Abstract
The shoot epidermis of land plants serves as a crucial interface between plants and the atmosphere: pavement cells protect plants from desiccation and other environmental stresses, while stomata facilitate gas exchange and transpiration. Advances have been made in our understanding of stomatal patterning and differentiation, and a set of ‘master regulatory’ transcription factors of stomatal development have been identified. However, they are limited to specifying stomatal differentiation within the epidermis. Here, we report the identification of an Arabidopsis homeodomain-leucine zipper IV (HD-ZIP IV) protein, HOMEODOMAIN GLABROUS2 (HDG2), as a key epidermal component promoting stomatal differentiation. HDG2 is highly enriched in meristemoids, which are transient-amplifying populations of stomatal-cell lineages. Ectopic expression of HDG2 confers differentiation of stomata in internal mesophyll tissues and occasional multiple epidermal layers. Conversely, a loss-of-function hdg2 mutation delays stomatal differentiation and, rarely but consistently, results in aberrant stomata. A closely related HD-ZIP IV gene, Arabidopsis thaliana MERISTEM LAYER1 (AtML1), shares overlapping function with HDG2: AtML1 overexpression also triggers ectopic stomatal differentiation in the mesophyll layer and atml1 mutation enhances the stomatal differentiation defects of hdg2. Consistently, HDG2 and AtML1 bind the same DNA elements, and activate transcription in yeast. Furthermore, HDG2 transactivates expression of genes that regulate stomatal development in planta. Our study highlights the similarities and uniqueness of these two HD-ZIP IV genes in the specification of protodermal identity and stomatal differentiation beyond predetermined tissue layers.
Keywords: Stomatal development, Epidermis, HD-ZIP IV proteins, Cell-fate specification, Arabidopsis
INTRODUCTION
Epidermis, the outermost cell layer of land plants, serves as an interface between plants and the surrounding environment. The shoot epidermis, which is derived from the L1 layer of the shoot apex, gives rise to specialized cell types - pavement cells, stomatal guard cells and trichomes - to optimize the balance between protection and gas exchange. The cuticulated pavement cells form a tightly sealed barrier that protects plants from desiccation, UV damage and pathogen entry, while the stomata act as valves for efficient gas exchange and transpiration (Dong and Bergmann, 2010; Javelle et al., 2011b; Pillitteri and Torii, 2012). Trichomes are appendages for herbivore protection, and, in some plants, accumulate and secrete defensive chemicals (Hülskamp, 2004; Serna and Martin, 2006).
The epidermal layer is specified during embryogenesis. In Arabidopsis, two closely related genes, A. thaliana MERISTEM LAYER1 (AtML1) and PROTODERMAL FACTOR2 (PDF2), redundantly specify epidermal identity; their double loss of function results in an embryo that is lethally devoid of a protoderm (Abe et al., 2003). AtML1 and PDF2 belong to a family of plant-specific homeodomain leucine zipper class IV (HD-ZIP IV) transcription factors (Mukherjee et al., 2009), many showing preferential expression in the epidermis (Ingram et al., 2000; Javelle et al., 2011a; Nakamura et al., 2006). The Arabidopsis genome contains 16 HD-ZIP IV (AtHD-ZIP IV) genes, of which at least nine exhibit expression in leaf epidermal cells (Nakamura et al., 2006; supplementary material Fig. S1). It is not known whether AtML1 and PDF2 are sufficient to specify epidermal fate. Some other HD-ZIP IV members are associated with differentiation of shoot epidermal cell types. For example, Arabidopsis GLABRA2 (GL2) promotes trichome differentiation (Rerie et al., 1994), and HDG11 and HDG12 regulate trichome branching (Nakamura et al., 2006). Microarray studies of maize plants overexpressing HD-ZIP IV gene, OUTER CELL LAYER1 (OCL1) identified target genes in lipid metabolism and cuticle biosynthesis, suggesting a role in pavement cell differentiation (Javelle et al., 2010). Likewise, Arabidopsis HDG1 overexpression upregulates epicuticular wax biosynthesis genes (Wu et al., 2011). Thus far, it is not known whether any HD-ZIP IV members promote differentiation of the remaining epidermal cell type: stomata.
Stomatal development occurs through a series of cell-state transitions. In Arabidopsis, a subpopulation of protodermal cells adopts the identity of a meristemoid mother cell (MMC) and initiates an asymmetric entry division that creates a meristemoid, a transiently amplifying stomatal lineage cell. The meristemoid reiterates asymmetric amplifying divisions but eventually differentiates into a guard mother cell (GMC), which divides symmetrically to form paired guard cells (GCs) that constitute a stoma (Dong et al., 2010; Pillitteri et al., 2012).
The stomatal cell-state transitions are specified by combinatorial activities of five basic helix-loop-helix (bHLH) proteins, SPEECHLESS (SPCH), MUTE, FAMA, SCREAM (SCRM, also known as ICE1) and SCRM2 (Kanaoka et al., 2008; MacAlister et al., 2007; Ohashi-Ito and Bergmann, 2006; Pillitteri et al., 2007). These bHLH proteins are necessary and sufficient for specifying each stomatal precursor state. Among them, MUTE and SCRM are capable of converting the aerial shoot epidermis into stomata when ectopically overexpressed or stabilized (Kanaoka et al., 2008; Pillitteri et al., 2008). By contrast, ectopic overexpressions of SPCH and FAMA confer excessive asymmetric entry divisions and the formation of guard cell-like cells, respectively (MacAlister et al., 2007; Ohashi-Ito and Bergmann, 2006; Pillitteri et al., 2007). Importantly, the ability of MUTE and SCRM to trigger stomatal differentiation is limited to the epidermis. Factors that enable stomatal differentiation beyond epidermal identity remain unknown.
Here, we report a meristemoid-enriched AtHD-Zip IV protein, HDG2, as a key component that links epidermal identity to stomatal differentiation. HDG2 ectopic expression confers striking ectopic differentiation of stomata in internal mesophyll tissues and occasional multiple epidermal layers. Conversely, the loss-of-function hdg2 mutant exhibits delayed meristemoid-to-GMC transition. Our work highlights the similarity and difference between HDG2 and its close relative AtML1 in the specification of protodermal identity, and their ability to override epidermal constraints of stomatal differentiation when ectopically expressed.
MATERIALS AND METHODS
Plant materials and growth conditions
Arabidopsis thaliana Columbia (Col) accession was used as wild type. Mutants used in the study were in the Col background. The following mutants and reporter lines have been reported previously: TMMpro::GUS-GFP (Nadeau and Sack, 2002), TMMpro::GUS (Shpak et al., 2005), EPF2pro::erGFP (Hara et al., 2009), AtML1pro::GUS (Uchida et al., 2012), SPCHpro::GUS and E994 (Pillitteri et al., 2007), GL2pro::GUS (Masucci et al., 1996) and At5g17710pro::nuc-3xVENUS (Roeder et al., 2012). T-DNA insertion lines: hdg2-2 (SALK_127828C); hdg2-3 (SALK_138646C); hdg2-4 (SALK_120064); atml1-3 (SALK_128172); atml1-4 (SALK_033408) and pdf2-2 (SALK_109425C) were obtained from the Arabidopsis Biological Resource Center (ABRC), and loss or disruption of their transcripts was confirmed (supplementary material Fig. S2). Seedlings and plants were grown as described previously (Pillitteri et al., 2011).
