Abstract
Objective
Multi-disciplinary and multi-site biomedical research programs frequently require infrastructures capable of enabling the collection, management, analysis, and dissemination of heterogeneous, multi-dimensional, and distributed data and knowledge collections spanning organizational boundaries. We report on the design and initial deployment of an extensible biomedical informatics platform that is intended to address such requirements.
Methods
A common approach to distributed data, information, and knowledge management needs in the healthcare and life science settings is the deployment and use of a service-oriented architecture (SOA). Such SOA technologies provide for strongly-typed, semantically annotated, and stateful data and analytical services that can be combined into data and knowledge integration and analysis “pipelines.” Using this overall design pattern, we have implemented and evaluated an extensible SOA platform for clinical and translational science applications known as the Translational Research Informatics and Data-management grid (TRIAD). TRIAD is a derivative and extension of the caGrid middleware and has an emphasis on supporting agile “working interoperability” between data, information, and knowledge resources.
Results
Based upon initial verification and validation studies conducted in the context of a collection of driving clinical and translational research problems, we have been able to demonstrate that TRIAD achieves agile “working interoperability” between distributed data and knowledge sources.
Conclusion
Informed by our initial verification and validation studies, we believe TRIAD provides an example instance of a lightweight and readily adoptable approach to the use of SOA technologies in the clinical and translational research setting. Furthermore, our initial use cases illustrate the importance and efficacy of enabling “working interoperability” in heterogeneous biomedical environments.
Keywords: Clinical research informatics, data access, data integration, data analysis, standards, workflow, socio-organizational issues
Introduction
The availability of scalable and extensible biomedical informatics platforms, such as those commonly associated with service oriented architectures (SOA), has been cited in numerous reports as being critical to the performance of efficient, timely, and high quality research in multi-site and multi-disciplinary settings (1–3). Such research programs frequently involve a broad variety of data and knowledge sources, stakeholders, and analytic resources (4–7). In this report we will describe the design, initial adoption, and future plans for a SOA-based biomedical informatics platform, known as the Translational Research Informatics and Data-management grid (TRIAD), developed as part of the activities of the Clinical and Translational Science Award (CTSA)-funded Center for Clinical and Translational Science (CCTS) at The Ohio State University (OSU). TRIAD leverages the caGrid middleware and extends it to support “working interoperability” and the needs of clinical and translational researchers. In addition, and as part of the TRIAD project, we have created a set of wrappers for widely used research data management tools in order to accelerate and facilitate TRIAD adoption as a platform for exchanging clinical and translational research data, information, and knowledge. We will also describe the successful use of TRIAD for biospecimen and patient cohort discovery as an exemplar deployment use case.
In an effort to make maximal reuse of available SOA components and best practices, as described above, TRIAD has been implemented as a derivative and extension of the caGrid middleware (2). TRIAD is designed to address a number of requirements related to the information management needs of domain-agnostic clinical and translational science projects (as opposed to the cancer-specific focus of caGrid), resulting in the following two major additions to the core caGrid architecture:
-
1.
openMDR, an implementation of an extensible knowledge management suite that extends the United Kingdom’s CancerGrid metadata repository (cgMDR) platform (8); and
-
2.
software components capable of publishing or consuming information via TRIAD that are contained in common clinical and translational research data management systems.
Background
The use of service-oriented architectures (SOA), such as those commonly referred to as “computational grids” or alternatively “grid computing,” has become an increasingly common means of enabling the exchange, management, analysis, and dissemination of distributed and heterogeneous biomedical data and knowledge spanning traditional internal and external organizational boundaries. It is important to note that such an approach is commonly referred to in the computational sciences as “controlled and coordinated resource sharing and resource use in dynamic, scalable virtual organizations” (9). From a historical perspective, grid computing has evolved from an initial focus on massively parallelized distributed data analysis applications to a contemporary focus on the federated discovery and retrieval of distributed data and knowledge (1, 3). For instance, the United State’s National Cancer Institute’s (NCI) Cancer Biomedical Informatics Grid (caBIG) (10) program uses a grid infrastructure named caGrid to provide a common, extensible, and collaborative platform for data and knowledge sharing across cancer researchers and institutions (2). Similarly, the United States National Center for Research Resources (NCRR) sponsored Biomedical Informatics Research Network (BIRN) (11) seeks to use a grid-based approach to establish a service-oriented data sharing framework capable of enabling the exchange of heterogeneous data collected via a variety of modalities, including data-intensive imaging technologies. More recent projects that use variants of these types of SOA approaches include:
-
1.
the Shared Health Research Information Network (SHRINE) (12) program (13); and
-
2.
