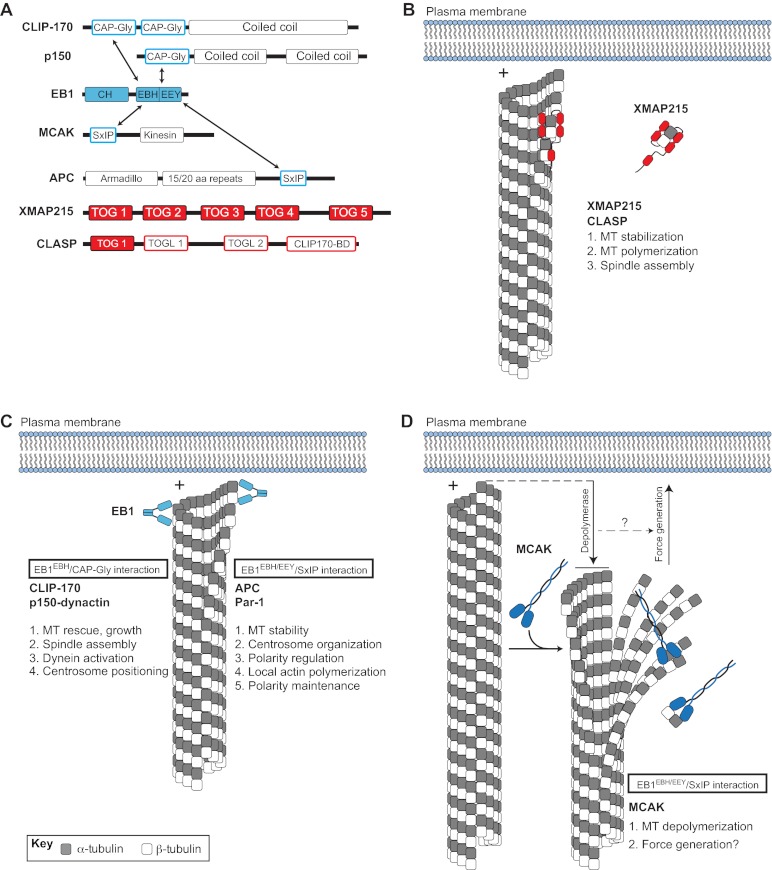
Fig. 2.
Plus-end binding proteins (+TIPs) contribute to microtubule dynamics and spindle orientation. (A) Domain architectures of various +TIPs. Arrows indicate direct interactions between +TIPs. (B) The XMAP215 and CLASP family of proteins (shown in red) are autonomously tracking +TIPs that regulate MT dynamics and spindle assembly. Interaction with microtubule (MT) plus-ends (+) occurs through direct binding of the XMAP215-CLASP TOG domain. (C) EB1 (shown in light blue) binds MT plus-ends through an N-terminal calponin homology (CH) domain and an acidic C-terminal EEY tripeptide motif. The EB1 homology domain (EBH) of EB1 also interacts with CAP-Gly domains found in CLIP-170 and p150-dynactin, thereby inducing their plus-end localization. CLIP-170 and p150-dynactin regulate MT dynamics, spindle assembly, centrosome positioning, and dynein activity. EB1 also binds a growing number of proteins containing the SxIP polypeptide motif, including APC and Par-1. The EBH/EEY domains directly bind the SxIP sequence and mediate plus-end tracking. Several members of the SxIP-containing family have been implicated in MT stability, centrosome/spindle orientation, actin polymerization, and polarity regulation/maintenance. (D) The MT depolymerase MCAK (shown in dark blue) also tracks to plus-ends through an EB1/SxIP interaction. MCAK-mediated depolymerization results in MT shortening and alteration in cortex-MT architecture (downward arrow), which may be coupled to cortical force generation, which is necessary for spindle orientation (upward arrow).