssd1-d2 | Trisha Davies |
| AY925 | As W303-1A but MATα | Kim Arndt |
| DEY132-1C | As AY925, but pph21::HIS3 | Evans and Stark (1997) |
| DEY10-2B | As AY925, but pph22::TRP1 | Evans and Stark (1997) |
| DEY132-2C | As AY925, but pph21::URA3 pph22::TRP1 | Evans and Stark (1997) |
| MMY09 | As W303-1A, but cna1::LEU2 cna2::URA3 | Trisha Davies |
| YJN519 | As W303-1A, but cnb1::LEU2 | Thomas Edlind |
| LFY3 | As W303-1A, but tot1Δ::TRP1 | This work |
| LFY4 | As W303-1A, but tot2Δ::TRP1 | This work |
| LFY5 | As W303-1A, but tot3Δ::TRP1 | Jablonowski et al. (2001c) |
| LFY6 | As W303-1A, but tot4Δ::TRP1 | Jablonowski et al. (2001c) |
| FY1679-08A | MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 GAL SSD1-v | Euroscarf, Frankfurt |
| DJY1t-a | As FY1679-08A, but TOT1-(c-myc)3::Sphis5+ | This work |
| FFY3t | As FY1679-08A, but TOT3-(c-myc)3::Sphis5+ | Frohloff et al. (2001) |
| FFY4t-a | As FY1679-08A, but TOT4-(c-myc)3::Sphis5+ | Fichtner et al. (2002a) |
| DJYS4H | As FY1679-08A, but SIT4-(HA)3::Sphis5+ | This work |
| DJYHS4 | As FY1679-08A, but TRP1::GAL1::(HA)3-SIT4 | This work |
| DJY8A-1H3 | As FY1679-08A, but TOT1-(HA)3::Sphis5 | Fichtner et al. (2003) |
| DJYT4H | As FY1679-08A, but TOT4-(HA)6::KITRP1 | This work |
| DJY2t1d-a | As FY1679-08A, but TOT2-(c-myc)3::Sphis5+ tot1Δ::KILEU2 | This work |
| FFY2/1dt | As FY1679-08A, but TOT2-(c-myc)3::Sphis5+ TOT1-(HA)6::KITRP1 | Fichtner et al. (2002a) |
| FFY3/1dt | As FY1679-08A, but TOT3-(c-myc)3::Sphis5+ TOT1-(HA)6::KITRP1 | Fichtner et al. (2002b) |
| FFY3/2dt | As FY1679-08A, but TOT3-(c-myc)3::Sphis5+ TOT2-(HA)6::KITRP1 | Fichtner et al. (2002a) |
| DJY3/2-d11 | As FFY3/2dt, but kti11Δ::KlLEU2 | This work |
| CY4029 | As W303-1A, but SSD1-v1 | Luke et al. (1996) |
| CY5224 | As CY4029, but sap185Δ::ADE2 sap190Δ::TRP1 | Luke et al. (1996) |
| CY5220 | As CY4029, but sap4Δ::LEU2 sap155Δ::HIS3 | Luke et al. (1996) |
| CY3938 | As CY4029, but sit4Δ::HIS3 | Luke et al. (1996) |
| DJY100 | As CY4029, but tot4Δ::KlLEU2 | This work |
| DJY101 | As CY4029, but sit4Δ::HIS3 tot4Δ::LEU2 | This work |
| DJY102 | As CY4029, but TOT1-(HA)6::KITRP1 | This work |
| DJY103 | As CY4029, but sit4Δ::HIS3 TOT1-(HA)6::KITRP1 | This work |
| DJY104 | As CY4029, but tot4Δ::LEU2 TOT1-(HA)6::KlTRP1 | This work |
| DJY105 | As CY4029, but sit4Δ::HIS3 tot4Δ::LEU2 TOT1-(HA)6::KITRP1 | This work |
| DJY107 | As CY3938, but TOT3-(c-myc)3::Sphis5+ TOT5-(HA)6::KITRP1 | This work |
| DJY108 | As CY4029, but TOT3-(c-myc)3::Sphis5+ TOT5-(HA)6::KITRP1 | This work |
| DJY109 | As CY3938, but TOT5-(c-myc)3::Sphis5+ TOT4-(HA)6::KITRP1 | This work |
| DJY110 | As CY4029, but TOT5-(c-myc)3::Sphis5+ TOT4-(HA)6::KITRP1 | This work |
| DJY111 | As CY3938, but TOT3-(c-myc)3::Sphis5+ TOT4-(HA)6::KITRP1 | This work |
| DJY112 | As CY4029, but TOT3-(c-myc)3::Sphis5+ TOT4-(HA)6::KITRP1 | This work |
| DJY113 | As CY3938, but TOT4-(HA)6::KITRP1 | This work |
| DJY114 | As CY4029, but TOT4-(HA)6::KITRP1 | This work |
| DJY115 | As CY5220, but sap4Δ::LEU2 sap155Δ::HIS3 TOT1-(HA)6::KITRP1 | This work |
| DJY116 | As CY4029, but sap185Δ::ADE2 sap190Δ::LEU2 TOT1-(HA)6::KITRP1 | This work |
Targeted Gene Disruptions and Epitope Tagging In Vivo
For PCR-mediated construction of defined kti11Δ, tot1Δ and tot2Δ null-alleles, the YDp-KlL plasmid carrying the K. lactis LEU2 marker and the YDpW deleter plasmid carrying S. cerevisae TRP1 was used (Berben et al., 1991; Frohloff et al., 2001) together with the following knockout primer pairs: FW-ko-KTI11 (5′-ACA TAC CAC GAC TGT AAG CAC ATC ATT TGT ACA ATA CAT TAC CAG CTG AAC GAC GGC CAG TGA ATT CCC GG-3′), RV-ko-KTI11 (5′-CTT TAT TTC TAT TTG TAT TCT CGA TCT AGC CTC TCA TCT TTA GGC AGC AGA GCT TGG CTG CAG GTC GAC GG-3′), FW-ko-TOT1 (5′-AGA AAC AGT ACA AAT GCC TAA TGG CTT ATG GTT GAA CAT GAC AAG AGT GGC GAC GGC CAG TGA ATT CCC GG-3′), RV-ko-TOT1 (5′-CAA TAT GAC TCT TAG GGA AAT CAT GAA TCT CTG GAA CAG GTA TTT CTG GGA GCT TGG CTG CAG GTC GAC GG-3′), FW-ko-TOT2 (5′-ATG GTG GAA TGT ATC ACT CCC GAA GCC ATT TTT ATA GGT GCT AAC AAG CAC GAC GGC CAG TGA ATT CCC GG-3′), and RV-ko-TOT2 (5′-CCT CAA TCT TGT AAT TTT GTC TGC TGG TGT TAT ATC CTC GTT TAG CTG CGA GCT TGG CTG CAG GTC GAC GG-3′). Leu+ or Trp+ transformants obtained on SC media were verified by polymerase chain reaction (PCR) by using the knockout primer pairs to amplify the foreign marker and ORF-specific primer pairs to check for proper integration.
Testing strain FY1679-08A for SSD1-v or ssd1-d allelism involved gene disruption of SIT4. To do so, we constructed a deletion cartridge in which SIT4 of YEpSIT4 (see above) was centrally disrupted by LEU2. The latter was inserted as a 1.8-kb BamHI segment of YDpL (Berben et al., 1991) into the single BglII site of SIT4. The disruption (sit4Δ::LEU2) was released by BamHI restriction and Leu+ transformants were verified to carry the sit4Δ::LEU2 disruption by using PCR and SIT4-A/B primers as described previously (Posas et al., 1991). All sit4Δ cells tested were found to display the characteristics of an SSD1-v background allowing SIT4 to be deleted with an accompanying slow growth phenotype (Sutton et al., 1991). Similarly, yeast strains harboring tot4Δ null-alleles (DJY100, DJY101, DJY103, and DJY104; Table 1) were obtained by transforming yeast cells with the pYF6 (kti12Δ/tot4Δ::LEU2) deletion construct (Butler et al., 1994) or by PCR-mediated gene disruption by using YDp-KlL (see above) and primers FW-ko-TOT4 (5′-AAA CTA AAC AGG CAA TTT AGT AAG AAG ATG CCA CTG GTG CTT TTT ACG GGC GAC GGC CAG TGA ATT CCC GG-3′) and RV-ko-TOT5 (5′-ATC TCA ATT CAA GTT TTT GTT AAG ATA ATC AGC GAA AAG CGG ACC GAT CCA GCT TGG CTG CAG GTC GAC GG-3′).
Tot proteins were C-terminally tagged with hemagglutinin (HA) and c-Myc epitopes by PCR by using tagging templates and S3/S2-primer pairs as described previously (Knop et al., 1999; Frohloff et al., 2001). For Sit4, C-terminal HA tagging used S3-primers (5′-TAA GAG AAT CCA CGG CAA ACC ATA ATA ATC AAA GAG CCG GCT ATT TCT TAC GTA CGC TGC AGG TCG AC-3′) and S2-primers (5′-ATT GTG AAA ATT ATT TTT ATT CGT CGA GTT AGG GAG GGC ATG CCG TCG TGA TCG ATG AAT TCG AGC TCG-3′) and PCR with plasmid pYM2 (Knop et al., 1999). HA tagging Sit4 at its N terminus involved PCR reactions by using pFA6a-TRP1-pGAL1-3HA (Longtine et al., 1998) and the following primers: F4-SIT4 (5′-TAT TAT TCT TCA GTC CCC TCC TCG CTC TTT TTA GAT TCG ACA TTA CAA GGG AAT TCG AGC TCG TTT AAA C-3′) and R3-SIT4 (5′-TGG CAT TTC TTT ATT GTT TCA AGC CAT TCG TCG GGG CCT CTA GAT ACC ATG CAC TGA GCA GCG TAA TCT G-3′). YCp vector pCB243 (LEU2 SIT4-HA) (Sutton et al., 1991) was kindly provided by Dr. M. Hall (Biozentrum Basel, Switzerland) and served as an additional source for a C-terminally HA-tagged Sit4 variant expressed from its native SIT4 promoter in strain CY3938 (sit4Δ; Table 1).
