Abstract
Tightly controlled DNA replication and RNA transcription are essential for differentiation and tissue growth in multicellular organisms. Histone chaperones, including the FACT (facilitates chromatin transcription) complex, are central for these processes and act by mediating DNA access through nucleosome reorganisation. However, their roles in vertebrate organogenesis are poorly understood. Here, we report the identification of zebrafish mutants for the gene encoding Structure specific recognition protein 1a (Ssrp1a), which, together with Spt16, forms the FACT heterodimer. Focussing on the liver and eye, we show that zygotic Ssrp1a is essential for proliferation and differentiation during organogenesis. Specifically, gene expression indicative of progressive organ differentiation is disrupted and RNA transcription is globally reduced. Ssrp1a-deficient embryos exhibit DNA synthesis defects and prolonged S phase, uncovering a role distinct from that of Spt16, which promotes G1 phase progression. Gene deletion/replacement experiments in Drosophila show that Ssrp1b, Ssrp1a and N-terminal Ssrp1a, equivalent to the yeast homologue Pob3, can substitute Drosophila Ssrp function. These data suggest that (1) Ssrp1b does not compensate for Ssrp1a loss in the zebrafish embryo, probably owing to insufficient expression levels, and (2) despite fundamental structural differences, the mechanisms mediating DNA accessibility by FACT are conserved between yeast and metazoans. We propose that the essential functions of Ssrp1a in DNA replication and gene transcription, together with its dynamic spatiotemporal expression, ensure organ-specific differentiation and proportional growth, which are crucial for the forming embryo.
Keywords: Ssrp1, Cell cycle, Hdac, Liver, Organogenesis, Zebrafish, Drosophila
INTRODUCTION
Expansion of progenitor pools and their tissue-specific differentiation are central for the formation of functional organs. Complex signalling cascades controlling organ growth and differentiation have been identified, but the associated basic cellular processes, such as regulation of DNA accessibility, in the developing embryo are still poorly understood. DNA accessibility, allowing the passage of RNA polymerase II and replication forks, is pivotal for coordinated gene expression and cell proliferation. Histone chaperones mediate DNA accessibility by structural destabilisation of nucleosomes and deconvolution of higher-order chromatin structure (Ransom et al., 2010). One histone chaperone, the FACT (facilitates chromatin transcription) complex consists of Spt16 (suppressor of Ty16; also known as Supt16h in zebrafish) and Ssrp1 (Structure specific recognition protein 1), which are both essential for nucleosome reorganisation and conserved across metazoans and plants (Formosa, 2008; Winkler and Luger, 2011). Metazoan Ssrp1 contains a C-terminal high mobility group box (HMGB), whereas its yeast homologue Pob3 solely encodes the N-terminal domains and the HMGB function is provided by the small independent protein Nhp6 (Brewster et al., 2001; Formosa et al., 2001; Stillman, 2010). Both yeast Spt16 and Pob3 are required for cell viability, whereas Nhp6 is not essential (Malone et al., 1991; Wittmeyer et al., 1999; Brewster et al., 2001), questioning the importance of the HMGB domain of Ssrp1 for FACT function in metazoans. In mice, Ssrp1 loss results in lethality around implantation (Cao et al., 2003). Consequently, insights into Ssrp1 and FACT functions during later stages of vertebrate embryogenesis are limited.
Here, we report that the FACT component Ssrp1a is essential for tissue differentiation and growth in vertebrate organogenesis. In the developing zebrafish liver and eye, zygotic loss of Ssrp1a results in impaired RNA transcription, defective S phase progression and cell death. Using cross-species rescue experiments, we examine the importance of the HMGB domain of Ssrp1a, as well as paralogous Ssrp1b, in metazoan organogenesis.
MATERIALS AND METHODS
Fish stocks
The following strains were bred under standard conditions (Westerfield, 2000): Tg(XIEef1a1:GFP)s854 (Field et al., 2003), clampeds819, clampedu428, cecyil: Tg(Efα:mKO2-zCdt1(1/190))rw0405b; Tg(Efα:mAG-zGem(1/100))rw0410h (Sugiyama et al., 2009) and tp53zdf1 (Berghmans et al., 2005) (for genotyping details, see Zebrafish International Resource Center).
Positional cloning of clampeds819
Meiotic mapping followed standard protocols (Geisler, 2002). Sibling and cmps819 mutant cDNAs were sequenced for ssrp1a, rad9a, zgc:113229, med19a and slc43a1a. For genotyping of homozygous cmps819 embryos, dCaps Finder 2.0 (http://helix.wustl.edu/dcaps/dcaps.html) was used to design primers creating a HinfI restriction site including the ssrp1as819 mutation. The following primers were used for PCR amplification from genomic DNA: forward 5′-AGCTCATTAGCACACAGATGG-3′ and reverse 5′-CAGTTCCCCAATTCCAGCCCTTGAC-3′. PCR amplification and subsequent HinfI digest from homozygous mutant, wild-type and heterozygous embryos confirmed the identified lesion (supplementary material Fig. S1). ‘MO-ssrp1a’ (MO-ATG-ssrp1a, 5′-CGTTAAACTCCAGAGTGTCTCCCAT-3′; Gene Tools) and standard ‘MO-control’ were injected into one-cell-stage wild-type or Tg(XIEef1a1:GFP)s854 zebrafish.
Zebrafish immunostaining and in situ hybridisation
Labelling was performed as described (Ober et al., 2006). The following antibodies were used: rabbit anti-Prox1 (1:1000; Chemicon), mouse anti-2F11 (1:1000; gift from J. Lewis, Cancer Research UK, London, UK), mouse anti-BrdU [1:20; Developmental Studies Hybridoma Bank (DSHB)] and rabbit anti-cleaved Caspase 3 (1:75; Cell Signaling Technology). Fluorescently conjugated secondary antibodies were obtained from Jackson Laboratories. Staining was visualised with a Zeiss LSM5Pascal Exciter confocal microscope. Images were processed and cell numbers determined with Volocity image analysis software (Improvision).