Microarray validation
Preparation and analysis of the meristemoid cell state microarray have been described previously (Pillitteri et al., 2011). Original, DNase I-treated RNA samples for all microarray genotypes were converted to cDNA using the iScript cDNA synthesis kit (Bio-Rad, Hercules, CA). Quantitative RT-PCR was performed as described previously (Pillitteri et al., 2011) and normalized using the ΔΔCq method (Livak and Schmittgen, 2001). RT-PCR was performed as described previously (Lee et al., 2012). See supplementary material Table S1 for a list of primer sequences.
Molecular cloning and generation of transgenic plants
The following constructs were generated for transgenic studies using Gateway cloning (Nakagawa et al., 2008; Nakagawa et al., 2007): pLJP249 (35S::HDG2), pKMP114 (HDG2pro::HDG2-GFP), pKMP145 (inducible HDG2), pKMP141 (inducible HDG2-SRDX), pKMP151 (inducible AtML1), pKMP139 (HDG2pro::nls-3xGFP). Plasmid pER8 (Zuo et al., 2000) was used for estradiol-inducible constructs. Generation and selection of transgenic plants were performed as described previously (Pillitteri et al., 2007). For all transgenic Arabidopsis lines, more than 20 T1 plants per construct were isolated and characterized for phenotypes. Among them, five or six lines were selected based on single insertion status inferred by the segregation of resistance genes and stability of the phenotype in subsequent generations. Estradiol-mediated induction of HDG2, HDG2-SRDX and AtML1 were as previously reported (Lee et al., 2012), and their induction was confirmed by RT-PCR using transgene-specific primers (see supplementary material Figs S3, S4). See supplementary material Table S1 for a list of primer sequences used for molecular cloning and supplementary material Table S2 for a list of plasmid constructs generated in this study.
Microscopy
Confocal laser scanning microscopy (CLSM) images were taken with either the Zeiss LSM700 (Thornwood, NY, USA) or the Leica SP5 (Solms, Germany). Cell peripheries were visualized with propidium iodide (PI: Molecular Probes, Carlsbad, CA), and GFP and PI signals were detected as described previously (Pillitteri et al., 2011). mPS-PI staining was performed as previously described (Truernit et al., 2008), with an ethanol treatment step of 2-3 minutes. Z-stack movies were produced using Quicktime 7 Pro (Apple, Cupertino, CA).
Molecular phylogenetic analyses
Full-length amino acid sequences of 16 AtHD-ZIP IV members, together with five AtHD-ZIP III members as outgroup, were aligned in MUSCLE (http://www.ebi.ac.uk/Tools/msa/muscle/). The model of evolution for the data set was determined by ProtTest version 2.4 (Abascal et al., 2005). The model selected under the Akaike Information Criterion (Akaike, 1974) was JTT+I+G+F.
Bayesian analyses were conducted in MrBayes version 3.1.2 (Huelsenbeck and Ronquist, 2001; Ronquist and Huelsenbeck, 2003) via the CIPRES Science Gateway version 3.1 (Miller et al., 2010) under the default priors. Three independent Markov Chain Monte Carlo (Yang and Rannala, 1997) analyses of 1 million generations were run under the default settings. Convergence was determined when the average standard deviation of split frequencies remained less than 0.01. The first 40% of trees were discarded before convergence. The remaining trees from each run were pooled to construct a 50% majority rule consensus tree and to obtain posterior probabilities. Graphical molecular phylogenetic tree was drawn using FigTree version 1.4 (http://tree.bio.ed.ac.uk/software/figtree/).
In situ hybridization
Wild-type seedlings were grown on sterile 1/2× Murashige and Skoog medium for 10 days. esHDG2 seedlings were grown for 5 days and transplanted on induction medium with 10 μM estradiol for 5 more days. The seedlings were harvested and fixed in FAA (50% ethanol, 10% formaldehyde, 5% acetic acid) for 1 hour under vacuum at room temperature and for 1 more hour at 4°C without vacuum. Samples were dehydrated through graded ethanol series then the solution was replaced with Histo-Clear (Cosmo Bio, Japan). The samples were embedded in paraffin wax (Paraplast, Sigma-Aldrich) and cut at 8 μm.
Partial HDG2 cDNA fragments (corresponding to exon 1 and exon 4) were amplified from pKMP144 using gene-specific primers and cloned into pCR4-TOPO vector (Invitrogen). A partial FDH cDNA fragment was amplified from first strand cDNA from 10-day-old Col seedlings using gene-specific primers and cloned into pCR4-TOPO vector. For primers and plasmids, see supplementary material Tables S1 and S2. These plasmids were used as templates for PCR amplification of in situ probes using M13 forward (-20) and M13 reverse primers. For probe synthesis, in vitro transcription and digoxigenin (DIG)-UTP labeling were carried out using MAXIscript T3 or T7 kit (Ambion, Life Technologies) according to manufacturer’s instructions. RNA polymerases used for probe synthesis were as follows: HDG2_exon1 (pMK370), antisense T3, sense T7; HDG2_exon4 (pMK371), antisense T3, sense T7; FDH_exon3 (pMK372), antisense T7, sense T3.
The sections were de-paraffinized in Histo-Clear and rehydrated through a graded ethanol series. They were treated with proteinase K (final 5 μg/ml) in ProK buffer [100 mM TRIS-C1 (pH 7.5), 50 mM EDTA)] at 37°C for 30 minutes and then washed twice with distilled water for 5 minutes each. They were subsequently treated with post-fixation solution [4% (w/v) paraformaldehyde in 0.1 M Na-P buffer (pH 7.2)] for 10 minutes at room temperature and then washed twice with distilled water for 5 minutes each. Samples were treated with TEA solution (0.5% acetic anhydride in 0.1 M triethanol amine) for 10 minutes at room temperature with stirring, washed twice in 2×SSPE for 5 minutes each and then dehydrated through a graded ethanol series. Sections were dried in a vacuum at room temperature for 1 hour, and subsequently incubated in a humid box at 45°C for 18 hours with hybridization buffer (200 μl per slide) containing the probes (0.1 μg/slide). The hybridization buffer consisted of 50% deionized formamide, 0.3 M NaC1, 10% dextran sulfate, 10 mM Tris-HCl (pH 7.5), 1 mM EDTA, 60 mM DTT, 1× Denhardt’s, 1 μg/μl Escherichia coli tRNA and 0.1 μg/μl polyA. For HDG2 detection, both exon 1 and exon 4 probes were mixed and used.
