Abstract
Dengue virus infection has become a global threat affecting around 100 countries in the world. Currently, there is no licensed antiviral agent available against dengue. Thus, there is a strong need to develop therapeutic strategies that can tackle this life threatening disease. RNA interference is an important and effective gene silencing process which degrades targeted RNA by a sequence specific process. Several studies have been conducted during the last decade to evaluate the efficiency of siRNA in inhibiting dengue virus replication. This review summarizes siRNAs as a therapeutic approach against dengue virus serotypes and concludes that siRNAs against virus and host genes can be next generation treatment of dengue virus infection.
Keywords: Dengue virus, Dengue haemorrhagic fever, RNA interference, Small interference RNA
1. Introduction
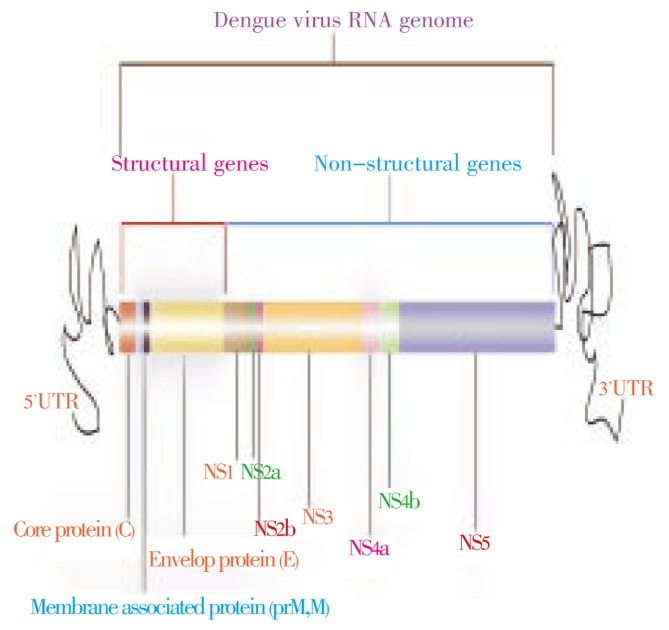
Dengue virus (DENV) infection is the wildest mosquito-borne infection affecting 2.5 billion people in tropical and sub-tropical regions of the world[1],[2]. DENV is a member of Flaviviridae family and is transmitted to humans by infected female Aedes genus, especially Aedes aegypti or Aedes albopictus[3],[4]. There are four serotypes of dengue virus (DENV 1-4) that manifest with similar symptoms[5],[6]. Two types of infections are caused by DENV, namely, primary infection and secondary infection. Primary infection causes acute febrile illness known as dengue fever. Secondary infection is more severe and results in hemorrhagic fever (DHF) or dengue shock syndrome (DSS)[7]. Both DHF and DSS can be fatal and can lead to death of the patients[8]. Thus, dengue is regarded as a life threatening fever. And there is a need to develop an efficient, low cost and safe vaccine that can target all the four serotypes of DENV. Life cycle of DENV begins with the attachment of DENV with specific receptors, this process is mediated by dengue envelop protein. After attachment, the viral particle is fused into acidic lysosomes through receptor mediated endocytosis. After that, the viral particle becomes uncoated and the RNA is released in host cell where it directs the synthesis of viral proteins. In only few hours after infection, tens of thousands of copies of viral molecules are produced from a single viral molecule, leading to cell damage and in severe cases to death. DENV is a positive stranded RNA virus having genome of 11 kb. It encodes a poly protein precursor that is cleaved to generate at least ten proteins[9]. These proteins include three structural proteins, namely core protein, a membrane associated protein, an envelope protein and seven nonstructural proteins. The genes are in order: 5′-CprM (M)-E-NS1-NS2A-NS2B-NS3-NS4A-NS4B-NS5-3′ (Figure 1)[10]. The increased expression of both extracellular and cytoplasmic pattern recognition receptors; retinoic acid inducible gene-I, melanoma differentiation associated gene-5 and Toll-like receptor-3 are collectively involved in initiating an effective IFN production against DENV[11]. Currently, there is no vaccination available against dengue infection. Due to four serotypes of DENV, developing vaccine against dengue has become quite challenging. There is a strong need to develop cost effective and less toxic anti-dengue therapies that can target all serotypes of dengue. Heterochromatin was discovered 75 years ago. Soon after, it was found that it had silence genes. This heterochromatin gives rise to small RNAs which directs the modification of proteins and DNA in heterochromatic repeats and transposable elements through RNA interference[12]. Double-stranded RNA-mediated interference (RNAi) is a simple and rapid method of silencing gene expression in a range of organisms[13]. RNAi and other silencing mechanisms act as a defense against incoming viruses and also play an important role in regulating cellular genes expression. Investigators proved RNAi as a promising approach to treat flavivirus infections in the host and to control flavivirus transmission by vector[14],[15].