SOA platforms being designed as part of the Nationwide Health Information Network (NHIN) (14) Decision Support System (15).
Within the scope of research programs that utilize these grid-based approaches there is a frequent need for mechanisms that aid in the discovery, query, and integration of heterogeneous data and knowledge resources such as data collected into electronic data capture systems, data warehouses, and project-specific databases. Often, this need is addressed by virtualizing a data source through a process of “wrapping” it with a standards-based service interface known as a façade. In the context of caGrid, this type of wrapper translates from a consistent data-source agnostic query language (i.e., Common Query Language or CQL) to a data-source specific query language (i.e., Structured Query Language or SQL, as is commonly used by relational database management systems). Individual wrappers are developed for each instance of a collection of heterogeneous data sources in order to provide a layer of homogeneity at the wrapper interface (2). A similar approach is taken in SHRINE with an aggregator-adapter model, where an aggregator service broadcasts individual queries to each adapter, which internally translates the query to a format that is compatible with the institution’s source database (22). In both cases, such wrappers and/or aggregators are used to enable this federation-façade approach to distributed or federated queries. A set of data resources are queried via a federation service that processes a given query and issues multiple component data queries to targeted data services via their wrapper interfaces. Subsequently, the federation service joins the data sets returned by the component queries. An example of such a federation service is provided in the Results section of this report.
System Rationale
Existing caGrid Design and Functionality
As previously introduced, our effort to adopt and adapt caGrid for use in the clinical and translational science domain is known as TRIAD. Our selection of caGrid as the foundation for the TRIAD project was motivated by an analysis of the four key aspects of caGrid’s design and functionality. These key aspects directly map to the requirements for data and knowledge sharing present in the contemporary clinical and translational research environment, as illustrated in ►Figure 1 and described in the following sub-sections.
Figure 1.
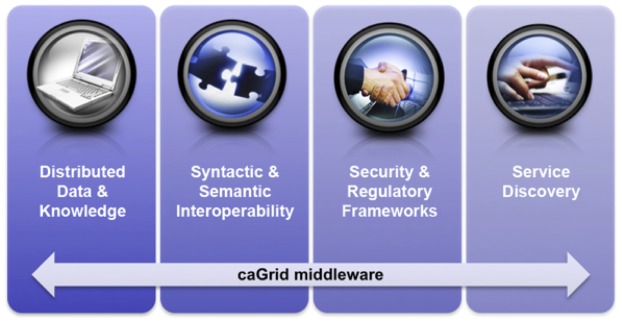
Overview of four caGrid design and key design and functional aspects that correspond to data and knowledge sharing requirements present in the contemporary clinical and translational research environment.
Distributed data and knowledge
caGrid’s basic functionality focuses upon enabling the exchange of data and knowledge, and providing access to analytical services in distributed settings. The current version of caGrid is built upon the Globus framework (16) and provides a robust SOA platform that supports the federation-wrapper model introduced earlier. The caGrid middleware is currently used in a variety of capacities at over 64 National Cancer Institute (NCI)-designated cancer centers, as well as many other basic science and clinical research organizations and consortia, such as the NCI-funded Chronic Lymphocytic Leukemia Research Consortium (17).
Syntactic and semantic interoperability
caGrid is designed to use a centrally curated, strongly-typed, predefined terminology managed and maintained by the NCI, with a specific focus on concepts used in the domain of cancer research. In caGrid, an emphasis is placed on enabling computable semantic interoperability between data and analytical services, wherein comprehensive modeling, in combination with centralized terminology and metadata management, allows for the inference of semantic interoperability by computational agents in an autonomous manner (18). In order to support computable semantic interoperability in caGrid data and analytical services, the caGrid platform provides for a full spectrum of terminology, metadata, and data model management components. These components include:
-
1.
the NCI Enterprise Vocabulary Service (EVS), a centrally curated terminology service;
-
2.
a Global Model Exchange (GME), which enables programmatic access to the logical data models employed by grid services; and
-
3.
a Cancer Data Standards Repository (caDSR), which provides access to metadata definitions that serve to link data models and terminologies in order to enable strong typing of resultant grid services.
In addition, caGrid provides a centralized index service for services to advertise metadata, including service-level semantics, in order to enable service discovery by data element definitions, semantic metadata, or other relevant identifying information.
