Significance
Synchronized activity among groups of interconnected cells is essential for diverse functions in the brain. Most studies on neuronal networks have been performed in the mature brain when chemical synapses have been established. However, less is known about networking during embryonic development. We have studied neural progenitors and found that they form gap junction-mediated small-world networks, which, via electrical depolarization, drive spontaneous calcium activity to stimulate cell proliferation. Our data underscore the critical role of intricate cell signaling during embryonic development and show that complex networks of immature cells exist in the brain before birth.
Keywords: calcium signaling, neural networks, neural progenitor cells, spontaneous activity, stem cells
Abstract
Coherent network activity among assemblies of interconnected cells is essential for diverse functions in the adult brain. However, cellular networks before formations of chemical synapses are poorly understood. Here, embryonic stem cell-derived neural progenitors were found to form networks exhibiting synchronous calcium ion (Ca2+) activity that stimulated cell proliferation. Immature neural cells established circuits that propagated electrical signals between neighboring cells, thereby activating voltage-gated Ca2+ channels that triggered Ca2+ oscillations. These network circuits were dependent on gap junctions, because blocking prevented electrotonic transmission both in vitro and in vivo. Inhibiting connexin 43 gap junctions abolished network activity, suppressed proliferation, and affected embryonic cortical layer formation. Cross-correlation analysis revealed highly correlated Ca2+ activities in small-world networks that followed a scale-free topology. Graph theory predicts that such network designs are effective for biological systems. Taken together, these results demonstrate that immature cells in the developing brain organize in small-world networks that critically regulate neural progenitor proliferation.
Cell-to-cell intercellular communication is essential for all multicellular organisms and permits exchange of cytosolic ions/molecules and information between cells. Such intercellular communication is frequently investigated in the adult brain, where cells form neural network circuits that constitute today’s notion of basic brain activity (1, 2). The relationship between network dynamics and topology has been studied using statistical physics and concepts from graph theory to further our general understanding of biological network systems (3, 4). Small-world and scale-free organizations are efficient and robust (5, 6) and found in diverse contexts, including the Internet, social sciences, transportation systems, and biology (7–9). In networks with small-world properties, clustering is high and path length is short, whereas scale-free networks display highly connected nodes, termed hubs. Coherent network activity has been previously reported at different stages of the developing embryonic brain (10, 11), before the formation of chemical synapses (12, 13).
Embryonic stem (ES) cells possess several unique features that make them an attractive tool for numerous purposes in regenerative medicine, including the development of novel treatments for different pathological conditions and neurodegenerative diseases such as Parkinson disease (14). ES cells are also ideal tools for studying very basic developmental processes, including neural cell proliferation and differentiation. Controlling these two processes, as well as how cells functionally integrate with each other and their environment, is critical to synchronize structure and function during nervous system development. Interestingly, stem cell-derived mature neurons can exhibit spontaneous (i.e., without externally applied stimuli) electrical and chemical activity (15, 16). This type of spontaneous activity has been shown to regulate developmental events, including axon outgrowth and path-finding, synaptic connectivity, and maturation of neuronal signaling properties (17–19). The mechanisms underlying spontaneous activity in developing neural tissue are not entirely understood.
Over the course of evolution, the calcium ion (Ca2+) has emerged as one of the most versatile intracellular second messengers, capable of controlling numerous cellular processes during development and in adulthood (20–23). Through biochemical mechanisms, the cytosolic concentration of Ca2+ can oscillate, providing a high degree of versatility to these signals (24). Interestingly, a number of studies report that spontaneous Ca2+ oscillations modulate important functions during brain development. For example, neuronal migration depends on the frequency of spontaneous Ca2+ fluctuations (25, 26), and hippocampal development depends on spontaneous nonsynaptic Ca2+ plateaus associated with recurrent burst discharges that are regulated by gap junctions (11). The channel-forming proteins in gap junctions allow direct communication between cells (27, 28) and regulate essential functions such as radial glia movement and differentiation in the ventricular zone (29), and adult neurogenesis in the dentate gyrus (30). However, the functional interplay between individual cells exhibiting spontaneous Ca2+ activity remains largely unknown.
In this study, we demonstrate that neural progenitor cells form functional networks that regulate cell proliferation, both in vitro and in vivo. Cross-correlation analysis revealed that these cell interactions are small-world networks that follow a scale-free topology. Neural progenitor cells are connected by connexin 43 (Cx43) gap junctions that facilitate correlated spontaneous Ca2+ activity. Interrupting gap junction-mediated cell-to-cell communication abolishes this Ca2+ activity and reduces the rate of proliferation.
Results
Calcium Signaling.
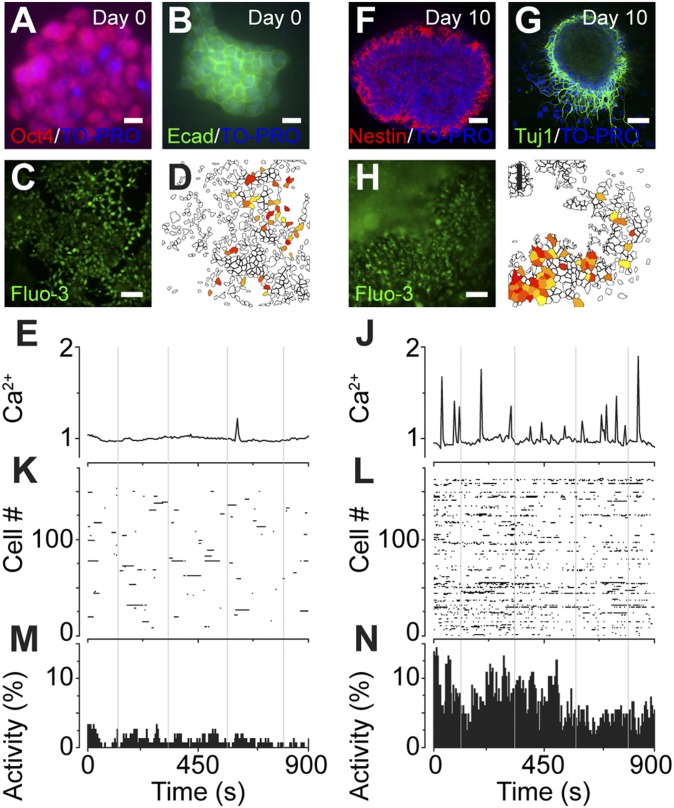
To acquire a dynamic and quantitative estimate of the activity of stem cells at different stages of neural differentiation, we conducted single-cell Ca2+ recordings in ES cell-derived neural progenitors. Before differentiation, ES cells expressed pluripotency markers Oct4 and E-cadherin (Ecad; Fig. 1 A and B). Time-lapse recordings of undifferentiated ES cells loaded with Fluo-3/AM (Fig. 1C) revealed sparse spontaneous Ca2+ signaling activity widely distributed spatially (Fig. 1 D and E; Fig. S1A). Neural differentiation of cells for 10 d induced neural progenitor marker nestin (Fig. 1F) and postmitotic marker βIII tubulin (TuJ1; Fig. 1G). Fluo-3/AM-loaded neural progenitor cells (Fig. 1H) demonstrated vivid spontaneous Ca2+ oscillations that were concentrated into small clusters of cells (Fig. 1 I and J; Fig. S1B). Raster plots of single-cell Ca2+ recordings from undifferentiated ES cells and neural progenitor cells (Fig. 1 K and L) showed a clear difference in overall Ca2+ activity between the two cell populations. Collapsing the raster plots into activity histograms further strengthened the difference between undifferentiated ES cells and neural progenitor cells (Fig. 1 M and N). The percentage of neural progenitor cells showing spontaneous Ca2+ activity from total single-cell recordings (n) and experiments (N) was 67.8 ± 5.7% (n = 1,105; N = 6). Fourier-transform spectral analysis revealed that active cells had a Ca2+ oscillatory frequency of 4.8 ± 0.8 mHz (period = 208 s, n = 1,105; N = 6). These results demonstrated that ES cells begin to exhibit spontaneous Ca2+ activity under neural differentiation conditions.
Fig. 1.
Neural progenitors derived from ES cells exhibit spontaneous Ca2+ activity. (A and B) Pluripotency markers Oct4 (A) and Ecad (B) were expressed in mouse ES cells on day 0. Nuclei were counterstained with TO-PRO-3. (Scale bars, 10 µm.) (C–E) ES cells on day 0 loaded with Fluo-3/AM (C) exhibited rare events of spontaneous Ca2+ activity. (D) Snapshot at 1 min of experiment. (Scale bar, 50 µm.) (E) Representative single cell Ca2+ trajectory. (F and G) Neural progenitor marker nestin (F) and TuJ1 (G) were expressed in mouse ES cells differentiated for 10 d. Nuclei were counterstained with TO-PRO-3. (Scale bars, 50 µm.) (H–J) Neural progenitor cells loaded with Fluo-3/AM (H) exhibited vivid clustered spontaneous Ca2+ activity. (I) Snapshot at 31 min of experiment. (Scale bar, 50 µm.) (J) Representative single cell Ca2+ trajectory. (K and L) Raster plots of ES cells (K) and neural progenitor cells (L) where each row represents a single-cell Ca2+ recording with peak activities marked with black dots. (M and N) Activity histograms of ES cells in K and neural progenitor cells in L depicting the percentage of active cells at each time point. See also Fig. S1.
