Abstract
The activity of the Notch pathway revolves around a CSL-class transcription factor, which recruits distinct complexes that activate or repress target gene expression. The co-activator complex is deeply conserved and includes the cleaved Notch intracellular domain (NICD) and Mastermind. By contrast, numerous CSL co-repressor proteins have been identified, and these are mostly different between invertebrate and vertebrate systems. In this study, we demonstrate that mammalian BEND6 is a neural BEN-solo factor that shares many functional attributes with Drosophila Insensitive, a co-repressor for the Drosophila CSL factor. BEND6 binds the mammalian CSL protein CBF1 and antagonizes Notch-dependent target activation. In addition, its association with Notch- and CBF1-regulated enhancers is promoted by CBF1 and antagonized by activated Notch. In utero electroporation experiments showed that ectopic BEND6 inhibited Notch-mediated self-renewal of neocortical neural stem cells and promoted neurogenesis. Conversely, knockdown of BEND6 increased NSC self-renewal in wild-type neocortex, and exhibited genetic interactions with gain and loss of Notch pathway activity. We recapitulated all of these findings in cultured neurospheres, in which overexpression and depletion of BEND6 caused reciprocal effects on neural stem cell renewal and neurogenesis. These data reveal a novel mammalian CSL co-repressor in the nervous system, and show that the Notch-inhibitory activity of certain BEN-solo proteins is conserved between flies and mammals.
Keywords: BEN domain, CBF1, CSL, Notch, Repression, Mouse, Drosophila
INTRODUCTION
The Notch pathway is an ancient cell communication system that is broadly utilized to determine cell fates and pattern tissues throughout the Metazoa (Lai, 2004). At its core are the transmembrane ligand and receptor pair, Delta and Notch, respectively. Their productive interaction induces Notch cleavage and nuclear translocation of the Notch intracellular domain (NICD). There, it functions as a transcriptional co-activator for a member of the CSL transcription factor family, so-named for human CBF1 (now known as RBPJ), Drosophila Suppressor of Hairless [Su(H)] and nematode LAG-1. A tripartite complex of NICD, CSL and Mastermind activates various target genes, often in a setting-specific manner. However, multiple genes encoding basic helix-loop-helix (bHLH) repressors figure prominently as conserved effectors of Notch signaling (Bailey and Posakony, 1995; Jarriault et al., 1995; Lecourtois and Schweisguth, 1995), and are activated in response to Notch signaling in diverse developmental settings.
Historically, studies of gene regulation by transcription factors in signaling pathways focused on targets that are ‘turned on’. However, growing attention has been paid to the fact that many signal-regulated transcription factors function as active repressors in the absence of pathway activity, referred to as ‘default repression’. Indeed, such dual transcriptional activity has been argued to be a fundamental attribute of signal-regulated transcription factors (Barolo and Posakony, 2002). This might serve to heighten the differential between ‘on’ and ‘off’ states of signaling, prevent spurious target gene activity, and/or help impose temporal precision on signaling.
Default repression by members of the conserved CSL transcription factor family is crucial for proper cell fate decisions mediated by Notch signaling. However, although many different CSL-interacting co-repressors have been defined in invertebrate and vertebrate systems, studies to date have not revealed strikingly conserved requirements for the various CSL co-repressors (Lai, 2002). For example, whereas Hairless is the major co-repressor for the Drosophila Su(H), acting as an adaptor protein that recruits both CtBP and Groucho repressor complexes (Morel et al., 2001; Barolo et al., 2002), mammalian Hairless proteins have not been identified. In mammals, SHARP (also known as MINT and SPEN) and CIR1 both bind the mammalian CSL protein CBF1 and recruit NcoR/SMRT (NcoR2) and histone deacetylase (HDAC) repressor complexes (Hsieh et al., 1999; Kuroda et al., 2003; Oswald et al., 2005). However, evidence that invertebrate SHARP and CIR1 proteins mediate substantial aspects of Notch target repression in vivo is limited. Recently, the histone demethylases KDM5A/Lid were reported as conserved members of CBF1-Su(H) co-repressor complexes that bind directly to these transcription factors (Moshkin et al., 2009; Liefke et al., 2010), although these chromatin factors have pleiotropic functions involving diverse DNA-binding partners such as Rb, Myc (also known as Dm in Drosophila) and PRC2 (Polycomb repressive complex 2) (Fattaey et al., 1993; Secombe et al., 2007; Pasini et al., 2008).
We recently characterized Drosophila Insensitive, a neural-specific CSL co-repressor that inhibits Notch and Su(H) target genes during multiple steps of peripheral neurogenesis (Duan et al., 2011). Because Insensitive contains a single BEN domain and lacks other motifs (save for a potential coiled-coil region), we refer to it as a ‘BEN-solo’ factor. This distinguishes it from other BEN-containing proteins that either have multiple copies of BEN and/or contain other characterized domains (Abhiman et al., 2008). Here, we analyzed the properties of the mammalian BEN-solo factor BEND6, yielding extensive parallels with Drosophila Insensitive. In vitro, BEND6 directly binds CBF1, opposes Notch-induced target transcriptional activation, inhibits Notch-maintained renewal of neural stem cells and promotes neural commitment. In vivo, BEND6 is expressed in a gradient along the embryonic neocortex with highest levels in differentiated neurons, and associates with Notch- and CBF1-regulated enhancers. We present multiple lines of evidence, using in vitro neurosphere assays and in vivo overexpression and knockdown manipulations in embryonic neocortex, that BEND6 is an endogenous inhibitor of Notch signaling that regulates neural stem cell (NSC) dynamics and neural differentiation.
MATERIALS AND METHODS
Molecular cloning
The mouse Bend6 (mBend6) open reading frame (ORF) was amplified and cloned into the EcoRI site of pCAG-IRES-GFP for in utero electroporation and into the EcoRI site of pMXIE (Hitoshi et al., 2002). The mBend6 ORF was also cloned into the pD/ENTR vector (Invitrogen) and transferred into pUAS-Myc using Gateway cloning. mBend6 short hairpin RNAs (shRNAs) were designed using PSICOLIGOMAKER 1.5. Annealed oligonucleotides were cloned into the HpaI and XhoI sites of the Lentiviral vector pLL3.7, which contains a separate CMV>DsRed marker. mBEND6 shRNA1 and shRNA2 effectively suppressed co-expressed mBEND6-V5 protein in HEK293T cells. mBEND6 shRNA3 failed to suppress mBEND6-V5 expression when used as 1:1 ratio (shRNA:mBEND6-V5) and had modest activity when used as 3:1 ratio. We used this relatively inefficient shRNA as a control in in vitro neurosphere assays, cell-pair assays and differentiation assays. Oligonucleotides are listed in supplementary material Table S1 and all plasmids were confirmed by sequencing.
Glutathione s-transferase (GST) pulldown assays
Constructs used in the GST-pulldown assay were amplified from Drosophila insv and human BEND6 (hBEND6) cDNAs with primers containing EcoRI and XhoI restriction sites (supplementary material Table S1), and cloned into the corresponding sites of pGEX5X-2. In vitro translation was performed using pET-His-Su(H) (gift of James Posakony, University of California, San Diego) or pCMV-SPORT6-CBF1 (ATCC-10437626).
GST and GST fusion proteins were expressed in Escherichia coli BL21 cells and purified on glutathione Sepharose beads (Sigma). 35S-labeled proteins were translated in vitro with TnT coupled reticulocyte lysate (Promega) and pre-cleared with GST-Sepharose (GE Healthcare). They were then incubated with GST-fusion proteins on GST Sepharose in NETN buffer (20 mM Tris pH 8.0, 100 mM NaCl, 1 mM EDTA, 0.5% NP40) for 1 hour. Sepharose beads were collected and washed four times in NETN before elution by boiling in loading buffer. Proteins were separated on 10% polyacrylamide-SDS, stained with Coomassie Blue, vacuum dried at 80°C, and exposed to a phosphorimager (Molecular Dynamics).
Luciferase assays
HeLa cells were cultured in Dulbecco’s Modified Eagle Medium (GIBCO) containing 10% fetal calf serum, 50 IU/ml penicillin and 50 μg/ml streptomycin. Transfections were performed in 96-well plates using Lipofectamine 2000 reagent (Invitrogen). For each well, 100 μl cells at 2×105/ml density were seeded 24 hours before transfection. The following DNA combinations were used: 40 ng 8xCBF1 firefly luciferase reporter (Zhou et al., 2000), 80 ng of pMXIE-Notch1ICD (Hitoshi et al., 2002) or pMXIE empty vector, 320 ng of pCMV-SPORT6-hBEND6 (American Type Culture Collection, MGC-30132) or 320 ng pBluescript plasmid for compensating the total amount of DNA. Renilla luciferase construct (10 ng) was used as internal control. Transfected cells were cultured for 24 hours before harvest. Luciferase activity was measured in cell lysates using Dual Luciferase Assays (Promega), and promoter activity was defined as the ratio between the firefly and Renilla luciferase activities.
Drosophila genetics
UAS-Myc-mBend6 was generated using standard P transformation (BestGene, Chino Hills). Other stocks used in rescue experiments were sca-Gal4, Hairless[E31] (from James Posakony), and UAS-Insv (Duan et al., 2011). Flies were cultured at 25°C.