Security and regulatory frameworks
In response to the need to exchange privileged and confidential data types in a variety of research settings, caGrid provides a set of core security and authentication/authorization services that are able to enforce policy-based data security and access controls. These services are collectively known as the GAARDS (Grid Authentication and Authorization with Reliably Distributed Services) platform. Included in the core services provided by GAARDS are the caBIG-developed Dorian, Grid Grouper, and NCI CBIIT (Center for Biomedical Informatics and Information Technology) Common Security Module (CSM). Using Dorian, user identities are managed and federated among grid participants. Grid Grouper enables individual grid services to be configured to ascribe varying levels of access to individual users based on their group membership. CSM enables the provisioning of finegrained data access policies for foundational relational data models that underlie grid data services.
Service Discovery
caGrid also provides a rich web portal interface, called the caGrid Portal, which enables users to securely discover data and analytical services, explore service metadata, execute queries using CQL, and leverage grid services published by caBIG participants.
Methods
Extending caGrid for TRIAD
When taken as a whole, the four factors described in the preceding sub-sections (distributed data and knowledge, syntactic and semantic interoperability, security and regulatory frameworks, service discovery) provide a basis for the design, deployment, and use of domain-agnostic SOA technologies in distributed clinical and translational research settings. However, there are a number of limitations of the current caGrid architectural model that must be addressed in order to adapt to the needs of such environments. In TRIAD, we have developed extensions to the caGrid middleware that provide for an extensible, standards-based, and secure data- and knowledge-sharing platform targeting domainagnostic use cases. The major differences between caGrid and TRIAD are outlined in ►Table 1 and defined within this section. A guiding principle relative to the design and deployment of TRIAD’s architecture is to enable an agile, efficient process of grid infrastructure configuration and utilization, as exemplified by the TRIAD data-service creation workflow illustrated in ►Figure 2.
Table 1.
Overview of the features of caGrid and TRIAD. Key differences between caGrid and TRIAD are listed in bold.
| Distributed Data and Knowledge | Syntatic and Semantic Interoperability | Security & Regulatory Frameworks | Socio-technical factors | Access to common translational research tools | Service Discovery & Exploration | |
|---|---|---|---|---|---|---|
| caGrid | Globus-based SOA platform |
EVS GME caDSR |
GAARDS CSM Dorian GridGrouper |
6 years of best practices | N/A | caGrid Portal |
| TRIAD | Globus-based SOA platform |
EVS GME openMDR |
GAARDS CSM Dorian GridGrouper |
Leverages existing caGrid knowledge | Wrappers for I2B2 REDCap caTissue | caGrid Portal modified for TRIAD |
Figure 2.
Overview of workflow culminating in the creation and use of TRIAD data services targeting underlying relational database constructs, involving the following major steps: 1) creation of UML models that map to existing relational data structures; 2) annotation of UML models using with semantic metadata; 3) the semi-automated generation of Java-based grid adapters using the caCORE SDK and Introduce toolkit; and 4) the implementation and deployment of resulting grid services using the TRIAD-specific instance of the caGrid middleware.
A major difference between the architecture of TRIAD and caGrid is the approach used to enable semantic interoperability and discovery. The semantic knowledge collection exposed by the TRIAD index service is cataloged, created, and curated using the openMDR repository and service, described below under “Support for working interoperbility.” OpenMDR allows a single institution to locally deploy, curate, and manage semantic knowledge and then enable this knowledge to be leveraged by multiple institutions in an ad-hoc and scalable way.
Another major differentiating feature of TRIAD is the addition of wrappers for common clinical and translational informatics research data management tools, such as i2b2 (Informatics for Integrating Biology and the Bedside) (19), the caTissue Suite (10), and REDCap (20). We discuss these wrappers in greater detail in “Facilitating access to widely adopted research data management tools.”
TRIAD’s architecture also provides a rich web portal interface built using the existing caGrid Portal software “stack” (21), which leverages the components of caGrid, in conjunction with openMDR, to allow users to securely discover new TRIAD data and analytical services, execute queries, and collaborate with other TRIAD users. This portal is highly extensible and provides an avenue for the rapid deployment of grid-enabled data-centric presentation layer components that meet the needs of motivating scientific use cases. This overall software architecture has been deployed at OSU for both internal and external use, employing a cost- and resource-efficient virtual server environment that supports the rapid re-use and deployment of service clusters by diverse sites and end-user communities. While we have deployed TRIAD and its individual services across a series of virtual machines in a virtual server environment that is running Red Hat Enterprise Linux and hosted on a high-performance, high-density blade server environment running VMware ESXi, if necessary, TRIAD can be successfully deployed on a single moderately powered computer.
In the remainder of this section, we describe our approaches to leveraging existing caGrid infrastructure and extending caGrid capabilities in order to support working interoperability and to facilitate access to widely used research data management tools as are prevalent in domain agnostic clinical and translational research use cases.