Network Activity.
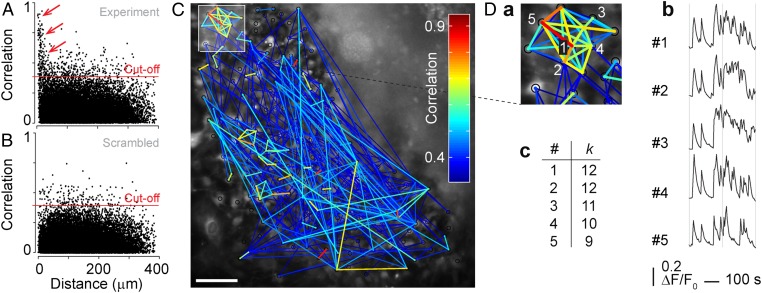
Time-lapse Ca2+ imaging indicated that neural progenitor cells in culture communicated with each other. This hypothesis was tested by cross-correlation analysis of Ca2+ recordings, in which cells act as nodes (4). The correlation coefficient and linear similarity were quantified between two single-cell Ca2+ recordings as one signal was shifted in time, using an algorithm implemented in MATLAB. Plotting correlation coefficients as a function of distance between cells (n = 1,105; N = 6) revealed a distinct group of cells (indicated by arrows) with high correlation coefficients and short intercellular distances (Fig. 2A). As a control, a scrambled set of data were generated by randomly shifting each individual cell signal in the time domain. The group of highly correlated cells was absent from the correlation plot of the scrambled data set (Fig. 2B). By calculating the mean of the 99th percentile of the scrambled data, a cutoff value of 0.39 ± 0.0012 was determined (Fig. 2B). Thus, two cells were defined as functionally connected if their correlation coefficient was greater than 0.39. A network plot of cell pairs with correlation coefficients above the cutoff showed small cluster formations of highly correlated cells (Fig. 2C). By contrast, no cell cluster formations were observed in a network plot of the scrambled data (Fig. S2A). Strongly correlated cells were closer to each other than weakly correlated cells (Fig. S2 B–F). Comparing five single-cell Ca2+ recordings within a cluster (Fig. 2 D, a) revealed distinct similarities in the shape of the Ca2+ traces (Fig. 2 D, b). Cells with a high number of connected cells k were suggested to function as hubs or trigger cells (Fig. 2 D, c).
Fig. 2.
Neural progenitor cells form functional Ca2+ signaling networks. (A and B) Correlation coefficients as a function of intercellular distance for experimental Ca2+ recordings (A) and scrambled data (B). Red lines indicate cutoff (0.39) equal to the mean value of the 99th percentile for scrambled data. Arrows indicate cells with high correlation and short intercellular distances. (C) Network plot of cross-correlation coefficients larger than cutoff (color coded as indicated) derived from Ca2+ recordings of neural progenitor cells. (Scale bar, 50 µm.) Cluster of cells (D, a), outlined in C, with strong intercellular correlation together with five single-cell Ca2+ traces (D, b). The number of connected cells is indicated with k (D, c). See also Fig. S2.
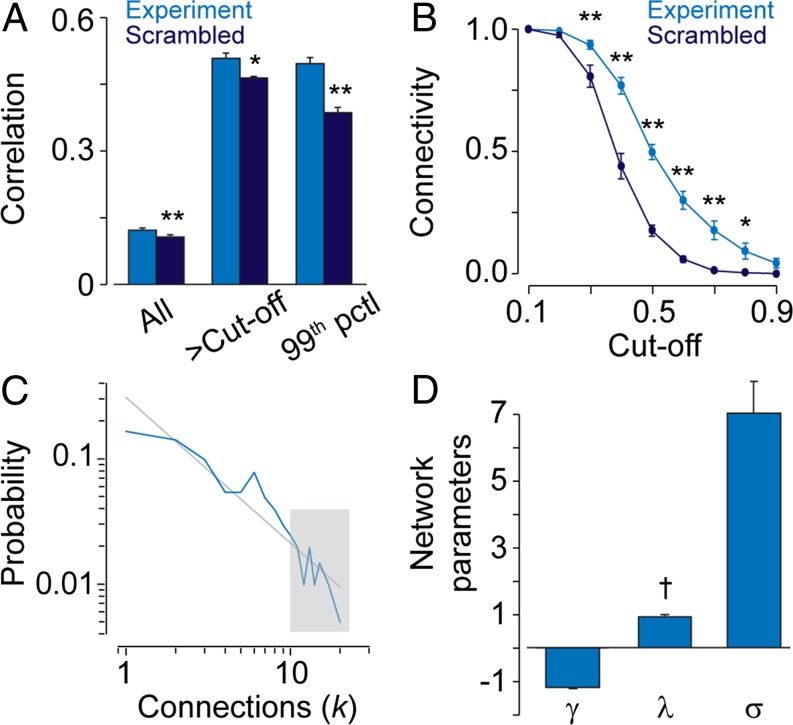
Additional analysis of the correlation coefficients of the experimental Ca2+ recordings and the scrambled data set showed significant differences when comparing all coefficients, only coefficients above the cutoff, and 99th-percentile coefficients (Fig. 3A). Plotting connectivity (defined in Materials and Methods) as a function of cutoff values revealed significantly higher connectivity in the experimental data set (Fig. 3B). The probability distribution P(k), where k is the number of connected cells (Fig. 2 D, c), for neural progenitor networks was heavy tailed and approximately linear on a log-log scale (Fig. 3C), indicating a scale-free network structure. Hubs are present in a scale-free network, in contrast to regular or random networks (6). The mean slope (γ = −1.23 ± 0.0489, N = 6), the mean shortest path length (λ = 0.974 ± 0.0559, N = 6), and the mean clustering value (σ = 7.68 ± 0.917, N = 6) together pointed toward a scale-free and small-world network (Fig. 3D; see Materials and Methods for further details). Small-world networks are suggested to pose an evolutionary advantage due to their increased robustness (6) that would enhance their impact. In conclusion, network analyses showed that neural progenitor cells were nodes in a functional network with scale-free and small-world features.
Fig. 3.
Neural progenitors organize in small-world networks that follow a scale-free topology. (A) Statistical analyses of correlation coefficients derived from experimental Ca2+ recordings and scrambled data. The entire data set (All), and filtered data sets greater than cutoff (>Cut-off) or the 99th percentile (99th pctl) are presented. (B) Connectivity as a function of cutoff values of experimental and scrambled data. (C) The probability distribution, P(k), plotted in a log-log scale with linear regression fit in gray. The gray box indicates highly connected cells (five cells shown in Fig. 2D). (D) Network parameters calculated from experimental Ca2+ recordings. Slope (γ) is a measure for scale-free networks, and path length (λ) and clustering (σ) are measures for small-world networks. Values are mean ± SEM; †P > 0.95, *P < 0.05, **P < 0.01. See also Fig. S2.
Signaling Mechanism.
