Abstract
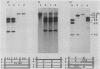
To examine the relationship between genetic and physical chromosome maps, we constructed a diploid strain of the yeast Saccharomyces cerevisiae heterozygous for 12 restriction site mutations within a 23-kilobase (5-centimorgan) interval of chromosome III. Crossovers were not uniformly distributed along the chromosome, one interval containing significantly more and one interval significantly fewer crossovers than expected. One-third of these crossovers occurred within 6 kilobases of the centromere. Approximately half of the exchanges were associated with gene conversion events. The minimum length of gene conversion tracts varied from 4 base pairs to more than 12 kilobases, and these tracts were nonuniformly distributed along the chromosome. We conclude that the chromosomal sequence or structure has a dramatic effect on meiotic recombination.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Angel T., Austin B., Catcheside D. G. Regulation of recombination at the his-3 locus in Neurospora crassa. Aust J Biol Sci. 1970 Dec;23(6):1229–1240. doi: 10.1071/bi9701229. [DOI] [PubMed] [Google Scholar]
- Clarke L., Carbon J. Isolation of a yeast centromere and construction of functional small circular chromosomes. Nature. 1980 Oct 9;287(5782):504–509. doi: 10.1038/287504a0. [DOI] [PubMed] [Google Scholar]
- Coleman K. G., Steensma H. Y., Kaback D. B., Pringle J. R. Molecular cloning of chromosome I DNA from Saccharomyces cerevisiae: isolation and characterization of the CDC24 gene and adjacent regions of the chromosome. Mol Cell Biol. 1986 Dec;6(12):4516–4525. doi: 10.1128/mcb.6.12.4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dicarprio L., Hastings P. J. Gene conversion and intragenic recombination at the SUP6 locus and the surrounding region in Saccharomyces cerevisiae. Genetics. 1976 Dec;84(4):697–721. doi: 10.1093/genetics/84.4.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T. Translocations Involving the Third and the Fourth Chromosomes of DROSOPHILA MELANOGASTER. Genetics. 1930 Jul;15(4):347–399. doi: 10.1093/genetics/15.4.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink G. R., Petes T. D. Gene conversion in the absence of reciprocal recombination. 1984 Aug 30-Sep 5Nature. 310(5980):728–729. doi: 10.1038/310728a0. [DOI] [PubMed] [Google Scholar]
- Fitzgerald-Hayes M., Clarke L., Carbon J. Nucleotide sequence comparisons and functional analysis of yeast centromere DNAs. Cell. 1982 May;29(1):235–244. doi: 10.1016/0092-8674(82)90108-8. [DOI] [PubMed] [Google Scholar]
- Fogel S., Mortimer R., Lusnak K., Tavares F. Meiotic gene conversion: a signal of the basic recombination event in yeast. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 2):1325–1341. doi: 10.1101/sqb.1979.043.01.152. [DOI] [PubMed] [Google Scholar]
- Gutz H. Site Specific Induction of Gene Conversion in SCHIZOSACCHAROMYCES POMBE. Genetics. 1971 Nov;69(3):317–337. doi: 10.1093/genetics/69.3.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hieter P., Pridmore D., Hegemann J. H., Thomas M., Davis R. W., Philippsen P. Functional selection and analysis of yeast centromeric DNA. Cell. 1985 Oct;42(3):913–921. doi: 10.1016/0092-8674(85)90287-9. [DOI] [PubMed] [Google Scholar]