Support for working interoperability
As introduced previously, in the existing caGrid architectural model, an emphasis is placed on enabling computable semantic interoperability between data and analytical services. Unfortunately, such an approach exhibits significant scalability problems when applied beyond a well-defined domain or scope of functionality (e.g., disease or organizational context), such as that encountered in the broad clinical and translational science arena. An alternative approach to computable semantic interoperability is what is known as working interoperability, where stakeholders negotiate and use context-relevant semantic models to enable the agile deployment and use of grid services that are tailored to a specific use case or requirement. Such an approach ideally retains the ability to harmonize or federate locally relevant semantic models with external standards or models when necessary to enable more systematic exchange as is required by a given use case (18, 22–23). Based upon our analysis of local, regional, and national-scale data and knowledge sharing requirements associated with the activities of the OSU CCTS and the CTSA consortium-at-large, we believe that such a working interoperability approach is both necessary and critical to successful system adoption and use.
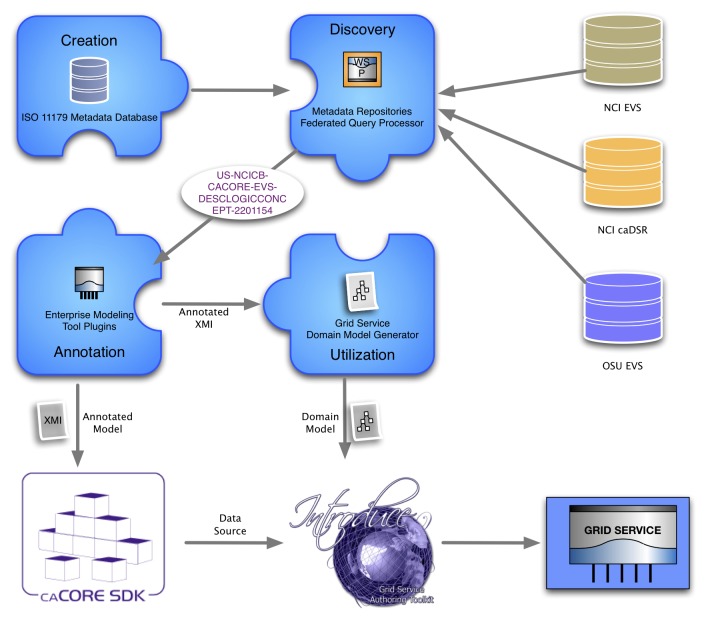
In order to facilitate a more flexible and real-time approach to supporting locally relevant semantic models, we have developed the openMDR platform (24), a suite of open-source tools that incorporates four major components, as is illustrated in ►Figure 3 and described below:
Figure 3.
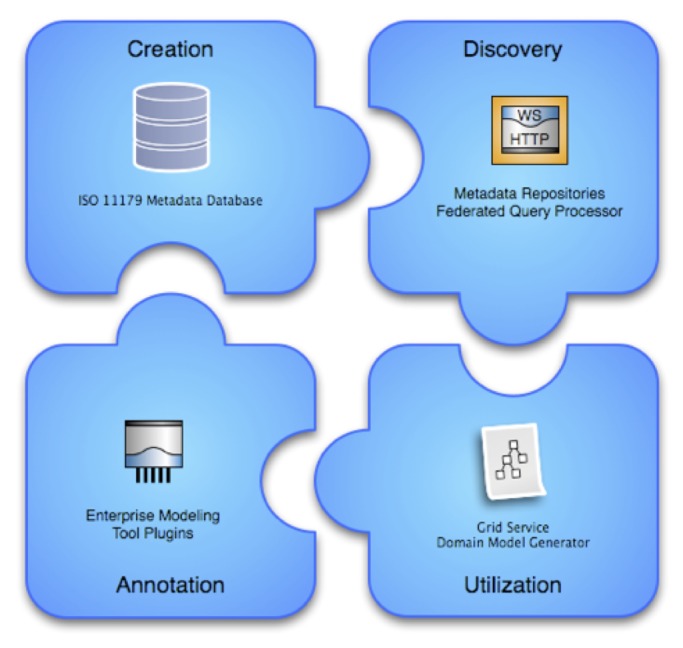
Overview of openMDR components, including an ISO 11179 compliant metadata database, enterprise modeling tools and annotation plug-ins, grid-service domain model generator, and federated metadata query processor.
-
1.
MDR Core is an ISO11179-compliant Metadata registry that stores domain specific semantic metadata for use by local institutions using an XML database (eXist) to store such information. MDR Core is capable of creating, storing, versioning, searching, and maintaining semantic metadata in the form of Common Data Elements (CDEs), accessed via a web-based user interface. The metadata stored and managed in the MDR Core is annotated with conceptual information from one or more local or remote LexEVS (25) instances.
