Abstract
Objective
Data from clinical care is increasingly being used for research purposes. The i2b2 platform has been introduced in some US research communities as a tool for data integration and querying by clinical users. The purpose of this project was to assess the applicability of i2b2 in Germany regarding use cases, functionality and integration with privacy enhancing tools.
Methods
A set of four research usage scenarios was chosen, including the transformation and import of ontology and fact data from existing clinical data collections into i2b2 v1.4 instances. Query performance was measured in comparison to native SQL queries. A setup and administration tool for i2b2 was developed. An extraction tool for CDISC ODM data was programmed. Interfaces for the TMF privacy enhancing tools (PID Generator, Pseudonymization Service) were implemented.
Results
Data could be imported in all tested scenarios from various source systems, including the generation of i2b2 ontology definitions. The integration of TMF privacy enhancing tools was possible without modification of the platform. Limitations were found regarding query performance in comparison to native SQL and certain temporal queries.
Conclusions
i2b2 is a viable platform for data query tasks in use cases typical for networked medical research in Germany. The integration of privacy enhancing tools facilitates the use of i2b2 within established data protection concepts. Entry barriers should be lowered by providing tools for simplified setup and import of medical standard formats like CDISC ODM.
Keywords: Medical Record Systems, Information Storage and Retrieval, Single-Source
Introduction
Increasing amounts of data are captured digitally during clinical routine care with the primary objective of supporting the care process. The need for making these data available for scientific reuse has been discussed extensively [1-3]. Thus, routine data could be beneficial throughout the whole research lifecycle, as a base for hypothesis generation, estimation of expected study cohort sizes, to enhance patient recruitment in ongoing studies and to reduce duplication of data entry, among other uses. Currently, the availability of these data for research purposes is limited, and projects trying to tap them are facing complex challenges: Data items are typically spread among disparate databases, including electronic medical records (EMR), laboratory and order entry systems or electronic data capture (EDC) systems for research, and have to be extracted and transformed into a common schema optimized for analysis. Records of the same patients across various systems have to be linked and de-duplicated to create added value. At the same time, data protection guidelines mandate the de-identification of patient data as well as strict access controls. Query interfaces have to be developed that allow clinical users to analyze complex datasets.
The “informatics for integrating biology and the bedside” project (i2b2) was funded by the NIH as one of seven National Centers for Biomedical Computing to provide a generic and scalable platform for the integration of clinical and research data [4, 5]. The i2b2 platform uses a modular approach that provides several “cells” to carry out queries, export and visualize result data as well as generate additional data points through further analysis, e.g. natural language processing [6, 7]. i2b2 has reached wide adoption in the United States and created an active user community [8-13]. i2b2 uses a generic Entity-Attribute-Value (EAV) database schema [14, 15] that facilitates rapid integration with additional data sources as well as a simplified user interface that allows clinicians to formulate complex Boolean queries. Limitations of this approach have been discussed, including restrictions in the formulation of queries containing post-coordination as well as certain temporal or aggregated expressions [15, 16].
The German Technology and Method Platform for Networked Medical Research (TMF) [17] is a non-profit organization financed by member institutions carrying out multicenter medical research. Its mission is to support researchers by identifying and providing solutions for organizational, legal and technical issues. In this context the TMF also evaluates established tools and platforms regarding their applicability for specific use cases in the German medical research context as well as their conformity to German data protection requirements.
The TMF sponsored an IT strategy project in 2009 in order to identify relevant tools and platforms to support networked medical research in the near future. Within this project, the authors’ objective was to assess the applicability of i2b2 within the German research context, including the identification of possible use cases and limitations, data protection issues and the integration with privacy enhancing tools developed by the TMF.
Methods
Installations of i2b2 v1.4 were carried out using both a preconfigured virtual machine (VM) as well as from scratch using source code provided on the i2b2 website [18]. Installations were set up on a VMware ESXTM virtualization platform (VMware Inc.) with a single virtual CPU and 1 GB RAM. The database was placed on a dedicated server (Sun Fire V440TM, Sun Inc-) with 4 CPUs (1.59 GHz) and 16 GB of RAM running Oracle 10gTM (Oracle Inc.). A setup and administration tool was developed within the project to simplify installation from source code as well the setup and loading of i2b2 project databases.