After hybridization, successive washing steps were performed as follows: twice in 4×SSC (for 7 minutes and 8 minutes, at 45°C), treated with RNase A (50 μg/ml) in RNase buffer [0.5 M NaC1, 10 mM Tris-HCl (pH 7.5), 5 mM EDTA] at 37°C for 30 minutes, three times in RNase buffer at 37°C for 5 minutes each, and twice in 0.5×SSC at 45°C for 20 minutes each. The slides were soaked twice with buffer 1 [150 mM NaC1, 100 mM Tris-HCl (pH 7.5)] at room temperature for 5 minutes each and then incubated with 0.5% blocking reagent (Boehringer) in buffer 1 for 30 minutes at room temperature. After removing the blocking reagent, the slides were briefly rinsed with buffer 1, followed by incubation with the diluted anti-digoxigenin-alkaline phosphatase (anti-DIG-AP) conjugate (1:1000) in buffer 1 containing 0.1% BSA and 0.25% Tween 20 for 60 minutes at room temperature. The slides were subsequently washed three times with buffer 1 for 10 minutes each with agitation and with buffer 3 [100 mM NaC1, 50 mM MgCl2, 100 mM Tris-HC1 (pH 9.5)] for 5 minutes, and then covered with buffer 3 containing 4.5 μl/ml nitroblue tetrazolium salt (NBT) solution and 3.5 μl/ml 5-bromo-4-chloro-3-indolyl phosphate toluidinium salt (BCIP) solution. After incubation at 37°C for 19 hours in the dark, the color reaction was stopped by immersing the slides in TE (pH 7.5). The sections were dehydrated through a graded ethanol series and then mounted in Eukitt (O. Kindler, Freiburg, Germany) (Kouchi and Hata, 1993).
Tissue sectioning
Plastic embedding and sectioning using Technovit 7100 (Heraeus Kulzer, Wehrheim, Germany) and Toluidine Blue O (TBO) staining were performed as described previously (Shpak et al., 2003). Histochemical staining for GUS activity was performed as described previously (Sessions et al., 1999). Images were taken with the Olympus FV1000 (Center Valley, PA, USA). To visualize GFP expression inside leaves, fresh tissue was embedded in 3% low-melt agarose. The blocks were cut into 150 μm sections using Vibratome Series 1000 (Ted Pella, Redding, CA, USA), stained with propidium iodide and imaged with the Zeiss LSM700 as above.
Quantitative analysis of stomatal phenotype
Stomatal index (SI; number of stomata/total number of stomata + non-stomatal epidermal cells×100), meristemoid index (MI; number of meristemoids/total number of stomata + non-stomatal epidermal cells×100) and stomatal-lineage index (SLI; number of stomata + meristemoids/total number of stomata + non-stomatal epidermal cells×100) were calculated using TBO-stained 10-day-old cotyledons as well as rosette leaves (fully expanded rosette leaf 4 from 6-week-old plants) as previously reported (Guseman et al., 2010).
Yeast one-hybrid assays and HDG2 autoactivation assays
The following plasmids were constructed and used in yeast one-hybrid assays: pCAB120 (Gal4AD-HDG2), pCAB121 (HAHR1-A box), pCAB122 (HAHR1-T box), pCAB123 (HAHR1-m box), pCAB124 (L1-C box), pCAB125 (L1-T box), pCAB126 (L1-m box), pCAB130 (Gal4AD-AtML1) and pCAB132 (Gal4AD-SCRM). Gal4AD-PDF2 was reported previously (Wu et al., 2011). SCRM was used as a negative control. DNA binding was tested using the Matchmaker One-Hybrid system (Clontech Laboratories) and the yeast expression vector pDEST22 (Invitrogen). Target binding sequences were created by annealing oligonucleotide pairs containing three tandemly repeated target-binding sequences (see supplementary material Table S3). Each target binding sequence was integrated into yeast strain YM4271 according to the manufacturer’s protocols. DNA binding ability was assayed by testing β-galactosidase activity using 2-nitrophenyl-β-D-galactopyranoside (ONPG; Sigma). All protocols were run according to manufacturer’s specifications. Yeast cell density and colorimetric change caused by ONPG were measured using the Victor3 V Plate Reader (Perkin Elmer) at 620 nm and 405 nm, respectively.
The following constructs were generated for HDG2 transactivation assays using the Matchmaker Gold system (Clontech Laboratories): pCAB127 (Gal4DB-HDG2) and pCAB128 (Gal4DB-HDG2-SRDX). Gal4DB-SCRM and Gal4DB-MUTE (Kanaoka et al., 2008) were used as positive and negative controls, respectively, of activation. These constructs were transformed into yeast strain AH109 according to manufacturer’s protocols. Activation was measured by testing for ONPG (Sigma) as a substrate. See supplementary material Tables S1, S2 for details about plasmid construction and oligo DNA sequences used.
Dual luciferase transactivation assay in planta
The following constructs were generated and used in transactivation assays in planta: pCS001 (pGREEN-0800-LUC), pCS003 (pGREEN-0800-LUC-TMMpro) and pCS004 (pGREEN-0800-LUC-MUTEpro). Agrobacterium carrying both effector and reporter constructs were infiltrated into 4- to 5-week old N. benthamiana as described previously (Hellens et al., 2005). Five to 7 days after infiltration, firefly luciferase (LUC) and Renilla luciferase (REN) were assayed using dual luciferase reagents (Promega) as previously described (Li et al., 2010). LUC and REN activity were measured using a Victor3 V Plate Reader. At least three biological replicates were measured for each sample. See supplementary material Tables S1, S2 for details about plasmid construction and oligo DNA sequences used.
Statistical analysis of promoter motifs
Enrichment of specific promoter motifs in epidermally expressed genes regulating stomata development (EPF1, EPF2, ERECTA, ERL1, ERL2, TMM, SPCH, MUTE, FAMA, SCRM/ICE1, SCRM2, HDG2, AtML1, POLAR, BASL, MYB88, FLP and SDD1) was tested using the ATHENA database (O’Connor et al., 2005). Up to 3 kb upstream of each gene was considered, with a cutoff at adjacent genes.
RESULTS
HDG2 is a meristemoid-expressed nuclear protein
We have previously performed a comparative transcriptomic analysis of seedlings specifically enriched in particular stomatal cell state: spch, in which epidermis is devoid of any stomatal-lineage cells (i.e. pavement cells only); scrm-D mute, which produces an epidermis solely composed of meristemoids and sister cells (known as stomatal lineage ground cells, SLGCs); and scrm-D, which produces a stomata-only epidermis (Pillitteri et al., 2011). Through that analysis, we identified HDG2, a member of the HD-ZIP IV family, as a meristemoid-enriched gene. HDG2 shows strikingly high expression in the meristemoid-enriched population (2069.7±200.8, mean±s.e.m.) with over 18-fold, 20-fold and 10-fold enrichment over wild type (114.6±4.76), spch (102.2±18.3) and scrm-D (200.5±9.02), respectively (Fig. 1A). Quantitative RT-PCR analysis confirmed specific and high HDG2 expression in scrm-D mute, with over 31-fold, 62-fold and 18-fold expression compared with wild type, spch and scrm-D, respectively (Fig. 1A).