Immunological Techniques
Detection of tagged proteins by anti-c-Myc (9E10) and anti-HA (3F10) (Roche Diagnostics, Mannheim, Germany) antibodies was as described previously (Frohloff et al., 2001). Elp1/Tot1 was immunodetected using anti-Elp1/Tot1 rabbit antiserum (provided by Dr. J. Svejstrup, London Research Institute, United Kingdom) essentially as described by Otero et al. (1999). Protein concentrations were determined using the method of Bradford (1976), and standardized protein loadings were controlled with a rabbit antibody directed against the α and β subunits of yeast Pfk1 (provided by Dr. J. Heinisch, University of Osnabrück, Osnabrück, Germany) and diluted 1:10,000 in standard Western studies. Immunoprecipitation was performed as described previously (Zachariae et al., 1996; Frohloff et al., 2001). Analysis of Tot1 phosphomodification by Western blots with anti-phosphoserine (Q5) and anti-phosphothreonine (Q7) antibodies followed essentially the manufacturer's manual (Phospho-Protein Purification Handbook August, 2002; QIAGEN, Hilden, Germany). Cell fractionation involving sucrose-ultracentrifugation was done as described by Kölling and Hollenberg (1994). Separation of enriched yeast cytoplasmic from nuclear fractions followed the protocol of Pereira et al. (1998). To exclude cross-contamination between the fractions, aliquots were analyzed in parallel by anti-Nop1, anti-RFA, and anti-Cdc19 antibodies, kind gifts from Drs. J. Aris (University of Florida, Gainesville, FL), B. Stillman, (Cold Spring Harbor Laboratory, Cold Spring Harbor, NY), and J. Thorner, (University of California, Berkeley, Berkeley, CA), at 1:2,000 (anti-Nop1), 1:5,000 (anti-RFA), and 1:10,000 (anti-Cdc19) dilutions. Detection of Sit4 phosphatase by a Sit4-specific monoclonal antibody (kindly provided by Dr. Y. Jiang, University of Pittsburgh, Pittsburgh, PA) rather than an immunoreactive epitope-tagged version used a 1:1,000 dilution (Wang et al., 2003).
RNA and Reverse Transcription (RT)-PCR Methods
Total RNA was isolated from equal amounts of SIT4 and sit4Δ strains cells by using the RNAeasy midi kit (QIAGEN) according to the manufacturer's recommendations. RT-PCR experiments involved equal amounts of total RNA (4 μg) with the RevertAid kit (MBI Fermentas) for 1 h at 42°C in 20-μl reaction volumes. After first-strand cDNA synthesis, 1/20 of the reaction was subjected to PCR (30 cycles) by using Taq polymerase (MBI Fermentas, Vilnius, Lithuania) and oligonucleotide primers (10 μM) to amplify fragments specific for Elongator subunits (TOT1-7) or histone (HHT1) and actin (ACT1) controls. These were as follows: HHT1 (5′-AGC AAG AAA GTC CAC TGG TG-3′ and 5′-GAA TGG CAG CCA AGT TGG TA-3′), ACT1 (5′-CTT CCG GTA GAA CTA CTG GT-3′ and 5′-CCT TAC GGA CAT CGA CAT CA-3′), TOT1/ELP1 (5′-CTT GGT GTA TGA AAC TCG CG-3′ and 5′-TTC TTA CCT CTG CCA GTA CC-3′), TOT2/ELP2 (5′-AAC CTG ATG AGA CTT CAG GC-3′ and 5′-CAA ACC TAA CAC AGG AAC GG-3′), TOT3/ELP3 (5′-TCA GTC CTT GTA CGA AGA CG-3′ and 5′-ATA AGC TCG ACC TGA TCT GG-3′), TOT4/KTI12 (5′-TCC GGT ATC AAC TTC ACT GC-3′ and 5′-CTT GTT CCG TTA CTT ACC CC-3′), TOT5/ELP5 (5′-TAT TGA CGC TAC GCA GAT GG-3′ and 5′-CTC CTC TTC TTG CTT AGT GG-3′), TOT6/ELP6 (5′-GAT GCT ACC TTC GTC AAC TC-3′ and 5′-TAC GTC CTT TGC AAA ACC GG-3′), and TOT7/ELP4 (5′-TTT GCA AAG GAG CTA CCT GG-3′and 5′-GGA AGC AAC AGT ACA ACC C-3′). RT-PCR products were analyzed by agarose gel electrophoresis: ACT1 (0.44 kb), HHT1 (0.32 kb), TOT1/ELP1 (0.49 kb), TOT2/ELP2 (0.57 kb), TOT3/ELP3 (0.53 kb), TOT4/KTI12 (0.47 kb), TOT5/ELP5 (0.5 kb), TOT6/ELP6 (0.5 kb), and TOT7/ELP4 (0.5 kb).
RESULTS
Nucleocytoplasmic Sit4 Mediates the Ceramide G1 Block Together with Tot4 and Elongator
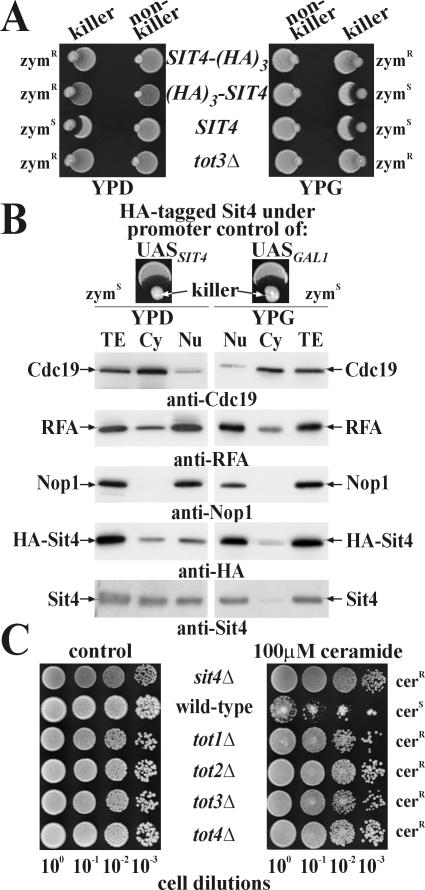
Having HA-tagged Sit4 at its C terminus, we observed zymocin resistance and wild-type cell viability atypical of sit4Δ SSD1-v cells, indicating a crucial role for Sit4's C terminus in zymocicity (Figure 1A; our unpublished data). Because this region contains a conserved PP2A motif (-YFL) whose carboxymethylation mediates subunit interaction (Evans and Hemmings, 2000; Wei et al., 2001), the tag may have interfered with formation of zymocin-relevant Sit4 subcomplexes. Consistent with this, an HA-tag incorporated at Sit4's C terminus upstream of this -YFL motif and expressed from single-copy vector pCB243 (Sutton et al., 1991) was fully capable of complementing zymocin resistance associated with the sit4Δ background of strain CY3938 (Table 1 and Figure 1B, top left). An N-terminal HA-tag under GAL1-promoter control yielded glucose-dependent zymocin protection with sit4Δ SSD1-v-like slow growth (Figure 1A; our unpublished data). Galactose restored normal growth and zymocin inhibition, indicating that (HA)3-Sit4 is functional (Figure 1A). On separating the cytoplasmic and nuclear fractions from galactose-grown (HA)3-SIT4 cells, Sit4 was found to be predominantly nuclear localized with a minor cytoplasmic pool (Figure 1B, right). Similarly, in sit4Δ cells, HA-tagged Sit4 expressed from its natural promotor on pCB243 (Sutton et al., 1991) showed up in both cytoplasmic and nuclear fractions (Figure 1B, left). Therefore, we conclude that contrary to data inferred from indirect immunofluorescence microscopy by using multicopy SIT4 cells (Sutton et al., 1991), Sit4 cannot be considered to be exclusively cytoplasmic, a notion supported by several independently observed nuclear Sit4 interactions (Arndt et al., 1989; Ho et al., 2002, Singer et al., 2003). As for its zymocin relevant function, it is noteworthy that comparably with balanced nucleocytoplasmic localization in the SIT4 wild-type promoter context (Figure 1B, top left), GAL1-promoter induced nuclear accumulation of Sit4 did not alter a yeast cell's vulnerability to the toxin complex (Figure 1B, top right). This supports a nuclear-associated role of Sit4 for zymocin's lethal action and is in line with fractionation data that recently reported Elongator to be preferentially found in the nucleus (Fichtner et al., 2003). Comparing functional linkage between Sit4, Elongator and Tot4, an Elongator partner protein (Fichtner et al., 2002a,b), we checked how tot1-4Δ and sit4Δ cells responded to C2-ceramide, another SIT4-dependent G1 cell cycle blocker (Nickels and Broach, 1996). Unlike wild-type cells and consistent with phenocopying zymocin protection, cells lacking Elongator, Tot4, or Sit4 efficiently resisted ceramide (Figure 1C). So, Elongator, Tot4, and Sit4 function together in cell cycle-related processes susceptible to antizymotics.
Figure 1.
Sit4 can be found in the nucleus and sit4Δ and Tot- mutants resist ceramide. (A) The Sit4 C terminus confers zymocin sensitivity. Killer eclipse assays used K. lactis killer (AWJ137) and nonkiller (NK40) strains (Table 1) and S. cerevisiae strains expressing wild-type Sit4 (FY1679-08A: SIT4) or Sit4 tagged with the HA epitope at either the C terminus (DJYS4H: SIT4-(HA)3), or at the N terminus (DJYHS4: (HA)3-SIT4) under GAL promoter control. Eclipse formation around the killer strain indicates zymocin sensitivity (zymS), whereas lack of inhibition indicates a resistant (zymR) response. The Elongator mutant LFY5 (tot3Δ) served as zymR control. Growth was on glucose (YPD) or galactose (YPG) medium. (B) Sit4 is nucleocytoplasmic. Total extracts (TE) from lysed CY3938 (sit4Δ) spheroplasts carrying pCB243 (UASSIT4::SIT4-HA) (Sutton et al., 1991) and DJYHS4 (UASGAL1::(HA)3-SIT4) spheroplasts grown in YPD (left) or YPG (right) were immunoprobed together with separated cytoplasmic (Cy) and nuclear (Nu) fractions by using Sit4- and HA-specific antibodies to detect the PP2A-type Sit4. Marker distribution used anti-Nop1 (nucleus), anti-RFA (cytoplasm and nucleus), and anti-Cdc19 (predominantly cytoplasm) antibodies. To test effects of promoter-dependent SIT4 expression on zymocin sensitivity, both promoter scenarios (UASSIT4 vs. UASGAL1) were assayed by killer eclipse assays (top; also see A). (C) Ceramide assay. Tenfold dilutions of S. cerevisiae strains (W303-1A: wild-type, CY3938: sit4Δ, LFY3: tot1Δ, LFY4: tot2Δ, LFY5: tot3Δ and LFY6: tot4Δ) were spotted on YPD plates without (control) and with ceramide. Sensitivity and resistance to ceramide are indicated (cerS and cerR, respectively).