The system was tested in 4 different usage scenarios chosen by the authors to represent typical use cases (not only) within the medical research entities organized in the TMF:
-
•
A: Query frontend for a local clinical data warehouse
-
•
B: Research database for local prostate cancer project
-
•
C: Research database for multicenter dermatologic research network
-
•
D: Research database for long term storage
For scenario A, a Clinical Data Warehouse (Cognos BITM, IBM Inc.) established at Erlangen University Hospital was integrated with i2b2. Metadata for diagnoses (German ICD 10 GM 2010) and procedures (German OPS 2010) were converted from star schema dimension tables into the i2b2 ontology format using the IBM Cognos DataManagerTM Extraction, Transformation and Loading (ETL) tool. Patient demographics and fact dataa [19] for diagnoses and procedures were transformed from warehouse fact tables into the i2b2 EAV representation using the same method. The TMF PID-Generator, a tool for pseudonymization and the robust linkage of patient demographic data [20-22] was integrated into the import process. Pseudonymization was carried out asynchronously to generate a patient list containing both identifying data and pseudonyms. The patient list was then joined to the demographic source data during conversion into the i2b2 format.
To verify query performance and capabilities within scenario A, a set of consecutive clinical selections were carried out in comparison of direct SQL requests on the clinical data warehouse (relational database schema) against requests through the i2b2 user interface (generic EAV database schema). Selection criteria were chosen by the authors to represent a stepwise approach of narrowing down a prospective study cohort, using data items available in the EHR. Both the clinical data warehouse and the i2b2 project resided on the same Oracle database server and contained the same number of patients and diagnosis/procedure codes. The query was started by selecting a set of female patients presenting in 2009 with a diagnosis of breast cancer (ICD10-GM [23] code C50). The selection was then further restricted by a procedure code for radiation therapy (OPS [24] code 8-52), a procedure code for surgical breast excision or resection (OPS code 5-87) and a procedure code for chemotherapy (OPS code 8-54). The dataset was then restricted to the patients aged 30-49 years at diagnosis. For this query, patient age at the beginning of each encounter was made available as a fact item in i2b2, whereas it was calculated at runtime for the native query. Finally, additional restrictions were added for the chemotherapy to have occurred before the surgical procedure and the radiation to have taken place after surgery. All queries were carried out 5 times consecutively and the average runtime was computed.
For scenario B, a prostate cancer documentation based on the Erlangen University hospital EMR system (SoarianTM, Siemens Inc.) was integrated with i2b2. Ontology metadata was automatically generated from the EMR forms definition tables using an Oracle PL/SQL script for conversion into the i2b2 ontology format. Fact data was extracted from the EMR system for script based conversion into the i2b2 EAV representation.
For scenario C, a locally developed EDC system used with the German Epidermolysis Bullosa Research Network [25] was integrated with i2b2. Ontology metadata was transferred manually into an ExcelTM (Microsoft Inc.) spreadsheet containing the concept and hierarchy followed by script-based conversion into the i2b2 ontology format.
For Scenario D the trial database of the Competence Network for Congenital Heart Defects (KN AHF) was integrated with i2b2. The trial database is based on the commercial EDC system SecuTrialTM (iAS GmbH). Data was exported using standard CDISC ODM (Operational Data Model) 1.2 and 1.3 export files [26, 27]. Ontology metadata was extracted and converted into SQL-statements suitable for populating the i2b2 ontology tables using a Java-based program developed within this project. Fact data was similarly extracted from the ODM files and converted into individual SQL statements for each patient. The TMF pseudonymization service (PSD), a tool for the reversible pseudonymization of medical research data [21, 22], was integrated into the import process. The integration consisted of Python programs importing the fact data SQL files generated by the ODM converter, sending them in XML form to the PSD web service, receiving the pseudonymized records from the PSD web service, parsing and storing them in SQL files ready for import into i2b2.
Results
The setup and administration tool for i2b2 developed in this project provided functions for the setup and initial configuration of a fully functional i2b2 server instance from source including the installation of required Linux packages. Administration functions covered the setup of i2b2 project instances including the configuration of related database schemas and i2b2 users. The tool was made available for public use on the TMF website [28] and the i2b2 Academic User Group (AUG) [29].