Fig. 1.
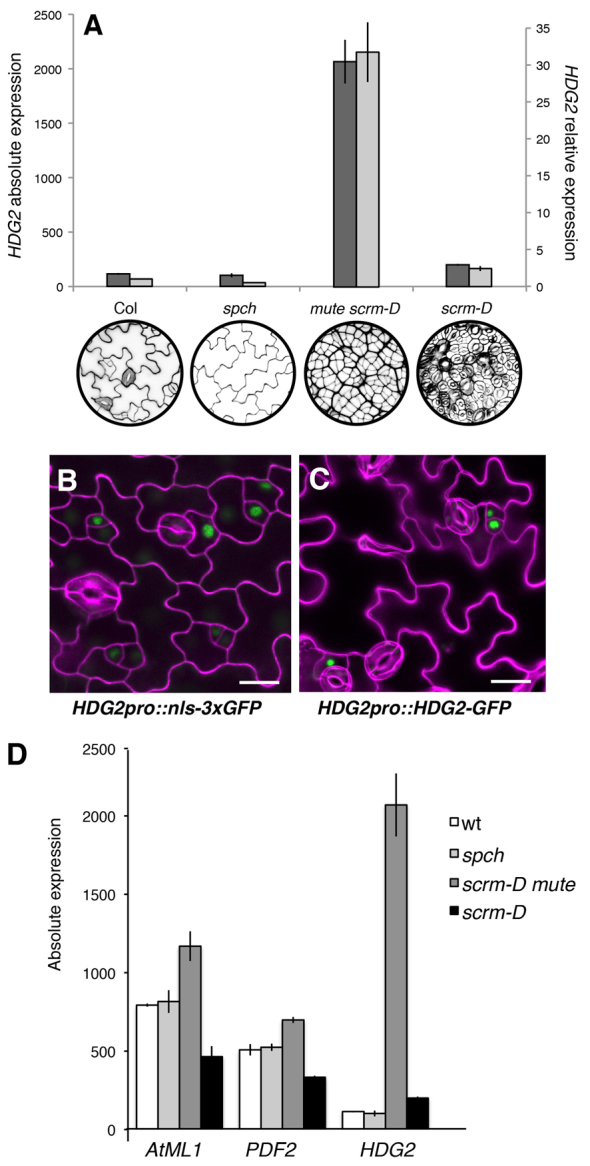
HDG2 is highly enriched in meristemoid population of stomatal cell lineages. (A) HDG2 absolute and relative expression levels among wild-type and stomatal mutants enriched in specific epidermal cell populations. Absolute expressions (dark gray) are from ATH1 microarray data; relative expressions (light gray) are from qRT-PCR analysis. Data are mean values of triplicates; error bars indicate s.e.m. Col, wt; spch, pavement-cell only; mute scrm-D, overwhelmingly enriched in meristemoids; scrm-D, stomata-only epidermis. Below each graph are confocal images of cotyledons from corresponding genotypes. (B) Stomatal-lineage accumulation of HDG2 transcriptional reporter (HDG2pro::nls-3xGFP) in seedling epidermis. (C) Stomatal-lineage accumulation of HDG2 translational reporter (HDG2pro::HDG2-GFP) in 10-day-old abaxial cotyledon epidermis. Scale bars: 20 μm. (D) Expression levels of AtML1 and PDF2 compared with HDG2 among wild-type and stomatal mutants.
We next examined the cell-type expression patterns of HDG2 in developing cotyledon and leaf epidermis. HDG2 transcriptional reporter (HDG2pro::nls-3xGFP) showed strong GFP signals in meristemoids and SLGCs (Fig. 1B). This is consistent with a previous report on HDG2pro::GUS (Nakamura et al., 2006). Furthermore, the HDG2 translational reporter (HDG2pro::HDG2-GFP) accumulated in the nuclei of meristemoids and SLGCs (Fig. 1C). The nuclear localization of HDG2-GFP accords with its predicted structure as a HD-ZIP IV transcription factor.
Our molecular phylogenetic analysis of 16 AtHD-ZIP IV family members using Bayesian algorithms shows that, similar to the neighbor-joining analysis by Nakamura et al. (Nakamura et al., 2006), HDG2 is most closely related to AtML1 and PDF2, two well-studied L1 layer (protoderm)-specific genes (Abe et al., 2003; Lu et al., 1996) (supplementary material Fig. S1). To exclude the possibility that high expression of HDG2 in the meristemoid-enriched population may simply be reflecting the protodermal character of the actively dividing scrm-D mute epidermis, we further surveyed the expression of AtML1 and PDF2 in stomatal cell-state mutants. Unlike HDG2, both AtML1 and PDF2 showed moderately high expression in all genetic backgrounds (Fig. 1D). The expression levels of AtML1 and PDF2 were only 1.5-fold and 1.4-fold higher in scrm-D mute compared with wild type, which are significantly less than the more than 18-fold increase in HDG2 (Fig. 1D). Combined, these results highlight the difference in HDG2 expression from that of AtML1 and PDF2, and establish HDG2 as a stomatal lineage-expressed nuclear-localized protein.
HDG2 overexpression confers differentiation of ectopic stomata in internal mesophyll tissues
To understand the function of HDG2, we first ectopically overexpressed HDG2. CaMV35Spro::HDG2 plants were infertile (data not shown). For this reason, we subsequently used an induction system (Zuo et al., 2000), which specifically induced the transgenes after estradiol application (supplementary material Figs S3, S4). Both CaMV35Spro::HDG2 and inducible HDG2 seedlings develop narrow, dark-green leaves, suggesting that HDG2 ectopic overexpression (HDG2-OX) affects leaf growth (supplementary material Fig. S3 and data not shown).