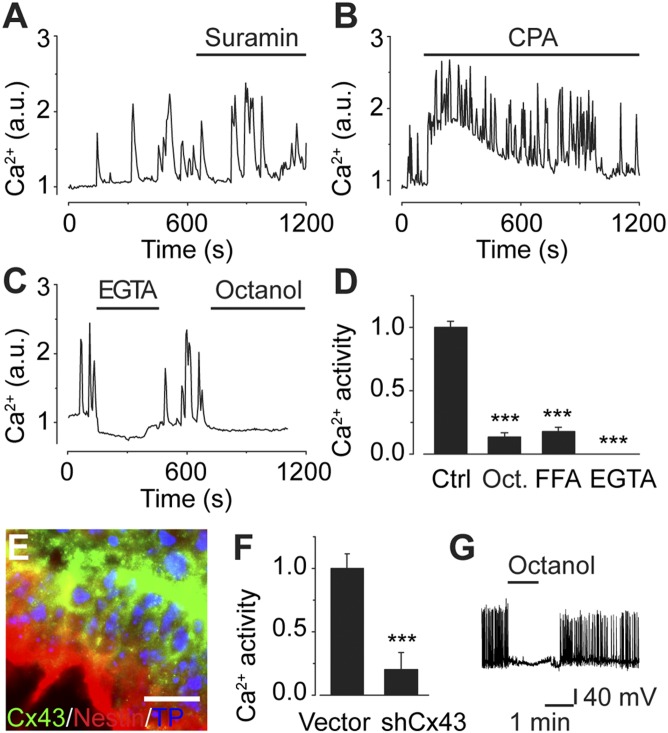
To elucidate the signaling mechanisms responsible for inducing spontaneous Ca2+ oscillations in neural progenitors, cells were challenged with different pharmacological blockers. The nonselective P2 ATP receptor antagonist suramin (100 µM) had no effect on spontaneous Ca2+ activity (Fig. 4A). Treatment with 10 U/mL hexokinase, an enzyme that consumes extracellular ATP by phosphorylation of glucose, had no effect on spontaneous Ca2+ activity (Fig. S3A), indicating that ATP release into the extracellular medium did not trigger the Ca2+ activity. Depleting internal Ca2+ stores with the endoplasmic reticulum Ca2+-ATPase blocker cyclopiazonic acid (CPA; 20 µM) produced the characteristic prolonged increase in cytosolic Ca2+ concentration, but neither CPA nor thapsigargin (1 µM; Fig. S3B) had any effect on spontaneous Ca2+ activity (Fig. 4B). Inhibiting phospholipase C with U73122 (5 μM; Fig. S3C) or voltage-gated sodium channel-induced action potentials with TTX (3 µM; Fig. S3D) also failed to stop spontaneous Ca2+ activity. Blocking neurotransmitter receptors for NMDA and AMPA with D(−)-2-amino-5-phosphonovaleric acid (D-AP5; 100 µM) or 2,3-dihydroxy-6-nitro-7-sulfamoyl-benzo[f]quinoxaline (NBQX; 50 µM), respectively, did not suppress spontaneous Ca2+ activity (Fig. S3 E and F).
Fig. 4.
Gap junctions and extracellular Ca2+ mediate spontaneous Ca2+ activity. (A and B) Neither ATP receptor inhibitor suramin (100 µM; A) nor sarco/endoplasmic reticulum Ca2+-ATPase inhibitor CPA (20 µM; B) inhibited spontaneous Ca2+ activity. (C) Eliminating extracellular Ca2+ (EGTA, 2 mM) or gap junction inhibitor octanol (1 mM) abolished Ca2+ activity. (D) Quantification of spontaneous Ca2+ activity following inhibition with octanol (Oct.), FFA (100 µM), or EGTA compared with control (Ctrl). (E) Immunocytochemistry of neural progenitor cells (day 8) stained for Cx43 and nestin. Nuclei were counterstained with TO-PRO-3 (TP). (Scale bar, 20 µm.) (F) Cx43 knock-down with shRNA (shCx43) significantly reduced the spontaneous Ca2+ activity, compared with empty vector (Vector). (G) Octanol (1 mM) blocked spontaneous electrical activity. Line under name of applied drug indicates time of drug administration. Values are mean ± SEM; ***P < 0.001. See also Fig. S3.
Cells were next subjected to a Ca2+-free extracellular medium containing EGTA (2 mM) to examine whether Ca2+ influx from the extracellular space was involved. Following this treatment, virtually all spontaneous Ca2+ activity was inhibited (Fig. 4 C and D). When Ca2+ was added back to the extracellular medium, the response was rapidly recovered (Fig. 4C). Adding Ni2+ (50 µM) to the medium also abolished the spontaneous Ca2+ activity (Fig. S3G), suggesting that extracellular Ca2+ entered the cell through plasma membrane voltage-dependent Ca2+ channels. Following EGTA treatment, the progenitor cells were exposed to the gap junction inhibitor octanol (1 mM; Fig. 4C). Gap junction inhibition with octanol or flufenamic acid (FFA; 100 µM) significantly abolished the Ca2+ activity (Fig. 4D). To verify that the effect of gap junction inhibition was not due to perturbed Ca2+ homeostasis, cells were shown to respond to ATP (10 µM; Fig. S3H) and glutamate (100 µM; Fig. S3I) following inhibition with octanol.
Cx43 is a major gap junction channel that has been implicated in neural development of the embryonic brain (29, 31–34). Expression of Cx43 in ES cell-derived neural progenitors expressing nestin was confirmed with immunocytochemistry (Fig. 4E) and real-time PCR (Fig. S3J). Cells immunostained for the neural progenitor marker doublecortin (DCX) were also positive for Cx43 (Fig. S3K). Gene silencing using shRNA reduced the level of Cx43 by 58 ± 1.6% (Fig. S3L) and significantly reduced spontaneous Ca2+ activity (20.1 ± 0.1%, n = 71; N = 9; Fig. 4F).
Taken together, these experiments demonstrated that influx of extracellular Ca2+ and Cx43 gap junction channels played an essential role for spontaneous Ca2+ activity in neural progenitor cells.
Electrical Connections.
Our data indicated that both extracellular Ca2+ and functional gap junctions were involved in the observed spontaneous Ca2+ activity. Whole-cell patch-clamp recordings were then carried out to test whether neural progenitor cells exhibited spontaneous electrical firing (membrane depolarization). Indeed, ES cell-derived neural progenitors showed spontaneous electrical activity that was inhibited by octanol (1 mM), Ni2+ (50 µM), and Cd2+ (0.1 mM; Fig. 4G; Fig. S3 M and N), respectively.
To investigate the interconnectivity between touching and nontouching progenitor cells, multielectrode patch-clamp recordings were applied (Fig. 5A). A current step injected in one cell induced depolarization in a neighboring cell, indicating electrical coupling mediated by gap junctions (Fig. 5 B, a). Blockade of gap junction by bath application of octanol (1 mM) blocked the electrotonic transmission between cells and increased the voltage response of injected cells due to the reduced membrane conductance (Fig. 5 B, b). Similar results with multipatch recordings were obtained using the gap junction inhibitors FFA (100 µM; Fig. S4A) and 18α-glycyrrhetinic acid (GA18α, 50 µM; Fig. S4B).
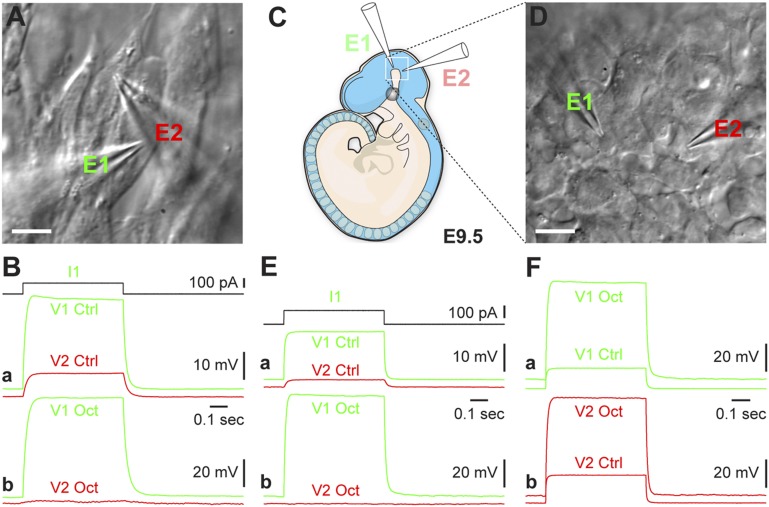
Fig. 5.
Neural progenitors establish gap junction-dependent electrical circuits in vitro and in vivo. (A) Two ES cell-derived neural progenitor cells simultaneously patched with electrode 1 (E1) and electrode 2 (E2). (Scale bar, 10 µm.) (B, a) Injecting a 100 pA-pulse (I1) into one cell (E1) revealed electrical coupling with a neighboring cell (E2). (B, b) Adding octanol (1 mM) to the medium blocked the electrical coupling between cells. Note the increased input resistance of the recorded cells, as evidenced by the larger voltage response in cell 1 (green traces: V1 control vs. V1 octanol). (C and D) In vivo patch clamp recordings of two separated neural progenitors in whole-mouse embryos on day E9.5 (C) using two electrodes E1 and E2 (D). (Scale bar, 10 µm.) (E, a) Injecting a 100 pA-pulse (I1) into one cell (E1) induced a voltage response in the neighboring cell (E2). (E, b) Bath application with octanol (1 mM) blocked transmission between cells. (F) Bath application of octanol (1 mM) increased the input resistance of recorded cells due to the blockade of gap junctions, as seen by the increased voltage response to a 100-pA current step in each of the cells. See also Fig. S4.