Sit4·Sap155 Is Dispensable for Elongator Expression and Elongator Complex Integrity
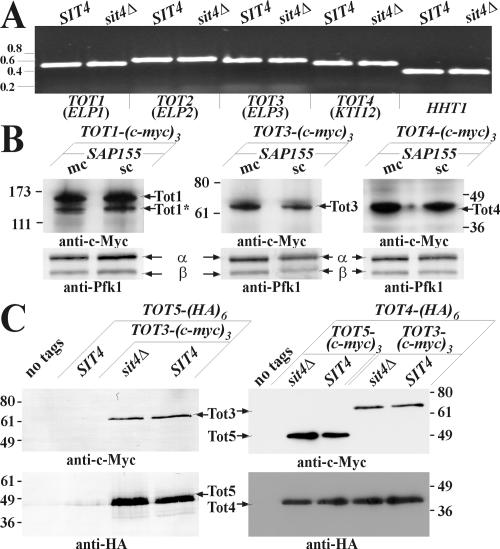
Because sit4 mutants display pol II transcription defects (Arndt et al., 1989), we checked whether sit4Δ cells affect Elongator gene expression, a condition predicted to induce zymocin resistance (Frohloff et al., 2001). We examined transcription of TOT1-7 by RT-PCR and found essentially identical mRNA levels for these Elongator subunits in SIT4 and sit4Δ cells (Figure 2A; our unpublished data). Comparison of Tot1-5 levels in multicopy SAP155 cells, which phenocopy loss of SIT4 function with regards to zymocin resistance (Jablonowski et al., 2001a), also failed to show an effect of Sit4·Sap155 on Elongator expression at the translational level. Thus, c-Myc-tagged Tot protein levels were hardly affected by excess Sap155 (Figure 2B; our unpublished data). Intriguingly, c-Myc-tagged Tot1 (Elp1), separated as two distinct forms (Figure 2B), confirming recent data that Tot1 may undergo proteolysis (Fichtner et al., 2003). Our failure to detect alterations in Elongator subunit expression prompted us to study whether Sit4 affects Elongator assembly, disruption of which protects from zymocin (Fichtner et al., 2002b; Frohloff et al., 2003). Coimmune precipitation between Elongator subunits in the presence or absence of Sit4 revealed that all the individual subunit interactions tested remained unaffected in sit4Δ cells; contacts between Tot3-Tot5, Tot4-Tot3, and Tot4-Tot5 (Figure 2C) were unaltered. In conclusion, zymocin-resistance of sit4Δ or multicopy SAP155 cells is not based on deregulation of Elongator subunit expression or interference with Elongator complex assembly or integrity.
Figure 2.
Elongator expression and complex assembly in correlation to SIT4 and SAP155. (A) Elongator gene transcription (TOT1-4) is unaffected by sit4Δ. RT-PCR was used to compare expression of TOT1-4 in CY4029 (SIT4) and CY3938 (sit4Δ) strains. Histone H3 (HHT1) served as control. Numbers refer to kilobases (Gene Ruler; MBI Fermentas). (B) Elongator expression is unaffected by multicopy SAP155. Identical amounts of protein extracts from c-Myc-tagged Elongator strains DJY1t-a (TOT1-(c-myc)3), FFY3t (TOT3-(cmyc)3) and FFY4t-a (TOT4-(c-myc)3) carrying multi-(mc) or single copy (sc) SAP155 were immunoprobed by 9E10 (anti-c-Myc). Loading was followed using anti-Pfk1 antibody (recognizing phosphofructokinase α and β subunits as indicated). Positions of c-Myc-tagged Tot proteins are marked by arrows. Tot1 separates (at least) in two forms, with the faster (*) being N-terminally truncated (Fichtner et al., 2003). (C). Loss of SIT4 does not affect Elongator assembly. Equal amounts of protein extracts obtained from wild-type S1T4 DJY108, DJY110, and DJY112 and sit4Δ DJY107, DJY109, and DJY111 strains expressing the indicated tagged Tot proteins were subjected to 9E10 (anti-c-Myc) immunoprecipitations. Detection of Tot3 and Tot5 used 9E10 (anti-c-Myc), monitoring HA-tagged Tot5 or Tot4 used 3F10 (anti-HA). Numbering (B and C) refers to kilodaltons of molecular markers (Invitrogen, Carlsbad, CA).
Elongator Is a Phospho-Complex
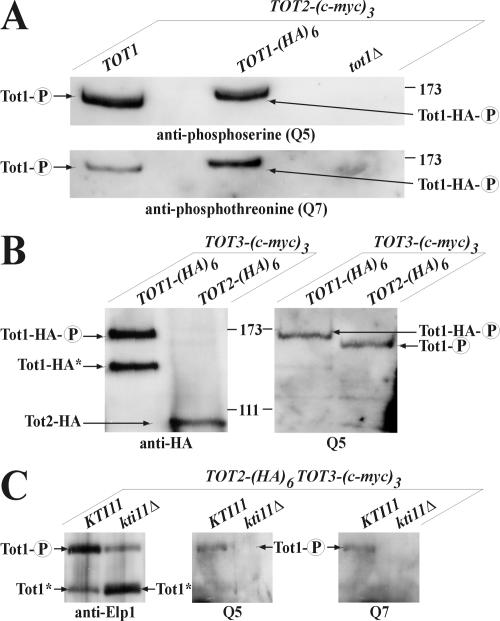
To test whether Sit4 rather acts posttranslationally, we asked whether Elongator is phosphomodified. Protein extracts obtained from TOT2-(c-myc)3 cells coexpressing TOT1-(HA)6, wild-type TOT1 or carrying a tot1Δ null-allele were subjected to immunoprecipitation followed by Western blots by using anti-phosphoserine (Q5) and anti-phosphothreonine (Q7) antibodies. Remarkably, Tot1 produced strong Q5/Q7 signals, which were lost in tot1Δ cells as expected (Figure 3A). Precipitates from TOT1-(HA)6 expressors produced a Q5/Q7-immunoreactive form of Tot1-HA that, compared with wild-type Tot1, showed an electrophoretic up-shift of ∼10 kDa, which is in line with the expected ∼9 kDa (HA)6-tag extension (Figure 3A). Immunoprecipitates obtained from TOT3-(c-myc)3 cells coexpressing TOT1-(HA)6 or TOT2-(HA)6 were analyzed by Western blots with 3F10 (anti-HA) and Q5 antibodies. Again, Q5 signals correlating to the slowest migrating Tot1-HA form (Figure 3B) were found. Wild-type Tot1 produced a Q5 signal down-shifted by ∼10 kDa relative to Tot1-HA as expected (Figure 3B). In contrast, an electrophoretically faster Tot1-HA* variant, down-shifted by ∼20 kDa, was not Q5-responsive (Figure 3B). Thus, Elongator is a phospho-complex, consisting of (at least) two Tot1 species that differ from each other by phosphorylation and by a ∼20-kDa mobility shift. Because the latter is likely to involve proteolytic truncation in a manner suppressed by Kti11, an Elongator partner protein vital for zymocin and diphtheria toxin sensitivity (Fichtner and Schaffrath, 2002; Fichtner et al., 2003; Liu and Leppla, 2003), we tested whether KTI11 affects Elongator phosphorylation. We immunoprecipitated Elongator from KTI11 or kti11Δ cells co-expressing TOT3-(c-myc)3 and TOT2-(HA)6 and probed the precipitates by using anti-Elp1/Tot1 (Otero et al., 1999), Q5, and Q7 antibodies. In kti11Δ cells, the Tot1 down-shift (∼20 kDa) was significantly pronounced as judged from a steep decrease of full-length Tot1 and a parallel increase of truncated Tot1 (Figure 3C). Markedly, kti11Δ cells no longer produced the Q5/Q7-signals corresponding to full-length Tot1 of KTI11 cells (Figure 3C). Thus, Tot1 phosphorylation is coupled to the full-length protein. Given the low levels of full-length Tot1 in kti11Δ cells, however, it remains to be seen whether Tot1 phosphorylation depends on KTI11 gene function or whether it evaded detection due to underrepresentation. Nonetheless, Kti11 is biochemically and genetically linked to Elongator, and kti11Δ cells induce TOT deficiency (Fichtner and Schaffrath, 2002; Fichtner et al., 2003).
Figure 3.