Chromatin immunoprecipitation (ChIP)-qPCR
Dissected embryonic day (E)13.5 mouse heads, HEK293T cells (with or without mBEND6-V5), or OT11 (Cbf1 knockout) cells transfected with CBF1 and/or mBEND6-V5, were homogenized and fixed in 1.8% formaldehyde. Chromatin in lysis buffer containing 0.1% SDS was sheared to ∼0.5 kb, then treated overnight with anti-mBEND6 antibody or pre-immune serum for E13.5 material, and with anti-V5 (Invitrogen) or mouse IgG (Jackson ImmunoResearch) for HEK293T cells and OT11 cells. For each immunoprecipitation, 0.5 ml supernatant containing sheared chromatin was pre-cleared overnight with 40 μl Gammabind G agarose (GE Healthcare) coated with BSA to remove background, then treated overnight with anti-BEND6 antibody or pre-immune serum. Precipitated complexes were washed and eluted, and cross-links were reversed overnight at 65°C. After treatment with proteinase K, DNA was purified using QIAprep Spin columns (Qiagen) and recovered in 50 μl elution buffer containing RNase A.
Real-time PCR was performed on a BIO-RAD CFX96 using Power SYBR Green (Applied Biosystems) using primers listed in supplementary material Table S1. PCR was performed on 1 μl template DNA in triplicate samples, and immunoprecipitated DNA was compared against standard curves from serial dilutions of input DNA. Values were plotted as fold enrichment normalized to pre-immune reference samples, with standard deviation of triplicate samples indicated.
Mice and in utero electroporation
We generated mouse BEND6 constructs in pCAG-IRES-GFP or pCAG-IRES-DsRed (Matsuda and Cepko, 2004) and sh-mBEND6 constructs in pll3.7-IRES-DsRed (Rubinson et al., 2003). Previously described plasmids included Notch-ICD (Weng et al., 2003), DN-MAML (Maillard et al., 2004) and Hes5>DsRed (Mizutani et al., 2007). In utero electroporation into CD-1 mice was performed as described (Saito and Nakatsuji, 2001; Saito, 2006).
Briefly, timed pregnant CD-1 mice with E13.5 embryos were anesthetized using isofluorane and the uterine horns were exposed. We microinjected ∼1 μl of plasmid DNA (1-3 μg/μl) spiked with Fast Green (Sigma) into the lateral ventricles of embryos using beveled and polished glass micropipettes (Drummond Scientific). Five 50-msecond pulses of 40-45 mV with 950-msecond intervals were delivered across the uterus with two 9-mm electrode paddles positioned on either side of the embryo head, with the negative paddle upon the ventricle containing injected DNA (BTX, ECM830). The embryos were kept moist by constant bathing with 37°C PBS (pH 7.4). As standard procedure, the uterus was placed back in the abdominal cavity, the wound surgically sutured and wound clips were placed covering the incision. The animals were placed in a 28°C incubator until they recovered and visibly resumed normal activity, and we monitored their health at 24 and 48 hours post-surgery. All animal procedures were approved by MSKCC Institutional Animal Care and Usage Committee (IACUC #09-04-005).
At appropriate time points, embryos were transcardially perfused with saline PBS followed by 4% paraformaldehyde (PFA). The perfused brains were dissected, fixed overnight and sectioned coronally (80-100 μm) using a vibratome (Leica Microsystems).
Immunochemistry and confocal imaging
Mouse brain sections were incubated for 1 hour at room temperature in blocking solution [10% fetal bovine serum (FBS), 0.3% Triton X-100], then in primary antibodies at 4°C overnight. Sections were washed three times for 20 minutes each in 0.3% Triton X-100 in PBS, then incubated with fluorophore-conjugated secondary antibodies for 2 hours at room temperature. Incubation with DAPI for 5 minutes and three 20-minute 0.1% Triton X-100 washes followed. Primary antibodies were: rabbit anti-mBEND6 (1:500; custom made by Strategic Diagnostics), rabbit anti-PAX6 (1:200; Covance), rabbit anti-TBR2 (1:200; Abcam), mouse anti-TUJ1 (1:200; Covance), chicken anti-GFAP (1:500; Millipore) and goat anti-SOX2 (1:150; R&D Systems). Secondary antibodies were: donkey anti-rabbit Alexa Fluor 488 (1:500; Invitrogen), donkey anti-goat Dylight 549 (1:1000; Jackson ImmunoResearch), donkey anti-chicken Dylight 649 (1:1000; Jackson ImmunoResearch) and donkey anti-mouse Dylight 549 (1:1000; Jackson ImmunoResearch).
Neural stem cell culture, self-renewal and differentiation assays
Mouse embryonic cortices were dissected and dissociated into single cell suspensions using a fire-polished Pasteur pipette. Cells were maintained in proliferation media (STEMCELL Technologies) supplemented with 20 ng/ml epidermal growth factor (EGF; Sigma) and 10 ng/ml fibroblast growth factor (FGF; Invitrogen), and transfected using Amaxa nucleofection. For self-renewal assays, neurospheres were dissociated using Accutase (Sigma) and seeded at very low density (0.5 cell/μl) in 96-well plates. After 4 days, neurospheres were quantified, then dissociated and re-plated at 0.5 cell/μl for the second passage; this was repeated for a third passage.
For differentiation assays, we dissociated neurospheres and plated 75,000 cells on Matrigel. These were cultured in media with FGF for 2 days (to permit neuronal survival), followed by 5 days in 2% FBS (to promote astrocyte maturation). Cells were then fixed for immunostaining and quantification. For the cell pair assay, cells overexpressing the different constructs were plated at 25 cells/μl in 24-well plates. Twenty-four hours later, cells were fixed in 4% PFA for 15 minutes. We performed immunostaining using anti-PAX6 (1:250; Covance), anti-TUJ1 (1:600; Covance) and anti-GFP (1:600; Abcam).
RESULTS
BEND6 is a conserved direct co-repressor for the Notch transcription factor CBF1
Based on our recent observation that the Drosophila BEN-solo factor Insensitive (Insv) is a direct co-repressor for Su(H) (Duan et al., 2011), we investigated whether any mammalian factor might serve a similar function. BEN proteins are not very well conserved (Abhiman et al., 2008), which obscures potential orthology relationships. For example, mammalian BEND6 and Insv bear only 6.7% overall identity, with their BEN domains exhibiting 33% identity. However, as BEND6 has a similar organization to Insv, i.e. it has a C-terminal BEN domain and a predicted N-terminal coiled-coil region (Fig. 1A), we investigated its properties further. Human and mouse BEND6 are 84% identical and we performed assays with both constructs in this study; their activities proved very similar.
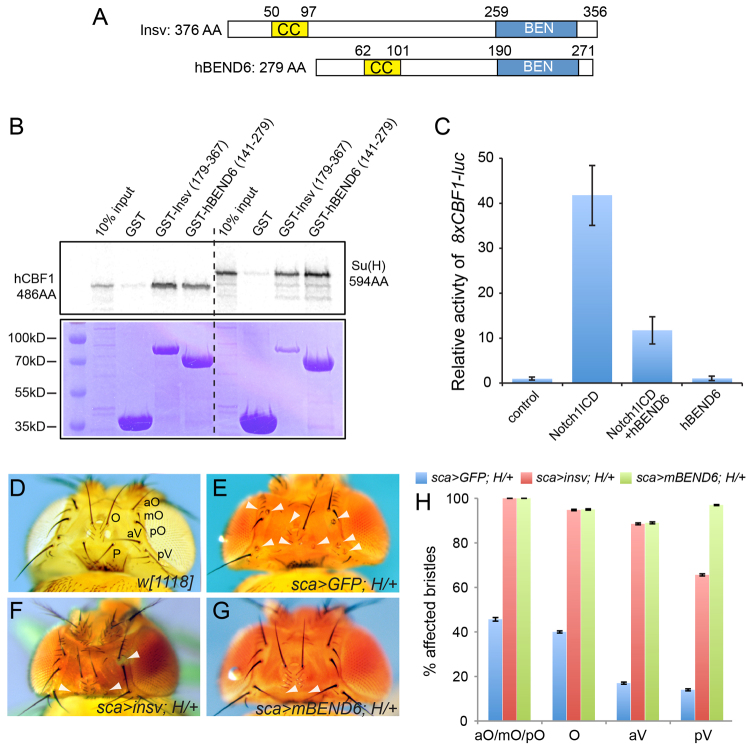
Fig. 1.
Properties of mammalian BEND6 proteins. (A) Similar domain structures of Drosophila Insensitive (Insv) and human BEND6. (B) GST pulldown assays demonstrate reciprocal cross-species interactions between the BEN domains of Insv and hBEND6 and the CSL proteins human CBF1 and Drosophila Su(H). (C) Expression of hBEND6 inhibits Notch1-ICD-mediated activation of 8xCBF1-luc in HEK293T cells. (D-G) Drosophila heads demonstrating suppression of Hairless (H) haploinsufficient phenotypes by BEN-solo factors. (D) Wild-type w[1118] head; macrochaete positions are labeled: orbital (O), anterior/medial/posterior orbital (aO/mO/pO), anterior/posterior vertical (aV/pV); postvertical (P). (E) Control H[E31]/+ head expressing UAS-GFP under control of the sca-Gal4 driver (sca>GFP) exhibits shaft-to-socket transformations (arrowheads) characteristic of H heterozygotes. (F,G) Expression of Insv (F) rescues many double sockets, as does expression of mouse BEND6 (G). (H) Quantification of shaft rescues. Error bars represent s.e.m.