Figure 1. Dengue virus genome.
DENV genome encodes 10 viral proteins in the order of 5′-CprM (M)-E-NS1-NS2A-NS2B-NS3-NS4A-NS4B-NS5-3′.
2. RNAi mechanism
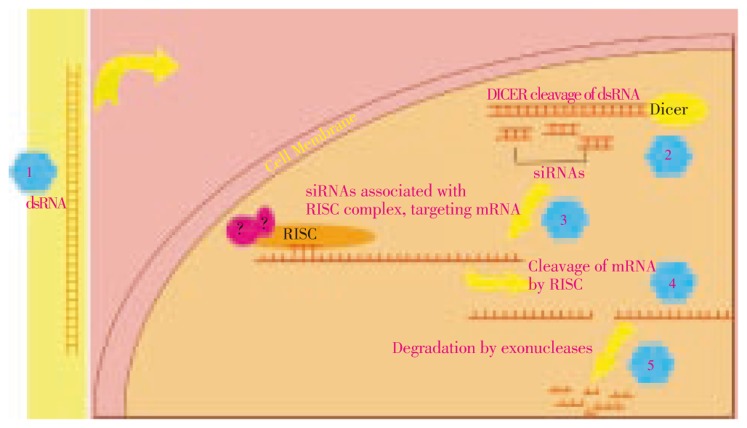
To date, several studies have undergone to understand the mechanisms of RNAi. This ancient antiviral mechanism has been discovered in Drosophila melanogaster, Caenorhabditis elegans, plants and human. Once a virus invades, dsRNA acts as a warning signal and starts the mechanism of RNAi which leads to the catalytic degradation of targeted gene's mRNA, thus silencing its expression. Majority of the RNA viruses generates dsRNA as a replication byproduct. Two mosquito species, Aedes aegypti and Anopheles gambiae can silence endogenous gene expression by dsRNA and thus have RNAi reponse similar to Drosophila melanogaster[16]. The first step of RNAi mechanism is the incorporation of dsRNA into the cytoplasm of cell. In cytoplasm it acts as a trigger to start the cascade of RNAi. Dicer enzyme (DICER), dsRNA specific endonuclease, cuts the dsRNA and generates the small interfering RNAs (siRNAs∼21-23 base pairs in length) pool. These siRNAs are double-stranded and include two nucleotides overhanging at both 3′ ends. These siRNAs are incorporated in Argonaut-containing RNA-Induced Silencing Complex (RISC). Now RISC is activated. It unwinds the two siRNA strands and keeps one strand acting as a RISC-targeting co-factor. After that, the RISC containing siRNA binds with target complementary mRNA. At the end, the RISC having endonucease activity causes a single-site cleavage of the target mRNA approximately in the middle of the siRNA binding region. Thus, the mRNA is destabilized and is degraded through natural endogenous mechanisms (Figure 2).
Figure 2. RNAi mechanism: dsRNA is degraded by DICER resulting in siRNAs. These siRNAs associates with RISC complex, targets and cleaves complementary mRNA.