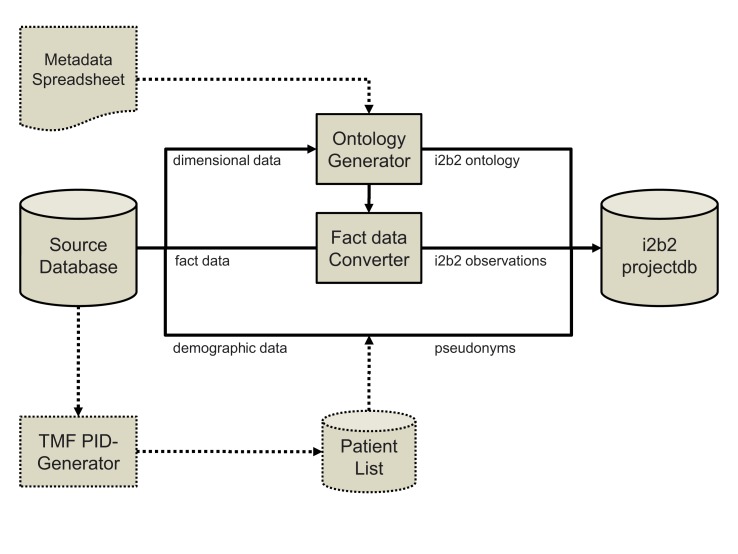
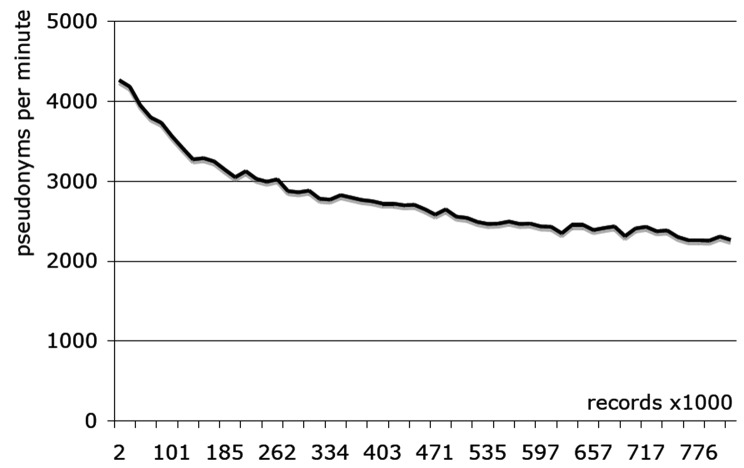
►Figure 1 shows the process established in scenarios A-C for the conversion and import of ontology, demographic and fact data into the i2b2 project database. Ontology metadata was extracted directly from source databases in scenarios A and B and was prepared manually for scenario C. ►Table 1 presents the number of patients, ontology and fact records imported in each scenario as well as loading times. The import process was tested both with and without PID generator integration for demographic data in scenario A. ►Figure 2 shows the performance of the PID generator against the number of coded demographic records. ►Table 2 shows the composition of the loading time for scenario D, including the pseudonymization service.
Fig. 1.
Ingest data flow for usage scenarios A-C
Table 1.
Content structure and loading times in i2b2 usage scenarios
| Scenario | Contents | Items | Concept Codes |
Patients | Records | Loading time (mm:ss) |
|---|---|---|---|---|---|---|
|
A Clinical Data Warehouse |
Demographics Diagnoses Procedures |
4 | 56,275 | 672,225 | 5,375,223 | 45:05 |
|
B Prostate Cancer Project |
Demographics Med. History Surgery Pathology Selected Lab |
46 | 232 | 121 | 2238 | 00:05 |
|
C Dermatology Research Network |
Demographics Med. History Skin status |
253 | 546 | 418 | 113,993 | 01:52 |
|
D Long-term research database |
Demographics Diagnoses Procedures MRT/US |
3195 | 94,117 | 143 | 54,534 | 33:44 |
Fig. 2.
Performance of TMF PID-Generator Pseudonymization tool
Table 2.