We prepared a GST fusion to the BEN domain of human BEND6, and tested for interaction with 35S-labeled hCBF1, the human Notch transcription factor. We detected specific pulldown by comparison with GST alone (Fig. 1B), suggesting that BEND6 is an Insv homolog. This was bolstered by cross-species interactions amongst metazoan BEN-solo proteins and CSL-class Notch transcription factors: the BEN domain of Insv could pull down both Drosophila Su(H) and human CBF1, and the BEN domain of hBEND6 performed similarly (Fig. 1B). These findings prompted functional tests of hBEND6. We utilized an 8xCBF1-luc reporter that was induced by Notch1-ICD (subsequently referred to as NICD) when transfected in HEK293T cells. Activation by NICD was diminished by co-transfection of an hBEND6 construct (Fig. 1C), indicating that BEND6 opposes the function of activated Notch.
Finally, we performed a strict test of the functional equivalence of Insv and BEND6 using an in vivo rescue assay. Among the many developmental settings of Notch signaling, one that is very sensitive is asymmetric division of the mechanosensory organ pIIA cell, which generates socket and shaft cells that are visible on the fly cuticle (Lai and Orgogozo, 2004). Heterozygosity of the major Drosophila Su(H) co-repressor encoded by Hairless results in a mild Notch gain-of-function phenotype (Bang et al., 1991), including socket-to-shaft transformations in mechanosensory organs that are particularly evident amongst head macrochaetes (Fig. 1D,E). This was rescued using sca-Gal4 to drive expression of Insv in the peripheral nervous system (Fig. 1F). We generated an inducible transgene of mouse Bend6, and observed that it similarly rescued H/+ double-socketed organs (Fig. 1G, quantified in 1H).
These biochemical, molecular and genetic tests suggest that, despite modest overall sequence similarity, mammalian BEND6 is a functional ortholog of Drosophila Insensitive and shares activity as a CSL co-repressor.
Mouse BEND6 is a nuclear protein with neural-restricted expression
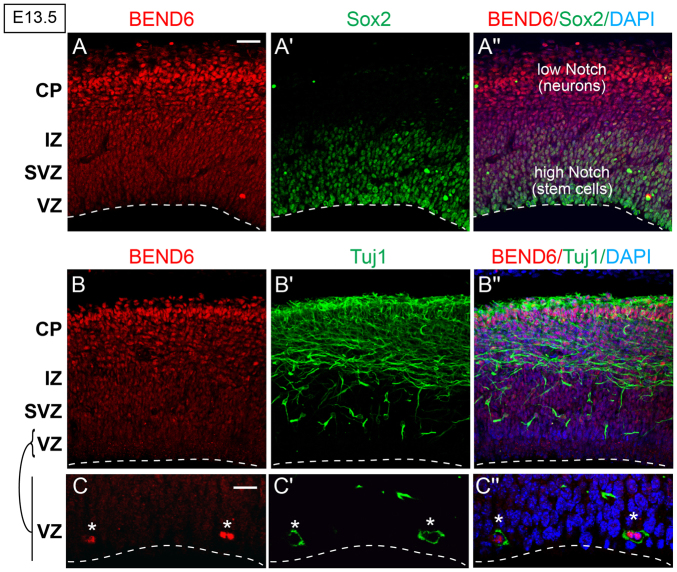
We generated a rabbit mBEND6 antibody and used it to assess mBEND6 expression. Peak periods of neocortical neurogenesis occur at E13-E14. During this time, radial glial cells (RGCs) predominantly divide asymmetrically to allow self-renewal and produce either a neuron or an intermediate progenitor cell (IPC); IPCs divide symmetrically to generate neurons (Malatesta and Götz, 2013). Neural stem cells (NSCs) and IPCs are located in the ventricular zone (VZ) and sub-VZ (SVZ), as revealed by expression of SOX2 (both NSCs and IPCs), PAX6 (NSCs) and TBR2 (EOMES - Mouse Genome Informatics) (IPCs). Neurons, as labeled by TUJ1 (TUBB3 - Mouse Genome Informatics), migrate radially from the VZ/SVZ, passing through the intermediate zone (IZ), ultimately to the cortical plate (CP).
We observed that mBEND6 is nuclear and expressed in a gradient along E13.5 neocortex, with lowest expression in the stem cell layers and highest expression in the differentiating neural layers towards the cortical plate (Fig. 2A). This gradient is opposite to SOX2 expression and to Notch activity, both of which are highest in the VZ and lowest in the CP (Fig. 2A-A″). Cells that express higher levels of mBEND6 in the IZ and CP were co-stained with the neuronal marker TUJ1 (Fig. 2B), suggesting their committed neuronal fate. Interestingly, a small population of VZ cells strongly co-stain for mBEND6 and TUJ1 (Fig. 2C). As these cells are likely to be neuronal daughter cells produced from RGC division, upregulation of mBEND6 might be an early marker for neural commitment. Notably, this characteristic also applies to Insensitive, which initiates early in the sensory organ precursor lineage and persists specifically in the cell destined to become the postmitotic neuron (Duan et al., 2011). Therefore, BEND6 not only shares molecular and functional properties with Insensitive, but it is deployed in an analogous spatial fashion.
Fig. 2.
Preferential expression of BEND6 expression in differentiating neurons in mouse embryonic neocortex. (A-C″) Images of E13.5 cortices immunostained with the antibodies against mBEND6 (A) and SOX2 (A′), or against mBEND6 (B) and TUJ1 (B′). Higher magnification images of mBEND6 and TUJ1 stainings in the VZ are shown in C-C″. A″-C″ show merged images with both markers and DAPI. Dashed lines indicate the luminal surface of the VZ. Scale bars: 50 μm in A-B″; 10 μm in C-C″.
We extended our assessment to E10, E18, postnatal day (P)10 and adult brains (supplementary material Fig. S1). Consistently, BEND6+ cells co-labeled with TUJ1 throughout all these stages, although not all TUJ1+ cells expressed mBEND6 protein. However, BEND6 was not co-expressed with GFAP, a typical astrocyte marker (supplementary material Fig. S1). Therefore, BEND6 appears to be restricted to a neuronal subpopulation and is not expressed in astrocytes.
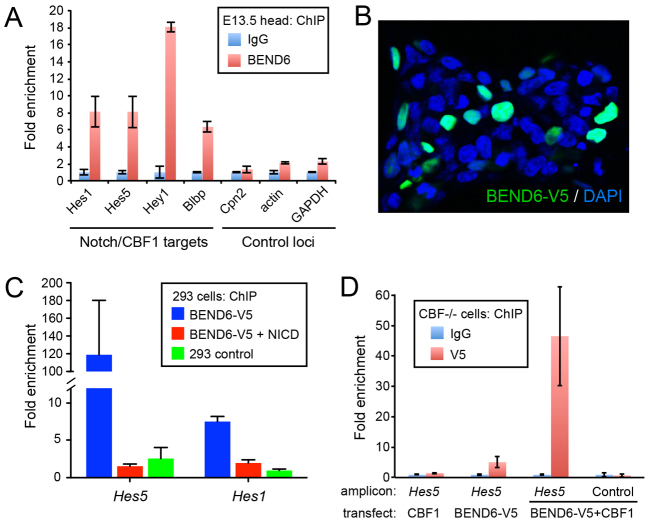
Endogenous BEND6 is localized to Notch- and CBF1-target enhancers
Drosophila Insensitive binds many Notch-regulated enhancers across the Enhancer of split-Complex [E(spl)-C] (Duan et al., 2011), including multiple bHLH repressor genes that are major conserved effectors of Notch signaling. We tested whether mouse BEND6 associates with Notch targets, by immunoprecipitating chromatin from dissected E13.5 heads with mBEND6 or IgG antibodies. BEND6 bound in vivo at regions bearing CBF1-binding sites in the Notch-regulated bHLH repressors Hes1, Hes5 and Hey1 (Fig. 3A), which are conserved Notch effectors (Jarriault et al., 1995; Cau et al., 2000; Maier and Gessler, 2000). Three control promoters, including Cpn2 (which is genomically adjacent to Hes1), actin and Gapdh, were not substantially bound by mBEND6 (Fig. 3A). Therefore, bHLH repressors are not only conserved Notch and CSL targets that execute Notch signaling, but also conserved targets of neural BEN-solo proteins in flies and mammals. We also observed specific binding of BEND6 to the Notch- and CBF1-regulated enhancer of brain lipid binding protein (Blbp; Fabp7 - Mouse Genome Informatics) (Vo et al., 2005) (Fig. 3A).
Fig. 3.
Chromatin immunoprecipitation (ChIP) of BEND6 to Notch-regulated enhancers. (A) ChIP-qPCR on E13.5 mouse head extracts show that BEND6 generates substantial signals in the vicinity of characterized CBF1-binding sites in multiple Notch targets, compared with control loci (note Cpn2 is the neighboring gene to Hes1). (B) mBEND6-V5 accumulates exclusively in nuclei of transfected HEK293T cells. (C) ChIP-qPCR of HEK293T cells transfected with NICD and/or mBEND6-V5. V5 immunoprecipitation was normalized to IgG immunoprecipitation. Robust ChIP signals in BEND6-V5-transfected cells were eliminated by introduction of NICD. (D) ChIP-qPCRs on Cbf1 knockout (-/-) cells transfected with the indicated CBF1 and/or BEND6-V5 plasmids. As before, the Hes5 amplicon contains the known CBF1 site, whereas the control amplicon is located 1 kb upstream from the CBF1 site. Mild mBEND6-V5 was observed at the Hes5 CBF1-binding site in cells expressing mBEND6-V5 alone, but this was greatly enhanced upon introduction of CBF1. Error bars represent s.e.m.