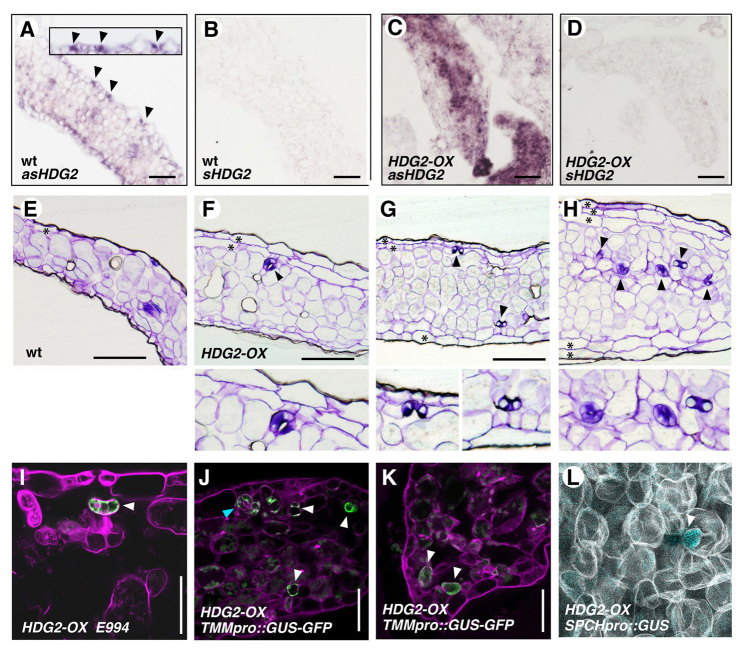
In situ hybridization analysis of total HDG2 transcripts showed strong expression in small epidermal cells (Fig. 2A), confirming the meristemoid-enriched expression seen in scrm-D mute seedlings, as well as in HDG2 GFP reporters (Fig. 1). By contrast, induced HDG2-OX resulted in strong signals through internal cotyledon and leaf tissues (Fig. 2C). Control sense probes showed no signal, indicating that the probes are highly specific (Fig. 2B,D).
Fig. 2.
Ectopic overexpression of HDG2 confers ectopic stomata within mesophyll tissues and formation of multiple epidermal layers. (A-D) In situ hybridization analysis of HDG2 expression in 10-day-old wild-type (A,B) and HDG2-OX (C,D) seedlings treated with HDG2 antisense (A,C) or sense (B,D) probes. In wild type, endogenous HDG2 shows stomatal-lineage expression (A, arrowheads). Inset shows an enlarged image. Scale bars: 40 μm. (E-H) Histological cross-sections of plastic-embedded 10-day-old cotyledons from wild-type (E) and transgenic plants overexpressing HDG2 (HDG2-OX), which are CaMV35S::HDG2 (F-H). In HDG2-OX, ectopic differentiation of stomata in internal tissues (arrowheads) is evident. Bottom insets indicate enlarged images. Multiple epidermal layers (asterisks) occasionally formed. Images are taken under the same magnification. Scale bars: 100 μm. (I-K) Live sections of HDG2-OX cotyledons expressing mature guard cell marker E994 (I) and stomatal-lineage marker TMMpro::GUS-GFP (J,K). GFP is detected in ectopic internal stomata (I, arrowhead) and internal stomatal-lineage cells (J,K, arrowheads). A mature ectopic internal stoma is also evident in J (cyan arrowhead). Scale bars: 20 μm. (L) Optical section of mPS-PI stained mesophyll layer from HDG2-OX cotyledon expressing SPCHpro::GUS (arrowhead).
Strikingly, histological sectioning revealed that HDG2-OX confers differentiation of stomata in internal tissues of cotyledons and true leaves. These internal stomata are correctly shaped: round and mirror-symmetric with paired GCs (Fig. 2E-H). Furthermore, the internal stomata express mature GC marker E994 (Fig. 2I), indicating that they possess molecular characteristics of stomata. The internal stomata phenotype was consistently observed in multiple T1 CaMV35Spro::HDG2 seedlings and in all independent inducible HDG2-OX lines analyzed in subsequent T2-T3 generations (Fig. 2; supplementary material Figs S3, S4).
To unravel whether HDG2 can activate earlier steps of stomatal differentiation, we next examined whether HDG2-OX triggers ectopic expression of earlier stomatal cell-lineage markers. As shown in Fig. 2J,K, a subpopulation of HDG2-OX mesophyll cells expresses TMMpro::GUS-GFP, which is normally expressed in early stomatal-lineage cells, such as meristemoids and SLGCs (Nadeau and Sack, 2002). The TMM-negative internal cells are round and have numerous chloroplasts (Fig. 2J,K), indicating that they probably retain mesophyll cell identity. We further detected the expression of SPCHpro::GUS, the earliest marker of stomatal cell lineages, in the mesophyll tissue (Fig. 2L). Based on these observations, we conclude that HDG2 is sufficient to induce ectopic stomatal identities.
Ectopic HDG2 overexpression can confer multiple epidermal layers and ectopic expression of epidermal markers
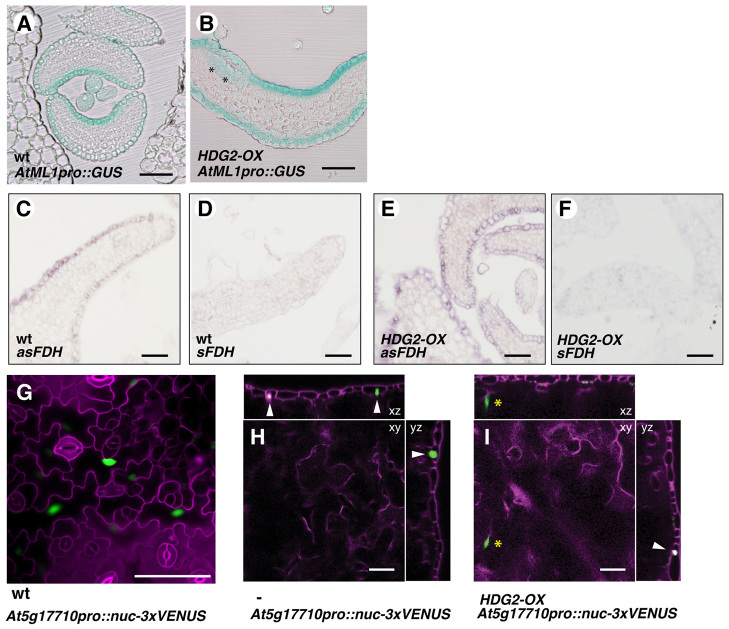
In some instances, we observed formation of multiple epidermal layers in HDG2-OX cotyledons and leaves (Fig. 2F-H). This prompted us to test whether HDG2 overexpression confers ectopic epidermal fate. Indeed, the L1-layer marker AtML1pro::GUS was occasionally detected in the internal tissues (Fig. 3A,B). We next examined the expression of the L1-layer-specific gene FIDDLEHEAD (FDH) (Pruitt et al., 2000) using in situ hybridization (Fig. 3C-F). As expected, strong FDH signals are detected in the epidermis of both wild-type and HDG2-OX seedlings (Fig. 3C,E); however, it was inconclusive whether FDH is ectopically expressed in the internal tissues of HDG2-OX seedlings (Fig. 3E). We therefore used an endoreduplicating epidermal cell marker (At5g17710pro::nuc-3xVENUS) (Roeder et al., 2012) to address this issue (Fig. 3G-I). The marker was originally identified for sepal epidermal ‘giant’ cells, but it also exhibits expression in large pavement cells in cotyledons and rosette leaves (Fig. 3G; data not shown). The At5g17710pro::nuc-3xVENUS signal was detected in occasional subepidermal nuclei in HDG2-OX seedlings (Fig. 3I). By contrast, we did not detect clear ectopic expression of a trichome marker in GL2pro::GUS seedlings nor trichome differentiation in internal tissues in HDG2-OX (data not shown). These results indicate that, in addition to stomatal differentiation, ectopic HDG2 could induce ectopic epidermal identities.