3. RNAi: next generation antiviral therapy?
RNAi, discovered in 1998, has provided new horizons for drug discovery and biological research. Since then, it has been emerging as a powerful tool for combating the most challenging diseases as cancer, genetic disorders and viral clinical targets[17]. However, this novel technology has several critical issues to progress towards human clinical trials. Until recent years, researches are being conducted to solve these issues[18]. RNAi involves three types of small non-coding RNAs: microRNAs, siRNAs and Piwi-interacting RNAs. Mammals have several thousand different Piwi-interacting RNAs produced from gene clusters of repetitive elements and more than 1000 different microRNAs regulated cellular gene expression programs to control diverse steps in cell development and physiology[19],[20]. RNAi plays an important role as a defense against viruses of invertebrates, but its contribution towards mammalian antiviral defense has been a matter of dispute. RNAi mechanism is conserved in mammals and siRNA introduction efficiently silences replication of viruses[19]. RNAi has become a widely used technique for genetic knock-outs and for studying gene function by reverse genetics[21]. Several experiments have used RNAi to assess the function of particular proteins and to discover enzymes/proteins involved in metabolic pathways[22]. After the recently described RNA-based strategies for gene inhibition in mammalian cells, RNAi has become a promising antiviral therapy[23]. Thus, RNAi not only can serve as an extremely powerful instrument for functional genomic analyses but also can be an important tool to develop highly specific dsRNA based gene-silencing therapeutics[24].
4. RNAi against DENV
RNAi is an exciting field of functional genomics that can silence viral genes[25]. This antiviral mechanism discovered in many eukaryotes and is a promising treatment for flaviviruses infections in hosts. However, RNAi against flaviviruses has received only a little study[26],[27]. To date, RNAi has been used against several human pathogens including human immunodeficiency virus type 1, hepatitis C virus, hepatitis B virus, poliovirus, influenza virus A and DENV[21]. These viruses possess ssRNA genomes and these genomes are exposed within cytoplasm and become potential targets for RNAi. This happens between viral RNA uncoating and viral replication[28]. Any variation in RNAi pathway can explain why some mosquitoes are competent vectors of arthropod borne viruses (arboviruses), but others are not. Existence of RNAi pathway in Aedes species has been evidenced. The first evidence is the interference of recombinant Sindbis viruses expressing a RNA fragment from a genetically unrelated dengue-2 virus (DENV-2) with DENV-2 replication in Aedes aegypti mosquitoes by a mechanism that is similar to silencing mechanism in plants. The second evidence is the interference of C6/36 (Aedes albopictus) cells transfected with dsRNA or siRNA derived from arbovirus genome with replication of homologous virus. The third evidence is the generation of virus resistance C6/36 cells from a hairpin DENV-2 specific RNA transcribed from a plasmid. All these evidence points towards the fact that Aedes species has RNAi similar to plants and other organisms[16]. Both innate and adaptive immune responses greatly influence the DENV infection, but only a little is known about the innate immune response of the mosquito vector Aedes aegypti to arbovirus infection. RNAi is not completely evaded by DENV-2 as silencing expression of dcr2, r2d2, or ago2 genes increase virus replication in vector and decrease the extrinsic incubation period of viral transmission. Sánchez-Vargas and collegues provided evidence that RNAi is an important modulator of mosquito infections[29]. AAV (adeno-associated virus)-siRNA infected DCs (dendritic cells) showed a dose dependent reduction in dengue infection. AAV-siRNA treated DCs also decreased the dengue induced apoptosis of DCs. Thus, it can be said that AAV-mediated siRNA delivery can reduce dengue infection and replication in humans[30]. More than 100 proteins of host factors involved in DENV replication have been screened through large-scale siRNA. These host factors serves as drug targets in drug designing. Only few host factors (proteases, glucosidases etc.) have been found through siRNA screens. Moreover, these screens could not identify