The activity of CBF1 on target genes switches depending on whether it is associated with co-repressor or co-activator molecules (Lai, 2004). We tested whether NICD could disrupt binding of BEND6 to a CBF1-regulated enhancer using the tissue culture model. A mouse BEND6 construct tagged at the C-terminus with V5 (mBEND6-V5) accumulated in the nucleus of transfected HEK293T cells (Fig. 3B). We prepared chromatin from V5 and IgG immunoprecipitates and analyzed them by qPCR for occupancy at Notch- and CBF1-regulated enhancers of Hes1 and Hes5. We observed enrichment of mBEND6-V5 by >100-fold at Hes5 and by more than sevenfold at Hes1 (Fig. 3C). The appropriate subcellular and chromatin localization of mBEND6-V5 indicated that the tag did not affect BEND6 activity. Strikingly, the enrichments at Hes1 and Hes5 were almost completely depleted by co-transfecting NICD along with mBEND6-V5 (Fig. 3C). Therefore, activated Notch displaces BEND6 from CBF1-target enhancers.
Finally, we tested whether CBF1 mediates chromatin localization of BEND6, using the Cbf1 knockout cell line OT11 (Kato et al., 1997). We compared binding of mBEND6 to the Hes5 promoter in OT11 cells transfected with CBF1 and/or mBEND6-V5. As expected, V5 antibody pulled down no DNA, unlike IgG from cells expressing CBF1 alone, and little binding was observed in OT11 cells transfected with mBEND6-V5. However, in cells expressing both CBF1 and mBEND6-V5, the association of mBEND6 to the Hes5 promoter increased to 40-fold above background (Fig. 3D). These data, along with the GST pulldown results, are consistent with the notion that CBF1 recruits BEND6 to target promoters for repression.
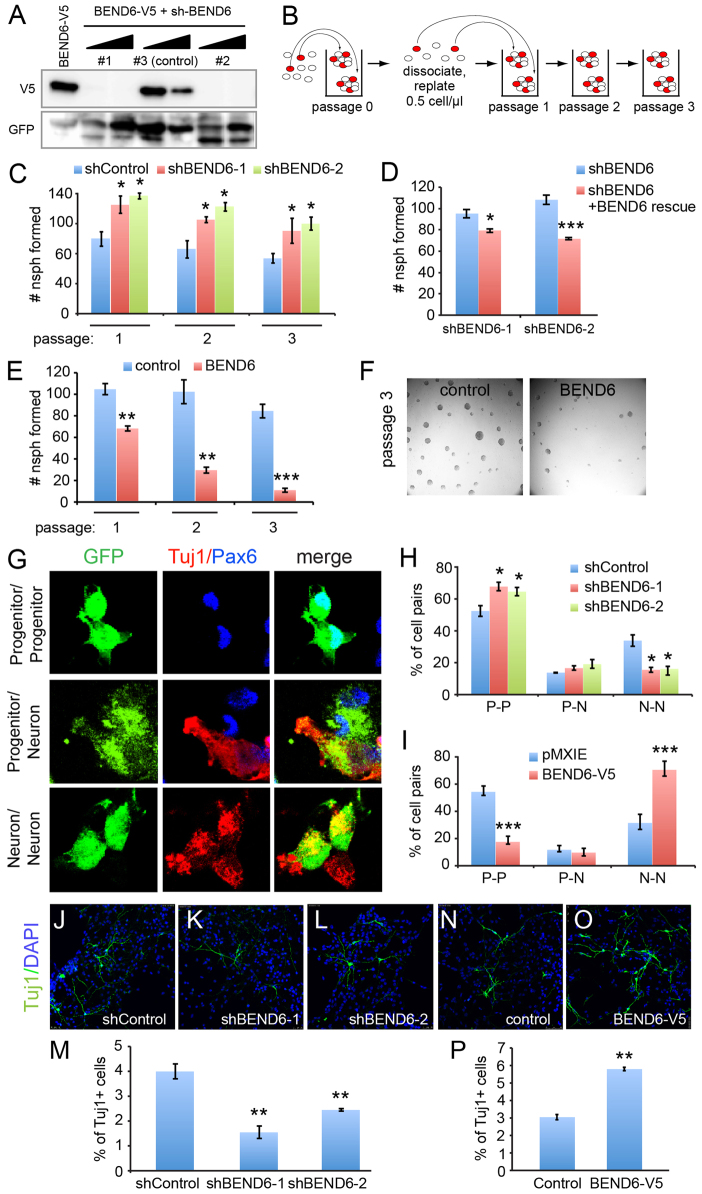
Mouse BEND6 inhibits Notch-mediated self-renewal of NSCs and promotes neurogenesis in neurospheres
Among its myriad developmental requirements, Notch signaling is crucial for maintaining the self-renewal of neural stem cells (Pierfelice et al., 2011). In neuroblasts, Notch signaling is downregulated, thereby permitting commitment to the neural fate. We tested whether BEND6, as a neural inhibitor of the Notch pathway, was able to influence neural stem cell self-renewal or differentiation. To do so, we used the mBEND6-V5 construct to evaluate several shRNAs for their knockdown capacity in transfected cells. We identified two shRNAs (shBEND6-1 and -2) that strongly and specifically suppressed mBEND6, and we utilized an ineffective sh-mBEND6 construct as a control (Fig. 4A).
Fig. 4.
Reciprocal effects of BEND6 gain and loss of function on neural stem cell self-renewal and neural differentiation in neurospheres. (A) Validation of mBEND6 shRNAs by co-transfection of HEK293T cells at 1:1 and 3:1 ratio with mBEND6-V5. sh-BEND6 constructs #1 and #2 were effective, whereas #3 was relatively inefficient and was subsequently used as a control. (B) Schematic of long-term neurosphere self-renewal assay. Red circles represent stem cells, which can self-renew and generate new neurospheres following dissociation. (C) Assays of neurospheres expressing control or active shRNAs against mouse Bend6 (n=4 animals/condition) shows that mBEND6 depletion increases self-renewal. (D) Effects of shBEND6-1 and shBEND6-2 were rescued by an shRNA-resistant BEND6 construct. (E). Enforced BEND6 expression induces progressive loss of neurosphere self-renewal (n=3 animals/condition). (F) Representative images of pMXIE- and mBEND6-V5-expressing neurospheres from passage 3. (G) Pair-cell assays illustrating the different outcomes of transfected (GFP+, green) progenitor cells undergoing symmetric or asymmetric division, as labeled by progenitor (P) marker PAX6 (blue) or neuronal (N) marker TUJ1 (red). (H) Quantification of P-P, P-N and N-N cell pairs from progenitor cells expressing shRNA control (196 cells from three animals) or mBEND6 shRNAs (201 cells from three animals). (I) Quantification of daughter cell pairs from pMXIE (235 cells from three animals) or mBEND6-V5 (213 cells from three animals) expressing progenitor cells. (J-L) Neurospheres expressing control or mBEND6 shRNAs were differentiated and stained for TUJ1 (green) and DAPI (blue). (M) Quantification of TUJ1+ cells shows that depletion of mBEND6 reduces neurogenesis (n=3 animals). (N,O) Differentiation of cells from neurospheres expressing pMXIE and mBEND6-V5. (P) Quantification of TUJ1+ cells shows that overexpression of mBEND6 increases neurogenesis (n=3 animals). All data are shown as mean±s.e.m. *P<0.05; **P<0.005; ***P<0.0005.
We isolated stem cell cultures by dissociating mouse embryonic cortices. Under proliferative conditions, these generate discrete neurospheres, the number of which provides an estimate of the stem cell content of the starting population. The capacity of these progenitor cells for long-term self-renewal can be demonstrated by serial rounds of dissociating neurospheres into single-cell suspensions, replating them, and allowing them to regenerate neurospheres (Fig. 4B). In this model, the loss of self-renewal factors or blockade of Notch pathway activity impairs the recovery of neurospheres over successive cycles of dissociation and regeneration.
We set up self-renewal assays using neurospheres transfected with control sh-mBEND6, and established a baseline for neurosphere self-renewal by plating cell suspensions at 0.5 cells/μl, and monitoring their self-renewal capacity over three serial rounds. We used these data to normalize the yield of companion neurospheres cultures expressing active sh-mBEND6. Knockdown of mBEND6 enhanced neurosphere self-renewal capacity by ∼30-40% relative to the shRNA control (Fig. 4C). We observed these effects with both on-target shRNAs, throughout several rounds of serial passaging. The effects of both sh-mBEND6 constructs were reversed by co-expression of a mBEND6 construct bearing point mutations that rendered it resistant to the shRNA, demonstrating specificity of the knockdown (Fig. 4D).
In reciprocal experiments, we analyzed cells derived from in vivo electroporated brains expressing control or mBEND6 constructs. We observed strong and progressive loss of their ability to maintain BEND6-expressing neurospheres (Fig. 4E), and these cultures were nearly extinguished by the third passage (Fig. 4F). The opposite effects from loss- and gain-of-function manipulations indicate that BEND6 inhibits self-renewal of neural stem cells.