Composition of scenario D loading time
| ODM->SQL | SQL->XML | PSEUD | XML->SQL | SQL->i2b2 | Total time |
|---|---|---|---|---|---|
| 01:25 | 00:03 | 13:15 | 00:03 | 18:57 | 33:44 |
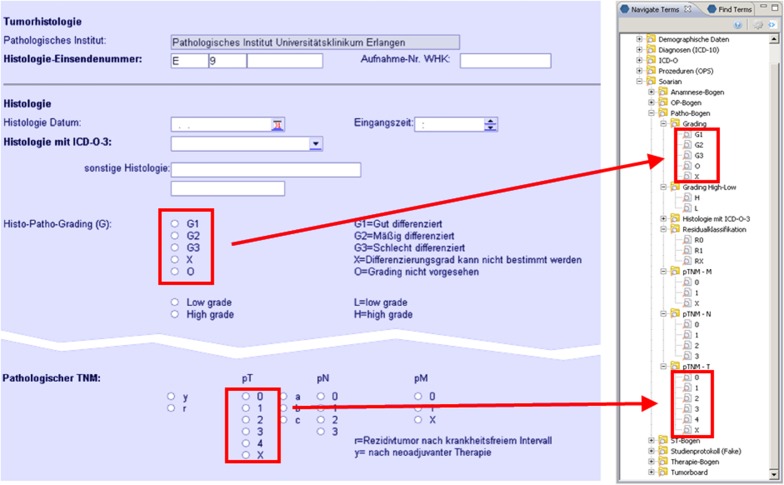
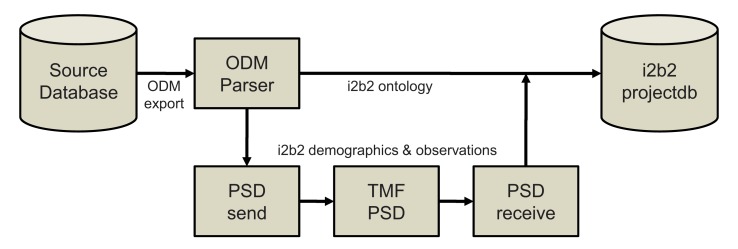
►Table 3 presents the record counts and runtimes from the SQL/i2b2 query comparison in scenario A. ►Figure 3 shows a screenshot of a prostate cancer EMR form from scenario B in comparison to an i2b2 ontology hierarchy extracted from its metadata. ►Figure 4 illustrates the import process established in scenario D for the extraction of ontology and fact data from ODM files and their subsequent processing through the TMF pseudonymization service.
Table 3.
Query performance and capabilities for scenario A
| Query Stage | Native SQL | i2b2 | ||
|---|---|---|---|---|
| Patients retrieved |
Runtime (sec) |
Patients retrieved |
Runtime (sec) |
|
| Female, Breast Cancer (ICD C50) in 2009 | 1081 | 9 | 1081 | 47 |
| + Radiation therapy (OPS 8-52) | 384 | 5 | 384 | 57 |
| + Surgical Breast excision/resection (OPS 5-87) | 194 | 8 | 194 | 39 |
| + Chemotherapy (OPS 8-54) | 55 | 5 | 55 | 49 |
| + age group 30-49 at diagnosis | 18 | 4 | 21 | 53 |
| + Chemotherapy pre & radiation post surgery | 10 | 4 | - | - |
Fig. 3.
Screenshots of prostate cancer EMR documentation form and corresponding i2b2 ontology elements: the left window shows the EMR system screen with a tumor histology form, the right window shows the corresponding generated ontology tree
Fig. 4.
Ingest workflow in scenario D with integration of TMF pseudonymization service: the ontology metadata (s. fig. 3 right window) is imported separately from the other data which is being de-identified in the lower branch using TMF tools.
Discussion
While the preconfigured i2b2 virtual machine download provides a quick method of experimenting with the platform, it poses restrictions regarding disk space, operating system updates and adherence to local IT guidelines. A dedicated installation from source should therefore be used for production environments. The setup and configuration from scratch, however, was complicated by dependencies on specific library versions and multiple interdependent configuration steps. Feedback gathered at two well-attended national i2b2 workshops indicated that the complex setup posed a serious obstacle to production use at several sites. By the creation of a dedicated setup and administration tool within this project it was possible to automate this process and reduce installation and configuration times to a few minutes.
In all tested usage scenarios it was possible to connect existing data sources with i2b2. The simple hierarchical structure of the i2b2 ontology allowed a direct conversion of standard star schema dimension tables from the clinical data warehouse. Metadata definitions extracted from the EMR could be structured by form, field and value levels which provide easy recognition for i2b2 users experienced with the EMR. For data sources without readily available structured metadata, a manual spreadsheet could be constructed containing a similar form/field/value hierarchy.
Pseudonymization tools developed to meet national data protection requirements could be integrated seamlessly into the import process. While performance of the TMF PID generator is fast (average >2000 records per minute), pseudonymization of the full demographic dataset in scenario A took more than 5 hours. By using asynchronous integration, this additional time burden can be separated from the import workflow itself. Additionally, only new or modified records have to be pseudonymized once the initial dataset has been processed.