innate immunity genes that act as a defense against DENV infection. Delivery of siRNA in patients is the biggest issue, and a proper delivery system is yet to be established[31]. It was proved by the presence of DENV-2 derived siRNA in RNA extracts from midguts of Carb77 and by RNAi pathway interruption that lead to the loss of resistance phenotype that DENV-2 resistance was caused by a RNAi response[32]. siRNA against dengue PreM gene was transfected in C6/36 cells, which then was attacked by DENV1 virus. After 7 d, the cell survival rate of transfected cells increased by 2.26 fold and virus RNA reduced by 97.54%. This finding provides evidence that siRNA can effectively inhibit dengue replication[33]. Mukherjee et al. indicated that DENV can replicate in Drosophila S2 cells and that the RNAi pathway plays a role in modulating DENV replication in these cells[26]. Down regulation of HSP60 in infected cells resulted in decreased viral load, RNA copy number and increased IFN-α level. Thus, elevated levels of HSP60 in infected cells may help in viral multiplication and can be a possible target to manage dengue infection[34]. Effects of genes can be temporarily silenced by using RNAi along with plasmid transfection technology with inducible vectors. siRNA is highly specific against target RNA. Therefore, siRNA is an important tool in understanding and discovering the function of gene[35]. siRNA mediated silencing of attachment receptor and clathrin-mediated endocytosis can inhibit DENV entry and multiplication in HepG2 cells, ultimately reducing the viral load. Thus, preventing dengue fever to develop into more severe forms (DHF/DSS)[36]. Ang et al. identified key cellular genes involved in processes of endocytosis and cytoskeletal dynamics, important for DENV infection. siRNA targeting genes associated with clathrin-mediated endocytosis inhibited DENV entry into Huh7 cells[37]. Villegas-Rosales et al. recently predicted three siRNAs that are able to silence four DENV genome serotypes by targeting NS4B and NS5 sequences[38]. siRNA in combination with endogenous RNAi processing machinery can help in prevention of severe dengue infection. DC-3 siRNA is a novel approach against different serotypes of dengue and thus can be helpful in developing new therapeutic regimens[39]. Korrapati et al. used replication defective human adenovirus type 5 vector for short-hairpin RNA delivery in cells to target conserved sites in viral genome. This short-hairpin RNA matures to cognate siRNA and can affectively inhibit antigen secretion, viral replication of all four dengue serotypes. All these researches and their significant results declare RNAi as a possible therapeutic strategy against DENV replication in vitro and in animal models[40],[41].
5. Conclusion
Arhtropod-borne viral diseases have become serious public health issue and novel methods are required to control pathogen transmission. DENV, because of its four serotypes, has become a challenging target to design effective drugs. Currently, there is no vaccination or antiviral drugs available against dengue. The only treatment against dengue is the preventive and supportive care. So there is a strong need to develop therapeutic approaches that can halt DENV replication and decrease viral load. RNAi is a sequence specific RNA degradation process that initiates with dsRNA. This pathway has lead to the use of siRNAs against several viral diseases, especially ssRNA genome containing viruses. Several researches have been conducted to use siRNA against DENV and many of them have provided results of effectively using siRNA against DENV replication. Hopefully, in future siRNA approach may provide a better and effective treatment that will lead to eradication of dengue infection.
Comments
Background
DENV is an arthropod-borne flaviviruses that causes dengue fever with significant morbidity and mortality in tropical and subtropical regions of the world. There are four serotypes of DENV (DENV types 1–4). Humans are main amplifying host of the virus and they circulate virus in their blood at approximately same time as they have fever, and other Aedes spp. mosquitoes may acquire the virus if they feed on an individual at this time. DENV being an RNA virus has great potential to be targeted through nucleotide based therapeutic options like RNAi. This antiviral mechanism is a promising treatment for flaviviruses including HCV and DENV.