Elongator is a phospho-complex. (A) 9E10 (anti-c-Myc) immunoprecipitates of strains FFY2t-a (TOT2-(c-myc)3), FFY2/1dt (TOT2-(c-myc)3 TOT1-(HA)6), and DJY2t1d-a (TOT2-(c-myc)3 tot1Δ) were subjected to 6% SDS-PAGE analysis and probed with Q5 and Q7 antibodies, respectively. Phosphoforms ( ) of Tot1 and Tot1-HA are shown by arrows. (B) Phosphorylation of Tot1 requires full-length protein. 9E10 (anti-c-Myc) immunoprecipitates from strains FFY3/1dt (TOT3-(c-myc)3 TOT1-(HA)6) and FFY3/2dt (TOT3-(c-myc)3 TOT2-(HA)6) were probed with 3F10 (anti-HA) and Q5 to detect Tot1-HA, Tot2-HA, and phosphoforms (
) of Tot1 and Tot1-HA are shown by arrows. (B) Phosphorylation of Tot1 requires full-length protein. 9E10 (anti-c-Myc) immunoprecipitates from strains FFY3/1dt (TOT3-(c-myc)3 TOT1-(HA)6) and FFY3/2dt (TOT3-(c-myc)3 TOT2-(HA)6) were probed with 3F10 (anti-HA) and Q5 to detect Tot1-HA, Tot2-HA, and phosphoforms ( ) of Tot1 and Tot1-HA. The truncated (∼20 kDa) Tot1-HA form is indicated (*) (also see Figure 2B). (C) Phosphorylation of Tot1 is supported by Elongator partner protein Kti11. 9E10 (anti-c-Myc) immunoprecipitates from the strains FFY3/2dt (KTI11) and DJY3/2-d11 (kti11Δ) coexpressing epitope-tagged Tot2 and Tot3 were probed with anti-Elp1/Tot1, Q5 and Q7 antibodies. Truncated Tot1 is indicated (*). Numbers (A and B) refer to protein marker sizes in kilodaltons (Invitrogen).
) of Tot1 and Tot1-HA. The truncated (∼20 kDa) Tot1-HA form is indicated (*) (also see Figure 2B). (C) Phosphorylation of Tot1 is supported by Elongator partner protein Kti11. 9E10 (anti-c-Myc) immunoprecipitates from the strains FFY3/2dt (KTI11) and DJY3/2-d11 (kti11Δ) coexpressing epitope-tagged Tot2 and Tot3 were probed with anti-Elp1/Tot1, Q5 and Q7 antibodies. Truncated Tot1 is indicated (*). Numbers (A and B) refer to protein marker sizes in kilodaltons (Invitrogen).
Dephosphorylation of Elongator Depends on Sit4·Sap185/190
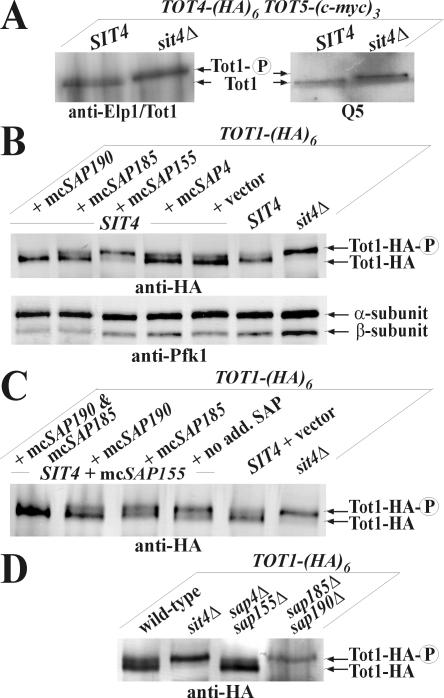
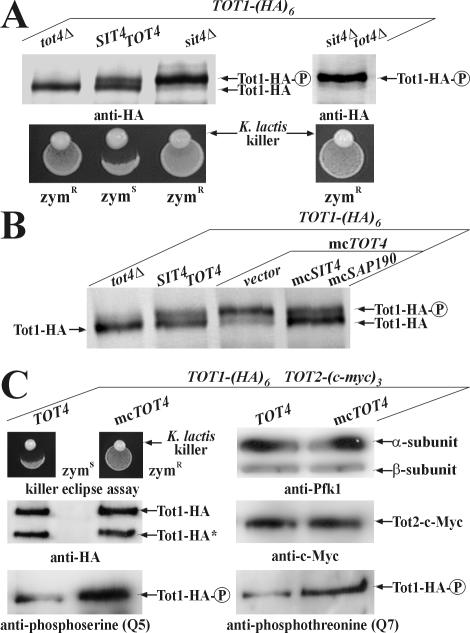
To examine the relationship between Tot1 phosphomodification and Sit4 function, Elongator was immunoprecipitated from a SIT4 strain and its isogenic sit4Δ knockout. Tot1 was identified using the anti-Elp1/Tot1 antibody (Figure 4A). Importantly, Tot1 isolated from sit4Δ cells electrophoretically migrated more slowly than Tot1 isolated from an equivalent SIT4 strain (Figure 4A). That this reflected phosphorylation of the slower Tot1 band is supported by Western blots by using the Q5 antibody on Elongator immunoprecipitates from the same SIT4 and sit4Δ strains. Again, whereas in sit4Δ cells, a more abundant, up-shifted phospho-Tot1 species accumulated (Figure 4A), SIT4 cells contained an electrophoretically faster and hypophosphorylated Tot1 form (Figure 4A) that may reflect a minor Sit4-insensitive Tot1 pool or a steady-state balance with phospho-Tot1 being underrepresented in SIT4 cells (Figure 6A). Collectively, loss of SIT4 suppresses Tot1 dephosphorylation. Among the Saps, Sit4-specific associators that act as Sit4 activators (Luke et al., 1996), multicopy SAP155 has been shown to confer zymocin resistance in a manner antagonized by excess Sap185 and Sap190 (Jablonowski et al., 2001a). To check whether high Sap155 out-titrates binding of Sap185/190 to Sit4, we compared Tot1-HA from sit4Δ cells and from SAP4, SAP155, SAP185, and SAP190 overexpressors. Analysis of total protein extracts revealed identical electrophoretic up-shifts of HA-tagged Tot1 produced from sit4Δ and multicopy SAP155 cells. Thus, consistent with phenocopying zymocin resistance of sit4Δ cells, excess Sap155 sustains phospho-Tot1 levels. In contrast, multicopy SAP4, SAP185, and SAP190 produced Tot1 patterns similar to SIT4 wild-type cells (Figure 4B). To test whether the effect of multicopy SAP155 on sustaining phospho-Tot1 levels could be counteracted by excess Sap185 and Sap190, we cointroduced multicopy SAP185, SAP190, or both into SAP155 overexpressors and investigated the electrophoretic behavior of Tot1-HA. Consistent with bypassing zymocin resistance, multicopy SAP190 and SAP185/190 suppressed the effect of multicopy SAP155 on Tot1 phosphorylation (Figure 4C). Thus, as judged from suppressing the electrophoretic up-shift of Tot1 produced in multicopy SAP155 cells, Sap185/190 specifically counteract Sap155 (Figure 4C). Contrary to sap4Δsap155Δ cells, the sap185Δsap190Δ double mutant up-regulated Tot1 phosphorylation (Figure 4D) and induced sit4Δ-like zymocin resistance (Figure 5A). Collectively, these data indicate that Tot1 dephosphorylation requires the Sit4 phosphatase in combination with Sap185 and/or Sap190.
Figure 4.
Sit4·Sap185/190 is specific for Tot1 dephosphorylation. (A) sit4Δ cells suppress Tot1 dephosphorylation. 9E10 (anti-c-Myc) Elongator immunoprecipitates from strains DJY110 (SIT4 TOT5-(cmyc)3 TOT4-(HA)6) and DJY109 (sit4Δ TOT5-(c-myc)3 TOT4-(HA)6) were probed with anti-Elp1/Tot1 and Q5 antibodies. The positions of hypo-(Tot1) and hyperphosphorylated Tot1 ( ) forms are shown. sit4Δ cells accumulate the up-shifted Tot1-
) forms are shown. sit4Δ cells accumulate the up-shifted Tot1- form. (B) sit4Δ or multicopy SAP155 cells suppress Tot1 dephosphorylation. Protein extracts from strains DJY103 (sit4Δ TOT1-(HA)6), DJY102 (SIT4 TOT1-(HA)6), and DJY102 carrying the indicated multicopy SAP constellations were standardized by anti-Pfk1 (see Figure 2B) and probed with 3F10 (anti-HA). (C) Suppressed Tot1 dephosphorylation by multicopy SAP155 is antagonized by excess Sap185/190. Protein extracts from indicated TOT-(HA)6 expressors were probed with 3F10 (anti-HA) to monitor Tot1-HA migration dependent on SAP copy number. (D) sap185Δsap190Δ cells phenocopy high phospho-Tot1 levels of sit4Δ cells. Strains DJY102 (SIT4), DJY103 (sit4Δ), DJY115 (sap4Δsap155Δ), and DJY116 (sap185Δsap190Δ) expressing TOT1-(HA)6 were analyzed as described above (B and C). Non- and phosphorylated forms (
form. (B) sit4Δ or multicopy SAP155 cells suppress Tot1 dephosphorylation. Protein extracts from strains DJY103 (sit4Δ TOT1-(HA)6), DJY102 (SIT4 TOT1-(HA)6), and DJY102 carrying the indicated multicopy SAP constellations were standardized by anti-Pfk1 (see Figure 2B) and probed with 3F10 (anti-HA). (C) Suppressed Tot1 dephosphorylation by multicopy SAP155 is antagonized by excess Sap185/190. Protein extracts from indicated TOT-(HA)6 expressors were probed with 3F10 (anti-HA) to monitor Tot1-HA migration dependent on SAP copy number. (D) sap185Δsap190Δ cells phenocopy high phospho-Tot1 levels of sit4Δ cells. Strains DJY102 (SIT4), DJY103 (sit4Δ), DJY115 (sap4Δsap155Δ), and DJY116 (sap185Δsap190Δ) expressing TOT1-(HA)6 were analyzed as described above (B and C). Non- and phosphorylated forms ( ) of Tot1-HA (B-D) are shown by arrows; vector control (B and C) denotes empty YEp24 vector used to clone the SAP genes in multicopy (Luke et al., 1996).
) of Tot1-HA (B-D) are shown by arrows; vector control (B and C) denotes empty YEp24 vector used to clone the SAP genes in multicopy (Luke et al., 1996).
Figure 6.