The query performance comparison (►Table 3) showed that the average runtime was generally five to tenfold slower in i2b2 than using native SQL queries. The data warehouse tables used for native queries are modeled in a relational schema optimized for reporting, making use of indices and optimizer hints to gain maximum performance. i2b2 in comparison uses a generic schema, putting all dimensional data in a single table and all fact data in one additional large table. Data segmentation in a relationally modeled schema allows the database to gain speed by having to access only those tables containing data relevant to the query. It should be evaluated whether i2b2 query performance can be optimized by partitioning the fact data table, modifying indices or adding database-specific optimizer hints. It should be noted that the creation of the native SQL statements for the queries required detailed knowledge of the database structure as well as complex SQL commands including subselects and optimizer hints. Using the i2b2 frontend, all queries were constructed graphically, regardless of the underlying table structures. A limitation of the performance tests could result from the setup of i2b2 in a virtual machine. Due to the size of the underlying dataset, however, the major part of the query runtime occurred on the database, which resided on a hardware server used also for the native SQL queries.
Regarding query results, the number of patients retrieved was identical between native SQL and i2b2 for the first 4 queries. Adding an age restriction in step 5 resulted in different patient counts. Further analysis revealed that i2b2 retrieved 3 additional patients because the age restriction was handled differently. In native SQL it was possible to combine the diagnosis and age restriction in the same statement. i2b2, however, treats each query item separately and combines them afterwards using Boolean operators. The age restriction was thus not applied in co-occurrence with the breast cancer diagnosis, but rather as a separate set of all patients aged 30-49, regardless of diagnosis. While this behavior is consistent with the i2b2 user interface, it imposes restrictions on queries referring to age at a specific event. As age is an important inclusion/exclusion criterion in many patient samples, i2b2 should be extended to allow restricting patient age in direct combination with other query items. The i2b2 development roadmap [30] states that in v1.6 it will be possible to filter query conditions to occur within the same encounter, which would remove this restriction. The final query step required the temporal combination of query items in the sense that they had to occur in a specific sequence over time. As it has been noted before [15], i2b2 does not provide functions to define this type of query, so this step could not be carried out in i2b2. The integrated timeline view could have been used to manually examine individual patient histories with regard to temporal conditions, but this approach would not be feasible for selecting records from a large dataset. Even though the addition of temporal constraints would be desirable, the increased complexity of the user interface should be balanced against overall usability. Alternatively, data could be preprocessed before importing to provide derived fact items containing the required temporal restrictions [15].
The implementation of a generic CDISC ODM parser and converter for i2b2 for scenario D allowed the automatic extraction of both ontology and fact data from a standardized format. As ODM is used widely e.g. in pharmaceutical trials [26], this tool could potentially be useful to many sites looking at analyzing their study datasets in i2b2. Metadata available in the ODM files was, however, not in all cases sufficient to automatically generate optimal ontology definitions: e.g. for numeric items the data type itself is defined in the ODM file, but there is no information about suitable intervals for displaying valid choices in the ontology window. i2b2 v1.4 did not provide a means of modifying ontology data, apart from importing a new ontology dataset changed outside of the system. It should be considered to implement an ontology editor that can be pre-populated with metadata from source systems. Users could then adapt the ontology where needed and add details that could not be derived from source systems. Ongoing efforts include using the Protégé ontology editor to create i2b2 ontologies [31] as well as the CTSA Health Ontology Mapper project [32]. Starting with v1.5 an ontology editor was added to the base i2b2 distribution as well.
The TMF pseudonymization service could be integrated into the import process by the addition of sending and receiving programs accessing the PSD web service in scenario D. No modifications of the i2b2 systems had to be carried out. Integration of the TMF Pseudonymization Service resulted in an overhead of 65% for additional processing during loading time (►Table 2). The Pseudonymization Service currently accepts only individual patient records, which made it necessary to individually for each patient extract from ODM, encode into XML, decode the pseudonymized record from XML and load them into the i2b2 database. Extending the Pseudonymization Service to allow batch processing should result in a relevant reduction of processing time. Also, records were loaded with individual SQL statements into the i2b2 database. In scenario A, much higher loading performance was achieved by using flat-file-based batch importing (Oracle SQL*Loader™), which could be implemented for the ODM/PSD pathway as well.
Role-based access controls became available in i2b2 v1.4 that can restrict user access to aggregated patient counts rather than individual, exportable records. This new feature can be used for a graduated approach, allowing a broader group of users to query a non-identified view of the database for relevant subsets and then request approval for full access. The platform, however, does not provide ways to generally restrict access to specific subsets (e.g. the patients of a single department). As a workaround, subsets can be extracted and copied into separate i2b2 project instances. When applied to large datasets, this approach would however greatly increase loading times as well as the complexity of database and user administration. The feasibility of adding fine-grained user permissions in i2b2 has been demonstrated within a translational medicine project [13] and should be considered for the general release.