An implication of these findings is that BEND6 might interfere with the division mode of progenitor cells, which normally regenerates a stem cell along with a differentiating cell. We assessed this using pair-cell assays (Shen et al., 2002; Szulwach et al., 2010), in which dissociated neurosphere cells are plated at low density and allowed to undergo at most a single division (24 hours after plating). At this point, the cultures are fixed and stained for a progenitor marker (PAX6) and a neural marker (TUJ1) to identify cell pairs that yield two progenitors, two neurons, or one of each (Fig. 4G). We observed that both sh-mBEND6 constructs caused ∼50% decrease in neuron-neuron pairs, with concomitant increase in progenitor-progenitor pairs (Fig. 4H). Conversely, ectopic BEND6 strongly decreased the proportion of progenitor-progenitor divisions, and increased the proportion of neuron-neuron pairs by 50% (Fig. 4I). These data reflect an endogenous contribution of mBEND6 to the control of progenitor division mode, suppressing progenitor self-renewal and favoring neural differentiation.
Finally, we analyzed the differentiation capacity of neurospheres expressing sh-mBEND6 or mBEND6. Consistent with the pair-cell assays, we observed that expression of either sh-mBEND6 construct decreased the yield of TUJ1+ cells by ∼50% compared with the control shRNA (Fig. 4J-L, quantified in 4M). By contrast, overexpression of mBEND6 doubled the proportion of TUJ1+ neurons upon differentiation of neurospheres (Fig. 4N,O, quantified in 4P). Overall, the reciprocal phenotypes from loss- and gain-of-function experiments support an endogenous Notch-inhibitory activity for mBEND6 that helps restrict NSC self-renewal and promote neurogenesis.
BEND6 inhibits NSC self-renewal and promotes neurogenesis in mouse embryonic neocortex
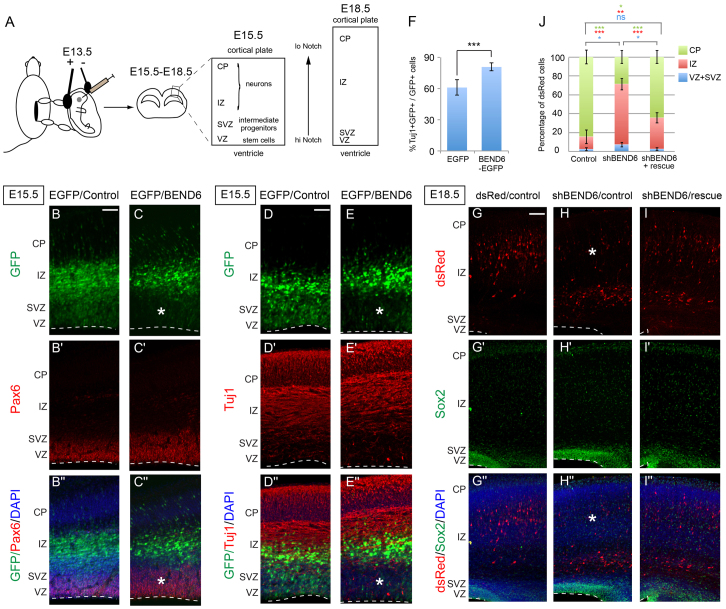
We sought to extend our findings to a genuine in vivo setting. The layered temporal progression of neocortical development facilitates investigation of the neural stem cell lineage. We used in utero electroporation to deliver control, mBEND6 or sh-mBEND6 plasmids into E13.5 ventricles, then returned these embryos to develop for additional days before processing for immunohistochemistry (Fig. 5A). During peak phases of neurogenesis at E13 and E14, RGCs in the VZ and intermediate progenitors in the SVZ divide actively to self-renew and generate neurons; progenitors in these layers are maintained by Notch activity. At later stages, progenitor cells are more committed to generate neuronal daughter cells. As the neocortex continues to develop, diminishing pools of progenitors and intermediate progenitors lead to thinning of the VZ/SVZ layers by E18, concomitant with expansion of the neuronal layers, the IZ and CP. The earlier time points are better suited for scoring effects on progenitor cell fate, whereas later time points are more appropriate for testing effects on neuronal cell fate and/or defects in neuronal migration (Fig. 5A).
Fig. 5.
Reciprocal effects of BEND6 gain and loss of function on neural stem cell self-renewal and neural differentiation in embryonic neocortex. (A) Schema for in utero electroporation and cellular analysis. Plasmids were in utero electroporated to the embryo neocortex at E13.5, and brain cortices were processed at E15.5 for staining of PAX6, GFP and DAPI (B,C) or TUJ1, GFP and DAPI (D,E) or at E18.5 for staining of dsRed, SOX2 and DAPI (G-I). (B-E″) Expression of BEND6 inhibits neural stem cell self-renewal. Constructs introduced were CAG-IRES-EGFP/CAG-IRES vector as control (B-B″,D-D″) or CAG-IRES-EGFP/mBEND6 (C-C″,E-E″). Ectopic expression of BEND6 forces cells to exit the PAX6+ VZ layer and leads to an increase in neuronal production as marked by TUJ1 staining. Asterisks indicate area in the VZ/SVZ where GFP+ cells are reduced in BEND6 overexpression. Dashed lines indicate the luminal surface of the VZ. (F) Quantification of the percentage of transfected cells that are TUJ1+ (control, 2934 cells from seven animals; BEND6, 2548 cells from seven animals). (G-I″) Knockdown of BEND6 attenuates neurogenesis/neuronal migration. Plasmids injected were pll3.7-dsRed/CAG-IRES vector as control (G-G″), shBEND6-dsRed/CAG-IRES vector (H-H″) or shBEND6-dsRed/mBEND6 rescue (I-I″). Retention of cells in the SOX2+ VZ/SVZ and IZ layers caused by sh-mBEND6 were rescued by co-injection of an shRNA-resistant mBEND6 rescue construct. Asterisks indicate area in the CP where dsRed+ cells are reduced in BEND6 knockdown. Dashed lines indicate the luminal surface of the VZ. (J) Quantification of the percentage of dsRed cells in different layers (dsRed/control, 724 cells from three animals; shBEND6/control, 362 cells from three animals; shBEND6/mBEND6 rescue, 747 cells from four animals). Data are shown as mean±s.e.m.; ns, not significant; *P<0.05; **P<0.005; ***P<0.0001. Scale bars: 100 μm in B-E″; 140 μm in G-I″.
In control electroporated brains, GFP+ cells were broadly distributed amongst the VZ, SVZ and IZ, with slightly greater accumulation in IZ (Fig. 5B). However, very few GFP+ cells reached the CP by E15.5, 2 days after electroporation. Under these conditions, a greater fraction of mBEND6-expressing cells (as marked by GFP) had exited the PAX6+ stem cell layer and were found in the IZ and CP (Fig. 5C). Furthermore, ectopic BEND6 promoted neural differentiation, as observed in an increase in GFP+/mBEND6+/TUJ1+ cells relative to control electroporated cells (Fig. 5D,E). Quantification indicated that mBEND6 expression increased TUJ1+ cells by ∼20% (Fig. 5F) and reduced GFP+ cells in the VZ/SVZ to half (40% GFP+/TBR2+ cells in control compared with 20% in mBEND6 electroporations; Fig. 6E). Therefore, ectopic BEND6 inhibited neural stem cell self-renewal and promoted neurogenesis, consistent with its activity as a Notch pathway antagonist.
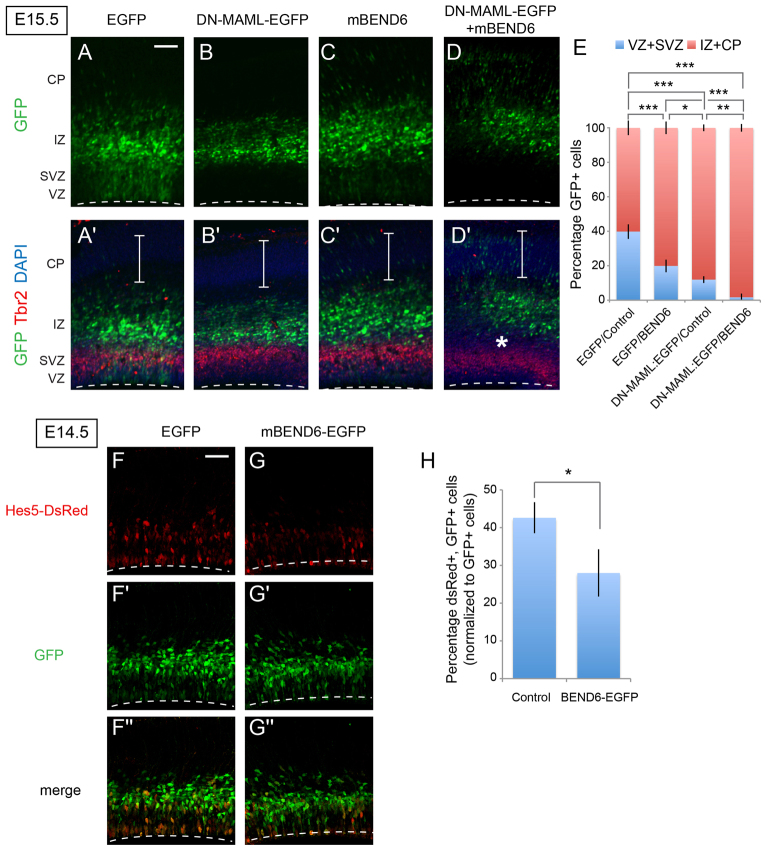
Fig. 6.