Cell-to-cell communication was subsequently examined in vivo in whole-mouse embryos on embryonic day 9.5 (E9.5). Embryos were loaded with Fluo-3/AM and visualized using a confocal microscope. Embryonic neural progenitors deep in the brain exhibited spontaneous Ca2+ activity (Movie S1). Analyzing networks in the in vivo experimental Ca2+ recordings together with a scrambled data set revealed functional connectivity (Fig. S2G). To test the octanol effect in E9.5 mice, we next performed multipatch recordings (Fig. 5 C and D), injecting a current step into one cell and detecting it in another cell, with or without inhibiting gap junctions. This experiment confirmed the in vitro data showing that octanol (1 mM) effectively blocked electrical coupling between neural progenitor cells (Fig. 5E). As in ES cell-derived progenitors, the input resistance of the recorded cells increased following octanol application (Fig. 5F).
These experiments not only confirmed the presence of functional gap junctions connecting neural progenitors in vitro and in vivo, but also demonstrated that octanol can efficiently block gap junctions formed between neural progenitors.
Cell Proliferation.
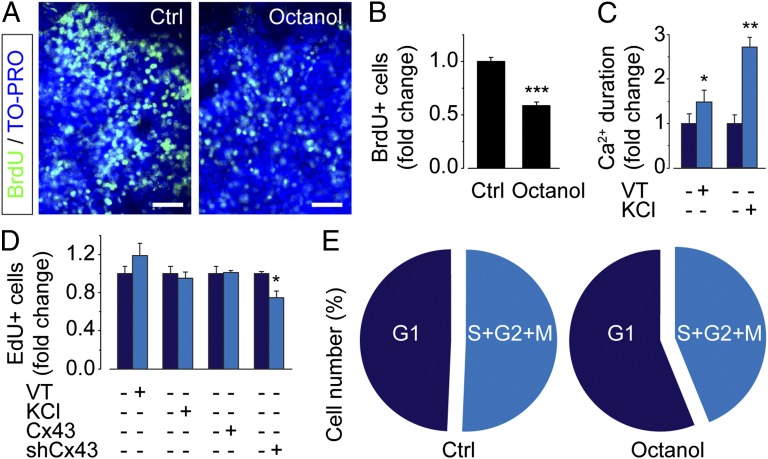
Cell proliferation and cell cycle progression in the embryonic brain are suggested to depend on cytosolic Ca2+ signaling (35). To examine the proliferative effect of spontaneous Ca2+ activity in ES cell-derived neural progenitors, cells were exposed to BrdU, incorporation of which reflects S-phase entry. Under control conditions, 34.2 ± 1.5% of total cells (n = 241,430; N = 23) were positive for BrdU (10 µM, 1 h; Fig. 6A). Significantly fewer BrdU- positive cells, 20.1 ± 1.1% (n = 111,887; N = 17), were observed after 6-h octanol treatment followed by 1-h BrdU incubation together with octanol (Fig. 6 A and B), compared with the normalized control. A significant reduction in BrdU-positive cells, 17.2 ± 2.2% (n = 63,948; N = 9), was also observed when voltage-dependent Ca2+ channels were blocked with Ni2+ (50 µM; Fig. S5A). This reduction in proliferation was not a toxic effect, because the number of undifferentiated ES cells was unaffected by octanol or Ni2+ (Fig. S5B). To test if elevated Ca2+ as such stimulated proliferation, cells were subjected to veratridine or high KCl to enhance Ca2+ signaling. Veratridine, through activation of sodium channels, and KCl depolarize the cell, causing voltage-dependent Ca2+ channels to open. Both veratridine (20 µM) and KCl (25 mM) significantly increased the duration of spontaneous Ca2+ oscillations (Fig. 6C) but failed to alter small-world network parameters as well as proliferation (Fig. 6D). Transfecting progenitors with Cx43-mCherry also failed to alter small-world networks and proliferation (Fig. 6D). Knocking down Cx43 specifically using shRNA significantly reduced the number of 5-ethynyl-2′-deoxyuridine (EdU)-positive neural progenitor cells (Fig. 6D). Applying flow cytometry measurements, we next examined the cell cycle phase of ES cell-derived neural progenitors treated with or without octanol. Cells stained with propidium iodide (PI) treated with octanol (1 mM) demonstrated more cells in G1 (56.1 ± 2.0%, N = 4) compared with control treated-cells (49.3 ± 2.1%, N = 4; Fig. 6E).
Fig. 6.
Inhibiting gap junction-dependent network circuits diminishes neural progenitor proliferation. (A) Immunocytochemical staining with BrdU and TO-PRO-3 of neural progenitor cells treated with control (Ctrl) or octanol (1 mM) for 7 h. (Scale bars, 50 µm.) (B) Quantification of BrdU-positive neural progenitors treated with octanol (1 mM) normalized to control. (C) Ca2+ duration in neural progenitors treated with veratridine (VT; 20 μM) or KCl (25 mM). (D) Quantification of EdU-positive neural progenitors treated with VT (20 μM) or KCl (25 mM) or transfected with Cx43-mCherry (Cx43) or shRNA/Cx43 (shCx43). (E) Flow cytometry measurements of cell cycle distribution using PI staining of control (Ctrl)- or octanol (1 mM)-treated neural progenitor cells. Values are mean ± SEM; *P < 0.05, **P < 0.01, ***P < 0.001. See also Fig. S5.
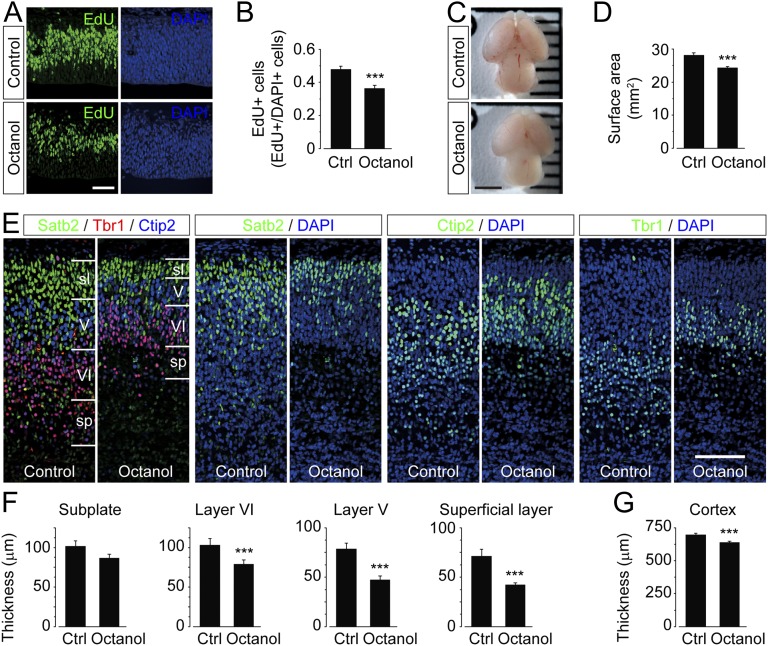
To examine how an acute pharmacological blockade of gap junctions affected proliferation and brain development in vivo, we i.p. injected octanol (0.5 mg/g body weight) into pregnant mice at E12.5. After 6 h, EdU was i.p. injected, and the pregnant mouse was killed after 1 h. Embryos were then removed, fixed, and sectioned with a cryostat. Measuring the amount of EdU-positive cells in the dorsolateral cortex in the central region of the brain hemisphere revealed a significant reduction of EdU-positive cells in octanol-treated animals (Fig. 7 A and B). Cell death was not significantly increased in octanol-injected animals at E12.5 (2.2 ± 0.5 vs. 3.7 ± 0.7 cells/field of view; Fig. S6A).
Fig. 7.
Inhibiting gap junction-dependent network circuits affects proliferation and cortical layer formation in the embryonic brain. (A) Immunohistochemistry images of the dorsolateral cortex in the central region of the brain hemisphere stained with EdU (green) following i.p. injection of control or octanol (0.5 mg/g body weight) at E12.5. Nuclei were counterstained with DAPI (blue). (Scale bar, 100 µm.) (B) Quantification of EdU-positive cells, measured as number of EdU- vs. DAPI-positive cells in sections from control (Ctrl)- or octanol-treated animals. (C and D) Images of brains (C) and calculations of brain surface area (D) from E17.5 animals treated with control or octanol (0.5 mg/g body weight) at E12.5. (Scale bar, 2 mm.) (E) Immunohistochemistry images of the dorsolateral cortex in the central region of the brain hemisphere at E17.5 following i.p. injection of control or octanol (0.5 mg/g body weight) at E12.5 stained for: Satb2, Tbr1, and Ctip2. Marked layers are superficial layer (sl), layer V (V), layer VI (VI), and subplate (sp). Nuclei were counterstained with DAPI (blue). (Scale bar, 100 µm.) (F and G) Thicknesses of layers: subplate, V, VI, and superficial layer (F) and overall cortical thickness (G) in E17.5 animals that were control- or octanol-injected at E12.5. Values are mean ± SEM; ***P < 0.001. See also Fig. S6.