The project was carried out using version 1.4 of i2b2, so functionality and performance improvements introduced in later versions could not be taken into account. The ETL process was carried out according to public documentation, so the effect of undocumented optimization techniques was not evaluatedb [33]. Performance comparisons were only carried out between i2b2 and native SQL on the same database, not in comparison to other query platforms.
Conclusions
It was demonstrated that i2b2 is a viable platform for data query tasks in use cases typical for networked medical research in Germany. The integration of TMF privacy enhancing tools for record linkage and pseudonymization was possible without modification of the platform or reduced performance, facilitating the use of i2b2 within established data protection concepts in Germany. In order to reach broad acceptance of the platform, entry barriers should be lowered further. This includes simplifying the complex setup process, which was achieved in this project by implementing a dedicated installation and administration tool not yet included in the original distribution. While data import from various sources was carried out successfully during the project, manual interventions were necessary in many cases, especially regarding the construction of valid ontology metadata. Support for extracting fact and ontology data from established standard formats could reduce the effort to set up and maintain a productive i2b2 installation. The ODM import tool developed in this project can be seen as a first step in this direction, as well as the recent addition of an ontology editor in v1.5 of i2b2. Known restrictions of the platform regarding temporal queries could be reproduced in this project. However, workarounds exist by preprocessing related data items, and temporal extensions have been announced to address this limitation in future versions of i2b2. While query performance of i2b2 was inferior in comparison to native SQL, it provides an intuitive graphical user interface that allows clinical users to construct complex queries without detailed knowledge of database structure and SQL optimization.
Overall, i2b2 proved to be a valuable addition to the tools curated by the TMF for German networked medical research. Several TMF member networks are currently in the process of setting up i2b2 installations.
Clinical Relevance Statement
Single-source strategies facilitate the re-use of data acquired in routine clinical care for research purposes. The availability of adequate tools for data integration and analysis positively impacts the cost-effectiveness, quality and timeliness of clinical research projects relying on such data.
Conflict of Interest
The authors have established in 08/2009 a memorandum of understanding with the i2b2 National Center for Biomedical Computing to collaborate on the further development, evaluation and dissemination of i2b2 in Germany.
Human Subject Research
The study was performed in compliance with the World Medical Association Declaration of Helsinki on Ethical Principles for Medical Research Involving Human Subjects.
Acknowledgments
This project was supported in part by the grant KFO179 “Biological Basis of Individual Tumor Response in Patients with Rectal Cancer” of the German Research Foundation (DFG) as well as by the grant Kompetenznetz Angeborene Herzfehler (Competence Network for Congenital Heart Defects) funded by the German Federal Ministry of Education and Research (BMBF), FKZ 01GI0210, and the grant Netzwerk Epidermolysis Bullosa (German Epidermolysis Bullosa Network) funded by the German Federal Ministry of Education and Research (BMBF), FKZ 01GM0831. The authors wish to thank Lars Reimann for his work on the integration of the TMF pseudonymization service, Steffen Zeiss, Roman Ostertag, Benedikt Schäffler and Christian Bauer for their work on the ODM import and Andreas Becker for his support on Clinical Data Warehouse integration.
Glossary
Abbreviations
- AUG
Academic User Group
- CDISC
Clinical Data Interchange Standards Consortium
- EAV
Entity-Attribute-Value
- EDC
Electronic Data Capture
- EMR
Electronic Medical Record
- ETL
Extraction, Transformation and Loading
- i2b2
Informatics for Integrating Biology and the Bedside
- ICD
International Classification of Diseases
- KN AHF
Competence Network for Congenital Heart Defects
- MRT
Magnetic Resonance Tomography
- NIH
National Institutes of Health
- ODM
Operational Data Model
- OPS
Operation and Procedure Codes
- PID
Patient Identifier
- PSD
Pseudonymization Service
- SQL
Structured Query Language
- TMF
Technology and Method Platform for Networked Medical Research
- US
Ultrasound
Footnotes
a Fact data are usually defined in Data Warehousing as business performance measurements which are typically numeric and additive. In Clinical Data Warehousing, fact data may also include textual concepts linked to a patient, which do not have to be numeric.