Ectopic BEND6 inhibits neural stem cell self-renewal via the Notch pathway. (A-D′) Images of mouse cortices that were electroporated in utero at E13.5 and processed at E15.5 for immunostaining to detect EGFP (green), TBR2 (red) and DAPI (blue). Constructs introduced were EGFP/CAG-IRES vector (A,A′), DN-MAML-EGFP/CAG-IRES vector (B,B′), EGFP/mBEND6 (C,C′) and DN-MAML-EGFP/mBEND6 (D,D′). Expression of DN-MAML forces cells to completely exit the VZ/SVZ into the IZ at this timeframe, and co-expression of mBEND6 pushes cells further to the CP. Asterisks indicate separation of GFP+ cells from the TBR2-labeled SVZ layer in BEND6 overexpression. Brackets indicate the CP. Dashed lines indicate the luminal surface of the VZ. (E) Quantification of the distribution of EGFP-expressing cells in the developing neocortex. (control, 4129 cells from seven animals; mBEND6, 3630 cells from seven animals; DN-MAML-EGFP/Control, 871 cells from four animals; DN-MAML-EGFP/mBEND6, 862 cells from three animals). Data are shown as mean±s.e.m.; *P<0.05; **P<0.005; ***P<0.0005. (F-G″) Images of cortices that were electroporated in utero at E13.5 and processed at E14.5 to detect GFP (green) and DsRed (red). Constructs introduced were Hes5>dsRed and EGFP (F-F″) or Hes5>dsRed and mBEND6-EGFP (G-G″). (H) Quantification of the percentage of dsRed cells versus EGFP cells in the developing neocortex. (control, 2504 cells from three animals; mBEND6, 3334 cells from four animals). Data are shown as mean±s.e.m.; *P<0.05. Scale bars: 100 μm in A-D′; 50 μm in F-G″.
We investigated next the consequences of BEND6 loss of function. We electroporated in utero two shRNAs or vector alone as control. By E18.5, the majority of dsRed+ cells migrate to the CP in control, but those expressing sh-mBEND6 were mostly retained in the IZ (Fig. 5G,H; see also similar results at E16.5 in Fig. 7A,B). We also observed that the frequency of dsRed+/sh-mBEND6 cells located in the VZ/SVZ was increased relative to control (Fig. 5G,H). To confirm that this phenotype was indeed due to knockdown of mBEND6, we generated a BEND6 rescue construct bearing mutations in the region targeted by sh-mBEND6. Co-electroporation of these constructs showed that shRNA-resistant BEND6 completely rescued defects in the VZ/SVZ and strongly suppressed the phenotype in the IZ (Fig. 5I). Quantifications of the sh-mBEND6 phenotype and its rescue are presented in Fig. 5J. We conclude that endogenous BEND6 represses NSC self-renewal and promotes neuronal differentiation/migration during neocortex development.
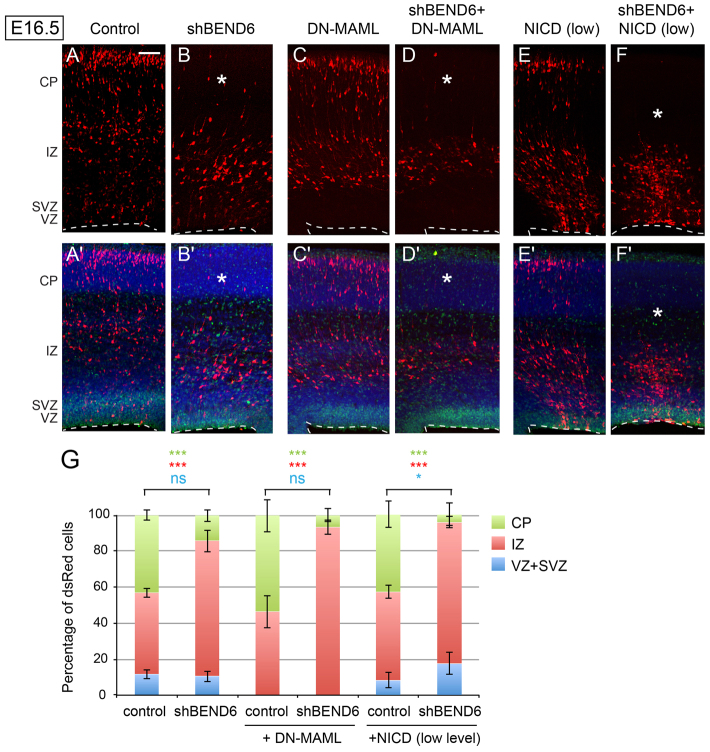
Fig. 7.
Endogenous BEND6 opposes N signaling in embryonic neocortex. (A-F′) Images of E16.5 mouse cortices expressing control (pll37-dsRed/CAG-IRES vector; A,A′) and shBEND6-dsRed/CAG-IRES vector (B,B′), DN-MAML/pll37-dsRed (C,C′), shBEND6-dsRed/DN-MAML (D,D′), NICD/pll37-dsRed (E,E′) or shBEND6-dsRed/NICD (F,F′). (B) At E16.5, shBEND6 had no effect on frequencies of cells in the VZ/SVZ, but increased the proportion of cells retained in the IZ. (C) DN-MAML forced cells to exit the VZ/SVZ and migrate to the IZ and CP. (D) Addition of shBEND6 and DN-MAML strongly reduced the proportion of cells in the CP. (E) A low level of NICD (1:10 to control or shBEND6) did not induce an obvious effect on cell fate/distribution. (F) The combination of shBEND6 and this low level of NICD depleted cells from the CP nearly completely, and caused most cells to be located in VZ/SVZ and IZ. Asterisks indicate area in the CP where dsRed+ cells are reduced in mBEND6 knockdown alone, or in combination with DN-MAML or NICD. Dashed lines indicate the luminal surface of the VZ. Scale bar: 100 μm. (G) Quantification of the percentage of dsRed cells in different layers (dsRed/control, 1632 cells from three animals; shBEND6/control, 885 cells from four animals; DN-MAMl/control, 671 cells from five animals; shBEND6/DN-MAML, 486 cells from six animals; NICD/control, 1373 cells from four animals; shBEND6/NICD, 717 cells from three animals). Data are shown as mean±s.e.m.; ns, not significant; *P<0.05; **P<0.005; ***P<0.0001.
BEND6 opposes Notch signaling to regulate NSC self-renewal
We wished to link the phenotypic influence of mBEND6 during NSC self-renewal to alteration of Notch signaling. Our tests in HEK293T cells, neurospheres and neocortex all presented a consistent picture that elevation of mBEND6 inhibits Notch signaling whereas knockdown of mBEND6 enhances Notch signaling. The effects of ectopic BEND6 resembled those of dominant-negative Mastermind-like (DN-MAML) (Maillard et al., 2004), the full-length counterpart of which is a co-activator for cleaved Notch. Using TBR2 as an SVZ marker, we observed that electroporated DN-MAML and mBEND6 both promoted exit of cells from the neural stem layer (Fig. 6A-C). Co-electroporation of mBEND6 and DN-MAML further promoted the differentiation of neural stem cells (Fig. 6D′, asterisk) and increased cortical migration of transfected cells, compared with either single electroporation (compare Fig. 6B′-D′, brackets, and quantification in 6E). The genetic enhancement of DN-MAML by mBEND6 was consistent with functional inhibition of Notch signaling by BEND6.
We tested this interpretation by analyzing a DsRed reporter under control of Hes5 regulatory sequences, including its Notch-responsive CBF1-binding sites (Mizutani et al., 2007). When electroporated in utero, the activity of Hes5>DsRed is restricted to the VZ and SVZ, where neural stem cells are actively renewing under Notch-mediated control. For these experiments, we restricted the analysis to E14.5, a time at which substantial mBEND6-overexpressing cells were still present in the VZ and SVZ where Hes5>DsRed is active. We observed that co-expression of mBEND6 resulted in a significant decrease in the expression of Hes5>DsRed, compared with co-expression of control GFP plasmid (Fig. 6F,G, quantified in 6H). These data support the notion that ectopic BEND6 inhibits NSC self-renewal by inhibiting Notch-mediated transcriptional activation.
Finally, we assessed mBEND6 knockdown for genetic interactions with the Notch pathway. As mentioned, expression of sh-mBEND6 delayed the migration of cells to the cortical plate (Fig. 7A,B), whereas introduction of DN-MAML accelerates this process concomitant with exit of NSCs from the VZ/SVZ (Fig. 7C). Co-electroporation in utero showed that sh-mBEND6 was unable to restore DN-MAML-expressing cells to the VZ/SVZ layers. Nevertheless, sh-mBEND6 suppressed the ability of cells expressing DN-MAML to reach the CP, so that almost all cells were maintained in the IZ (Fig. 7D). Therefore, depletion of endogenous BEND6 partially counteracted the consequences of expressing DN-MAML.