Balanced de-/phospho-Tot1 ratios involve opposing effects of Tot4 and Sit4. (A) tot4Δ cells enhance Tot1 dephosphorylation. Standardized protein extracts from indicated TOT1-(HA)6 expressing strains DJY102 (SIT4 TOT4), DJY103 (sit4Δ), DJY104 (tot4Δ), and DJY105 (sit4Δtot4Δ) were probed with 3F10 (anti-HA). The positions of non- and phosphorylated ( ) Tot1-HA forms are shown. (B) Excess Tot4 shifts electrophoretic mobility of Tot1, a situation antagonized by excess Sit4·Sap190. Standardized protein extracts from strain DJY102 (SIT4 TOT4) expressing TOT1-(HA)6 and maintaining multicopy (mc) TOT4, SIT4, and SAP190 genes as indicated were probed with 3F10 (anti-HA). As a control served DJY104 (tot4Δ) (see A). (C) mcTOT4 intensifies Tot1 phosphorylation. 9E10 (anti-c-Myc) immunoprecipitates from strain FFY2/1dt (TOT1-(HA)6 TOT2-(c-myc)3) maintaining single copy or mcTOT4 were probed with 3F10 (anti-HA), 9E10 (antic-Myc), Q5 (anti-phosphoserine) and Q7 (anti-phosphothreonine) antibodies to detect Tot1-HA, Tot2-c-Myc and phospho-Tot1-HA (
) Tot1-HA forms are shown. (B) Excess Tot4 shifts electrophoretic mobility of Tot1, a situation antagonized by excess Sit4·Sap190. Standardized protein extracts from strain DJY102 (SIT4 TOT4) expressing TOT1-(HA)6 and maintaining multicopy (mc) TOT4, SIT4, and SAP190 genes as indicated were probed with 3F10 (anti-HA). As a control served DJY104 (tot4Δ) (see A). (C) mcTOT4 intensifies Tot1 phosphorylation. 9E10 (anti-c-Myc) immunoprecipitates from strain FFY2/1dt (TOT1-(HA)6 TOT2-(c-myc)3) maintaining single copy or mcTOT4 were probed with 3F10 (anti-HA), 9E10 (antic-Myc), Q5 (anti-phosphoserine) and Q7 (anti-phosphothreonine) antibodies to detect Tot1-HA, Tot2-c-Myc and phospho-Tot1-HA ( ). Before immunoprecipitation, extracts were standardized by anti-Pfk1 to follow content of Pfk1 α and β subunits (Figure 2B). Tot1-HA truncation is indicated (*). For zymocin read-outs (A and C), see Figure 1A.
). Before immunoprecipitation, extracts were standardized by anti-Pfk1 to follow content of Pfk1 α and β subunits (Figure 2B). Tot1-HA truncation is indicated (*). For zymocin read-outs (A and C), see Figure 1A.
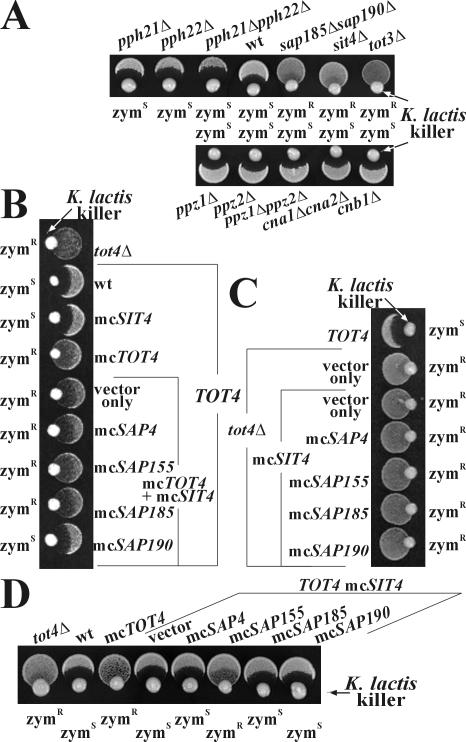
Figure 5.
Zymocicity strictly depends on Sit4·Sap185/190 and high copy TOT4 zymocin resistance is suppressed by Sit4·Sap190. (A) K. lactis killer eclipse assays (see Figure 1A) involved S. cerevisiae strains LFY5 (tot3Δ), AY925 (wild-type (wt)), and phosphatase mutants CY3938 (sit4Δ), CY5224 (sap185Δsap190Δ), DEY132-1C (pph21Δ), DEY10-2B (pph22Δ), DEY132-2C (pph21Δpph22Δ), EDN75 (ppz1Δ), JA103 (ppz2Δ), EDN76 (ppz1Δppz2Δ), MMY09 (cna1Δcna2Δ), and YJN519 (cnb1Δ). (B) Eclipse assays involving resistant control DYJ100 (tot4Δ) and wild-type TOT4 strain CY4029 carrying multicopy (mc) TOT4, SIT4, and/or SAP genes. (C) Eclipse assays involving mcSIT4/SAP genes maintained in strain DYJ100 (tot4Δ) and compared with wild-type TOT4 CY4029 cells. (D) Eclipse assays of zymocin-resistant strain DYJ100 strain (tot4Δ) and wild-type CY4029 (TOT4) cells carrying the indicated mcSIT4/SAP genes. As a high copy control served mcTOT4. For phenotypic zymocin readouts, see Figure 1A. Vector controls (B-D) refer to empty YEplac181, YEp24, and YEplac112 plasmids used to clone the SIT4, SAP, and TOT4 genes, respectively (Butler et al., 1994; Luke et al., 1996; Jablonowski et al., 2001c).
Sit4·Sap190 Opposes Zymocin Resistance Induced by Multicopy TOT4
Because defects in several other Ser/Thr phosphatases (pph21Δ, pph22Δ, pph21Δpph22Δ, ppz1Δ, ppz2Δ, ppz1Δppz2Δ, pph3Δ, and ppg1Δ), including calcineurin (cna1Δcna2Δ and cnb1Δ), neither affected phospho-Tot1 levels nor zymocin sensitivity (Figure 5A; our unpublished data), dephosphorylation of Elongator is specifically linked to the PP2A-type phosphatase Sit4. To study a possible role in Elongator regulation, we compared the relationship between Sit4, Tot4, and the lethal response toward zymocin. Tot4, a potential Elongator-pol II up-loader, is able to protect cells against zymocin either when overexpressed or removed from the cell (Frohloff et al., 2001, 2003; Fichtner et al., 2002a,b). Whereas multicopy TOT4 cells and tot4Δ cells show similar zymocin resistance, multicopy SIT4 on its own has no effect (Figure 5B). However, we found that when multicopy SIT4 was cointroduced into multicopy TOT4 cells, zymocin protection associated with high copy TOT4 was slightly antagonized in the presence of extra Sit4 (Figure 5B). This effect was significantly enhanced by cointroducing multicopy SAP190, which reinstated wild-type zymocin sensitivity to the multicopy SIT4 TOT4 strain (Figure 5B). The action is specific because SAP4, SAP155, and SAP185 remained ineffective in comparison with SAP190 (Figure 5B). tot4Δ-associated resistance, however, was not opposed by multicopy SIT4 SAP190 (Figure 5C), nor was normal zymocin sensitivity of single copy TOT4 cells affected by multicopy SIT4 SAP190 (Figure 5D). In a single copy TOT4 strain, only multicopy SIT4 SAP155 suppressed zymocin sensitivity (Figure 5D), most likely as a result of outcompeting Sap185/190 from Sit4 binding and augmenting Tot1 phosphorylation (Figure 4B). In conclusion, suppression of excess Tot4 by elevated Sit4·Sap190 implies competition between Tot4 and Sit4·Sap190 for a shared factor (TOT/Elongator) and opposing effects on Tot1 phosphorylation.
Tot4 Antagonizes Sit4-dependent Dephosphorylation of Elongator
To correlate genetic suppression between Sit4 and Tot4 on zymocin action (Figure 5B) with Tot1 dephosphorylation, we analyzed Tot1-HA expressed from sit4Δ, tot4Δ, tot4Δsit4Δ, TOT4 SIT4 wild-type and multicopy TOT4 cells by using the anti-HA antibody in Western blots. Although zymocin-resistant sit4Δ cells exclusively reproduced high phospho-Tot1 levels (Figure 6A), sensitive TOT4 SIT4 wild-type cells expressed a balanced de-/phospho-Tot1 ratio with phospho-Tot1 being slightly underrepresented (Figure 6A). Conversely, in zymocin-resistant tot4Δ cells, phospho-Tot1 was abolished, whereas a zymocin-resistant tot4Δsit4Δ double mutant phenocopied the phospho-Tot1 levels present in single sit4Δ cells (Figure 6A). Thus, removal of Sit4 or Tot4 affect Tot1 phosphomodification in opposite ways, and tot4Δ cells intensify Tot1 dephosphorylation. In line with TOT4 influencing Elongator phosphorylation, zymocin resistant-multicopy TOT4 cells up-regulated phospho-Tot1 levels compared with balanced de-/phosphorylation of zymocin-sensitive wild-type TOT4 cells (Figure 6B). Remarkably, stimulation of Tot1 phosphorylation by excess Tot4 as detected by intensified Q5/Q7-immunoreactivity of Tot1 in Elongator immunoprecipitations (Figure 6C) did not interfere with Elongator complex assembly or subunit interaction: stoichiometric coimmunoprecipitation of Elongator subunits Tot1 and Tot2 was unaffected by multicopy TOT4 cells (Figure 6C). Consistent with genetic suppression and functional antagonism, excess Sit4·Sap190 enhanced Tot1 dephosphorylation in multicopy TOT4 cells, reinstated Tot1's wild-type phospho-balance (Figure 6B), and reinstalled zymocin sensitivity (Figure 5B). Thus, a fine-tuned balance of Tot1 de-/phosphomodification that is critical for Elongator's TOT function is mutually controlled by SIT4 and TOT4, and our data provide evidence to suggest that locking Tot1 into either a hyper- or hypophosphorylated state is detrimental to Elongator function and protects against the G1 cell cycle block by zymocin.