Research frontiers
Several studies has been conducted worldwide describing the efficiency of RNAi silencing of DENV, in the current review, authors summarize the updated knowledge about the research on the use of RNAi against DENV and human genes.
Related reports
DENV is one of the first animal viruses that could be efficiently inhibited by RNAi. Like other flaviviruses, DENV generates intracellular dsRNA as an intermediate of their replication, which may induce RNAi in the host cells. Wu et al. (2010) reported synthetic siRNA against the DENV-1 membrane glycoprotein precursor gene effectively inhibited DENV-1 viral RNA replication and increased C6/36 cell survival rate. Recently, Alhoot et al. (2012) reported silencing the attachment receptor and clathrin-mediated endocytosis using siRNA could inhibit DENV entry and multiplication into HepG2 cells.
Innovations and breakthroughs
The article reviewed all the available literature on the use of RNAi as an exciting field of treatment against DENV infection.
Applications
The only available treatment against dengue infection is preventive and supportive medicine. Keeping this in mind, it is a significant piece of information regarding the RNAi, DENV infection and its possible treatment targeting both DENV genes and human genes.
Peer review
This is a good study in which the authors summarize all the available information and suggest that RNAi could be used as therapy in future against DENV infection.
Footnotes
Foundation Project: Supported by Higher Education Commission (HEC) with Grant # PM-IPFP/HRD/HEC/2012/2770.
Conflict of interest statement: We declare that we have no conflict of interest.
References
- 1.Guzman MG, Halstead SB, Artsob H, Buchy P, Farrar J, Gubler DJ, et al. et al. Dengue: a continuing global threat. Nat Rev Microbiol. 2010;8(Suppl 12):S7–S16. doi: 10.1038/nrmicro2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Halstead SB. Dengue. Lancet. 2007;370:1644–1655. doi: 10.1016/S0140-6736(07)61687-0. [DOI] [PubMed] [Google Scholar]
- 3.Beatty ME, Stone A, Fitzsimons DW, Hanna JN, Lam SK, Vong S, et al. et al. Best practices in dengue surveillance: a report from the Asia-Pacific and Americas Dengue Prevention Boards. PLoS Negl Trop Dis. 2010;4(11):e890. doi: 10.1371/journal.pntd.0000890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fatima Z, Idrees M, Bajwa MA, Tahir Z, Ullah O, Zia MQ, et al. et al. Serotype and genotype analysis of dengue virus by sequencing followed by phylogenetic analysis using samples from three mini outbreaks-2007-2009 in Pakistan. BMC Microbiol. 2011;11:200. doi: 10.1186/1471-2180-11-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ross TM. Dengue virus. Clin Lab Med. 2010;30:149–160. doi: 10.1016/j.cll.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang E, Ni H, Xu R, Barrett AD, Watowich SJ, Gubler DJ, et al. et al. Evolutionary relationships of endemic/epidemic and sylvatic dengue viruses. J Virol. 2000;74:3227–3234. doi: 10.1128/jvi.74.7.3227-3234.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guzman MG, Kouri G. Dengue: an update. Lancet Infect Dis. 2002;2:33–42. doi: 10.1016/s1473-3099(01)00171-2. [DOI] [PubMed] [Google Scholar]
- 8.WHO . 2nd ed. Geneva: WHO; 1997. Dengue haemorrhagic fever: diagnosis, treatment, prevention and control. [Google Scholar]