Fig. 3.
Ectopic overexpression of HDG2 induces some epidermal reporters. (A,B) Histological cross-sections of emerging rosette leaves from 10-day-old wild-type (wt: A) and HDG2-OX seedlings expressing AtML1pro::GUS (B). Ectopic GUS expression (asterisks) can be seen in B. Scale bars: 100 μm. (C-F) In situ hybridization analysis of FDH expression in 10-day-old wild-type (C,D) and HDG2-OX (E,F) seedlings treated with HDG2 antisense (C,E) or sense (D,F) probes. Scale bars: 40 μm. (G) At5g17710pro::nuc-3xVENUS in wild-type rosette leaf abaxial epidermis showing specific signals in large pavement cells. Scale bar: 50 μm. (H,I) Confocal optical sections (xy, xz and yz planes) of 10-day-old HDG2-OX cotyledon expressing At5g17710pro::nuc-3xVenus, without (H) or with (I) HDG2 induction. Asterisk, GFP in subepidermal mesophyll nucleus. Arrowheads, GFP epidermal nuclei (co-stained with propidium iodide). Scale bars: 50 μm.
Optical sectioning of intact cotyledons reveals architecture of ectopic internal stomata
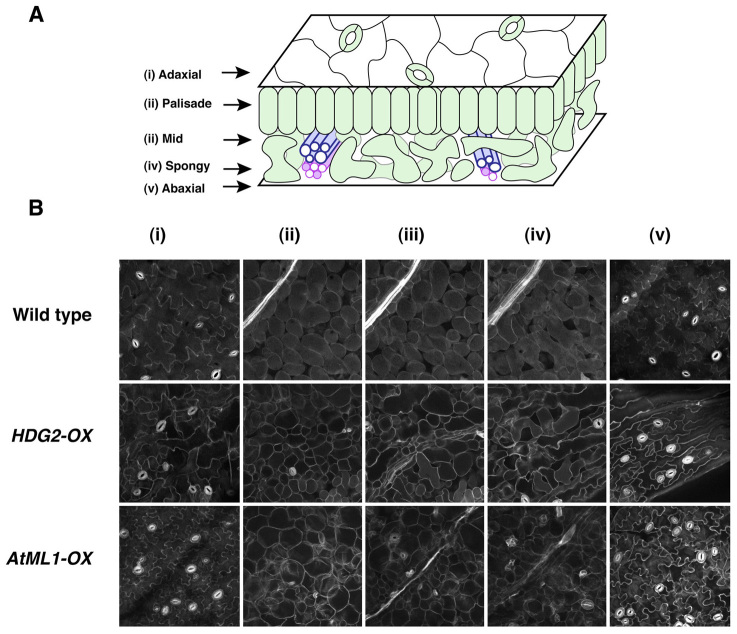
To demonstrate conclusively that internal stomata were not artifacts of tissue sectioning, we took advantage of the mPS-PI staining protocol (Truernit et al., 2008). This technique enabled us to section optically through whole undamaged cotyledons to document the exact locations of internal stomata (Fig. 4; supplementary material Movies 1-3). As expected, optical scanning of wild-type cotyledons showed characteristic layers of adaxial epidermis, palisade mesophyll, midvein, spongy mesophyll and abaxial epidermis [Fig. 4A,Bi-v (top row); supplementary material Movie 1]. By contrast, optical scanning of HDG2-OX cotyledons revealed internal stomata in both palisade and spongy mesophyll layers [Fig. 4Bii-iv (middle row); supplementary material Movie 2]. The palisade mesophyll layer was thicker and tightly packed with smaller mesophyll cells (supplementary material Movie 2).
Fig. 4.
Optical serial sectioning through intact cotyledons by mPS-PI staining reveals ectopic internal stomata. (A) Cartoon of a cotyledon indicating locations of each tissue: (i) adaxial epidermis; (ii) palisade mesophyll; (iii) mid-vein region; (iv) spongy mesophyll; (v) abaxial epidermis. Stomata are found in both adaxial and abaxial epidermis in Arabidopsis. (B) mPS-PI stained optical serial sections of locations (i-v) in wild-type (top); induced ectopic HDG2 overexpression (middle: HDG2-OX) and induced ectopic AtML1 overexpression (bottom: AtML1-OX). Images were taken under the same magnification. Scale bar: 20 μm. See supplementary material Movies 1-3 for z-stack 3D sections.
Because HDG2-OX induces ectopic AtML1 expression in mesophyll tissues (Fig. 3B) and because HDG2 and AtML1 are closely related, we tested whether they confer similar ectopic effects. As there is no previous report on AtML1 overexpression studies, we generated an estradiol-inducible AtML1 overexpressor (AtML1-OX) and analyzed the phenotypes of six independent transgenic lines (Fig. 4B; supplementary material Figs S3, S4). Like HDG2-OX, AtML1-OX seedlings from all lines developed cotyledons with striking, internal stomatal in mesophyll tissues [Fig. 4Bii-iv (bottom); supplementary material Movie 3]. These results unambiguously demonstrate that ectopic HDG2 overexpression results in the differentiation of stomata in internal tissues and suggest that HDG2 and AtML1 can act interchangeably for ectopic stomatal differentiation.
Loss of function in HDG2 results in delayed progression of stomatal development, which is enhanced by additional loss of AtML1
Our stomatal cell state-specific transcriptome analysis highlighted expression differences between HDG2 and AtML1. HDG2 is highly specific to the meristemoid-enriched population (scrm-D mute) whereas AtML1 is expressed throughout the epidermal mutant genotypes (spch, scrm-D mute and scrm-D) (Fig. 1D). Nevertheless, both HDG2-OX and AtML1-OX triggered ectopic, internal stomatal differentiation (Figs 3, 4). This raises the issue of whether the endogenous HDG2 plays a specific role in stomatal differentiation. To address this, we investigated the hdg2 loss-of-function phenotype. A previous report on the systematic characterization of the Arabidopsis HD-ZIP IV family members did not reveal any phenotype associated with the hdg2 T-DNA knockout mutation (Nakamura et al., 2006). We therefore predicted that hdg2 phenotype, if any, would be less obvious.