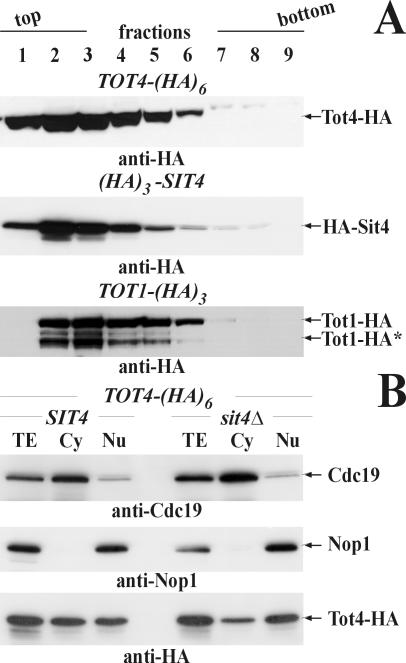
Tot1, Tot4, and Sit4 Cofractionate and sit4Δ-Nuclei Retain Tot4
Consistent with Sit4-dependent Tot1 dephosphorylation being under Tot4 control, sucrose gradient cell fractionations demonstrated that HA-tagged Sit4 and Tot4 essentially comigrated (Figure 7A). A strikingly similar fractionation pattern was produced by HA-tagged Tot1, and peak fractions coincided with maximal levels of either Tot4 or Sit4 (Figure 7A, lanes 2 and 3). This implies both a physical and functional relationship among Sit4, Tot4, and Elongator. Intriguingly, although the Tot4·Elongator contact does not require SIT4 function (our unpublished data), Sit4 seems to impact the subcellular distribution of Tot4 itself. In contrast to SIT4 cells, which showed a balanced distribution of cytoplasmic and nuclear Tot4 pools, Tot4 was preferentially retained in sit4Δ-nuclei with a parallel drop in the cytoplasmic pool (Figure 7B). Provided this was due to prolonged pol II·Elongator association, Sit4 may act following the assembly of the HAT-productive holo-Elongator (Winkler et al., 2002) by recycling the potential up-loader Tot4. So, Sit4 likely plays a nuclear role for Elongator's TOT function and zymocin-resistant sit4Δ cells may have an Elongator·pol II up-loading defect.
Figure 7.
Sit4, Tot4, and Tot1 cofractionate and Tot4 is retained in sit4Δ-nuclei. (A) Cell fractionation. Tot4-HA, HA-Sit4, and Tot1-HA were detected in sucrose-gradient fractions (lanes 1-9 from a total of 14) from strains DJY114 (TOT4-(HA)6), DJYHS4 ((HA)3-SIT4), and DJY8A-1H3 (TOT1-(HA)3) by using 3F10 (anti-HA). Their positions are indicated by arrows. The truncated Tot1-HA form (see Figure 3B) is indicated (*). (B) Tot4 is retained in sit4Δ-nuclei. Fractions of DJY114 (SIT4) or DJY113 (sit4Δ) cells expressing TOT4-(HA)6 were probed using 3F10 (anti-HA) and compared with nuclear and cytoplasmic marker proteins Nop1 and Cdc19, respectively (see Figure 1C). Their positions are indicated by arrows.
DISCUSSION
Dephosphorylation of Elongator Subunit 1 Depends on Sit4·Sap185/190
Like Elongator mutants, cells lacking the phosphatase Sit4 or its associators Sap185 and Sap190 survive the K. lactis zymocin (Frohloff et al., 2001; Jablonowski et al., 2001a,c). In line with a link to pol II Elongator, SIT4 was originally identified as a transcriptional suppressor, and sit4 mutations combined with pol II defects are synthetically lethal (Arndt et al., 1989). Our findings that sit4Δ and totΔ cells equally well resist the ceramide-induced G1 block (Nickels and Broach, 1996) (Figure 1C) reinforce functional linkage between Sit4 and pol II Elongator in processes required for progression through G1. Our finding that directly tagging Sit4's C-terminal -YFL motif elicits zymocin resistance (Figure 1A) may be interpreted that this PP2A-type domain known to mediate subunit interaction enables Sit4 to form subcomplexes with zymocin-relevant associators (Evans and Hemmings, 2000; Wei et al., 2001). Consistently, here we underscore that Sit4 associators Sap4, Sap155, Sap185, and Sap190 act in two functional subfamilies (Sap4/155 and Sap185/190). Only combined loss of Sap185 and Sap190 leads to zymocin resistance (Figure 5B), whereas sap4Δsap155Δ cells and triple sap deletion strains still containing either Sap185 or Sap190 are zymocin sensitive (our unpublished data). As for Elongator, a phospho-complex (Figure 3), dephosphorylation of its largest subunit (Tot1/Elp1) is shown here to depend on Sit4·Sap185 and/or Sit4·Sap190 (Figure 4). Intriguingly, SAP185 cannot effectively replace SAP190 in suppressing multicopy TOT4 zymocin resistance (Figure 5B). With regard to zymocin action, Sap190 thus seems to be the more potent member of the Sap185/190 subfamily. Contrary to sap185Δsap190Δ, sap4Δsap155Δ cells have normal phospho-Tot1 levels (Figure 4D). However, multicopy SAP155 induces zymocin resistance with high phospho-Tot1 levels similar to sit4Δ and sap185Δsap190Δ cells (Figure 4, B and D). Because this effect is antagonized by multicopy SAP185 or SAP190 (Figure 4C), excess Sap155 is likely to abduct Sit4 from its TOT-relevant associators Sap185/190, a notion consistent with the observation that Sap155 is particularly important for the TOR pathway (Jacinto et al., 2001). This adds to our proposal that despite a shared requirement for SIT4, there is hardly any evidence for Sit4-mediated TOT-TOR cross talk (Jablonowski et al., 2001a). In line, excess Sap185/190 opposes Sap155's ability to remove Sit4 and reinstates wild-type Tot1 phospho-balance and zymocin sensitivity. Collectively, besides other dephosphorylation events related to the functioning of the TOR pathway (Crespo and Hall, 2002), Sit4·Sap185/190 promotes Elongator dephosphorylation and conditions the zymocin G1 block.
Unlike suppression of multicopy TOT4 zymocin resistance, Sit4·Sap190 fails to antagonize zymocin protection of tot4Δ cells (Figure 5C). Thus, suppression requires Tot4 to be physically present. This reinforces the TOT relevant role of Sit4 and implies that Sit4 competes with or opposes Tot4 (for a shared Elongator substrate). Consistently, phospho-Tot1 levels are entirely abolished in tot4Δ cells, whereas excess Tot4 intensifies Tot1 phosphorylation (Figure 6). This indicates that Sit4 may be kept in check by Tot4 inhibition. Based on its capability to occupy promoter DNA and to associate with pol II and Elongator, Tot4 has been proposed to play a regulatory role, possibly as a candidate pol II-Elongator up-loader (Fichtner et al., 2002a,b; Frohloff et al., 2003). Our findings that Sit4, Tot4, and Tot1 cofractionate (Figure 7A) and that sit4Δ cells increase, whereas tot4Δ abolish Tot1 phosphorylation (Figures 4A and 6A) suggest Elongator loading via Tot4 may require SIT4. Accordingly, high Sit4·Sap190 levels could correct deregulated loading due to excess Tot4 by enhancing dephosphorylation of Tot1 (or increasing Tot4 recycling). Consistently, Tot4 is retained in sit4Δ nuclei (Figure 7B), a scenario that may reflect a loading or recycling defect. Together with the findings that high phospho-Tot1 levels typical of sit4Δ cells remain unaltered in sit4Δtot4Δ cells (Figure 6A) and that Tot4 does not relate to any known protein kinase, Tot4 is unlikely to act as a Sit4-antagonizing Elongator kinase.
Despite the elusive nature of an Elongator kinase, we report here on a supporting activity of Kti11 on Tot1 phosphorylation (Figure 3C). Kti11, recently identified as Elongator partner protein and zymocin and diphtheria toxin sensitivity factor, also interacts with components of the diphthamide synthesis pathway crucial for bacterial toxins to ADP-ribosylate elongation translation factor EF2. kti11Δ cells lack EF2 ADP-ribosyl acceptor activity, survive zymocin, and accumulate a truncated Tot1 form shown here to be phosphorylation-resistant (Fichtner and Schffrath, 2002; Fichtner et al., 2003; Liu and Leppla, 2003). If Elongator phosphoacceptor sites are identifiable by comparative mass spectrometry on SIT4, sit4Δ, and kti11Δ cells and whether Elongator links up to diphthamidisation via Kti11 are intriguing questions presently under investigation.
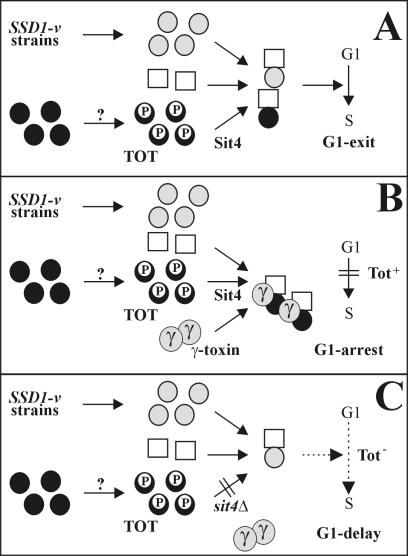
A Model for Sit4's Role in the G1 Block by K. lactis Zymocin
A model (Figure 8) that combines a yeast cell's requirement for Sit4 and Elongator to become blocked in G1 by zymocin's γ-toxin subunit can be conceived. In the absence of γ-toxin, either an alternative pathway (in SSD1-v strains) or TOT/Elongator dephosphorylation dependent on Sit4 can activate a key cell cycle regulator needed for G1 exit (Figure 8A). In the presence of γ-toxin, this key activator is sequestered in combination with TOT, the possible Sit4 substrate, regardless of whether the alternative SSD1-v pathway is normally sufficient and hence Sit4 dispensable (Figure 8B). Eventually, this leads to inactivation of the crucial activator, preventing START execution and G1 exit. If cells fail to dephosphorylate Tot1 (sit4Δ) (Figure 8C) or do not assemble Elongator (totΔ), they express TOT deficiency that no longer permits γ-toxin to hijack Elongator (Figure 8C). As a result, sit4Δ and totΔ cells resist γ-toxin at the expense of a G1 delay due to an Elongator defect. Consequently, reduced activation by the alternative SSD1-v pathway alone may cause a cell cycle delay. The findings that totΔ, sit4Δ, and ssd1Δ cells are each cell cycle affected support these predictions (Sutton et al., 1991; Frohloff et al., 2001). The model's key activator ought to physically contact TOT/Elongator. As judged from SSD1-v-dependent pol II stimulation, copurification between Elongator and pol II and assistance of pol II transcription through chromatin by the Elongator HAT, we propose pol II as this activator (Stettler et al., 1993; Otero et al., 1999; Winkler et al., 2001, 2002; Kim et al., 2002). In line, pol II mutations can induce a G1 block per se (Sugaya et al., 2001), suggesting that zymocin-dependent Elongator intoxication and pol II limitation may prevent G1 exit. Indeed, zymocin affects pol II performance and CTD phosphorylation (Frohloff et al., 2001; Jablonowski and Schaffrath, 2002). The findings that phospho-CTD stabilizes Elongator·pol II association, that CTD phosphatase modulation alters the zymocin response and that a Tot1 NLS truncation yields resistance, suggest that zymocin requires nuclear Elongator pools to physically contact pol II (Otero et al., 1999; Jablonowski et al., 2001c; Kitamoto et al., 2002, Fichtner et al., 2003).