We also tested for genetic interactions with NICD. Considering that activated Notch is very powerful molecule, and that NICD could displace BEND6 from chromatin in transfection experiments (Fig. 3C), we titrated the amount of NICD plasmid to 1:10 with control dsRed or sh-mBEND6. Under this condition, NICD barely changed cell fate in DsRed control electroporations (Fig. 7E); although some sections showed a mild increase in VZ/SVZ-retained cells, this was not significant overall (Fig. 7G). However, co-electroporation of this low amount of NICD with sh-mBEND6 had a synergistic effect on the capacity of cells to be retained as VZ/SVZ neural stem cells or intermediate progenitors, along with strong depletion of cells from the cortical plate (Fig. 7F, compare with 7B,E). In other words, NICD is more potent in cells depleted of BEND6. Quantification of all of these interactions is shown in Fig. 7G. Altogether, these results provide compelling evidence that endogenous BEND6 antagonizes Notch pathway activity during embryonic neurogenesis.
DISCUSSION
Conserved function of neural BEN-solo proteins as CSL co-repressors
Until recently, the BEN domain was known only as a protein domain of unknown function. Nevertheless, its presence in several chromatin factors suggested a role in nuclear gene regulation (Abhiman et al., 2008). Notably, several BEN-containing proteins have been characterized as transcriptional repressors, including Drosophila Mod(mdg4) (Cai and Levine, 1997) and Insensitive (Duan et al., 2011), and vertebrate BANP (also known as SMAR1) (Rampalli et al., 2005; Pavithra et al., 2009; Sreenath et al., 2010), NAC1 (also known as NACC1) (Korutla et al., 2005; Korutla et al., 2007) and BEND3 (Sathyan et al., 2011). NAC1 and Mod(mdg4) proteins also contain a BTB/POZ domain, a protein interaction motif that in certain cases recruits histone deacetylase co-repressor complexes (Huynh and Bardwell, 1998; Wong and Privalsky, 1998; Rampalli et al., 2005). In addition, some BEN proteins that lack other recognizable domains, including BANP (which contains a single BEN domain) and BEND3 (which contains four BEN domains), also recruit histone deacetylases (Rampalli et al., 2005; Sathyan et al., 2011). Collectively, these findings indicate general attributes of BEN superfamily proteins in transcriptional repression.
We recently performed the first in vivo characterization of a BEN-solo protein, and showed that Drosophila Insensitive is a new co-repressor for the Notch transcription factor Su(H) (Duan et al., 2011). Our present work identifies striking functional similarities between Insv and the mammalian BEN-solo factor BEND6, despite their limited overall sequence homology. Working with both human and mouse BEND6 proteins, we establish the following shared characteristics of Drosophila and mammalian neural BEN-solo proteins. (1) They are nuclear proteins with restricted expression in the nervous system, and are upregulated specifically in cell types known to have downregulated Notch signaling. (2) They exhibit cross-species capacity to interact directly with the CSL transcription factors Su(H) and CBF1. (3) They share the capacity to inhibit transcriptional reporters of Notch pathway activity, both in cultured cell systems and in intact animals. (4) They antagonize Notch-mediated cell identities in the nervous system and reciprocally promote neurogenesis. (5) They associate with key Notch and CSL target genes, and their occupancy of Notch-regulated enhancers of multiple bHLH-repressor effector genes is conserved across animals. Altogether, the striking molecular and functional parallels of Drosophila Insv and mammalian BEND6 contrast with the general observation of molecular divergence in the well-characterized CSL co-repressors (Lai, 2002).
Endogenous requirement of BEND6 in the mammalian central nervous system
As is the case with Drosophila Insv (Duan et al., 2011), overexpression of BEND6 in a setting of its endogenous expression induces phenotypes that suggest decreased Notch signaling. In vitro, overexpression of mouse BEND6 reduced the self-renewal capacity of neurospheres and promoted neural differentiation. Using in utero electroporation to deliver expression constructs into the neocortex, we also observed that BEND6 induced the exit of cells from the stem cell layers and promoted their differentiation into neurons.
We performed reciprocal tests to assess the endogenous requirement of BEND6 in neurogenesis. Evidence from neurospheres showed that knockdown of BEND6 increased NSC self-renewal, reduced neural differentiation and suppressed the incidence of neuron-neuron division mode. Moreover, in vivo knockdown in neocortical cells increased the proportion of cells in the IZ at the expense of cells in the CP (by E16.5), and mildly increased the frequency of NSCs in the VZ/SVZ layers (by E18.5). These phenotypes support the notion that endogenous BEND6 contributes to NSC cell fate decisions and/or influences neuronal migration.
The insensitive mutant displays only mild Notch gain-of-function phenotypes on its own, but exhibits strong genetic interactions with the Notch pathway. For example, simply removing one copy of the Notch antagonist Hairless from insensitive mutants results in the complete failure to specify hundreds of shaft cells across the body surface, concomitant with their transformation into socket cells (Duan et al., 2011); i.e. a Notch gain-of-function phenotype. Similarly, we find that phenotypes induced by knockdown of BEND6 showed strong genetic interactions with manipulations of mammalian Notch signaling, such that sh-mBEND6 effectively suppressed the activity of DN-MAML and strongly enhanced the activity of NICD in neocortex development. On this basis, we infer that these phenotypes are at least partly due to enhancement of Notch signaling in BEND6-depleted cells, consistent with our findings from the various cell systems.
Along with our observation that mammalian BEND6 is able to rescue the gain-of-function defect seen in Drosophila Hairless/+ bristle lineages, similar to the capacity of Insv (Duan et al., 2011), the myriad shared properties of Insv and BEND6 strongly support the view that neural BEN-solo proteins have conserved roles in restricting Notch pathway activity. Finally, the high level of BEND6 present in specified neurons deserves consideration. Drosophila Insensitive is similarly an early marker of neurons in the mechanosensory lineage, but it is switched off before canonical neural markers, such as Elav, are expressed. By contrast, mouse BEND6 is maintained at a robust level throughout the cortical layer and remains co-expressed with canonical neural markers, such as TUJ1. Therefore, BEND6 might serve some function in neural differentiation or function. We note that interactions between reelin and notch were reported to control neuronal migration in cortex (Hashimoto-Torii et al., 2008), and this might potentially be influenced by BEND6. Alternatively, BEND6 might serve a Notch-independent function in committed neurons. The work described in this study provides a foundation that motivates the generation and analysis of Bend6 knockout mice, as well as searches for additional BEN proteins that might influence Notch signaling and/or neurogenesis.
Supplementary Material
Acknowledgments
We thank Diane Hayward, James Posakony, Tasuku Honjo and Rafi Kopan for reagents used in this study.
Footnotes
Funding
Q.D. was supported by a fellowship from the Swedish Research Council, C.A.A. was supported by a NYSTEM postdoctoral fellowship. Work in E.C.L.’s group was supported by the Burroughs Wellcome Fund; the Starr Cancer Consortium (I3-A139); and the National Institute of General Medical Sciences of the National Institutes of Health [1R01-NS074037]. Deposited in PMC for release after 12 months.