The effect of reduced proliferation on the anatomy of brains subjected to octanol multiple days (E12.5–E17.5) in utero was next examined. Brains from control and octanol-injected animals (0.5 mg/g body weight, E12.5) showed a significant reduction in surface area when octanol was injected (Fig. 7 C and D). Cortical layer thickness was next examined using immunohistochemistry staining for Satb2, Tbr1, and Ctip2 to indicate cells in the superficial layers; the deep layers V and VI; and the subplate (Fig. 7E). Quantification of the thickness of the superficial layer, indicated by Satb2-positive and Ctip2-negative cells; layer V, indicated by Ctip2-positive and Tbr1-negative cells; layer VI, indicated by high-density Tbr1-positive cells; and subplate, indicated by the low density of Tbr1-positive cells, revealed greater differences for the more outermost layers (Fig. 7F). A significant decrease in the overall cortical thickness in E17.5 animals was observed when octanol was injected in utero at E12.5 (Fig. 7G; Fig. S6B). No significant cell death was also observed at E17.5 when octanol was injected at E12.5 (5.4 ± 0.8 vs. 6.6 ± 1.3 cells/field of view; Fig. S6C).
Thus, our in vitro and in vivo experiments indicate that spontaneous Ca2+ activity facilitated by gap junctions is involved in cellular processes that stimulate proliferation of neural progenitor cells. Blocking small-world networks with octanol in utero significantly reduced cortical thickness in the embryonic brain.
Discussion
Spontaneous Ca2+ activity in the developing nervous system has been reported previously (36). For example, nuclear migration in ventricular zone precursors (33) and radial glia proliferation (37) are regulated by spontaneous Ca2+ waves driven by ATP release through gap junctions/hemichannels. Also, grafted neural stem cells exhibit spontaneous Ca2+ activity that depends on gap junctions (38). Moreover, different patterns of spontaneous activity has been reported in regions of the developing visual cortex (39), and amplitude and frequency of Ca2+ oscillations were correlated positively with the rate of neuronal migration (25, 26). Our findings show that the spontaneous Ca2+ activities in each cell are not uncorrelated, independent cell signals, but rather highly ordered signaling events creating small-world networks that follow a scale-free topology. Such functional network designs are effective for biological systems (5, 6). Indeed, our data show that these networks are essential for neural progenitor proliferation.
Our in vitro and in vivo experiments demonstrate the existence of gap junction-mediated networks that regulate neural progenitor cell proliferation early in development. We hypothesize that depolarizing currents are involved in network activity, in which excess of positive charges travel to neighboring cells through gap junctions, causing depolarization and influx of Ca2+ through voltage-dependent Ca2+ channels. This hypothesis is supported by the following observations. The network activity, measured as spontaneous Ca2+ oscillations, was inhibited by gap junction blockers, blockade of Ca2+ channels in the plasma membrane with Ni2+, or removal of extracellular Ca2+. Interestingly, a few progenitors remained active after blocking gap junctions, indicating the presence of trigger or hub cells, as previously reported (17, 40). Our mathematical analyses revealed cells with node degrees (k) up to 13 (i.e., a cell functionally connected to 13 other cells), which make them plausible hub cell candidates. Functional coupling between cells was confirmed with multielectrode electrophysiology, by injecting a current in one cell and simultaneously recording the current in neighboring cells. These functional interactions were disconnected when gap junctions were inhibited. Furthermore, spontaneous activity was detected when measuring cytosolic Ca2+ concentration and plasma membrane potential. However, the frequency of the electrical signal was many times faster than the Ca2+ signal, indicating an intricate relationship between the two signals. We also noticed that the onset of spontaneous Ca2+ activity coincided with the time-point when differentiating ES cells began responding with Ca2+ increase to depolarization. Thus, these findings demonstrate concert actions of different ion transporters and signaling systems that together are responsible for building functional networks of neural progenitor cells.
Our findings show that neural progenitor cells during early embryonic development were forming small-world networks that followed a scale-free topology. Neurons in the developing mouse hippocampus (P5–P7) also connect in a scale-free network with GABAergic neurons acting as hubs, giving the total cell population a semisynchronized Ca2+ activity pattern (41). The Barabasi and Albert (8) mathematical model shows that scale-free networks emerge by adding new nodes to an existing network in a “rich get richer” fashion. Furthermore, many real-world networks have been shown to possess small-world properties—e.g., in newborn human brains (42) and cultured neurons (43). Small-world networks are characterized by a mean clustering value (Materials and Methods) larger than random networks but of the same magnitude as regular networks. The mean shortest path-length in a small-world network is similar to a random network, but smaller than a regular network. According to the Watts and Strogatz (44) model, small-world networks can be generated by rewiring a low number of links in a regular network, thereby creating shortcuts that decrease the shortest path length but preserve the high clustering value. Although the functional networks formed by neural progenitor cells presented here are rather small compared with other networks, they appear to follow both scale-free and small-world characteristics; as such, they are resistant to the death of individual nodes in preserving network activity, enable effective information flow, and provide structure for correlated/synchronized activity (4). Interestingly, we present unique data showing network structures during early embryonic development, and hypothesize that the scale-free network structure emerges by preferably adding new nodes (cell division assessed by S-phase entry) to already existing highly connected nodes (cells expressing gap junctions and possessing spontaneous Ca2+ activity). This hypothesis is consistent with our finding of decreased proliferation upon blocking gap junctions and abolishing spontaneous Ca2+ activity.
Ca2+ signaling is known to modulate several important processes during neural development. For example, specification of neuronal phenotype is regulated by spontaneous Ca2+ signaling (45). Borodinsky et al. (45) suggested that suppression of Ca2+ activity during the course of development increases excitatory neurons, whereas enhancement results in more inhibitory neurons. The onset of GABAergic phenotype and neurite outgrowth during neural stem cell differentiation is modulated by local and global spontaneous Ca2+ events (16). Spontaneous Ca2+ waves also affect the cell cycle in radial glial cells (34). These reports and others (46, 47) are in line with our observations of spontaneous Ca2+ activity in ES cell-derived neural progenitor cells and in whole animals. Our further scrutiny of these signals revealed that spontaneous Ca2+ activity is not random, but involves correlated and coherent activity. Network activity is driven by gap junctions and stimulates proliferation because pharmacological inhibition or gene silencing of Cx43 suppresses cell proliferation. Off-target effects of pharmacological inhibition of gap junctions, however, cannot be ruled out because these drugs have been reported to affect, for example, T-type Ca2+ channels (48). In vivo studies of Cx43 have been hampered by two difficulties: (i) Cx43 knockout mice display heart defects and die at birth (49), and (ii) developmental compensation among the 20 different gap junction genes (27) is hard to assess. These problems were circumvented by application of shRNA against Cx43 in vitro in ES cell-derived neural progenitor cells. The fact that overexpressing Cx43 had no detectable effect on Ca2+ activity, networking and proliferation was likely due to saturated endogenous Cx43. Interestingly, a conditional knockout of Cx43 in GFAP-expressing cells results in reduced size of the hippocampus, cortex, and cerebellum in postnatal animals, but the signaling mechanism is unknown (32). Our data showing that inhibition of network activity critically reduces progenitor proliferation in the neocortex demonstrates a plausible signaling pathway. Further strengthening this notion are the results showing no effect on networks and proliferation when the spontaneous Ca2+ activity as such was enhanced.
The data presented here support earlier studies reporting that gap junctions are more abundant in the embryonic brain (37, 50), when most proliferation takes place. We hypothesize that small-world networks built by gap junctions can form functional stem cell and progenitor niches (51). Further knowledge about how gap junctions, Ca2+ signaling, and small-world networks regulate proliferation may find applications to the fields of stem cell biology and regenerative medicine. Modulating small-world Ca2+ signaling networks may permit either expansion of progenitor pools or inhibit excessive proliferation in stem cell preparations, thus increasing throughput and control over phenotypically defined cells for applications in drug screening/development or future cell replacement therapies.
Materials and Methods
Embryonic Stem Cells.
Differentiation of mouse ES cells (R1) toward neural progenitors was carried out as described (52, 53). The Ying et al. (53) protocol was applied for Cx43 knock-down experiments, and the Barberi et al. (52) protocol was applied elsewhere.
Embryo Preparations.
Embryos from C57BL6 mice on E9.5 were loaded with Fluo-4/AM (Invitrogen) using a bulk-loading double-incubation protocol described elsewhere (54). Briefly, Fluo-4/AM was diluted in an artificial cerebrospinal fluid (ACSF) solution containing (in mM) 125 NaCl, 25 NaHCO3, 1.25 NaH2PO4, 2 CaCl2, 1 MgCl2, 20 D-glucose, and 5 KCl to final concentrations of 500 µM and 5 µM. First the embryos were loaded in the high-concentration Fluo-4/AM for 15 min at 37 °C in 5% (vol/vol) CO2 and then in the low-concentration Fluo-4/AM for 30 min. Embryos were then washed and covered with Matrigel (BD Biosciences). Ca2+ measurements of neural progenitors in whole embryos were carried out in ACSF bubbled with 5% (vol/vol) CO2 at 37 °C using a heat-controlled chamber (Warner Instruments) with a Carl Zeiss LSM510 META confocal laser scanning microscope at 0.1-Hz sampling frequency.