The following probes were used for in situ hybridisation: ath5 (also known as atoh7) (Masai et al., 2000), ceruloplasmin (Korzh et al., 2001), ccne (Shkumatava et al., 2004), foxa1 (Odenthal and Nüsslein-Volhard, 1998), group specific component (Noël et al., 2010), mdm2 (Chen et al., 2005), p21 [cyclin-dependent kinase inhibitor 1a (cdkn1a)] (Chen et al., 2005), pcna (Leung et al., 2005) and ptf1 (Lin et al., 2004).
BrdU incorporation assay
Embryos were incubated for 30 minutes at 28.5°C in 10 mM 5-bromo-deoxyuridine (BrdU) with 15% DMSO in embryo medium (5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2, 0.33 mM MgSO4) and prior to immunolabelling were treated with 2 M HCl for one hour at room temperature.
Global RNA labelling
Embryos were incubated in 10 mM 5-ethynyluridine (EU; Invitrogen) with 15% DMSO in embryo medium for one hour at 28.5°C. Click-iT detection was performed following manufacturer’s instructions (Invitrogen).
Mitotic index analysis
Foregut-containing tissue was isolated manually in Hank’s balanced salt solution with 5% fetal calf serum on ice and passaged through a 40 μm filter. Single-cell suspensions were stained with Vybrant DyeCycle Ruby (Invitrogen) and analysed by fluorescence-activated cells sorting (FACS) (Calibur, Becton Dickinson). Raw data were analysed by FlowJo software.
Drosophila molecular biology, genetics and histology
Zebrafish genes were cloned into a modified pKC26 UAS vector containing an attB site and a 3′ V5 epitope-tag sequence. Plasmids were inserted into the attP site-containing locus VIE-260b to generate transgenic lines (for details, see Hadjieconomou et al., 2011). Genetic crosses are summarised in supplementary material Table S2. Dissected tissues were processed as described (Hadjieconomou et al., 2011) and labelled using mouse mAb24B10 (1:75; DSHB), rat anti-Elav (1:50; DSHB), rabbit anti-PH3 (1:100; Millipore/Upstate), rabbit anti-aPKC ς (1:100; sc-216, Santa Cruz) and mouse anti-V5 (1:500; Invitrogen) and fluorescently labelled F(ab′)2 fragments from Jackson ImmunoResearch Laboratories as secondary antibodies. Images were collected with a Bio-Rad/Zeiss Radiance2100 or a Leica TCS SP5II confocal microscope. Samples for scanning electron microscopy were prepared using standard procedures (Cheyette et al., 1994) and imaged with a JEOL 35CF microscope.
qPCR
Quantitative PCR (qPCR) was performed using primer sets for ssrp1a and ssrp1b (ssrp1a1F 5′-CCTCATCCTCCTGTTCTCCAAA-3′, ssrp1aR 5′-TCTCCACCTCATCTTCGCTCAT-3′; ssrp1bF 5′-TCCGGCTCTCTCTATGAGATGG-3′, ssrp1bR 5′-TGTGATCTTCCTGTTGACCAAGG-3′; rpl13F, 5′-TCTGGAGGACTGTAAGAGGTATGC-3′; rpl13R, 5′-AGACGCACAATCTTGAGAGCAG-3′). mRNA was extracted from five to ten embryos and processed. qPCR (60°C annealing temperature) was carried out using ABsolute SYBR Green Mix (Thermo) on the ABI Prism 7000 RTPCR Detection system. Expression values were normalized using rpl13 and compared using Student’s t-test. Unless otherwise indicated, three independent experiments were assessed with three samples each.
RESULTS AND DISCUSSION
The clampeds819 mutation disrupts organ growth and differentiation
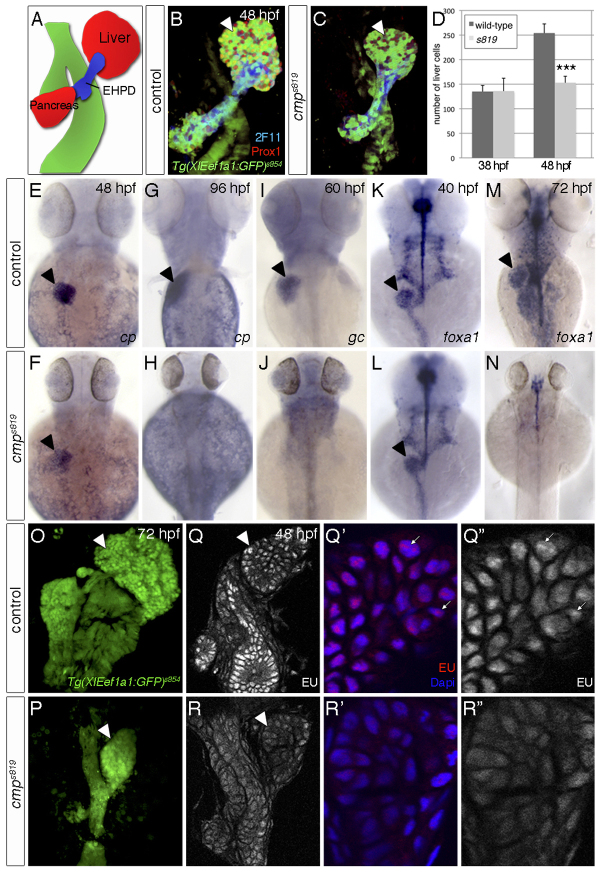
The clampeds819 (cmps819) mutant was previously identified in a forward genetic screen for factors essential for endodermal organogenesis (Ober et al., 2006). cmps819 embryos display liver and pancreas hypoplasia at 48 hours post-fertilisation (hpf; Fig. 1A-D), and smaller fins, head and eyes from 40 hpf (Fig. 2B-E). Liver progenitors arise from the foregut endoderm and aggregate into the liver bud (Field et al., 2003). Concomitant with liver-bud outgrowth, hepatoblasts begin to differentiate into functional hepatocytes and biliary epithelial cells. From about 50 hpf, the liver grows rapidly. Quantification of hepatic progenitors in cmps819 mutants using Prox1 expression showed a 40% reduction at 48 hpf (Fig. 1B-D). ceruloplasmin (cp), which encodes a plasma protein in differentiating hepatoblasts (Korzh et al., 2001), is expressed in cmps819 mutant livers at 48 hpf, but is not maintained at 96 hpf (100%, n=18 and n=10, respectively; Fig. 1E-H). In addition, group specific component (gc), marking late hepatocyte differentiation (Noël et al., 2010), was not detected in cmps819 mutants (100%, n=15; Fig. 1I-J). Hence, hepatic differentiation fails to progress in cmps819 mutants. Moreover, pan-endodermal expression of the transcription factor foxa1 in cmps819 mutants is similar to wild type at 40 hpf (100%, n=12; Fig. 1K,L), but undetectable at 72 hpf (100%, n=9; Fig. 1M,N). This is due to defects in transcription and not tissue loss, as Tg(XIEef1a1:GFP)s854 labels the digestive system in controls and cmps819 mutants at 72 hpf (100%, n=10; Fig. 1O,P). In wild-type embryos, EU labelling of newly synthesised RNA (Jao and Salic, 2008) was detected in all nuclei with higher levels in transcriptionally active sites at 48 hpf (Fig. 1Q-Q″). These foci were largely absent in cmps819 mutants and overall EU incorporation levels were lower (Fig. 1R-R″), indicating a general defect in RNA transcription. Consistently, actively transcribed genes with a short mRNA half-life, such as those encoding cyclins (Ohtani et al., 1995), represent some of the first genes exhibiting mRNA transcription defects (supplementary material Fig. S4L,M). Altogether, this suggests that cmps819 affects a key factor mediating gene transcription in the differentiating zebrafish liver.