-
2.
MDR Query is an API and TRIAD grid service that supports query and retrieval of semantic metadata from multiple repositories across the grid, including MDR Core, as well as the caDSR where applicable.
-
3.
MDR Plugin is an Enterprise Architect (EA) (26) plug-in that enables a knowledge engineer or information architect to search for and retrieve relevant semantic metadata represented as CDEs using the MDR Query service, and utilizes such search results to semantically annotate UML models that will in turn be used to create TRIAD grid services.
-
4.
MDR Domain Model Generator is a tool used by a grid service developer to generate a TRIAD-compatible domain model from a semantically annotated UML model. This component provides caGrid↔TRIAD compatibility by generating caGrid specific service metadata, an artifact that is required to create a semantically annotated caGrid data service. The TRIAD and caGrid tooling leverages this domain model to enable semantic discovery of data services across the grid by way of registration to the grid index service. This allows users of the grid to locate new and relevant sources of information and to subsequently query such resources using relevant domain models and semantics.
Facilitating access to widely adopted research data management tools
With the advent of the national CTSA initiative in the United States, there has been a marked consolidation of informatics platforms and data management tools targeting the biomedical research domain. For example, the i2b2 (19), caTissue Suite (10), and REDCap (20) applications are widely used by institutions throughout the United States to facilitate research data warehousing, biospecimen management, and electronic data capture requirements. These tools are either open-source or freely available via technology transfer mechanisms, and have large-scale adopter and adapter communities. Given the prevalence of these applications, as part of the TRIAD project we have developed a set of “wrappers” that enable “turn key” integration of such applications with the TRIAD data and knowledge-sharing infrastructure. As an example case of such efforts, the integration of i2b2 with TRIAD via the aforementioned wrapper provides institutions with an efficient way to execute integrative queries spanning research registry data stored in an i2b2 instance and complementary clinical phenotype and biomolecular data stored in other enterprise or research systems. Of note, i2b2-based TRIAD data services are able to perform queries that are defined in terms of semantic information provided by the Health Ontology Mapper (27), bridging the gap between TRIAD and i2b2 metadata management systems.
Results
Exemplar deployment use case: biospecimen and patient cohort discovery
It is common for basic, clinical, and translational researchers to use clinical and histo-pathology information to define the characteristics of tissues of interest, identify conforming samples in a tissue bank, and then go on to generate additional bio-molecular data that can be associated with such samples and correlate resultant analysis with their clinical phenotype(s). However, such operation are often complicated by the distribution and heterogeneity of the data needed to satisfy such information needs. For example, at The Ohio State University Medical Center (OSUMC), data that is pertinent to such activities exist in a variety of data resources and information systems, such as:
-
1.
multiple biospecimen management systems, such as caTissue Suite instances, which are used to store data related to the procurement, processing, and storage of tissue samples in biospecimen repositories; and
-
2.
an enteprise-wide clinical data warehouse (known locally as the Information Warehouse or IW) that contains secondary use data derived from over 70 enterprise information systems.
At present, researchers seeking to access these types of resources to identify and annotate tissue samples must complete multiple data request forms, obtain permission to access data from the Institutional Review Board (IRB), execute their queries (often involving the participation of a dedicated data analyst), and then manually integrate the resulting data sets using any number of conventional software applications (e.g., statistical analysis packages, spreadsheets, etc.). As a specific instance of these types of challenges and potential solution to them, we have focused our initial deployment and evaluation of TRIAD on a reocurring use case at OSUMC that involves the integrative query of two OSUMC based data resources:
-
1.
a caTissue Suite instance associated with the OSU Comprehensive Cancer Center’s (OSUCCC) Biospecimen & Biorepository Resource (BBR) initiative (a universal bio-specimen collection and secondary clinical data use protocol executed at the point-of-care for OSUCCC patients); and
-
2.
a “data mart” containing phenotypic data pertaining to patients who have agreed to participate in the aforementioned BBR, populated from the contents of the OSUMC Information Warehouse (IW).
At the initiation of this project, there were no direct mechanisms or tools that allowed for the query and real-time integration of data from these two resources in order to answer questions such as “How many patients do we have where ‘XYZ’ occurs within a year of diagnosis?” (wherein ‘XYZ’ might be a clinical diagnosis or treatment). This current discontinuity between such data and knowledge resources means that researchers must submit individual requests for data or tissue to biospecimen management staff and IW staff, who in turn manually query the results from both biospecimen management software systems and the data warehouse. The researcher then is required to match the individual result sets obtained to narrow down the specific population and associated tissue inventory information. The process is time consuming and takes approximately three days to reduce 3000 results to about 100 correct answers. The time and energy expended in this process does not lend itself to rapid cohort discovery, nor exploratory queries intended to establish the potential feasibility of a given hypothesis or study design.