Pregnant C57BL6 mice at E12.5 were i.p. injected with octanol (0.5 mg/g body weight; Sigma-Aldrich) in peanut oil (Sigma-Aldrich) or peanut oil only. For proliferation analysis, mice were injected with EdU (50 μg/g mouse body weight; Invitrogen) in PBS after 6 h of treatment with octanol, then 1 h later, mice were killed. Embryos were fixed overnight in 4% (wt/vol) paraformaldehyde (PFA), dehydrated in 30% sucrose, frozen, and cryostat sectioned with 14-μm thickness. For layer formation analysis, octanol was injected at E12.5, and embryos were fixed with 4% (wt/vol) PFA at E17.5. Before sectioning, whole-brain pictures were taken using a digital camera (PowerShot G12; Canon) that was mounted on a stereo microscope (Carl Zeiss Stemi 2000-C). Brain surface area (2D) was measured using Photoshop CS5 (Adobe). Experiments were performed using at least six embryos from three litters for each condition.
All experiments were approved by Stockholm’s Ethical Committee North (ethical approval no. 256/10).
Reagents.
Reagents and concentrations, unless otherwise stated, were as follows: CPA (20 μM; Sigma-Aldrich), D-AP5 (100 µM; Sigma-Aldrich), EGTA (2 mM; Sigma-Aldrich), FFA (100 µM; Tocris), GA18α (50 µM; Sigma-Aldrich), glutamate (100 µM; Tocris), hexokinase (10 U/mL; Calbiochem), NBQX (50 µM; Tocris), 1-octanol (1 mM; Sigma-Aldrich), suramin (100 µM; Tocris), TTX (3 µM; Tocris), thapsigargin (1 mM; Sigma-Aldrich), U73122 (5 µM; Sigma-Aldrich), and veratridine (20 µM; Tocris).
Calcium Imaging.
Cells were loaded with the Ca2+-sensitive fluorescence indicator Fluo-3/AM (5 µM; Invitrogen) at 37 °C for 30 min in a Krebs-Ringer buffer containing (in mM) 119.0 NaCl, 2.5 KCl, 2.5 CaCl2, 1.3 MgCl2, 1.0 NaH2PO4, 20.0 Hepes (pH 7.4), and 11.0 dextrose. Ca2+ measurements were carried out in Krebs-Ringer buffer at 37 °C using a heat-controlled chamber (Warner Instruments) with a cooled electron-multiplying charged-coupled device Cascade II:512 camera (Photometrics) mounted on an inverted microscope (Carl Zeiss) equipped with an 25× 0.8 N.A. lens (Carl Zeiss) (55). Excitation at 495 nm was assessed with a filter wheel (Sutter Instrument) at sampling frequency 0.2–1 Hz. MetaFluor (Molecular Devices) was used to control all devices and to analyze acquired images. The overall Ca2+ activity was determined using CellProfiler and MATLAB (MathWorks Inc.). Single-cell traces were digitized by applying a threshold of 10%, assigning all time points above threshold to 1 and all other points to 0. An active cell had two Ca2+ peaks or more above the threshold. Peak duration was quantified by calculating the area under the curve for normalized time-traces divided by the total signal duration and number of peaks per second. Fourier-transform spectral analysis of Ca2+ oscillations was performed as described elsewhere (56). Drugs were bath applied.
Electrophysiology.
Whole-cell patch recordings were obtained from progenitor cells (after 7–14 d of differentiation from ES cells) and whole embryos (E9.5) at room temperature. Cells were visualized using IR differential interference contrast microscopy (Carl Zeiss). Recorded cells were selected visually, and for multielectrode patch-clamp recordings, up to four neighboring cells with lateral somatic distances less than 100 µm were simultaneously recorded. Recordings were amplified using 700B amplifiers (Molecular Devices), filtered at 2 kHz, digitized (5–20 kHz) using ITC-18 (InstruTech), and acquired using Igor Pro (WaveMetrics). Patch pipettes were pulled with a Flaming/Brown micropipette puller (P-97; Sutter Instruments) and had an initial resistance of 5–10 MΩ, containing (in mM) 110 K-gluconate, 10 KCl, 10 Hepes, 4 Mg-ATP, 0.3 GTP, and 10 phosphocreatine. Recordings were performed in current-clamp mode, with access resistance compensated throughout the experiments. Recordings were discarded when access resistance increased beyond 35 MΩ. Electrical coupling between cells was tested by injection of depolarizing and hyperpolarizing current steps into each cell while recording the responses in the neighboring cells and averaging at least 20 repetitions.
Network Analysis.
Graph theory and cross-correlation analysis were performed using the following functions implemented in MATLAB (MathWorks Inc.):
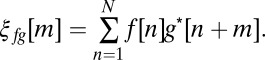 |
A correlation matrix was produced with the indices i and j running for all cells. Zero time-lag (m = 0) was used everywhere, except for calculations of full duration at half maximum for time lags. A scrambled data set (fscrambled) was created to test the correlation significance by translating signals (f) starting at random time points t of total N in a cyclic fashion, thereby conserving the activity of the data set:
A pair of cells was defined as connected if the correlation coefficient was above 0.39 (mean 99th percentile for scrambled signals). Once all links between nodes in the network had been defined, graph theory was used to calculate the network connectivity. The probability distribution, P(k), gives the probability that a selected cell has exactly k connections. P(k) was obtained by counting the number of nodes N(k) with k = 1, 2, 3… connections and divided by the total number of cells. Connectivity was defined as the number of cell pairs with a correlation coefficient larger than cutoff divided by the total number of pairs of cells.
Scale-free networks are characterized by a power law P(k) ∝ k−γ, and when plotting P(k) in a log-log scale, the data can be fitted to a linear function using the least-squares method (applying the maximum-likelihood estimation led to similar results) (57). Networks were considered to have scale-free properties if 1 < γ < 3. The small-world network properties were assessed by calculating the mean clustering value C and the mean shortest path-length L of the network using the MatlabBGL software package written for MATLAB. Next, these values were compared with the corresponding values Crand and Lrand for the randomized data, preserving the exact same level of connectivity and probability distribution. All columns were randomly translated in a Monte Carlo simulation with 1,000 iterations starting at the random row i in a cyclic fashion, as described for fscrambled. The path length (λ = L/Lrand) and clustering (σ = C/Crand) values were calculated, and networks were considered to have small-world properties if λ ∼ 1 and σ >> 1.
Immunostaining.
Immunocytochemical staining was performed according to a standard protocol using 20-min fixation in 4% (wt/vol) PFA. After blocking with 5% (vol/vol) normal goat serum, cells were incubated with primary antibodies: BrdU (1/200, rat; Abcam), Cx43 (1/100, mouse; BD Biosciences), doublecortin (1/500, goat; Santa Cruz), Ecad (1/400, goat; Santa Cruz), nestin (1/1000, rabbit; Chemicon), Oct4 (1/300, rabbit; Santa Cruz), or TuJ1 (1/200, mouse; Chemicon) for 1 h and then with Alexa fluorescent secondary antibodies (1:1,000; Molecular Probes) for 1 h, together with 0.25% Triton. Nuclei were stained with TO-PRO-3 (1:200; Molecular Probes) for 5 min. Proliferation assays were performed using BrdU (10 µM; Sigma) or EdU (10 µM; Invitrogen) 1 h before fixation. The ratio of BrdU- or EdU-positive cells and total cells (TO-PRO-3 positive) were quantified using ImageJ (National Institutes of Health, Washington, DC). Images were collected with confocal microscopes (Carl Zeiss LSM 5 Exciter or Olympus FluoView FV1000).
Immunohistochemistry was performed on brain sections washed with PBS + 0.05% Triton ×100 (PBS-Tx) and then blocked with blocking reagent (PerkinElmer) in PBS for 1 h at room temperature (RT). Sections were incubated overnight at 4 °C with primary antibodies: cleaved Caspase-3 (1/500, rabbit; Cell Signaling Technology), Ctip2 (1/200, rat; Abcam), Satb2 (1/100, mouse; Abcam), and Tbr1 (1/200, rabbit; Abcam). After washing with PBS-Tx three times, brain sections were incubated for 1 h at RT with secondary antibodies: goat anti-mouse Alexa 488, goat anti-rabbit Alexa 555, and goat anti-rat Alexa 647 (all from Invitrogen). Nuclei were stained with DAPI (5 μg/mL; Molecular Probes) for 1 h. For EdU visualization, we used Click-iT EdU Alexa Fluor 488 Imaging Kit (Invitrogen). Images were collected with a confocal microscope (Olympus FluoView FV1000).