Fig. 1.
cmps819 controls endodermal differentiation and global transcription. (A) Schematic of endodermal organs. (B,C) Confocal projections of endodermal Tg(XlEef1a1:GFP)s854, hepatic and pancreatic Prox1 and 2F11 in the extrahepatopancreatic ducts (EHPD) show liver and pancreas hypoplasia in cmps819 mutants. (D) Prox1-positive hepatoblasts are significantly reduced in cmps819 embryos at 48 hpf; data show mean±s.e.m., ***P=1.2-5. (E-H) At 48 hpf, cp is expressed in cmps819 livers (E,F), but undetectable at 96 hpf (G,H). (I,J) gc expression is absent in cmps819 embryos at 60 hpf. (K-N) foxa1 is expressed throughout the cmps819 endoderm at 40 hpf (K,L) and is absent at 72 hpf, whereas residual foxa1 is detectable in neural tissues (M,N). (O,P) Tg(XIEef1a1:GFP)s854 labels the digestive system in sibling and cmps819 embryos at 72 hpf. (Q-R″) EU incorporation reveals reduced RNA transcription in the digestive system of cmps819 mutants at 48 hpf. Using the same confocal settings, magnifications of representative sections show fewer EU-positive foci (arrows) in cmps819 livers compared with controls. A-C,O-R″ are ventral views; E-N are dorsal views; all show anterior to the top. Arrowheads indicate liver.
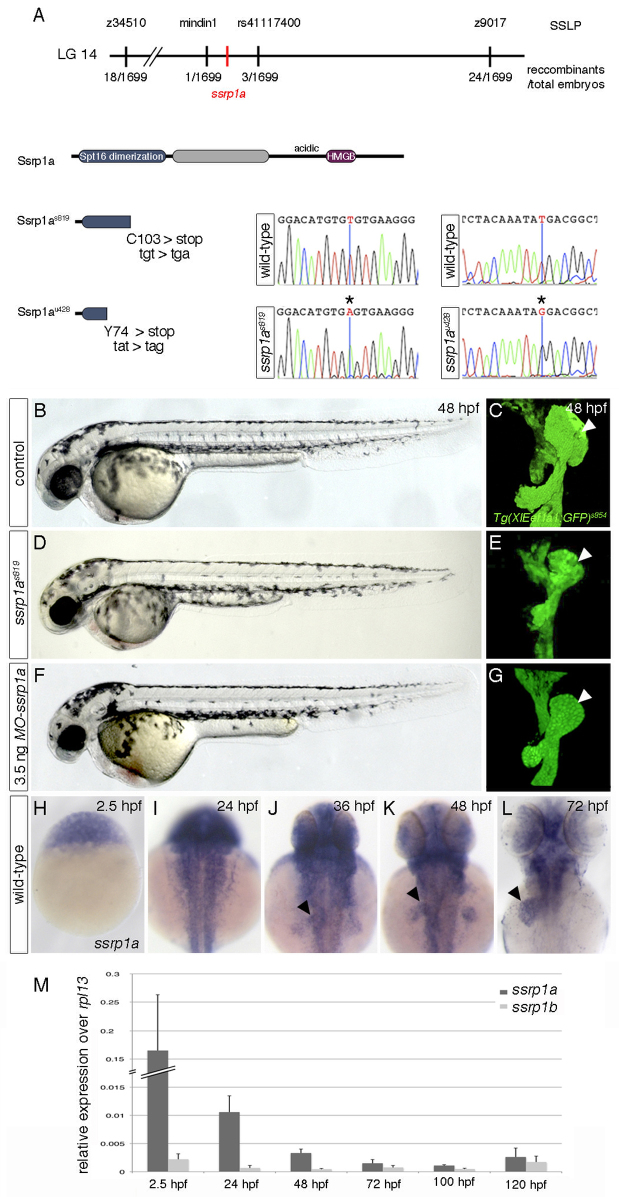
Fig. 2.
ssrp1a is disrupted in cmp mutants. (A) Genetic map of the cmp-containing region. s819- and u428-lesions in ssrp1a are shown at the nucleotide and protein level. (B-G) MO-ssrp1a-injected embryos (F,G) phenocopy cmps819 defects (D,E). Lateral views show smaller heads and eyes in mutant (D) and MO-ssrp1a embryos (F) compared with controls (B). Confocal projections of Tg(XIEef1a1:GFP)s854 highlight liver (arrowheads) and pancreas hypoplasia in cmps819 (E), and MO-ssrp1a injected embryos (G) compared with controls (C); ventral views, anterior to the top. (H-L) ssrp1a expression in wild-type embryos between 2.5 and 72 hpf. Dorsal views, anterior to the top. After 24 hpf, ssrp1a is enriched in the liver (arrowheads), eyes and fins. (M) qPCR analyses show dynamic ssrp1a and ssrp1b expression levels between 2.5 and 120 hpf. Error bars represent s.d.