As part of ongoing efforts to support clinical and translational research, as well as personalized healthcare delivery, OSUMC has undertaken an effort to connect these disparate sources of information. The goal of this project is to facilitate the rapid discovery of phenotypically characterized cohorts of patients and tissue samples only to users who have the privileges to access the data. To address this challenge, the TRIAD core infrastructure components have been used to rapidly create TRIAD specific data services for the IW and targeted biospecimen managements systems that seek to expose a consistent and meaningful query interface to such data resources. A TRIAD-compatible Federated Query Processor (FQP) engine capable of performing arbitrary joins (i.e., queries that do not have to adhere to a predefined set of allowable join operations) between data points across the various data services (as above) was developed and implemented. Of note, the query and join operations supported by the FQP technologies employed in this capacity are expressed using an XML-based object-oriented query laguage known as DCQL (Distributed caGrid Query Language), which is a derivative of the caGrid foundations used in TRIAD. Further collaboration with stakeholders led to the development of a set of meaningful and reusable query patterns relevant to the biospecimen research community, which was then used to optimize federated query patterns being applied to the preceding data resources. Finally, a web-based cohort discovery and data query interface was built to communicate with underlying TRIAD Security framework, data services, and FQP so that researchers could quickly and securely identify phenotypically characterized deidentified or partially-identified patient and tissue cohorts. Considering that the implementation of the system requires data instance level security where clinical researchers access data from multiple locations, the TRIAD/caGrid security framework including Dorian, Credential Delegating Service (CDS), Grid Trust Service (GTS), and Grid Grouper provides a robust infrastructure for authenticating, authorizing, managing, and federating user identities in this environment.
Clinical researchers can now use the TRIAD-based biospecimen and patient cohort discovery system to:
-
1.
Build queries from available data resources such as IW and caTissue data services,
-
2.
Select different sets of attributes for which the data is desired,
-
3.
Constrain the query by providing conditions for the data type and values of the attributes,
-
4.
Perform complex Boolean operations between different attributes,
-
5.
See a summary of the query at any point of time during the query building process, and
-
6.
Execute and view results of the newly built query.
As mentioned above, such grid tooling and wrappers are then used to deliver a portal-based interface that delivers a simple and rapid means of discovering new cohorts. An overview of the specific design and implementation pattern being used for this use case is provided in ►Figure 4.
Figure 4.
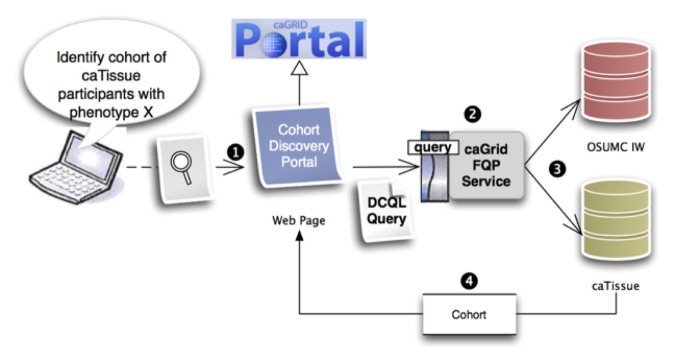
Overview of tissue cohort discovery tool implementation, in which: 1) end users pose a query via a cohort discovery portal built as a derivative of the caGRID portal platform; 2) that query is distributed and executed using Distributed Common Query Language (DCQL) via a TRIAD-specific instance of the caGrid-developed Federated Query Processor (FQP); 3) the ensuing source-specific queries, as specified via the initial DCQL statement and related semantic metadata and object modes, is executed against source systems; and 4) aggregate cohort-specific result sets are communicated to the portal interface and presented to the end user from FQP. In this example instance, phenotype data is being retrieved from the OSUMC IW, and biospecimen management data is being retrieved from a project-specific instance of caTissue Suite.
At the time of submission, this use case has been fully executed at OSUMC. TRIAD is successfully being used to manage data and knowledge related to over 20,000 distinct biospecimens and a corpus of greater than 3 million related clinical phenotype records derived from our institution’s electronic health record.
Discussion
In the preceding sections, we have described the overall state of knowledge and practice in the domain of SOA informatics platforms as applied to biomedical research, and how we have applied such principles to adopt and adapt the caGrid middleware in order to create an extensible clinical and translational research data and knowledge sharing platform known as TRIAD. This work has largely been motivated by the need to enhance the ability of caGrid to support working interoperability in complex use cases and to minimize barriers to adoption and utilization of the resultant informatics platform. In addition, we have also described an exemplar use case in which TRIAD is actively being deployed and evaluated to demonstrate the possible applications for such a platform.