Cell Cycle Analysis.
Differentiating neural progenitor cells were gently dissociated using TrypLE express (Invitrogen), collected, and fixed in 70% (vol/vol) ethanol. Samples were rehydrated in PBS, filtered through a 40-µm cell strainer, and stained with anti-nestin conjugated Alexa Fluor 647 antibody (1/6, mouse; Invitrogen), PI (1 mg/mL), and RNaseA (0.1 mg/mL) following the manufacturer’s protocols. Cells were analyzed with a FACSort flow cytometer (BD Biosciences). Background fluorescence was measured using unlabeled cells. Cell cycle was analyzed only in nestin-positive cells, and the percentage of G1-, S-, and G2-phase cells were calculated with FlowJo software (Tree Star Inc.).
shRNA and Plasmids.
The short hairpin of Cx43 was cloned in a pSuper plasmid, and the plasmid was transfected at day 6 of differentiation using Lipofectamine 2000 (Invitrogen) following the manufacturer’s protocol. Cells were prepared for experiments 48 h after transfection. The knock-down was validated in mouse embryonic NIH 3T3 fibroblasts. Total RNA were extracted using an RNeasy kit (Qiagen), and reverse transcription was conducted with a SuperScript II reverse transcriptase (Invitrogen) according to the manufacturer’s protocol. Real-time PCR primers were as follows: Cx43 forward TTTGACTTCAGCCTCCAAGG, reverse CCATGTCTGGGCACCTCT, TATA-box binding protein forward GGGGAGCTGTGATGTGAAGT, reverse CCAGGAAATAATTCTGGCTCA. The 2−ΔCt values were used to calculate the relative expression levels and are given as mean ± SEM.
Cx43-mCherry was obtained from the human ORFeome Library using a Gateway LR Clonase II reaction (Invitrogen) into a custom Gateway destination vector and transfected at day 5 of differentiation using Lipofectamine 2000 (Invitrogen). After 48 h, Ca2+ recordings and proliferation assays were performed as described above.
Statistical Analyses.
Data are presented as means ± SEM of a minimum of three experiments, unless indicated otherwise. Student t test and one-way analysis of variance with a Bonferroni’s post hoc test were used, and significance was accepted at †P > 0.95, *P < 0.05, **P < 0.01, or ***P < 0.001.
Supplementary Material
Acknowledgments
The authors thank Drs. Michael J. Berridge, Nicolas Fritz, Hiromi Hiyoshi, Marie Karlsson, and Nicholas C. Spitzer for thoughtful discussions and comments on the manuscript. mCherry Gateway destination vector was contributed by Dr. Arnold Hayer. This study was supported by the Swedish Research Council Grants 2005-6682, 2008-2811, 2009-3364, 2010-4392, 2011-3116, and 2011-3318 (P.U. and E.A.), Swedish Foundation for Strategic Research (P.U. and E.A.), Linnaeus Center in Developmental Biology for Regenerative Medicine (DBRM) (P.U. and E.A.), the Knut and Alice Wallenberg Foundation Grant to Center of Live Imaging of Cells at Karolinska (CLICK) Institutet (P.U.), the Royal Swedish Academy of Sciences (P.U.), European Commission Project: Dopamine Development and Parkinson's Disease Genes (E.A.), Hjärnfonden Grant FO2011-0300 (P.U.), the Åke Wiberg Foundation (P.U. and G.S.), the Magnus Bergvall Foundation (P.U. and G.S.), Jeanssons Stiftelser (P.U. and G.S.), Karolinska Institutet's Thematic Center in Stem cells and Regenerative Medicine (E.A.), the Fredrik and Ingrid Thuring Foundation (P.U.), the Stockholm Brain Institute (G.S.), Swedish Research Council Postdoctoral Fellowship 2009-546 (S.M.), the Wenner-Gren Foundation (S.K.), Japan Society for the Promotion of Science through the Strategic Young Researcher Overseas Visits Program for Accelerating Brain Circulation (S.K.), and the Swedish Society for Medical Research (S.M. and P.U.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1220179110/-/DCSupplemental.
References
- 1.McCulloch W, Pitts W. A logical calculus of the ideas immanent in nervous activity. Bull Math Biophys. 1943;5:115–133. [PubMed] [Google Scholar]
- 2.Dehaene S, Changeux JP. Experimental and theoretical approaches to conscious processing. Neuron. 2011;70(2):200–227. doi: 10.1016/j.neuron.2011.03.018. [DOI] [PubMed] [Google Scholar]
- 3.Albert R, Barabasi AL. Statistical mechanics of complex networks. Rev Mod Phys. 2002;74(1):47–97. [Google Scholar]
- 4.Feldt S, Bonifazi P, Cossart R. Dissecting functional connectivity of neuronal microcircuits: Experimental and theoretical insights. Trends Neurosci. 2011;34(5):225–236. doi: 10.1016/j.tins.2011.02.007. [DOI] [PubMed] [Google Scholar]
- 5.Barkai N, Leibler S. Robustness in simple biochemical networks. Nature. 1997;387(6636):913–917. doi: 10.1038/43199. [DOI] [PubMed] [Google Scholar]
- 6.Barabási AL, Oltvai ZN. Network biology: Understanding the cell’s functional organization. Nat Rev Genet. 2004;5(2):101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 7.Amaral LA, Scala A, Barthelemy M, Stanley HE. Classes of small-world networks. Proc Natl Acad Sci USA. 2000;97(21):11149–11152. doi: 10.1073/pnas.200327197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barabasi AL, Albert R. Emergence of scaling in random networks. Science. 1999;286(5439):509–512. doi: 10.1126/science.286.5439.509. [DOI] [PubMed] [Google Scholar]
- 9.Guimerà R, Mossa S, Turtschi A, Amaral LA. The worldwide air transportation network: Anomalous centrality, community structure, and cities’ global roles. Proc Natl Acad Sci USA. 2005;102(22):7794–7799. doi: 10.1073/pnas.0407994102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Owens DF, Kriegstein AR. Patterns of intracellular calcium fluctuation in precursor cells of the neocortical ventricular zone. J Neurosci. 1998;18(14):5374–5388. doi: 10.1523/JNEUROSCI.18-14-05374.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crépel V, et al. A parturition-associated nonsynaptic coherent activity pattern in the developing hippocampus. Neuron. 2007;54(1):105–120. doi: 10.1016/j.neuron.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 12.Peinado A, Yuste R, Katz LC. Extensive dye coupling between rat neocortical neurons during the period of circuit formation. Neuron. 1993;10(1):103–114. doi: 10.1016/0896-6273(93)90246-n. [DOI] [PubMed] [Google Scholar]
- 13.Williams RW, Herrup K. The control of neuron number. Annu Rev Neurosci. 1988;11:423–453. doi: 10.1146/annurev.ne.11.030188.002231. [DOI] [PubMed] [Google Scholar]
- 14.Arenas E. Towards stem cell replacement therapies for Parkinson’s disease. Biochem Biophys Res Commun. 2010;396(1):152–156. doi: 10.1016/j.bbrc.2010.04.037. [DOI] [PubMed] [Google Scholar]
- 15.Ban J, et al. Embryonic stem cell-derived neurons form functional networks in vitro. Stem Cells. 2007;25(3):738–749. doi: 10.1634/stemcells.2006-0246. [DOI] [PubMed] [Google Scholar]
- 16.Ciccolini F, et al. Local and global spontaneous calcium events regulate neurite outgrowth and onset of GABAergic phenotype during neural precursor differentiation. J Neurosci. 2003;23(1):103–111. doi: 10.1523/JNEUROSCI.23-01-00103.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blankenship AG, Feller MB. Mechanisms underlying spontaneous patterned activity in developing neural circuits. Nat Rev Neurosci. 2010;11(1):18–29. doi: 10.1038/nrn2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spitzer NC. Electrical activity in early neuronal development. Nature. 2006;444(7120):707–712. doi: 10.1038/nature05300. [DOI] [PubMed] [Google Scholar]
- 19.Wong RO, Chernjavsky A, Smith SJ, Shatz CJ. Early functional neural networks in the developing retina. Nature. 1995;374(6524):716–718. doi: 10.1038/374716a0. [DOI] [PubMed] [Google Scholar]
- 20.Berridge MJ, Bootman MD, Roderick HL. Calcium signalling: Dynamics, homeostasis and remodelling. Nat Rev Mol Cell Biol. 2003;4(7):517–529. doi: 10.1038/nrm1155. [DOI] [PubMed] [Google Scholar]