clamped mutations disrupt an orthologue of Ssrp1
Meiotic recombination mapping placed the cmps819 mutation on linkage group 14 in a 340-kb genomic region between markers mindin1 (spon2a) and rs411174000 (primers amplifying a ∼160 bp product in an intron of ccng1: F: 5′-GCATAAAACGTCTCTGTGTGC-3′; R: 5′-GCAGAGAGGA GATGAGGAAAC-3′) (Fig. 2A). Sequencing of five candidate genes within this region revealed a T-to-A base pair substitution at position 309 of ssrp1a in cmps819 mutants, resulting in a premature stop codon and, thus, a truncated protein of 102 amino acids (aa), terminating halfway in the N-terminal Spt16-dimerisation domain (Fig. 2A). The cmps819 lesion was confirmed in mutants by a derived cleaved amplified polymorphic sequences (dCaps) assay (supplementary material Fig. S1A). Moreover, the mutant u428 failed to complement cmps819 and contained a T-to-G base pair substitution at aa 221 in ssrp1a (R. Young, H. Stickney, T. Hawkins, F. Cavodeassi, G. Gestri, S. Wilson and E.A.O., unpublished; Fig. 2A). Both cmp mutations are predicted to produce functionally inactive proteins, compromising FACT complex formation and function (Keller and Lu, 2002).
Knockdown of Ssrp1a by injection of morpholino antisense oligonucleotides (MOs) into Tg(XIEef1a1:GFP)s854 embryos phenocopied cmp mutants, with ∼80% of embryos showing digestive system hypoplasia (n=9) and 90% displaying smaller eyes, head and fins (n=126; Fig. 2B-G). Consistent with the RNA transcription defects in cmp mutants, previous studies implicated Ssrp1 function in transcription (Orphanides et al., 1999; Belotserkovskaya et al., 2003; Saunders et al., 2003; Bai et al., 2010). Based on these validations, the cmp mutants are referred to as ssrp1as819 and ssrp1au428. All experiments were performed with ssrp1as819, as both alleles display comparable phenotypes.
ssrp1a is expressed ubiquitously in the early embryo and from ∼36 hpf in proliferative tissues, such as the liver, eyes and fins (Fig. 2H-L). Quantitative RT-PCR (qPCR) analysis revealed high maternal ssrp1a mRNA levels (2.5 hpf), whereas zygotic expression is much lower (>3 hpf; Fig. 2M). Inhibiting translation of maternal ssrp1a mRNA by MO injection into Tg(XIEef1a1:GFP)s854 embryos does not produce stronger phenotypes compared with ssrp1as819 mutants (Fig. 2B-G), suggesting that maternal Ssrp1a protein compensates for Ssrp1a loss during early development. Alternatively, the zebrafish Ssrp1 paralogue Ssrp1b (81% identity) might act redundantly (supplementary material Fig. S1B), as it is expressed throughout development, albeit at up to 98% lower levels than ssrp1a (Fig. 2M).
Ssrp1a promotes cell cycle progression in the digestive system and eye
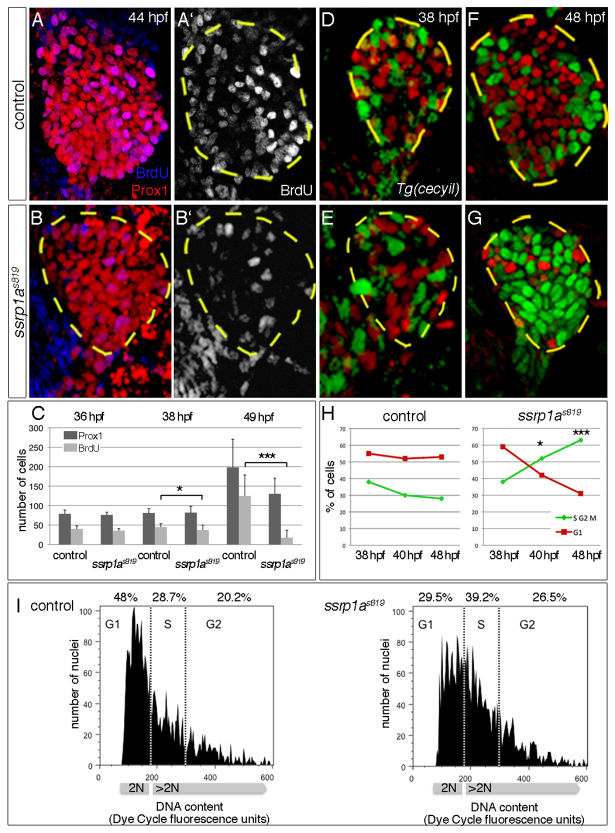
Hepatic growth arrests in ssrp1as819 mutants at ∼48 hpf. BrdU incorporation experiments revealed that at 36 hpf proliferation rates in ssrp1s819 mutants (n=10) are similar to those in controls (n=13), but are reduced by ∼10% at 38 hpf and by 87% at 49 hpf (n≥10; Fig. 3A-C). This indicates that Ssrp1a promotes hepatic cell divisions. To determine the Ssrp1a-dependent cell cycle phase, mutant and sibling livers were analysed in the transgenic cecyil background labelling S/G2/M phase cells in green and G1 phase cells in red (Sugiyama et al., 2009). At 38 hpf, 55-60% hepatoblasts are found in G1 phase and ∼40% in S/G2/M phase in controls and ssrp1as819 mutants (Fig. 3D,E,H). This distribution changes significantly in ssrp1as819 mutants by 48 hpf, with 30% of cells in G1 phase and 62% in S/G2/M phase (n≥7; Fig. 3F-H), indicating that Ssrp1a-deficient hepatoblasts accumulate in S/G2/M phase. Next, we examined single cell suspensions from the foregut domain of Tg(XIEef1a1:GFP)s854 embryos using FACS. Measuring the DNA content at 42-44 hpf revealed 18.5% fewer Ssrp1a-deficient endodermal cells in G1 phase and, conversely, 10.5% more in S phase compared with wild type (Fig. 3I). This indicates that Ssrp1a is required for S phase progression during development, consistent with in vitro results reporting Ssrp1 functions in DNA elongation (Abe et al., 2011).