- Hinnen A., Hicks J. B., Fink G. R. Transformation of yeast. Proc Natl Acad Sci U S A. 1978 Apr;75(4):1929–1933. doi: 10.1073/pnas.75.4.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst D. D., Fogel S., Mortimer R. K. Conversion-associated recombination in yeast (hybrids-meiosis-tetrads-marker loci-models). Proc Natl Acad Sci U S A. 1972 Jan;69(1):101–105. doi: 10.1073/pnas.69.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinks-Robertson S., Petes T. D. High-frequency meiotic gene conversion between repeated genes on nonhomologous chromosomes in yeast. Proc Natl Acad Sci U S A. 1985 May;82(10):3350–3354. doi: 10.1073/pnas.82.10.3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keil R. L., Roeder G. S. Cis-acting, recombination-stimulating activity in a fragment of the ribosomal DNA of S. cerevisiae. Cell. 1984 Dec;39(2 Pt 1):377–386. doi: 10.1016/0092-8674(84)90016-3. [DOI] [PubMed] [Google Scholar]
- Lambie E. J., Roeder G. S. Repression of meiotic crossing over by a centromere (CEN3) in Saccharomyces cerevisiae. Genetics. 1986 Nov;114(3):769–789. doi: 10.1093/genetics/114.3.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin J. C., Woolford J. L., Jr Molecular cloning and analysis of the CRY1 gene: a yeast ribosomal protein gene. Nucleic Acids Res. 1983 Jan 25;11(2):403–420. doi: 10.1093/nar/11.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meselson M. S., Radding C. M. A general model for genetic recombination. Proc Natl Acad Sci U S A. 1975 Jan;72(1):358–361. doi: 10.1073/pnas.72.1.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer R. K., Schild D. Genetic map of Saccharomyces cerevisiae, edition 9. Microbiol Rev. 1985 Sep;49(3):181–213. doi: 10.1128/mr.49.3.181-213.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Weaver T. L., Szostak J. W. Fungal recombination. Microbiol Rev. 1985 Mar;49(1):33–58. doi: 10.1128/mr.49.1.33-58.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PONTECORVO G., KAFER E. Genetic analysis based on mitotic recombination. Adv Genet. 1958;9:71–104. [PubMed] [Google Scholar]
- Palzkill T. G., Oliver S. G., Newlon C. S. DNA sequence analysis of ARS elements from chromosome III of Saccharomyces cerevisiae: identification of a new conserved sequence. Nucleic Acids Res. 1986 Aug 11;14(15):6247–6264. doi: 10.1093/nar/14.15.6247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponticelli A. S., Schultz D. W., Taylor A. F., Smith G. R. Chi-dependent DNA strand cleavage by RecBC enzyme. Cell. 1985 May;41(1):145–151. doi: 10.1016/0092-8674(85)90069-8. [DOI] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Stahl F. W., Stahl M. M., Malone R. E., Crasemann J. M. Directionality and nonreciprocality of Chi-stimulated recombination in phage lambda. Genetics. 1980 Feb;94(2):235–248. doi: 10.1093/genetics/94.2.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strathern J. N., Newlon C. S., Herskowitz I., Hicks J. B. Isolation of a circular derivative of yeast chromosome III: implications for the mechanism of mating type interconversion. Cell. 1979 Oct;18(2):309–319. doi: 10.1016/0092-8674(79)90050-3. [DOI] [PubMed] [Google Scholar]
- Symington L. S., Fogarty L. M., Kolodner R. Genetic recombination of homologous plasmids catalyzed by cell-free extracts of Saccharomyces cerevisiae. Cell. 1983 Dec;35(3 Pt 2):805–813. doi: 10.1016/0092-8674(83)90113-7. [DOI] [PubMed] [Google Scholar]
- Szostak J. W., Orr-Weaver T. L., Rothstein R. J., Stahl F. W. The double-strand-break repair model for recombination. Cell. 1983 May;33(1):25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- Warmington J. R., Anwar R., Newlon C. S., Waring R. B., Davies R. W., Indge K. J., Oliver S. G. A 'hot-spot' for Ty transposition on the left arm of yeast chromosome III. Nucleic Acids Res. 1986 Apr 25;14(8):3475–3485. doi: 10.1093/nar/14.8.3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston F., Chumley F., Fink G. R. Eviction and transplacement of mutant genes in yeast. Methods Enzymol. 1983;101:211–228. doi: 10.1016/0076-6879(83)01016-2. [DOI] [PubMed] [Google Scholar]