b The i2b2 Wiki made available in October 2010 mentions a previously undocumented “total_num” column in the i2b2 ontology table, which is used to optimize query performance
References
- 1.Prokosch HU, Ganslandt T. Perspectives for medical informatics. Reusing the electronic medical record for clinical research. Methods Inf Med 2009; 48(1): 38-44 PMid:19151882 [PubMed] [Google Scholar]
- 2.Ohmann C, Kuchinke W. Future developments of medical informatics from the viewpoint of networked clinical research. Interoperability and integration. Methods Inf Med 2009; 48(1): 45-54 PMid:19151883 [PubMed] [Google Scholar]
- 3.Kush R, Alschuler L, Ruggeri R, Cassells S, Gupta N, Bain L, et al. Implementing Single Source: the STARBRITE proof-of-concept study. J Am Med InformAssoc 2007; 14(5): 662-673 doi:10.1197/jamia.M2157PMid:17600107 PMCid:1975790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murphy SN, Mendis ME, Berkowitz DA, Kohane I, Chueh HC. Integration of clinical and genetic data in the i2b2 architecture. AMIA Annu Symp Proc 2006: 1040 PMid:17238659 PMCid:1839291 [PMC free article] [PubMed] [Google Scholar]
- 5.Murphy SN, Weber G, Mendis M, Gainer V, Chueh HC, Churchill S, et al. Serving the enterprise and beyond with informatics for integrating biology and the bedside (i2b2). J Am Med InformAssoc 2010; 17(2): 124-130 doi:10.1136/jamia.2009.000893PMid:20190053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mendis M, Wattanasin N, Kuttan R, Pan W, Philips L, Hackett K, et al. Integration of Hive and cell software in the i2b2 architecture. AMIA Annu Symp Proc 2007: 1048 PMid:18694146 [PubMed] [Google Scholar]
- 7.Mendis M, Phillips LC, Kuttan R, Pan W, Gainer V, Kohane I, et al. Integrating outside modules into the i2b2 architecture. AMIA Annu Symp Proc 2008: 1054 PMid:18999021 [PubMed] [Google Scholar]
- 8.Goryachev S, Sordo M, Zeng QT. A suite of natural language processing tools developed for the I2B2 project. AMIA Annu Symp Proc 2006: 931 PMid:17238550 PMCid:1839726 [PMC free article] [PubMed] [Google Scholar]
- 9.Gainer V, Hackett K, Mendis M, Kuttan R, Pan W, Phillips LC, et al. Using the i2b2 hive for clinical discovery: an example. AMIA Annu Symp Proc 2007: 959 PMid:18694059 [PubMed] [Google Scholar]
- 10.Uzuner O. Second i2b2 workshop on natural language processing challenges for clinical records. AMIA Annu Symp Proc 2008: 1252-1253 [PubMed] [Google Scholar]
- 11.Heinze DT, Morsch ML, Potter BC, Sheffer RE., Jr.Medical i2b2 NLP smoking challenge: the A-Life system architecture and methodology. J Am Med Inform Assoc. 2008; 15(1): 40-43 doi:10.1197/jamia.M2438 PMid:17947621 PMCid:2274871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Childs LC, Enelow R, Simonsen L, Heintzelman NH, Kowalski KM, Taylor RJ. Description of a rule-based system for the i2b2 challenge in natural language processing for clinical data. J Am Med Inform Assoc 2009; 16(4): 571-575 doi:10.1197/jamia.M3083 PMid:19390103 PMCid:2705261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Szalma S, Koka V, Khasanova T, Perakslis ED. Effective knowledge management in translational medicine. J Transl Med 2010; 8(1): 68 doi:10.1186/1479-5876-8-68 PMid:20642836 PMCid:2914663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nadkarni PM, Brandt C. Data extraction and ad hoc query of an entity-attribute-value database. J Am Med InformAssoc 1998; 5(6): 511-527 PMid:9824799 PMCid:61332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deshmukh VG, Meystre SM, Mitchell JA. Evaluating the informatics for integrating biology and the bedside system for clinical research. BMC Med Res Methodol 2009; 9: 70 doi:10.1186/1471-2288-9-70 PMid:19863809 PMCid:2779809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meystre SM, Deshmukh VG, Mitchell J. A clinical use case to evaluate the i2b2 Hive: predicting asthma exacerbations. AMIA Annu Symp Proc 2009; 2009: 442-446 [PMC free article] [PubMed] [Google Scholar]