Fig. 3.
Ssrp1a promotes cell cycle progression. (A-C) BrdU incorporation is reduced in ssrp1as819 mutant livers from 38 hpf onwards (outlined). Error bars represent s.d. (D-G) Transgenic cecyil expression marks G1 phase cells in red and S/G2/M phase cells in green. (H) Quantification of hepatoblast proliferation shows a reversed distribution in controls and ssrp1as819 mutants at 48 hpf. (I) FACS analysis of 42-44 hpf foregut endoderm shows an increase of cells in S phase and a decrease of those in G1 phase in ssrp1as819 mutants. A-B′,D-G are confocal projections of ventral views; all anterior to the top. *P<0.05, ***P<0.0005, determined by unpaired Student’s t-test.
Our data indicate that Ssrp1a-deficient cells accumulate in S phase, probably owing to more cells entering or stalling in S phase. A role for Ssrp1a during DNA synthesis, rather than at subsequent checkpoints prior to mitosis, is corroborated by fewer cells incorporating BrdU, and the intact, not polyploid appearance of nuclei in ssrp1as819 livers (Fig. 1Q′,R′). FACT interacts with the minichromosome maintenance (MCM) helicase in initiation and elongation of DNA synthesis (Gambus et al., 2006; Tan et al., 2006) and, consistent with this, cell cycle defects in ssrp1as819 mutants resemble the ones in Mcm5-depleted zebrafish embryos (Ryu et al., 2005). Intriguingly, Spt16-depleted yeast exhibits a G1 phase delay, pointing to specific functions of either FACT component (Morillo-Huesca et al., 2010). Similarly, human SSRP1 and SPT16 (SUPT16H) can regulate largely overlapping, but also distinct targets (Li et al., 2007).
To determine the specificity of different chromatin-remodelling factors on organ growth, we examined hdac1s436 mutants exhibiting similar liver size defects (Noël et al., 2008). Hdac1 mediates chromatin compaction and cell cycle progression from G1 to S phase (Yamaguchi et al., 2010), raising the possibility that its loss might counterbalance ssrp1as819 phenotypes. cecyil-based cell cycle analysis at 60 hpf showed that in hdac1s436 mutants 68% of cells are in G1 phase and 15% in S/G2/M phase (supplementary material Fig. S2A-E), suggesting that Hdac1 promotes cell cycle progression in liver organogenesis. Ssrp1a knockdown in hdac1s436 mutants partially rescues both individual phenotypes, with 26% cells in S/G2/M phase and 55% in G1 phase (supplementary material Fig. S2A-E), whereas neither organ size, nor hepatic gene expression improves (supplementary material Fig. S2F; data not shown). This is unexpected because in yeast impaired Ssrp1/Pob3 and Hdac/Rpd3(L) functions partially rescue gene transcription and cell viability compared with sole loss of Pob3 (Formosa et al., 2001). This suggests that both factors control different targets mediating cell viability in yeast and zebrafish liver growth.
Compared with the liver, Ssrp1a is required earlier in retina differentiation and proliferation (∼36 hpf; supplementary material Fig. S3), indicating similar, but temporally distinct, requirements for zygotic Ssrp1a in a tissue-specific manner. Liver and eye progenitors lacking Ssrp1a undergo apoptosis following S phase defects (supplementary material Fig. S3K-M and Fig. S4). Thus, in ssrp1as819 mutants, incomplete DNA synthesis may trigger a DNA-damage response. Indeed, increased Tp53-target gene expression (Vogelstein et al., 2000) (supplementary material Fig. S4H-K) and a partial rescue of cell death in ssrp1a;tp53 mutants (supplementary material Fig. S4F,G) showed that Tp53-dependent and -independent signalling is activated in Ssrp1a-deficient embryos. Human FACT activates TP53 following DNA damage (Keller et al., 2001) and in Ssrp1-deficient mice apoptosis is solely mediated by Tp53-independent pathways (Cao et al., 2003). By contrast, our data in zebrafish suggest that cell death is not mediated exclusively via Tp53, but might include alternative pathways, highlighting the complexity of the molecular mechanisms following replication stress in different vertebrates.
Full-length ssrp1a and ssrp1b, as well as ssrp1a1-518, can compensate for the loss of Drosophila Ssrp
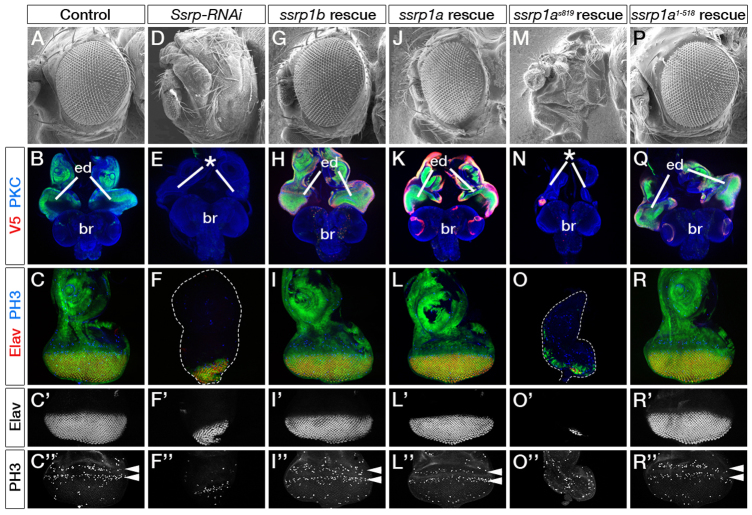
To elucidate the role of the HMGB domain, we performed cross-species rescue experiments using the Drosophila eye. The fly genome has one Ssrp gene (Shimojima et al., 2003), which encodes an HMGB-containing protein, similar to zebrafish Ssrp1a and Ssrp1b (supplementary material Fig. S1). Knockdown of Ssrp in the fly eye using a UAS-SsrpRNAi transgene caused severe defects (Fig. 4D-F″; supplementary material Fig. S6B,B′). Male pharate adults failed to eclose, and lacked eyes, antennae and head capsules, whereas 5.5% of females hatched, and displayed partial head capsules with absent or small eyes. Consistently, eye-antennal imaginal discs of third instar larvae were significantly smaller. Within the eye field, probably because of incomplete knockdown, few cells were phospho-Histone H3 (PH3) positive, and thus mitotically active, and few expressed Elav and mAb24B10 as indicators of photoreceptor (R-cell) differentiation. Upon overexpression of zebrafish full-length ssrp1b, full-length ssrp1a, ssrp1as819 or HMGB-deficient ssrp1a1-518, males and females hatched with fully developed heads and eyes, and proliferation and differentiation of eye-antennal imaginal discs proceeded normally (supplementary material Figs S5-S7 and Table S1). Strikingly, when Drosophila SsrpRNAi