- 9.Munoz-Jordan JL, Sanchez-Burgos GG, Laurent-Rolle M, Garcia-Sastre A. Inhibition of interferon signaling by dengue virus. Proc Natl Acad Sci USA. 2003;100:14333–14338. doi: 10.1073/pnas.2335168100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Umareddy I, Pluquet O, Wang QY, Vasudevan SG, Chevet E, Gu F. Dengue virus serotype infection specifies the activation of the unfolded protein response. Virol J. 2007;4:91. doi: 10.1186/1743-422X-4-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nasirudeen AM, Wong HH, Thien P, Xu S, Lam KP, Liu DX. RIG-I, MDA5 and TLR3 synergistically play an important role in restriction of dengue virus infection. PLoS Negl Trop Dis. 2011;5:e926. doi: 10.1371/journal.pntd.0000926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lippman Z, Martienssen R. The role of RNA interference in heterochromatic silencing. Nature. 2004;431:364–370. doi: 10.1038/nature02875. [DOI] [PubMed] [Google Scholar]
- 13.Agrawal N, Dasaradhi PV, Mohmmed A, Malhotra P, Bhatnagar RK, Mukherjee SK. RNA interference: biology, mechanism, and applications. Microbiol Mol Biol Rev. 2003;67:657–685. doi: 10.1128/MMBR.67.4.657-685.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fire A. RNA-triggered gene silencing. Trends Genet. 1999;15:358–363. doi: 10.1016/s0168-9525(99)01818-1. [DOI] [PubMed] [Google Scholar]
- 15.Szweykowska-Kulinska Z, Jarmolowski A, Figlerowicz M. RNA interference and its role in the regulation of eucaryotic gene expression. Acta Biochim Pol. 2003;50:217–229. [PubMed] [Google Scholar]
- 16.Sanchez-Vargas I, Travanty EA, Keene KM, Franz AW, Beaty BJ, Blair CD, et al. et al. RNA interference, arthropod-borne viruses, and mosquitoes. Virus Res. 2004;102:65–74. doi: 10.1016/j.virusres.2004.01.017. [DOI] [PubMed] [Google Scholar]
- 17.Haasnoot J, Westerhout EM, Berkhout B. RNA interference against viruses: strike and counterstrike. Nat Biotechnol. 2007;25:1435–1443. doi: 10.1038/nbt1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martínez MA. United Kingdom: Caister Academic Press; 2010. RNA interference and viruses: Current innovations and future trends. [Google Scholar]
- 19.Jeang KT. RNAi in the regulation of mammalian viral infections. BMC Biol. 2012;10:58. doi: 10.1186/1741-7007-10-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haasnoot J, Berkhout B. RNA interference: its use as antiviral therapy. Handb Exp Pharmacol. 2006:117–150. doi: 10.1007/3-540-27262-3_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haasnoot PC, Cupac D, Berkhout B. Inhibition of virus replication by RNA interference. J Biomed Sci. 2003;10:607–616. doi: 10.1159/000073526. [DOI] [PubMed] [Google Scholar]
- 22.Zentella R, Yamauchi D, Ho TH. Molecular dissection of the gibberellin/abscisic acid signaling pathways by transiently expressed RNA interference in barley aleurone cells. Plant Cell. 2002;14:2289–2301. doi: 10.1105/tpc.003376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang QC, Nie QH, Feng ZH. RNA interference: antiviral weapon and beyond. World J Gastroenterol. 2003;9:1657–1661. doi: 10.3748/wjg.v9.i8.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shuey DJ, McCallus DE, Giordano T. RNAi: gene-silencing in therapeutic intervention. Drug Discov Today. 2002;7:1040–1046. doi: 10.1016/s1359-6446(02)02474-1. [DOI] [PubMed] [Google Scholar]
- 25.Mahmood ur R, Ali I, Husnain T, Riazuddin S. RNA interference: the story of gene silencing in plants and humans. Biotechnol Adv. 2008;26:202–209. doi: 10.1016/j.biotechadv.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 26.Mukherjee S, Hanley KA. RNA interference modulates replication of dengue virus in Drosophila melanogaster cells. BMC Microbiol. 2010;10:127. doi: 10.1186/1471-2180-10-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu X, Hong H, Yue J, Wu Y, Li X, Jiang L, et al. et al. Inhibitory effect of small interfering RNA on dengue virus replication in mosquito cells. Virol J. 2010;7:270. doi: 10.1186/1743-422X-7-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uchil PD, Satchidanandam V. Architecture of the flaviviral replication complex. Protease, nuclease, and detergents reveal encasement within double-layered membrane compartments. J Biol Chem. 2003;278:24388–24398. doi: 10.1074/jbc.M301717200. [DOI] [PubMed] [Google Scholar]