- 17.TMF e.V. TMF Homepage [Internet]Berlin (Germany)2010[updated 11/19/2010; cited 11/28/2010]; Available from: http://www.tmf-ev.de/ [Google Scholar]
- 18.i2b2 NCBC i2b2 Software Download. [Internet]Boston (MA): Partners Healthcare; 2010[updated 11/10/2010; cited 11/28/2010]; Available from: https://www.i2b2.org/software/ [Google Scholar]
- 19.Kimball R, Ross M. The Data Warehouse Toolkit: John Wiley & Sons; 2002 [Google Scholar]
- 20.Faldum A, Pommerening K. An optimal code for patient identifiers. Comput Methods Programs Biomed 2005; 79(1): 81-88 doi:10.1016/j.cmpb.2005.03.004 PMid:15888350 [DOI] [PubMed] [Google Scholar]
- 21.Pommerening K, Reng M. Secondary use of the EHR via pseudonymisation. Studies in Health Technology and Informatics 2004; 103: 441-446 PMid:15747953 [PubMed] [Google Scholar]
- 22.Helbing K, Demiroglu SY, Rakebrandt F, Pommerening K, Rienhoff O, Sax U. A Data Protection Scheme for Medical Research Networks. Review after Five Years of Operation. Methods Inf Med 2010; 49(5). PMid:20644898 [DOI] [PubMed] [Google Scholar]
- 23.DIMDI International Classification of Diseases (ICD10) with German Modifications. [Internet]Cologne (Germany): German Institute of Medical Documentation and Information (DIMDI); 2010[updated 09/27/2010; cited 11/28/2010]; Available from: http://www.dimdi.de/static/de/klassi/diagnosen/icd10/ [Google Scholar]
- 24.DIMDI German Procedure Codes (OPS). [Internet]Cologne (Germany): German Institute of Medical Documentation and Information (DIMDI); 2010[updated 09/27/2010; cited 11/28/2010]; Available from: http://www.dimdi.de/static/de/klassi/prozeduren/ops301/ [Google Scholar]
- 25.Klein A, Prokosch HU, Muller M, Ganslandt T. Experiences with an interoperable data acquisition platform for multi-centric research networks based on HL7 CDA. Methods Inf Med 2007; 46(5): 580-585 PMid:17938783 [DOI] [PubMed] [Google Scholar]
- 26.Kuchinke W, Wiegelmann S, Verplancke P, Ohmann C. Extended cooperation in clinical studies through exchange of CDISC metadata between different study software solutions. Methods Inf Med 2006; 45(4): 441-446 PMid:16964363 [PubMed] [Google Scholar]
- 27.CDISC Operational Data Model (ODM). [Internet]Austin, TX: Clinical Data Interchange Standards Consortium; 2010[cited 11/28/2010]; Available from: http://www.cdisc.org/odm/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.TMF e.V. TMF Forum (registration required) [Internet]Berlin (Germany)2010[updated 11/19/2010; cited 11/28/2010]; Available from: http://www.tmf-ev.de/Forum.aspx [Google Scholar]
- 29.i2b2 NCBC i2b2 Academic Users Group. [Internet]Boston (MA)2010[cited 11/28/2010]; Available from: http://www.i2b2aug.org/ [Google Scholar]
- 30.i2b2 NCBC i2b2 Roadmap Release 1.6. [Internet]Boston (MA)2010[updated 10/05/2010; cited 11/28/2010]; Available from: https://community.i2b2.org/wiki/display/roadmap/Release+1.6 [Google Scholar]
- 31.Tokyo Medical and Dental University Japanese i2b2 database development project in TMDU. [Internet]Tokyo (Japan)2010[updated 10/27/2010; cited 11/28/2010]; Available from: http://bioomix.tmd.ac.jp/disease/i2b2/ [Google Scholar]
- 32.Wynden RW MG, Sim I, Gabriel D, Casale M, Carini S, Hastings S, Ervin D, Tu S, Gennari JH, Anderson N, Mobed K, Lakshminarayanan P, Massary M, Cucina RJ. Ontology Mapping and Data Discovery for the Translational Investigator. AMIA Summit on Clinical Research Informatics; San Francisco: 2010 [PMC free article] [PubMed] [Google Scholar]
- 33.i2b2 NCBC Optimizing Query Performance with the Ontology Total_Num field. [Internet]Boston (MA)2010[updated 10/12/2010; cited 11/28/2010]; Available from: https://community.i2b2.org/wiki/x/h4AW [Google Scholar]