Figure 8.
Model for the role of Sit4 in the cell cycle arrest imposed by K. lactis zymocin's γ-toxin subunit. In the absence of γ-toxin (A), the SSD1-v pathway ( ) and Sit4-dependent dephosphorylation of the toxin-target (TOT) substrate (
) and Sit4-dependent dephosphorylation of the toxin-target (TOT) substrate ( ) activate a key cell cycle regulator (□) required for processes preSTART. In the presence of γ-toxin (B), the critical protein can be sequestered and inactivated in combination with Sit4's dephosphorylated TOT substrate (•) to induce the G1 arrest (Tot+), irrespective of whether the alternative, Sit4-dispensable SSD1-v pathway would be sufficient. Failure to dephosphorylate TOT (C) no longer permits inactivation of the crucial regulator and causes sit4Δ cells to resist γ-toxicity (Tot-) at the expense of being G1 cell cycle delayed. The as yet elusive Elongator-specific kinase is marked by?.
) activate a key cell cycle regulator (□) required for processes preSTART. In the presence of γ-toxin (B), the critical protein can be sequestered and inactivated in combination with Sit4's dephosphorylated TOT substrate (•) to induce the G1 arrest (Tot+), irrespective of whether the alternative, Sit4-dispensable SSD1-v pathway would be sufficient. Failure to dephosphorylate TOT (C) no longer permits inactivation of the crucial regulator and causes sit4Δ cells to resist γ-toxicity (Tot-) at the expense of being G1 cell cycle delayed. The as yet elusive Elongator-specific kinase is marked by?.
Because neither assembly of Elongator nor its interaction with Tot4 are Sit4 sensitive (Figure 2C), Sit4 is likely to act after assembly of the Elongator HAT complex. Previous data demonstrated that Elongator HAT productivity conditions TOT proficiency (Frohloff et al., 2001; Winkler et al., 2001). TOT deficiency due to up-regulated (sit4Δ, sap185Δsap190Δ, and high copy SAP155 or TOT4) (Figure 4) or down-regulated (tot4Δ) phospho-Tot1 levels (Figure 6, A and B) points to HAT regulation by a fine-tuned Tot1 phospho-balance (via SIT4 and/or TOT4). Mutual control of Tot1 de-/phosphomodification by SIT4 and TOT4 suggests that phosphorylation of Tot1 does not seem to act as a functional Elongator “on/off” switch; thus, locking Tot1 into either hyper- or hypophosphorylated states impairs Elongator function as assessed by Tot- phenotypes of multicopy SAP155 and TOT4 cells as well as sit4Δ, tot4Δ, sit4Δtot4Δ, and sap185Δsap190Δ mutants. Possibly, Elongator's TOT function is supported by alternating de-/phosphorylation cycles. Whether Tot1 dephosphorylation depends directly or indirectly on Sit4 remains open. Although seemingly inconsistent with the cytoplasmic distribution deduced from immunolocalization studies on multicopy SIT4 cells (Sutton et al., 1991), our data that Sit4 is nucleocytoplasmically localized (Figure 1B) suggest a significant nuclear Sit4 pool. Consistently, sit4 mutations combined with nuclear ubiquitin or pol II defects are lethal, Sit4 physically interacts with Ctk1, a nuclear pol II CTD kinase, and with Hrr25/Kti14, a predominantly nuclear casein kinase I that copurifies with Sap185 and Sap190 (Arndt et al., 1989; Ho et al., 2002; Singer et al., 2003). Intriguingly, like Tot- Elongator mutants and sit4Δ cells, kinase-minus kti14 mutants survive zymocin (Mehlgarten and Schaffrath, 2003). A possible role of Kti14 as a Sit4 antagonist relevant to Elongator's TOT function and phosphomodification is under investigation.
Acknowledgments
We thank Drs. J. Ariño, J. Aris, K. Arndt, T. Davies, T. Edlind, M. Hall, J. Heinisch, Y. Jiang, B. Stillman, J. Svejstrup, and J. Thorner for providing antibodies, plasmids, and yeast strains. R.S. was supported by a Deutsche Forschungsgemeinschaft grant (Scha 750/2).
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03-10-0750. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-10-0750.
References
- Arndt, K.T., Styles, C.A., and Fink, G.R. (1989). A suppressor of a HIS4 transcriptional defect encodes a protein with homology to the catalytic subunit of protein phosphatases. Cell 56, 527-537. [DOI] [PubMed] [Google Scholar]
- Berben, G., Dumont, J., Gilliquet, V., Bolle, P.A., and Hilger, F. (1991). The YDp plasmids: a uniform set of vectors bearing versatile gene disruption cassettes for Saccharomyces cerevisiae. Yeast 7, 475-477. [DOI] [PubMed] [Google Scholar]
- Bradford, M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248-254. [DOI] [PubMed] [Google Scholar]
- Butler, A.R., O'Donnell, R.W., Martin, V.J., Gooday, G.W., and Stark, M.J. (1991a). Kluyveromyces lactis toxin has an essential chitinase activity. Eur. J. Biochem. 199, 483-488. [DOI] [PubMed] [Google Scholar]
- Butler, A.R., White, J.H., and Stark, M.J. (1991b). Analysis of the response of Saccharomyces cerevisiae cells to Kluyveromyces lactis toxin. J. Gen. Microbiol. 137, 1749-1757. [DOI] [PubMed] [Google Scholar]
- Butler, A.R., White, J.H., Folawiyo, Y., Edlin, A., Gardiner, D., and Stark, M.J. (1994). Two Saccharomyces cerevisiae genes which control sensitivity to G1 arrest induced by Kluyveromyces lactis toxin. Mol. Cell. Biol. 14, 6306-6316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespo, J.L., and Hall, M.N. (2002). Elucidating TOR signaling and rapamycin action: lessons from Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 66, 579-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross, F.R. (1990). Cell cycle arrest caused by CLN gene deficiency in Saccharomyces cerevisiae resembles START-I arrest and is independent of the mating pheromone signalling pathway. Mol. Cell. Biol. 10, 6482-6490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Torre-Ruiz, A.M., Torres, J., Ariño, J., and Herrero, E. (2002). Sit4 is required for proper modulation of the biological functions mediated by Pkc1 and the cell integrity pathway in Saccharomyces cerevisiae. J. Biol. Chem. 277, 33468-33676. [DOI] [PubMed] [Google Scholar]
- Di Como, C.J., and Arndt, K.T. (1996). Nutrients, via the Tor proteins, stimulate the association of Tap42 with type 2A phosphatases. Genes Dev. 10, 1904-1916. [DOI] [PubMed] [Google Scholar]
- Evans, D.R., and Stark, M.J. (1997). Mutations in the Saccharomyces cerevisiae type 2A protein phosphatase catalytic subunit reveal roles in cell wall integrity, actin cytoskeleton organization and mitosis. Genetics 145, 227-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, D.R., and Hemmings, B.A. (2000). Mutation of the C-terminal leucine residue of PP2Ac inhibits PR55/B subunit binding and confers supersensitivity to microtubule destabilization in Saccharomyces cerevisiae. Mol. Gen. Genet. 264, 425-432. [DOI] [PubMed] [Google Scholar]
- Fernandez-Sarabia, M.J., Sutton, A., Zhong, T., and Arndt, K.T. (1992). SIT4 protein phosphatase is required for the normal accumulation of SWI4, CLN1, CLN2 and HCS26 RNAs during late G1. Genes Dev. 6, 2417-2428. [DOI] [PubMed] [Google Scholar]
- Fichtner, L., Frohloff, F., Bürkner, K., Larsen, M., Breunig, K.D., and Schaffrath, R. (2002a). Molecular analysis of KTI12/TOT4, a Saccharomyces cerevisiae gene required for Kluyveromyces lactis zymocin action. Mol. Microbiol. 43, 783-791. [DOI] [PubMed] [Google Scholar]
- Fichtner, L., Frohloff, F., Jablonowski, D., Stark, M.J., and Schaffrath, R. (2002b). Protein interactions within Saccharomyces cerevisiae Elongator, a complex essential for Kluyveromyces lactis zymocicity. Mol. Microbiol. 45, 817-826. [DOI] [PubMed] [Google Scholar]
- Fichtner, L., and Schaffrath, R. (2002). KTI11 and KTI13, Saccharomyces cerevisiae genes controlling sensitivity to G1 arrest induced by Kluyveromyces lactis zymocin. Mol. Microbiol. 44, 865-875. [DOI] [PubMed] [Google Scholar]
- Fichtner, L., Jablonowski, D., Schierhorn, A., Kitamoto, H.K., Stark, M.J., and Schaffrath, R. (2003). Elongator's toxin-target (TOT) function is NLS-dependent and suppressed by post-translational modification. Mol. Microbiol. 49, 1297-1307. [DOI] [PubMed] [Google Scholar]
- Frohloff, F., Fichtner, L., Jablonowski, D., Breunig, K.D., and Schaffrath, R. (2001). Saccharomyces cerevisiae Elongator mutations confer resistance to the Kluyveromyces lactis zymocin. EMBO J. 20, 1993-2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohloff, F., Jablonowski, D., Fichtner, L., and Schaffrath, R. (2003). Subunit communications crucial for the functional integrity of the yeast RNA polymerase II Elongator (γ-toxin target (TOT)) complex. J. Biol. Chem. 278, 956-961. [DOI] [PubMed] [Google Scholar]