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.087502/-/DC1
References
- Abhiman S., Iyer L. M., Aravind L. (2008). BEN: a novel domain in chromatin factors and DNA viral proteins. Bioinformatics 24, 458–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey A. M., Posakony J. W. (1995). Suppressor of hairless directly activates transcription of enhancer of split complex genes in response to Notch receptor activity. Genes Dev. 9, 2609–2622 [DOI] [PubMed] [Google Scholar]
- Bang A. G., Hartenstein V., Posakony J. W. (1991). Hairless is required for the development of adult sensory organ precursor cells in Drosophila. Development 111, 89–104 [DOI] [PubMed] [Google Scholar]
- Barolo S., Posakony J. W. (2002). Three habits of highly effective signaling pathways: principles of transcriptional control by developmental cell signaling. Genes Dev. 16, 1167–1181 [DOI] [PubMed] [Google Scholar]
- Barolo S., Stone T., Bang A. G., Posakony J. W. (2002). Default repression and Notch signaling: Hairless acts as an adaptor to recruit the corepressors Groucho and dCtBP to Suppressor of Hairless. Genes Dev. 16, 1964–1976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai H. N., Levine M. (1997). The gypsy insulator can function as a promoter-specific silencer in the Drosophila embryo. EMBO J. 16, 1732–1741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cau E., Gradwohl G., Casarosa S., Kageyama R., Guillemot F. (2000). Hes genes regulate sequential stages of neurogenesis in the olfactory epithelium. Development 127, 2323–2332 [DOI] [PubMed] [Google Scholar]
- Duan H., Dai Q., Kavaler J., Bejarano F., Medranda G., Negre N., Lai E. C. (2011). Insensitive is a novel corepressor of Suppressor of Hairless and regulates Notch-mediated cell fate decisions in the peripheral nervous system. EMBO J. 30, 3120–3133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fattaey A. R., Helin K., Dembski M. S., Dyson N., Harlow E., Vuocolo G. A., Hanobik M. G., Haskell K. M., Oliff A., Defeo-Jones D., et al. (1993). Characterization of the retinoblastoma binding proteins RBP1 and RBP2. Oncogene 8, 3149–3156 [PubMed] [Google Scholar]
- Hashimoto-Torii K., Torii M., Sarkisian M. R., Bartley C. M., Shen J., Radtke F., Gridley T., Sestan N., Rakic P. (2008). Interaction between Reelin and Notch signaling regulates neuronal migration in the cerebral cortex. Neuron 60, 273–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitoshi S., Alexson T., Tropepe V., Donoviel D., Elia A. J., Nye J. S., Conlon R. A., Mak T. W., Bernstein A., van der Kooy D. (2002). Notch pathway molecules are essential for the maintenance, but not the generation, of mammalian neural stem cells. Genes Dev. 16, 846–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh J. J., Zhou S., Chen L., Young D. B., Hayward S. D. (1999). CIR, a corepressor linking the DNA binding factor CBF1 to the histone deacetylase complex. Proc. Natl. Acad. Sci. USA 96, 23–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh K. D., Bardwell V. J. (1998). The BCL-6 POZ domain and other POZ domains interact with the co-repressors N-CoR and SMRT. Oncogene 17, 2473–2484 [DOI] [PubMed] [Google Scholar]
- Jarriault S., Brou C., Logeat F., Schroeter E. H., Kopan R., Israel A. (1995). Signalling downstream of activated mammalian Notch. Nature 377, 355–358 [DOI] [PubMed] [Google Scholar]
- Kato H., Taniguchi Y., Kurooka H., Minoguchi S., Sakai T., Nomura-Okazaki S., Tamura K., Honjo T. (1997). Involvement of RBP-J in biological functions of mouse Notch1 and its derivatives. Development 124, 4133–4141 [DOI] [PubMed] [Google Scholar]
- Korutla L., Wang P. J., Mackler S. A. (2005). The POZ/BTB protein NAC1 interacts with two different histone deacetylases in neuronal-like cultures. J. Neurochem. 94, 786–793 [DOI] [PubMed] [Google Scholar]
- Korutla L., Degnan R., Wang P., Mackler S. A. (2007). NAC1, a cocaine-regulated POZ/BTB protein interacts with CoREST. J. Neurochem. 101, 611–618 [DOI] [PubMed] [Google Scholar]
- Kuroda K., Han H., Tani S., Tanigaki K., Tun T., Furukawa T., Taniguchi Y., Kurooka H., Hamada Y., Toyokuni S., et al. (2003). Regulation of marginal zone B cell development by MINT, a suppressor of Notch/RBP-J signaling pathway. Immunity 18, 301–312 [DOI] [PubMed] [Google Scholar]
- Lai E. C. (2002). Keeping a good pathway down: transcriptional repression of Notch pathway target genes by CSL proteins. EMBO Rep. 3, 840–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai E. C. (2004). Notch signaling: control of cell communication and cell fate. Development 131, 965–973 [DOI] [PubMed] [Google Scholar]
- Lai E. C., Orgogozo V. (2004). A hidden program in Drosophila peripheral neurogenesis revealed: fundamental principles underlying sensory organ diversity. Dev. Biol. 269, 1–17 [DOI] [PubMed] [Google Scholar]
- Lecourtois M., Schweisguth F. (1995). The neurogenic suppressor of hairless DNA-binding protein mediates the transcriptional activation of the enhancer of split complex genes triggered by Notch signaling. Genes Dev. 9, 2598–2608 [DOI] [PubMed] [Google Scholar]
- Liefke R., Oswald F., Alvarado C., Ferres-Marco D., Mittler G., Rodriguez P., Dominguez M., Borggrefe T. (2010). Histone demethylase KDM5A is an integral part of the core Notch-RBP-J repressor complex. Genes Dev. 24, 590–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier M. M., Gessler M. (2000). Comparative analysis of the human and mouse Hey1 promoter: Hey genes are new Notch target genes. Biochem. Biophys. Res. Commun. 275, 652–660 [DOI] [PubMed] [Google Scholar]
- Maillard I., Weng A. P., Carpenter A. C., Rodriguez C. G., Sai H., Xu L., Allman D., Aster J. C., Pear W. S. (2004). Mastermind critically regulates Notch-mediated lymphoid cell fate decisions. Blood 104, 1696–1702 [DOI] [PubMed] [Google Scholar]
- Malatesta P., Götz M. (2013). Radial glia - from boring cables to stem cell stars. Development 140, 483–486 [DOI] [PubMed] [Google Scholar]
- Matsuda T., Cepko C. L. (2004). Electroporation and RNA interference in the rodent retina in vivo and in vitro. Proc. Natl. Acad. Sci. USA 101, 16–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani K., Yoon K., Dang L., Tokunaga A., Gaiano N. (2007). Differential Notch signalling distinguishes neural stem cells from intermediate progenitors. Nature 449, 351–355 [DOI] [PubMed] [Google Scholar]
- Morel V., Lecourtois M., Massiani O., Maier D., Preiss A., Schweisguth F. (2001). Transcriptional repression by suppressor of hairless involves the binding of a hairless-dCtBP complex in Drosophila. Curr. Biol. 11, 789–792 [DOI] [PubMed] [Google Scholar]
- Moshkin Y. M., Kan T. W., Goodfellow H., Bezstarosti K., Maeda R. K., Pilyugin M., Karch F., Bray S. J., Demmers J. A., Verrijzer C. P. (2009). Histone chaperones ASF1 and NAP1 differentially modulate removal of active histone marks by LID-RPD3 complexes during NOTCH silencing. Mol. Cell 35, 782–793 [DOI] [PubMed] [Google Scholar]
- Oswald F., Winkler M., Cao Y., Astrahantseff K., Bourteele S., Knöchel W., Borggrefe T. (2005). RBP-Jkappa/SHARP recruits CtIP/CtBP corepressors to silence Notch target genes. Mol. Cell. Biol. 25, 10379–10390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D., Hansen K. H., Christensen J., Agger K., Cloos P. A., Helin K. (2008). Coordinated regulation of transcriptional repression by the RBP2 H3K4 demethylase and Polycomb-Repressive Complex 2. Genes Dev. 22, 1345–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavithra L., Mukherjee S., Sreenath K., Kar S., Sakaguchi K., Roy S., Chattopadhyay S. (2009). SMAR1 forms a ternary complex with p53-MDM2 and negatively regulates p53-mediated transcription. J. Mol. Biol. 388, 691–702 [DOI] [PubMed] [Google Scholar]
- Pierfelice T., Alberi L., Gaiano N. (2011). Notch in the vertebrate nervous system: an old dog with new tricks. Neuron 69, 840–855 [DOI] [PubMed] [Google Scholar]
- Rampalli S., Pavithra L., Bhatt A., Kundu T. K., Chattopadhyay S. (2005). Tumor suppressor SMAR1 mediates cyclin D1 repression by recruitment of the SIN3/histone deacetylase 1 complex. Mol. Cell. Biol. 25, 8415–8429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinson D. A., Dillon C. P., Kwiatkowski A. V., Sievers C., Yang L., Kopinja J., Rooney D. L., Zhang M., Ihrig M. M., McManus M. T., et al. (2003). A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nat. Genet. 33, 401–406 [DOI] [PubMed] [Google Scholar]
- Saito T. (2006). In vivo electroporation in the embryonic mouse central nervous system. Nat. Protoc. 1, 1552–1558 [DOI] [PubMed] [Google Scholar]
- Saito T., Nakatsuji N. (2001). Efficient gene transfer into the embryonic mouse brain using in vivo electroporation. Dev. Biol. 240, 237–246 [DOI] [PubMed] [Google Scholar]
- Sathyan K. M., Shen Z., Tripathi V., Prasanth K. V., Prasanth S. G. (2011). A BEN-domain-containing protein associates with heterochromatin and represses transcription. J. Cell Sci. 124, 3149–3163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secombe J., Li L., Carlos L., Eisenman R. N. (2007). The Trithorax group protein Lid is a trimethyl histone H3K4 demethylase required for dMyc-induced cell growth. Genes Dev. 21, 537–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q., Zhong W., Jan Y. N., Temple S. (2002). Asymmetric Numb distribution is critical for asymmetric cell division of mouse cerebral cortical stem cells and neuroblasts. Development 129, 4843–4853 [DOI] [PubMed] [Google Scholar]
- Sreenath K., Pavithra L., Singh S., Sinha S., Dash P. K., Siddappa N. B., Ranga U., Mitra D., Chattopadhyay S. (2010). Nuclear matrix protein SMAR1 represses HIV-1 LTR mediated transcription through chromatin remodeling. Virology 400, 76–85 [DOI] [PubMed] [Google Scholar]
- Szulwach K. E., Li X., Smrt R. D., Li Y., Luo Y., Lin L., Santistevan N. J., Li W., Zhao X., Jin P. (2010). Cross talk between microRNA and epigenetic regulation in adult neurogenesis. J. Cell Biol. 189, 127–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo N., Klein M. E., Varlamova O., Keller D. M., Yamamoto T., Goodman R. H., Impey S. (2005). A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis. Proc. Natl. Acad. Sci. USA 102, 16426–16431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng A. P., Nam Y., Wolfe M. S., Pear W. S., Griffin J. D., Blacklow S. C., Aster J. C. (2003). Growth suppression of pre-T acute lymphoblastic leukemia cells by inhibition of notch signaling. Mol. Cell. Biol. 23, 655–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong C. W., Privalsky M. L. (1998). Components of the SMRT corepressor complex exhibit distinctive interactions with the POZ domain oncoproteins PLZF, PLZF-RARalpha, and BCL-6. J. Biol. Chem. 273, 27695–27702 [DOI] [PubMed] [Google Scholar]
- Zhou S., Fujimuro M., Hsieh J. J., Chen L., Miyamoto A., Weinmaster G., Hayward S. D. (2000). SKIP, a CBF1-associated protein, interacts with the ankyrin repeat domain of NotchIC to facilitate NotchIC function. Mol. Cell. Biol. 20, 2400–2410 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.