We are currently conducting a series of socio-technical analyses in order to better understand the factors that may influence the successful adoption of TRIAD as a platform for clinical and translational research by the community. We anticipate that these activities will yield an understanding of best practices, policies, and procedures that can serve to address or inform responses to potential barriers to the adoption of TRIAD and their resultant workflow implications.
It should be noted that there are a number of limitations to the TRIAD project as described in this report, including:
-
1.
the number of current demonstration projects for the use of TRIAD are still somewhat limited;
-
2.
additional work must be performed to build highly-usable and configurable end-user facing interface applications that can serve as the basis for TRIAD-based data and knowledge integration and analysis “pipelines; and
-
3.
the technology “stack” used by TRIAD is closely tied to the activities of the NCI’s caBIG program, thus creating a number of external dependencies.
At a high level, it can be argued that our reliance on and extensions to the caGrid middleware as well as our choice of a working interoperability model for semantics and metadata management, may result in TRIAD inheriting many of the challenges associated with the adoption and use of caGrid, as well as a propensity for scalability concerns surrounding large volumes of distributed data. However, we believe that the incremental improvements to caGrid enabled by the TRIAD project, combined with the increased flexibility in terms of distributed and locally-relevant metadata management afforded by the use of openMDR, provides a value proposition that is capable of addressing the previously referenced barriers to adoption. Given the national scale adoption and use of TRIAD in a variety of use cases by numerous academic health centers and research organizations, we continue to explore and refine such approaches and to evaluate the resultant ability to readily adopt such a SOA platform in a timely and resource efficient manner. Such activities should ultimately enable a more comprehensive understanding of the challenges and opportunities inherent to these types of technologies in the biomedical research domain, allowing for empirically validated conclusions to be drawn concerning their usability, efficacy, and overall value relative to advances in health and life sciences research.
Given the aforementioned challenges, our future work as part of the TRIAD project will include:
-
1.
the execution and reporting of a wider variety of TRIAD deployment use cases;
-
2.
the design, deployment, and testing of a number of easily configurable end-user facing “knowledge discovery portals” that will enable data discovery, query, and pipeline/workflow execution by end-users; and
-
3.
the further refinement of the core caGrid/TRIAD middleware “stack” to enhance ease of deployment and support, including the creation of “appliance” or “cloud” deployment components and related best practices.
All of the preceding activities will be conducted with a major goal of reducing external dependencies relative to the core TRIAD middleware and increasing community-based contributions to the foundational TRIAD software architecture and its constituent components.
Conclusion
The availability of scalable and extensible biomedical informatics platforms, such as TRIAD, is central to the conduct of modern clinical and translational research programs (28–30). We believe that the dissemination of TRIAD and associated best practices will serve to greatly enhance the clinical and translational science capacity of the United States’ CTSA consortium, as well as the broader health and life science research communities. Furthermore, this effort represents a significant opportunity to overcome inherent information-centric translational barriers between basic science and clinical research (T1) as well as between clinical research and clinical practice or public health (T2) (4, 5). Ultimately, we hope to develop a broad-based consortium of TRIAD adopters and invite members of the informatics community with interests in such efforts to utilize and contribute to the TRIAD project web site and wiki (31).
Clinical Relevance Statement
The work presented will enable researchers to leverage existing clinical databases to support a wide variety of research objectives. An example of such an objective could include the creation of large-scale federated data repositories in order to conduct population-based outcomes research studies and to generate new knowledge in support of evidence-based practice.
Conflict of Interest
The authors have no conflicts of interest related to the content or findings reported in this manuscript.
Protection of Human Subjects
All human subject research conducted during the course of the study described in this manuscript was approved by the The Ohio State University Institutional Review Board, and in accordance with the World Medical Association Declaration of Helsinki on Ethical Principles for Medical Research Involving Human Subjects.
Acknowledgments
The project described in this manuscript was supported by award number U54RR024384 from the National Center for Research Resources. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Center For Research Resources or the National Institutes of Health.