- Gietz, D., St. Jean, A., Woods, R.A., and Schiestl, R.H. (1992). Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20, 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz, D., and Sugino, A. (1988). New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenised yeast genes lacking six-base pair restriction sites. Gene 74, 527-534. [DOI] [PubMed] [Google Scholar]
- Ho, Y., et al. (2002). Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415, 180-183. [DOI] [PubMed] [Google Scholar]
- Jablonowski, D., and Schaffrath, R. (2002). Saccharomyces cerevisiae RNA polymerase II is affected by Kluyveromyces lactis zymocin. J. Biol. Chem. 277, 26276-26280. [DOI] [PubMed] [Google Scholar]
- Jablonowski, D., Butler, A.R., Fichtner, L., Gardiner, D., Schaffrath, R., and Stark, M.J. (2001a). Sit4p protein phosphatase is required for sensitivity of Saccharomyces cerevisiae to Kluyveromyces lactis zymocin. Genetics 159, 1479-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablonowski, D., Fichtner, L., Martin, V.J., Klassen, R., Meinhardt, F., Stark, M.J., and Schaffrath, R. (2001b). Saccharomyces cerevisiae cell wall chitin, the Kluyveromyces lactis zymocin receptor. Yeast 18, 1285-1299. [DOI] [PubMed] [Google Scholar]
- Jablonowski, D., Frohloff, F., Fichtner, L., Stark, M.J., and Schaffrath, R. (2001c). Kluyveromyces lactis zymocin mode of action is linked to RNA polymerase II function via Elongator. Mol. Microbiol. 42, 1095-1106. [DOI] [PubMed] [Google Scholar]
- Jacinto, E., Guo, B., Arndt, K.T., Schmelzle, T., and Hall, M.N. (2001). TIP41 interacts with TAP42 and negatively regulates the TOR signaling pathway. Mol. Cell 8, 1017-1026. [DOI] [PubMed] [Google Scholar]
- Kim, J-H., Lane, W.S., and Reinberg, D. (2002). Human Elongator facilitates RNA polymerase II transcription through chromatin. Proc. Natl. Acad. Sci. USA 99, 1241-12461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishida, M., Tokunaga, M., Katayose, Y., Yajima, H., Kawamura-Watabe, A., and Hishinuma, F. (1996). Isolation and genetic characterization of pGKL killer-insensitive mutants (iki) from Saccharomyces cerevisiae. Biosci. Biotechnol. Biochem. 60, 798-801. [DOI] [PubMed] [Google Scholar]
- Kitamoto, H.K., Jablonowski, D. Nagase, J., and Schaffrath, R. (2002). Defects in yeast RNA polymerase II transcription elicit hypersensitivity to G1 arrest induced by Kluyveromyces lactis zymocin. Mol. Genet. Genomics 268, 49-55. [DOI] [PubMed] [Google Scholar]
- Knop, M., Siegers, K., Pereira, G., Zachariae, W., Winsor, B., Nasmyth, K., and Schiebel, E. (1999). Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast 15, 963-972. [DOI] [PubMed] [Google Scholar]
- Kobor, M.S., and Greenblatt, J. (2002). Regulation of transcription elongation by phosphorylation. Biochim. Biophys. Acta 1577, 261-275. [DOI] [PubMed] [Google Scholar]
- Kölling, R., and Hollenberg, C.P. (1994). The ABC-transporter Ste6 accumulates in the plasma membrane in a ubiquitinated form in endocytosis mutants. EMBO J. 13, 3261-3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, S., and Leppla, S.H. (2003). Retroviral insertional mutagenesis identifies a small protein required for synthesis of diphthamide, the target of bacterial ADP-ribosylating toxins. Mol. Cell 12, 603-613. [DOI] [PubMed] [Google Scholar]
- Longtine, M.S., McKenzie, A 3rd, Demarini, D.J., Shah, N.G., Wach, A., Brachat, A., Philippsen, P., and Pringle, J.R. (1998). Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953-961. [DOI] [PubMed] [Google Scholar]
- Luke, M.M., Della Seta, F., Di Como, C.J., Sugimoto, H., Kobayashi, R., and Arndt, K.T. (1996). The SAP, a new family of proteins, associate and function positively with the SIT4 phosphatase. Mol. Cell. Biol. 16, 2744-2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehlgarten, C., and Schaffrath, R. (2003). Mutant casein kinase I (Hrr25p/Kti14p) abrogates the G1 cell cycle arrest induced by Kluyveromyces lactis zymocin in budding yeast. Mol. Genet. Genomics 269, 188-196. [DOI] [PubMed] [Google Scholar]
- Munoz, I., Simon, E., Casals, N., Clotet, J., and Ariño, J. (2003). Identification of multicopy suppressors of cell cycle arrest at the G1-S transition in Saccharomyces cerevisiae. Yeast 20, 157-169. [DOI] [PubMed] [Google Scholar]
- Nasmyth, K., and Dirick, L. (1991). The role of SWI4 and SWI6 in the activity of G1 cyclins in yeast. Cell, 66, 995-1013. [DOI] [PubMed] [Google Scholar]
- Nickels, J.T., and Broach, J.R. (1996). A ceramide-activated protein phosphatase mediates ceramide-induced G1 arrest of Saccharomyces cerevisiae. Genes Dev. 10, 382-394. [DOI] [PubMed] [Google Scholar]
- Otero, G., Fellows, J., Li, Y., de Bizemont, T., Dirac, A.M., Gustafsson, C.M., Erdjument-Bromage, H., Tempst, P., and Svejstrup, J.Q. (1999). Elongator, a multisubunit component of a novel RNA polymerase II holoenzyme for transcriptional elongation. Mol. Cell 3, 109-118. [DOI] [PubMed] [Google Scholar]
- Pereira, G., Knop, M., and Schiebel, E. (1998). Spc98p directs the yeast γ-tubulin complex into the nucleus and is subject to cell cycle-dependent phosphorylation on the nuclear side of the spindle pole body. Mol. Biol. Cell 9, 775-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posas, F., Clotet, J., and Ariño, J. (1991). Saccharomyces cerevisiae gene SIT4 is involved in the control of glycogen metabolism. FEBS Lett. 279, 341-345. [DOI] [PubMed] [Google Scholar]
- Schaffrath, R., and Breunig, K.D. (2000). Genetics and molecular physiology of the yeast Kluyveromyces lactis. Fungal Genet. Biol. 30, 173-190. [DOI] [PubMed] [Google Scholar]
- Sherman, F. (1991). Getting started with yeast. Methods Enzymol. 194, 3-21. [DOI] [PubMed] [Google Scholar]
- Sikorski, R.S., and Hieter., P. (1989). A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer, T., Haefner, S., Hoffmann, M., Fischer, M., Ilyina, J., and Hilt, W. (2003). Sit4 phosphatase is functionally linked to the ubiquitin-proteasome system. Genetics 164, 1305-1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark, M.J. (1998). Protein phosphorylation and dephosphorylation. In: The Metabolism and Molecular Physiology of Saccharomyces cerevisiae, ed. J.R. Dickinson and M. Schweizer, London: Taylor & Francis, Ltd., London, 209-276.
- Stark, M.J., and Boyd, A. (1986). The killer toxin of Kluyveromyces lactis: characterization of the toxin subunits and identification of the genes which encode them. EMBO J. 5, 1995-2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stettler, S., Chiannilkulchai, N., Hermann-Le Denmat, S., Lalo, D., Lacroute, F., Sentenac, A., and Thuriaux, P. (1993). A general suppressor of RNA polymerase I, II and III mutations in Saccharomyces cerevisiae. Mol. Gen. Genet. 239, 169-176. [DOI] [PubMed] [Google Scholar]
- Sugaya, K., Sasanuma, S., Cook, P.R., and Mita, K. (2001). A mutation in the largest (catalytic) subunit of RNA polymerase II and its relation to arrest of the cell cycle in G(1) phase. Gene 274, 77-81. [DOI] [PubMed] [Google Scholar]
- Sutton, A., and Freiman, R. (1997). The Cak1p protein kinase is required at G1/S and G2/M in the budding yeast cell cycle. Genetics 147, 57-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton, A., Immanuel, D., and Arndt, K.T. (1991). The SIT4 protein phosphatase functions in late G1 for progression into S phase. Mol. Cell. Biol., 11, 2133-2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, H., Ashby, D.G., Moreno, C.S., Ogris, E., Yeong, F.M., Corbett, A.H., and Pallas, D.C. (2001). Carboxymethylation of the PP2A catalytic subunit in Saccharomyces cerevisiae is required for efficient interaction with the B-type subunits Cdc55p and Rts1p. J. Biol. Chem. 276, 1570-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H., Wang, X., and Jiang, Y. (2003). Interaction with Tap42 is required for the essential function of Sit4 and type 2A phosphatases. Mol. Biol. Cell 14, 4342-4351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler, G.S., Kristjuhan, A., Erdjument-Bromage, H., Tempst, P., and Svejstrup, J.Q. (2002). Elongator is a histone H3 and H4 acetyltransferase important for normal histone acetylation levels in vivo. Proc. Natl. Acad. Sci. USA 99, 3517-3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler, G.S., Petrakis, T.G., Ethelberg, S., Tokunaga, M., Erdjument-Bromage, H., Tempst, P., and Svejstrup, J.Q. (2001). RNA polymerase II elongator holoenzyme is composed of two discrete subcomplexes. J. Biol. Chem. 276, 32743-32749. [DOI] [PubMed] [Google Scholar]
- Zachariae, W., Shin, T.H., Galova, M., Obermaier, B., and Nasmyth, K. (1996). Identification of subunits of the anaphase-promoting complex of Saccharomyces cerevisiae. Science 274, 1201-1204. [DOI] [PubMed] [Google Scholar]