was co-expressed with zebrafish ssrp1b, ssrp1a or ssrp1a1-518, the defects caused by Ssrp knockdown were substantially rescued (Fig. 4G-L″,P-R″; supplementary material Figs S6, S7). The majority of adult females and males hatched and displayed normal heads and eyes (supplementary material Table S1), apart from males rescued by ssrp1b and ssrp1a1-518, which exhibited slightly rough eyes. Proliferation and R-cell differentiation in eye-antennal discs were indistinguishable from controls. Hence, zebrafish ssrp1b, ssrp1a and ssrp1a1-518 can substitute for Drosophila Ssrp. By contrast, truncated ssrp1as819 did not rescue Drosophila Ssrp knockdown phenotypes (Fig. 4M-O″; supplementary material Fig. S6H,H′ and Table S1), indicating that it is neither functional nor acts as a dominant-negative fragment. Importantly, these findings argue that in zebrafish, Ssrp1b could perform all Ssrp1 functions, but probably fails to replace zygotic Ssrp1a owing to low expression levels. Moreover, because Ssrp1a1-518 can replace Drosophila Ssrp, our data indicate that the C-terminal HMGB-domain is dispensable in vivo. This is consistent with similar observations in vitro (Abe et al., 2011), and the fact that the yeast homologue Pob3 lacking an endogenous HMGB-domain interacts with the HMGB proteins Nhp6a and Nph6b (Wittmeyer et al., 1999). Also, other proteins can compensate for their function, as yeast lacking both factors are viable (Costigan et al., 1994). Likewise, Ssrp1a1-518 could interact with other HMGB proteins to carry out FACT functions, as zebrafish and Drosophila genomes each contain at least four HMGB-containing polypeptides (Ragab et al., 2006) (Ensembl Zv9; http://www.ensembl.org/Danio_rerio/Info/Index). Our in vivo findings suggest that, despite the presence of an endogenous HMGB domain, Drosophila, and probably metazoans in general, have maintained the molecular components for Pob3-like FACT function. Hence, the basic mechanism of histone reorganisation between unicellular and multicellular organisms may be conserved independently of gene fusion/separation events associated with Ssrp1 evolution.
Fig. 4.
ssrp1b, ssrp1a and ssrp1a1-518 substitute Drosophila Ssrp function in the eye. (A-R″) For full genotypes and sample numbers, see supplementary material Table S1. (A,D,G,J,M,P) Scanning electron micrographs of leftward-facing adult female Drosophila heads. (B,E,H,K,N,Q) Confocal images of female third instar larval eye-antennal discs (ed) and brains (br) labelled with anti-PKC (blue). GFP signals indicate the areas of transgene expression, and V5 labelling (red) the presence of Danio rerio Ssrp1b-V5, Ssrp1a-V5, Ssrp1as819-V5 or Ssrp1a1-518-V5. Anterior is up. (C-C″,F-F″,I-I″,L-L″,O-O″,R-R″) Eye-antennal discs and single channels of eye fields labelled with anti-Elav (red or white), visualising R-cell differentiation, and anti-PH3 (blue or white), monitoring two mitotic waves (arrowheads). (A-C″) Controls show adult eyes with a regular array of ommatidia and eye-imaginal discs with differentiated R-cells and two waves of dividing cells. (D-F″) Drosophila Ssrp knockdown causes the loss of adult eyes, and eye-antennal discs are significantly smaller (asterisk in E). (G-R″) ssrp1b (G-I″), ssrp1a (J-L″) and ssrp1a1-518 (P-R″) expression rescues eye defects caused by SsrpRNAi. Expression of ssrp1as819 (M-O″) fails to rescue. Asterisk in N indicates eye-antennal disc defects. (F,O) Dashed lines outline eye-antennal discs.
Ssrp1a has essential functions in basic cellular processes, including DNA synthesis and gene transcription. We propose that the spatiotemporal regulation of ssrp1a expression and the aforementioned functions are pivotal for achieving the appropriate rate of cell division and growth of each tissue, ensuring the correct shape, size and organ proportions in the developing embryo. Three lines of evidence indicate that maternal and zygotic Ssrp1a function in a specific rather than ubiquitous fashion in zebrafish embryogenesis. First, the onset of zygotic defects in ssrp1as819 mutants is tissue specific. Phenotypes in the eye precede those in the liver but at the same stage no phenotypes are apparent in the somites, despite an earlier phase of extensive proliferation (Sugiyama et al., 2009), suggesting that these dynamics cannot be explained solely by protein turnover associated with replication. This is consistent with specific requirements for Ssrp1a in plant development (Lolas et al., 2010). Second, zygotic ssrp1a-expression domains are dynamic over time and often spatially restricted, including presumptive progenitor populations. This is in line with histone chaperones performing functions central to the properties of progenitor populations. Indeed, zebrafish with germ cells lacking maternal and zygotic Ssrp1a develop into sterile males, indicative of essential Ssrp1a functions in germ cell formation (K.K. and E.A.O., unpublished). Third, the developmental defects in ssrp1a mutants differ significantly in their timely appearance and severity from those in mutants carrying lesions in other genes performing similar fundamental cellular functions (Ryu et al., 2005).
In summary, our study uncovers essential functions of Ssrp1a in ensuring coordinated replication and RNA transcription in vertebrate embryos, underscoring the complex interplay between chromatin state and gene expression programmes during organ differentiation and growth.