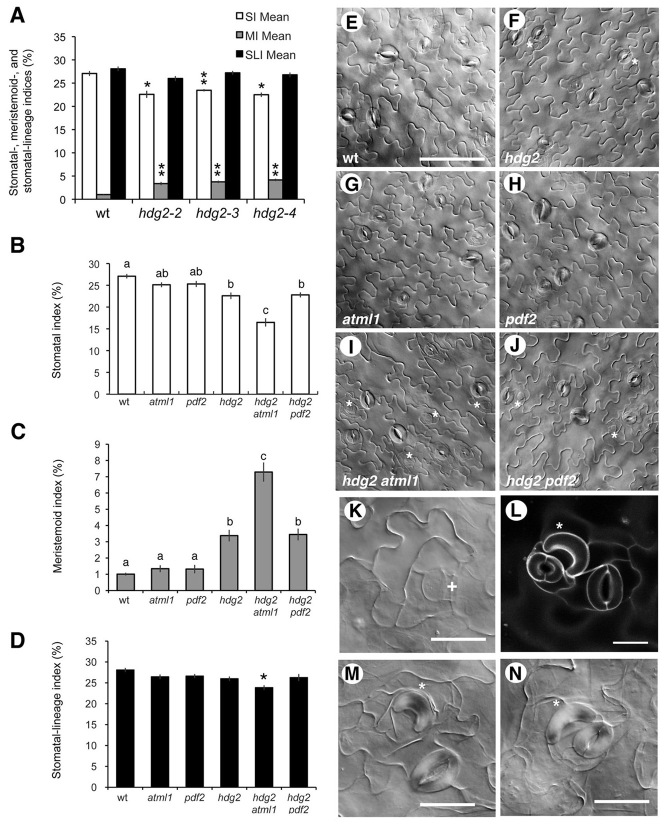
Careful examination of developing (10-day-old) hdg2 cotyledons revealed increased numbers of meristemoids, which are not commonly seen in wild type at this stage (Fig. 5). For quantitative analysis of stomatal phenotypes, we used three T-DNA insertion alleles of HDG2: hdg2-2, hdg2-3 and hdg2-4 (supplementary material Fig. S2). Extensive RT-PCR analysis detected aberrant, fused T-DNA-HDG2 transcripts of partial size, indicating that none of these alleles produce functional HDG2 transcripts (supplementary material Fig. S2D). Quantitative analysis showed that all three hdg2 alleles exhibit statistically significant reduction in stomatal index (SI) (Fig. 5A). By contrast, the number of meristemoids per epidermal cell, hereafter defined as meristemoid index (MI), was significantly higher in hdg2-2, hdg2-3 and hdg2-4 than in wild type (Fig. 5A). As total numbers of stomata and stomatal precursors, defined here as stomatal-lineage index (SLI), remained unchanged from wild type (Fig. 5A), hdg2 seedlings are delayed in progression of stomatal differentiation rather than producing more meristemoids. All three hdg2 alleles showed statistically indistinguishable phenotypes (Tukey’s HSD test, non-significant), strongly supporting that the phenotype is caused by the loss of function in HDG2.
Fig. 5.
Stomatal development defects in hdg2 mutants and higher-order mutants in closely related HD-ZIP IV genes. (A) Stomatal index (SI; white bars), meristemoid index (MI; grey bars) and stomatal-lineage index (SLI; black bars) of 10-day-old abaxial cotyledons from wild-type (wt) and three independent hdg2 T-DNA insertion alleles: hdg2-2, hdg2-3 and hdg2-4. SLI is defined here as the sum of SI and MI. See supplementary material Fig. S1 for RT-PCR analysis. Data are means (n=8); error bars indicate s.e.m. *P<0.0005; **P<0.0001 (two-tailed Student’s t-test of each hdg2 allele against wild type). Tukey’s HSD did not reveal statistical difference of SI, MI and SLI among three hdg2 alleles. (B) SI of 10-day-old abaxial cotyledons from wild type, atml1, pdf2, hdg2, hdg2 atml1 and hdg2 pdf2. hdg2-2 and atml1-3 alleles were used for the analysis. Data are means (n=8); error bars indicate s.e.m. Total numbers of stomata counted: 680 (wild type), 639 (atml1), 673 (pdf2), 630 (hdg2), 550 (hdg2 atml1) and 620 (hdg2 pdf2). (C) MI of the six genotypes described above. Data are means; error bars indicate s.e.m. Total numbers of meristemoids counted: 25 (wild type), 34 (atml1), 35 (pdf2), 93 (hdg2), 243 (hdg2 atml1) and 94 (hdg2 pdf2). (D) SLI of the six genotypes described above. Data are mean; error bars indicate s.e.m. Total numbers of cells counted: 2516 (wild type), 2542 (atml1), 2661 (pdf2), 2786 (hdg2), 3331 (hdg2 atml1) and 2661 (hdg2 pdf2). For B,C, genotypes with non-significant phenotypes were grouped together with a letter (Tukey’s HSD test after one-way ANOVA). For D, only one genotype was significantly different from others (Tukey’s HSD test; P<0.01). (E-J) Representative DIC images of 10-day-old abaxial cotyledons from wild type (E), hdg2 (F), atml1 (G), pdf2 (H), hdg2 atml1 (I) and hdg2 pdf2 (J). Asterisks indicate meristemoids. Images were taken under the same magnification. Scale bar: 100 μm. (K-N) Aberrant stomatal complexes found in hdg2. (K) Arrested stomatal precursor after extensive asymmetric amplifying divisions from 30-day-old cotyledon. (L) Confocal image of a stomatal complex with a single GC from 10-day-old abaxial cotyledon. (M,N) Singular GCs from 30-day-old cotyledon. Asterisks indicate singular GCs; + indicates arrested stomatal precursor. Scale bars: in L, 20 μm; in K,M,N, 25 μm.
We next examined the possible involvement of AtML1 and PDF2 in stomatal development. As shown in Fig. 5B-D,G,H, the SI, MI and SLI of atml1 and pdf2 cotyledon abaxial epidermis were not significantly different from those of wild type. Interestingly, introduction of an atml1 mutation into hdg2 dramatically enhanced MI and reduced SI. By contrast, pdf2 mutation had no such effects (Fig. 5B-D).
To understand the fate of delayed stomatal precursors, we observed fully mature, senescing cotyledon epidermis (30 dpg). In hdg2, some arrested stomatal precursors surrounded by excessive SLGCs were observed (Fig. 5K). Stomatal precursors were never observed in wild-type cotyledons at this age (not shown). Rarely, yet consistently (∼1-2 per cotyledon), some stomatal complexes resulted only in singular GCs (Fig. 5L-N).
Like cotyledons, rosette leaf epidermis of hdg2 exhibits statistically significant increase in MI with concomitant decrease in SI (supplementary material Fig. S5). In hdg2 rosette leaf abaxial epidermis, meristemoids occasionally failed to differentiate into stomata (supplementary material Fig. S5D). The phenotype was exaggerated by the additional atml1 mutation, and excessive numbers of SLGCs were seen in the hdg2 atml1 rosette leaf epidermis, implying extended asymmetric cell division state due to delayed differentiation (supplementary material Fig. S5D).
Although the cotyledon epidermis is formed during embryogenesis, the rosette leaf epidermis is derived from the L1 layer of the shoot apical meristem. Our results suggest that HDG2 promotes progression of stomatal differentiation in both types of organ, and that AtML1, but not PDF2, shares a redundant role in controlling the timing of meristemoid differentiation.