References
- 1.Foster I. Service-oriented science. Science. 2005; 308(5723): 814–817 [DOI] [PubMed] [Google Scholar]
- 2.Oster S, Langella S, Hastings S, Ervin D, Madduri R, Phillips J, et al. caGrid 1.0: An enterprise grid infrastructure for biomedical research. JAMIA. 2008; 15(2): 138–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buetow K. An infrastructure for interconnecting research institutions. Drug Discovery Today. 2009; 14(11–12): 605–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Embi PJ, Payne PR. Clinical research informatics: challenges, opportunities and definition for an emerging domain. J Am Med Inform Assoc 2009; 16(3): 316–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sung NS, Crowley WF, Jr, Genel M, Salber P, Sandy L, Sherwood LM, et al. Central challenges facing the national clinical research enterprise. JAMA 2003; 289(10): 1278–1287 [DOI] [PubMed] [Google Scholar]
- 6.Butte AJ. Translational bioinformatics: coming of age. J Am Med Inform Assoc 2008; 15(6): 709–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zerhouni EA. Translational research: moving discovery to practice. Nature 2007; 81(1): 126–128 [DOI] [PubMed] [Google Scholar]
- 8.UK Cancer Grid [cited 2011 February 14]; Available from: http://www.cancergrid.org
- 9.Foster I, Kesselman CST. The anatomy of the grid: enabling scalable virtual organizations. International Journal of High Performance Computing Applications 2001; 15(3): 200–222 [Google Scholar]
- 10.caBIG Program [cited 2011 Februrary 14]; Available from: https://cabig.nci.nih.gov/
- 11.BIRN Project [cited 2011 February 14]; Available from: http://www.birncommunity.org/
- 12.SHRINE Project [cited 2011 February 14]; Available from: http://catalyst.harvard.edu/shrine/
- 13.Weber GM, Murphy SN, McMurry AJ, MacFadden D, Nigrin DJ, Churchill S, et al. The Shared Health Research Information Network (SHRINE): A prototype federated query tool for clinical data repositories. JAMIA 2009; 16: 624–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.NHIN Project [cited 2011 February 14]; Available from: http://healthit.hhs.gov
- 15.Kawamoto K, Lobach DF, Willard HF, Ginsburg GS. A national clinical decision support infrastructure to enable the widespread and consistent practice of genomic and personalized medicine. BMC Medical Informatics and Decision Making 2009; 9(16). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Globus Project [cited 2011 February 14]; Available from: http://www.globus.org/
- 17.CLL Research Consortium [cited 2011 February 14]; Available from: http://cll.ucsd.edu
- 18.Park J, Ram S. Information systems interoperability: What lies beneath? ACM Transactions on Information Systems 2004; 22(4): 595–632 [Google Scholar]
- 19.i2b2 Project [cited 2011 February 14]; Available from: https://http://www.i2b2.org/
- 20.REDCap Project [cited 2011 February 14]; Available from: http://project-redcap.org/
- 21.caBIG. caGrid Portal Bethesda MA: National Cancer Institute; 2010[cited 2010 July 22, 2010]; Available from: http://cagrid.org/display/portal/Home [Google Scholar]
- 22.Borlawsky T, Dhaval R, Hastings S, Payne PR. Development of an agile knowledge engineering framework in support of multi-disciplinary translational research. 2009 AMIA Translational Bioinformatics Summit; San Francisco: American Medical Informatics Association; 2009 [PMC free article] [PubMed] [Google Scholar]
- 23.Foster I, Kesselman C. The Grid 2: Blueprint for a new computing infrastructure. 2nd ed.New York: Morgan Kaufman; 2003 [Google Scholar]
- 24.openMDR Project [cited 2011 February 14]; Available from: http://cagrid.org/display/MDR
- 25.Pathak J, Solbrig HR, Buntrock JD, Johnson TM, Chute CG. LexGrid: A framework for representing, storing, and querying biomedical terminologies from simple to sublime. JAMIA 2009; 16(3): 305–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Enterprise Architect 8. SPARX Systems; 2010 [Google Scholar]
- 27.Health Ontology Mapper Project [cited 2011 February 14]; Available from: http://www.healthontologymapper.org/home
- 28.Brandt CA, Argraves S, Money R, Ananth G, Trocky NM, Nadkarni PM. Informatics tools to improve clinical research study implementation. Contemporary clinical trials 2006; 27(2): 112–122 [DOI] [PubMed] [Google Scholar]
- 29.Chung TK, Kukafka R, Johnson SB. Reengineering clinical research with informatics. J Investig Med 2006; 54(6): 327–333 [DOI] [PubMed] [Google Scholar]
- 30.Payne PR, Johnson SB, Starren JB, Tilson HH, Dowdy D. Breaking the translational barriers: the value of integrating biomedical informatics and translational research. J Investig Med 2005; 53(4): 192–200 [DOI] [PubMed] [Google Scholar]
- 31.TRIAD Project [cited 2011 February 14]; Available from: http://www.triadcommunity.org.