Supplementary Material
Acknowledgments
We thank the NIMR aquatics team for fish care; D. Stainier, H. Field and P. Dong for joining forces for the LiverplusScreen; and A. Miyawaki, A. Sakaue-Sawano, J. Fischer, T. Hawkins, G. Kassiotis, D. Wilkinson, the Bloomington Drosophila Stock Center and the DSHB (University of Iowa) for reagents or discussions.
Footnotes
Funding
This work was funded by the Medical Research Council [U117581329 to K.K., D.S. and E.A.O.; and U117581332 to H.A. and I.S.]. Deposited in PMC for release after 6 months.
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.093583/-/DC1
References
- Abe T., Sugimura K., Hosono Y., Takami Y., Akita M., Yoshimura A., Tada S., Nakayama T., Murofushi H., Okumura K., et al. (2011). The histone chaperone facilitates chromatin transcription (FACT) protein maintains normal replication fork rates. J. Biol. Chem. 286, 30504–30512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai X., Kim J., Yang Z., Jurynec M. J., Akie T. E., Lee J., LeBlanc J., Sessa A., Jiang H., DiBiase A., et al. (2010). TIF1gamma controls erythroid cell fate by regulating transcription elongation. Cell 142, 133–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belotserkovskaya R., Oh S., Bondarenko V. A., Orphanides G., Studitsky V. M., Reinberg D. (2003). FACT facilitates transcription-dependent nucleosome alteration. Science 301, 1090–1093 [DOI] [PubMed] [Google Scholar]
- Berghmans S., Murphey R. D., Wienholds E., Neuberg D., Kutok J. L., Fletcher C. D., Morris J. P., Liu T. X., Schulte-Merker S., Kanki J. P., et al. (2005). tp53 mutant zebrafish develop malignant peripheral nerve sheath tumors. Proc. Natl. Acad. Sci. USA 102, 407–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewster N. K., Johnston G. C., Singer R. A. (2001). A bipartite yeast SSRP1 analog comprised of Pob3 and Nhp6 proteins modulates transcription. Mol. Cell. Biol. 21, 3491–3502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao S., Bendall H., Hicks G. G., Nashabi A., Sakano H., Shinkai Y., Gariglio M., Oltz E. M., Ruley H. E. (2003). The high-mobility-group box protein SSRP1/T160 is essential for cell viability in day 3.5 mouse embryos. Mol. Cell. Biol. 23, 5301–5307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Ruan H., Ng S. M., Gao C., Soo H. M., Wu W., Zhang Z., Wen Z., Lane D. P., Peng J. (2005). Loss of function of def selectively up-regulates Delta113p53 expression to arrest expansion growth of digestive organs in zebrafish. Genes Dev. 19, 2900–2911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheyette B. N., Green P. J., Martin K., Garren H., Hartenstein V., Zipursky S. L. (1994). The Drosophila sine oculis locus encodes a homeodomain-containing protein required for the development of the entire visual system. Neuron 12, 977–996 [DOI] [PubMed] [Google Scholar]
- Costigan C., Kolodrubetz D., Snyder M. (1994). NHP6A and NHP6B, which encode HMG1-like proteins, are candidates for downstream components of the yeast SLT2 mitogen-activated protein kinase pathway. Mol. Cell. Biol. 14, 2391–2403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field H. A., Ober E. A., Roeser T., Stainier D. Y. (2003). Formation of the digestive system in zebrafish. I. Liver morphogenesis. Dev. Biol. 253, 279–290 [DOI] [PubMed] [Google Scholar]
- Formosa T. (2008). FACT and the reorganized nucleosome. Mol. Biosyst. 4, 1085–1093 [DOI] [PubMed] [Google Scholar]
- Formosa T., Eriksson P., Wittmeyer J., Ginn J., Yu Y., Stillman D. J. (2001). Spt16-Pob3 and the HMG protein Nhp6 combine to form the nucleosome-binding factor SPN. EMBO J. 20, 3506–3517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambus A., Jones R. C., Sanchez-Diaz A., Kanemaki M., van Deursen F., Edmondson R. D., Labib K. (2006). GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat. Cell Biol. 8, 358–366 [DOI] [PubMed] [Google Scholar]
- Geisler R. (2002). Mapping and cloning. In Zebrafish: A Practical Approach (ed. Nusslein-Volhard C., Dahm R.), pp. 175–212 Oxford: Oxford University Press; [Google Scholar]
- Hadjieconomou D., Rotkopf S., Alexandre C., Bell D. M., Dickson B. J., Salecker I. (2011). Flybow: genetic multicolor cell labeling for neural circuit analysis in Drosophila melanogaster. Nat. Methods 8, 260–266 [DOI] [PubMed] [Google Scholar]
- Jao C. Y., Salic A. (2008). Exploring RNA transcription and turnover in vivo by using click chemistry. Proc. Natl. Acad. Sci. USA 105, 15779–15784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller D. M., Lu H. (2002). p53 serine 392 phosphorylation increases after UV through induction of the assembly of the CK2.hSPT16.SSRP1 complex. J. Biol. Chem. 277, 50206–50213 [DOI] [PubMed] [Google Scholar]
- Keller D. M., Zeng X., Wang Y., Zhang Q. H., Kapoor M., Shu H., Goodman R., Lozano G., Zhao Y., Lu H. (2001). A DNA damage-induced p53 serine 392 kinase complex contains CK2, hSpt16, and SSRP1. Mol. Cell 7, 283–292 [DOI] [PubMed] [Google Scholar]
- Korzh S., Emelyanov A., Korzh V. (2001). Developmental analysis of ceruloplasmin gene and liver formation in zebrafish. Mech. Dev. 103, 137–139 [DOI] [PubMed] [Google Scholar]
- Leung A. Y., Leung J. C., Chan L. Y., Ma E. S., Kwan T. T., Lai K. N., Meng A., Liang R. (2005). Proliferating cell nuclear antigen (PCNA) as a proliferative marker during embryonic and adult zebrafish hematopoiesis. Histochem. Cell Biol. 124, 105–111 [DOI] [PubMed] [Google Scholar]
- Li Y., Zeng S. X., Landais I., Lu H. (2007). Human SSRP1 has Spt16-dependent and -independent roles in gene transcription. J. Biol. Chem. 282, 6936–6945 [DOI] [PubMed] [Google Scholar]