- 29.Sanchez-Vargas I, Scott JC, Poole-Smith BK, Franz AW, Barbosa-Solomieu V, Wilusz J, et al. et al. Dengue virus type 2 infections of Aedes aegypti are modulated by the mosquito's RNA interference pathway. PLoS Pathog. 2009;5:e1000299. doi: 10.1371/journal.ppat.1000299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang W, Singam R, Hellermann G, Kong X, Juan HS, Lockey RF, et al. et al. Attenuation of dengue virus infection by adeno-associated virus-mediated siRNA delivery. Genet Vaccines Ther. 2004;2:8. doi: 10.1186/1479-0556-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Canard B. Antiviral Research and development against dengue virus. 2012 [Online] Available from: www.who.int/tdr/research/ntd/dengue/dengue_full_length_report.pdf. [Accessed on 10 August, 2012]. [Google Scholar]
- 32.Franz AW, Sanchez-Vargas I, Adelman ZN, Blair CD, Beaty BJ, James AA, et al. et al. Engineering RNA interference-based resistance to dengue virus type 2 in genetically modified Aedes aegypti. Proc Natl Acad Sci USA. 2006;103:4198–4203. doi: 10.1073/pnas.0600479103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yue JY, Wu XW, Wu YJ, Li XZ, Jiang LY, Li QY, et al. et al. Study on the inhibitory effect of RNA interference on replication of dengue virus. Bing Du Xue Bao. 2010;26:373–378. [PubMed] [Google Scholar]
- 34.Padwad YS, Mishra KP, Jain M, Chanda S, Karan D, Ganju L. RNA interference mediated silencing of Hsp60 gene in human monocytic myeloma cell line U937 revealed decreased dengue virus multiplication. Immunobiology. 2009;214:422–429. doi: 10.1016/j.imbio.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 35.Ashfaq UA, Yousaf MZ, Aslam M, Ejaz R, Jahan S, Ullah O. siRNAs: potential therapeutic agents against hepatitis C virus. Virol J. 2011;8:276. doi: 10.1186/1743-422X-8-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alhoot MA, Wang SM, Sekaran SD. RNA interference mediated inhibition of dengue virus multiplication and entry in HepG2 cells. PLoS One. 2012;7:e34060. doi: 10.1371/journal.pone.0034060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ang F, Wong AP, Ng MM, Chu JJ. Small interference RNA profiling reveals the essential role of human membrane trafficking genes in mediating the infectious entry of dengue virus. Virol J. 2010;7:24. doi: 10.1186/1743-422X-7-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Villegas-Rosales1 PM, Méndez-Tenorio A, Ortega-Soto E, Barrón L B. Bioinformatics prediction of siRNAs as potential. Bioiinformation. 2012;8:519–522. doi: 10.6026/97320630008519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stein DA, Perry ST, Buck MD, Oehmen CS, Fischer MA, Poore E, et al. et al. Inhibition of dengue virus infections in cell cultures and in AG129 mice by a small interfering RNA targeting a highly conserved sequence. J Virol. 2011;85:10154–10166. doi: 10.1128/JVI.05298-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Korrapati AB, Swaminathan G, Singh A, Khanna N, Swaminathan S. Adenovirus delivered short hairpin RNA targeting a conserved site in the 5′ non-translated region inhibits all four serotypes of dengue viruses. PLoS Negl Trop Dis. 2012;6:e1735. doi: 10.1371/journal.pntd.0001735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Rij RP, Andino R. The silent treatment: RNAi as a defense against virus infection in mammals. Trends Biotechnol. 2006;24:186–193. doi: 10.1016/j.tibtech.2006.02.006. [DOI] [PubMed] [Google Scholar]