- 21.Webb SE, Miller AL. Ca2+ signaling and early embryonic patterning during the blastula and gastrula periods of zebrafish and Xenopus development. Biochim Biophys Acta. 2006;1763(11):1192–1208. doi: 10.1016/j.bbamcr.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 22.Komuro H, Rakic P. Modulation of neuronal migration by NMDA receptors. Science. 1993;260(5104):95–97. doi: 10.1126/science.8096653. [DOI] [PubMed] [Google Scholar]
- 23.Desfrere L, et al. Na,K-ATPase signal transduction triggers CREB activation and dendritic growth. Proc Natl Acad Sci USA. 2009;106(7):2212–2217. doi: 10.1073/pnas.0809253106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uhlén P, Fritz N. Biochemistry of calcium oscillations. Biochem Biophys Res Commun. 2010;396(1):28–32. doi: 10.1016/j.bbrc.2010.02.117. [DOI] [PubMed] [Google Scholar]
- 25.Komuro H, Rakic P. Intracellular Ca2+ fluctuations modulate the rate of neuronal migration. Neuron. 1996;17(2):275–285. doi: 10.1016/s0896-6273(00)80159-2. [DOI] [PubMed] [Google Scholar]
- 26.Yacubova E, Komuro H. Stage-specific control of neuronal migration by somatostatin. Nature. 2002;415(6867):77–81. doi: 10.1038/415077a. [DOI] [PubMed] [Google Scholar]
- 27.Söhl G, Maxeiner S, Willecke K. Expression and functions of neuronal gap junctions. Nat Rev Neurosci. 2005;6(3):191–200. doi: 10.1038/nrn1627. [DOI] [PubMed] [Google Scholar]
- 28.Yeager M, Harris AL. Gap junction channel structure in the early 21st century: facts and fantasies. Curr Opin Cell Biol. 2007;19(5):521–528. doi: 10.1016/j.ceb.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Elias LA, Kriegstein AR. Gap junctions: Multifaceted regulators of embryonic cortical development. Trends Neurosci. 2008;31(5):243–250. doi: 10.1016/j.tins.2008.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kunze A, et al. Connexin expression by radial glia-like cells is required for neurogenesis in the adult dentate gyrus. Proc Natl Acad Sci USA. 2009;106(27):11336–11341. doi: 10.1073/pnas.0813160106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bruzzone R, Dermietzel R. Structure and function of gap junctions in the developing brain. Cell Tissue Res. 2006;326(2):239–248. doi: 10.1007/s00441-006-0287-0. [DOI] [PubMed] [Google Scholar]
- 32.Wiencken-Barger AE, Djukic B, Casper KB, McCarthy KD. A role for Connexin43 during neurodevelopment. Glia. 2007;55(7):675–686. doi: 10.1002/glia.20484. [DOI] [PubMed] [Google Scholar]
- 33.Liu X, Hashimoto-Torii K, Torii M, Ding C, Rakic P. Gap junctions/hemichannels modulate interkinetic nuclear migration in the forebrain precursors. J Neurosci. 2010;30(12):4197–4209. doi: 10.1523/JNEUROSCI.4187-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu X, Sun L, Torii M, Rakic P. Connexin 43 controls the multipolar phase of neuronal migration to the cerebral cortex. Proc Natl Acad Sci USA. 2012;109(21):8280–8285. doi: 10.1073/pnas.1205880109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weissman TA, Riquelme PA, Ivic L, Flint AC, Kriegstein AR. Calcium waves propagate through radial glial cells and modulate proliferation in the developing neocortex. Neuron. 2004;43(5):647–661. doi: 10.1016/j.neuron.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 36.Gu X, Olson EC, Spitzer NC. Spontaneous neuronal calcium spikes and waves during early differentiation. J Neurosci. 1994;14(11 Pt 1):6325–6335. doi: 10.1523/JNEUROSCI.14-11-06325.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Montoro RJ, Yuste R. Gap junctions in developing neocortex: A review. Brain Res Brain Res Rev. 2004;47(1-3):216–226. doi: 10.1016/j.brainresrev.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 38.Jäderstad J, et al. Communication via gap junctions underlies early functional and beneficial interactions between grafted neural stem cells and the host. Proc Natl Acad Sci USA. 2010;107(11):5184–5189. doi: 10.1073/pnas.0915134107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Siegel F, Heimel JA, Peters J, Lohmann C. Peripheral and central inputs shape network dynamics in the developing visual cortex in vivo. Curr Biol. 2012;22(3):253–258. doi: 10.1016/j.cub.2011.12.026. [DOI] [PubMed] [Google Scholar]
- 40.Yuste R, Nelson DA, Rubin WW, Katz LC. Neuronal domains in developing neocortex: Mechanisms of coactivation. Neuron. 1995;14(1):7–17. doi: 10.1016/0896-6273(95)90236-8. [DOI] [PubMed] [Google Scholar]
- 41.Bonifazi P, et al. GABAergic hub neurons orchestrate synchrony in developing hippocampal networks. Science. 2009;326(5958):1419–1424. doi: 10.1126/science.1175509. [DOI] [PubMed] [Google Scholar]
- 42.Fransson P, Aden U, Blennow M, Lagercrantz H. The functional architecture of the infant brain as revealed by resting-state fMRI. Cereb Cortex. 2011;21(1):145–154. doi: 10.1093/cercor/bhq071. [DOI] [PubMed] [Google Scholar]
- 43.Downes JH, et al. Emergence of a small-world functional network in cultured neurons. PLOS Comput Biol. 2012;8(5):e1002522. doi: 10.1371/journal.pcbi.1002522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Watts DJ, Strogatz SH. Collective dynamics of ‘small-world’ networks. Nature. 1998;393(6684):440–442. doi: 10.1038/30918. [DOI] [PubMed] [Google Scholar]
- 45.Borodinsky LN, et al. Activity-dependent homeostatic specification of transmitter expression in embryonic neurons. Nature. 2004;429(6991):523–530. doi: 10.1038/nature02518. [DOI] [PubMed] [Google Scholar]
- 46.Garaschuk O, Linn J, Eilers J, Konnerth A. Large-scale oscillatory calcium waves in the immature cortex. Nat Neurosci. 2000;3(5):452–459. doi: 10.1038/74823. [DOI] [PubMed] [Google Scholar]
- 47.Ben-Ari Y. Developing networks play a similar melody. Trends Neurosci. 2001;24(6):353–360. doi: 10.1016/s0166-2236(00)01813-0. [DOI] [PubMed] [Google Scholar]
- 48.Juszczak GR, Swiergiel AH. Properties of gap junction blockers and their behavioural, cognitive and electrophysiological effects: Animal and human studies. Prog Neuropsychopharmacol Biol Psychiatry. 2009;33(2):181–198. doi: 10.1016/j.pnpbp.2008.12.014. [DOI] [PubMed] [Google Scholar]
- 49.Reaume AG, et al. Cardiac malformation in neonatal mice lacking connexin43. Science. 1995;267(5205):1831–1834. doi: 10.1126/science.7892609. [DOI] [PubMed] [Google Scholar]
- 50.Arumugam H, Liu X, Colombo PJ, Corriveau RA, Belousov AB. NMDA receptors regulate developmental gap junction uncoupling via CREB signaling. Nat Neurosci. 2005;8(12):1720–1726. doi: 10.1038/nn1588. [DOI] [PubMed] [Google Scholar]
- 51.Moore KA, Lemischka IR. Stem cells and their niches. Science. 2006;311(5769):1880–1885. doi: 10.1126/science.1110542. [DOI] [PubMed] [Google Scholar]
- 52.Barberi T, et al. Neural subtype specification of fertilization and nuclear transfer embryonic stem cells and application in parkinsonian mice. Nat Biotechnol. 2003;21(10):1200–1207. doi: 10.1038/nbt870. [DOI] [PubMed] [Google Scholar]
- 53.Ying QL, Stavridis M, Griffiths D, Li M, Smith A. Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat Biotechnol. 2003;21(2):183–186. doi: 10.1038/nbt780. [DOI] [PubMed] [Google Scholar]
- 54.Peterlin ZA, Kozloski J, Mao BQ, Tsiola A, Yuste R. Optical probing of neuronal circuits with calcium indicators. Proc Natl Acad Sci USA. 2000;97(7):3619–3624. doi: 10.1073/pnas.97.7.3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Malmersjö S, et al. Ca2+ and cAMP signaling in human embryonic stem cell-derived dopamine neurons. Stem Cells Dev. 2010;19(9):1355–1364. doi: 10.1089/scd.2009.0436. [DOI] [PubMed] [Google Scholar]
- 56.Uhlén P. Spectral analysis of calcium oscillations. Sci STKE. 2004;2004(258):pl15. doi: 10.1126/stke.2582004pl15. [DOI] [PubMed] [Google Scholar]
- 57.Clauset A, Shalizi CR, Newman MEJ. Power-law distributions in empirical data. SIAM Rev. 2009;51(4):661–703. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.