- Lin J. W., Biankin A. V., Horb M. E., Ghosh B., Prasad N. B., Yee N. S., Pack M. A., Leach S. D. (2004). Differential requirement for ptf1a in endocrine and exocrine lineages of developing zebrafish pancreas. Dev. Biol. 270, 474–486 [DOI] [PubMed] [Google Scholar]
- Lolas I. B., Himanen K., Gronlund J. T., Lynggaard C., Houben A., Melzer M., Van Lijsebettens M., Grasser K. D. (2010). The transcript elongation factor FACT affects Arabidopsis vegetative and reproductive development and genetically interacts with HUB1/2. Plant J. 61, 686–697 [DOI] [PubMed] [Google Scholar]
- Malone E. A., Clark C. D., Chiang A., Winston F. (1991). Mutations in SPT16/CDC68 suppress cis- and trans-acting mutations that affect promoter function in Saccharomyces cerevisiae. Mol. Cell. Biol. 11, 5710–5717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masai I., Stemple D. L., Okamoto H., Wilson S. W. (2000). Midline signals regulate retinal neurogenesis in zebrafish. Neuron 27, 251–263 [DOI] [PubMed] [Google Scholar]
- Morillo-Huesca M., Maya D., Muñoz-Centeno M. C., Singh R. K., Oreal V., Reddy G. U., Liang D., Géli V., Gunjan A., Chávez S. (2010). FACT prevents the accumulation of free histones evicted from transcribed chromatin and a subsequent cell cycle delay in G1. PLoS Genet. 6, e1000964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noël E. S., Casal-Sueiro A., Busch-Nentwich E., Verkade H., Dong P. D., Stemple D. L., Ober E. A. (2008). Organ-specific requirements for Hdac1 in liver and pancreas formation. Dev. Biol. 322, 237–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noël E. S., Reis M. D., Arain Z., Ober E. A. (2010). Analysis of the Albumin/alpha-Fetoprotein/Afamin/Group specific component gene family in the context of zebrafish liver differentiation. Gene Expr. Patterns 10, 237–243 [DOI] [PubMed] [Google Scholar]
- Ober E. A., Verkade H., Field H. A., Stainier D. Y. (2006). Mesodermal Wnt2b signalling positively regulates liver specification. Nature 442, 688–691 [DOI] [PubMed] [Google Scholar]
- Odenthal J., Nüsslein-Volhard C. (1998). fork head domain genes in zebrafish. Dev. Genes Evol. 208, 245–258 [DOI] [PubMed] [Google Scholar]
- Ohtani K., DeGregori J., Nevins J. R. (1995). Regulation of the cyclin E gene by transcription factor E2F1. Proc. Natl. Acad. Sci. USA 92, 12146–12150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orphanides G., Wu W. H., Lane W. S., Hampsey M., Reinberg D. (1999). The chromatin-specific transcription elongation factor FACT comprises human SPT16 and SSRP1 proteins. Nature 400, 284–288 [DOI] [PubMed] [Google Scholar]
- Ragab A., Thompson E. C., Travers A. A. (2006). High mobility group proteins HMGD and HMGZ interact genetically with the Brahma chromatin remodeling complex in Drosophila. Genetics 172, 1069–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ransom M., Dennehey B. K., Tyler J. K. (2010). Chaperoning histones during DNA replication and repair. Cell 140, 183–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu S., Holzschuh J., Erhardt S., Ettl A. K., Driever W. (2005). Depletion of minichromosome maintenance protein 5 in the zebrafish retina causes cell-cycle defect and apoptosis. Proc. Natl. Acad. Sci. USA 102, 18467–18472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders A., Werner J., Andrulis E. D., Nakayama T., Hirose S., Reinberg D., Lis J. T. (2003). Tracking FACT and the RNA polymerase II elongation complex through chromatin in vivo. Science 301, 1094–1096 [DOI] [PubMed] [Google Scholar]
- Shimojima T., Okada M., Nakayama T., Ueda H., Okawa K., Iwamatsu A., Handa H., Hirose S. (2003). Drosophila FACT contributes to Hox gene expression through physical and functional interactions with GAGA factor. Genes Dev. 17, 1605–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shkumatava A., Fischer S., Müller F., Strahle U., Neumann C. J. (2004). Sonic hedgehog, secreted by amacrine cells, acts as a short-range signal to direct differentiation and lamination in the zebrafish retina. Development 131, 3849–3858 [DOI] [PubMed] [Google Scholar]
- Stillman D. J. (2010). Nhp6: a small but powerful effector of chromatin structure in Saccharomyces cerevisiae. Biochim. Biophys. Acta 1799, 175–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama M., Sakaue-Sawano A., Iimura T., Fukami K., Kitaguchi T., Kawakami K., Okamoto H., Higashijima S. I., Miyawaki A. (2009). Illuminating cell-cycle progression in the developing zebrafish embryo. Proc. Natl. Acad. Sci. USA 106, 20812–20817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan B. C., Chien C. T., Hirose S., Lee S. C. (2006). Functional cooperation between FACT and MCM helicase facilitates initiation of chromatin DNA replication. EMBO J. 25, 3975–3985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B., Lane D., Levine A. J. (2000). Surfing the p53 network. Nature 408, 307–310 [DOI] [PubMed] [Google Scholar]
- Westerfield M. (2000). The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio rerio). Eugene, OR: University of Oregon Press; [Google Scholar]
- Winkler D. D., Luger K. (2011). The histone chaperone FACT: structural insights and mechanisms for nucleosome reorganization. J. Biol. Chem. 286, 18369–18374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmeyer J., Joss L., Formosa T. (1999). Spt16 and Pob3 of Saccharomyces cerevisiae form an essential, abundant heterodimer that is nuclear, chromatin-associated, and copurifies with DNA polymerase alpha. Biochemistry 38, 8961–8971 [DOI] [PubMed] [Google Scholar]
- Yamaguchi T., Cubizolles F., Zhang Y., Reichert N., Kohler H., Seiser C., Matthias P. (2010). Histone deacetylases 1 and 2 act in concert to promote the G1-to-S progression. Genes Dev. 24, 455–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.