HDG2 is a transcriptional activator that binds to L1 and HAHR1 boxes
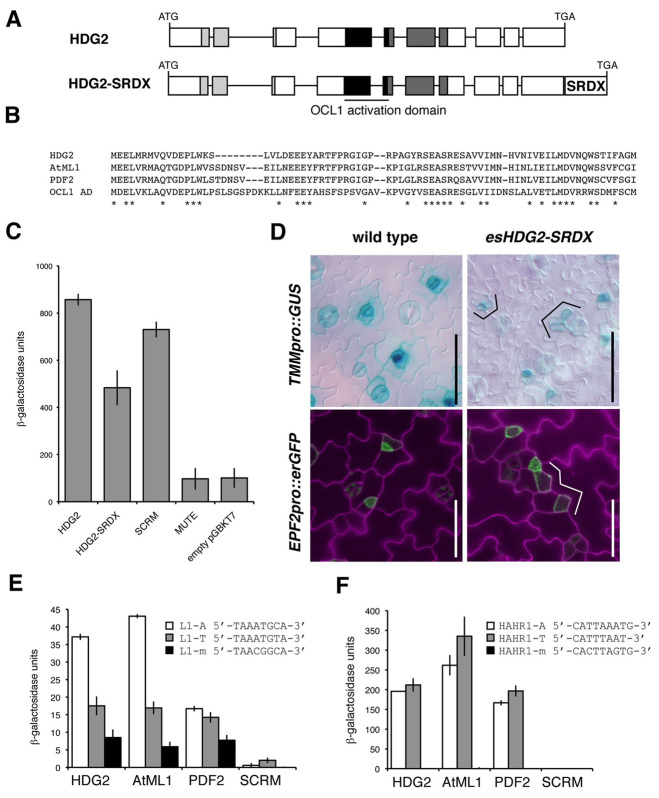
To gain insight into the mode of action of HDG2, we first tested whether HDG2 activates transcription, and, if so, whether forceful conversion of HDG2 into a transcriptional repressor mimics its loss-of-function effects (Fig. 6). HDG2 shares a sequence similarity with the known activation domain of the maize HD-ZIP IV protein OCL1 (Depège-Fargeix et al., 2011) (Fig. 6A,B). Indeed, HDG2 fused to Gal4 DNA-binding domain strongly transactivated the reporter in yeast at an equivalent level to known transcriptional activator SCRM/ICE1 (Fig. 6C), indicating that HDG2 is a transcriptional activator.
Fig. 6.
HDG2 is a transcriptional activator that can bind to both L1 box and HAHR1 box. (A) The HDG2 gene, from translational initiation codon to termination codon. Boxes indicate exons; lines indicate introns; light-gray boxes indicate homeodomain; dark-gray boxes indicate START domain, which also contains a transcriptional activation domain of maize OCL1 (black boxes). HDG2-SRDX has additional 12 amino acids (LDLDLELRLGFA) for forced gene repression. (B) Sequence similarity of HDG2 with AtML1, PDF2 and OCL1 activation domain region. (C) Transcriptional autoactivation of reporter lacZ gene in yeast by HDG2. Addition of SRDX reduces autoactivation. SCRM and MUTE, a known transcriptional activator and repressor, respectively (Kanaoka et al., 2008), were used as positive and negative controls. (D) Induced overexpression of HDG2-SRDX confers an epidermal phenotype with delayed stomatal precursors, resulting in expanded SLGC-like cells (brackets). Shown are 10-day-old cotyledon abaxial epidermis of wild type (left) and esHDG2-SRDX (right) expressing TMMpro::GUS (top) and EPF2pro::erGFP (bottom). Scale bars: 50 μm. (E) Yeast one-hybrid analysis for activation of lacZ reporter possessing 5′ upstream L1-A-box (white bars), L1-T-box (gray bars) or L1-m-box (black bars) by HDG2, AtML1 or PDF2. SCRM, a known E-box binder (Chinnusamy et al., 2003), was used as a negative control. Bars represent mean values of triplicates. Error bars indicate s.e.m. (F) Yeast one-hybrid analysis for activation of lacZ reporter possessing 5′ upstream HAHR1-A-box (white bars), HAHR1-T-box (gray bars) or HAHR1-m-box (black bars), respectively, by HDG2, AtML1 or PDF2. SCRM was used as a negative control. Bars represent mean value of triplicates. Error bars indicate s.e.m.
C-terminal addition of the 12 amino acid repressor domain SRDX is known to convert plant TFs to dominant repressors (Hiratsu et al., 2003). The addition of the SRDX sequence significantly decreased HDG2 reporter transactivation in yeast (Fig. 6C). To test whether HDG2-SRDX mimics loss of function of HDG2 in vivo, we generated transgenic Arabidopsis carrying inducible HDG2-SRDX (Fig. 6D; supplementary material Fig. S3). The HDG2-SRDX induction conferred delayed progression of meristemoids and patches of SLGCs (Fig. 6D). Expression of stomatal-lineage markers TMMpro::GUS and EPF2::erGFP supports the stomatal-lineage origins of these aberrant cells (Fig. 6D).
HD-ZIP IV-family proteins, including AtML1 and PDF2, show in vitro binding to the pseudopalindromic sequences known as the L1 box [5′-TAAATG(C/T)A-3′] and HAHR1 box [5′-CATT(A/T)AATG-3′] (Abe et al., 2003; Abe et al., 2001; Tron et al., 2001). Previous genome-wide study of AtHD-ZIP proteins did not include DNA-binding assays of HDG2 (Nakamura et al., 2006). We therefore performed yeast one-hybrid analysis to test whether HDG2 binds to these DNA sequences (Fig. 6E,F). The reporter assay shows that HDG2 binds to both L1-A (5′-TAAATGCA-3′) and L1-T (5′-TAAATGTA-3′) as strongly as AtML1 (Fig. 6E). HDG2, AtML1 and PDF2 all induced strong lacZ reporter activity when fused with the HAHR1-T (5′-CATTTAATG-3′) and HAHR1-A (5′-CATTAAATG-3′) boxes, with AtML1 inducing the highest HAHR1-reporter activity (Fig. 6F). These three HD-ZIP IV proteins did not induce reporter activity when key DNA residues of the L1 and HAHR1 boxes were mutated (L1-m and HAHR1-m; Fig. 6E,F), confirming the sequence specificity of binding. Similarly, a control bHLH protein, SCRM/ICE1, which has been shown to bind in vitro to an E-box element (CANNTG) (Chinnusamy et al., 2003), did not induce any reporter activity (Fig. 6E,F). These results suggest that HDG2 acts as a transcriptional activator that binds to L1 and HAHR1 boxes. The similar in vitro binding of HDG2 and AtML1 to these cis-regulatory elements further emphasizes the possibility that they share common downstream targets.
HDG2 transactivates promoters of stomatal development genes in planta
To address the molecular connection of HDG2, an L1/HAHR1-binding transcriptional activator, with known regulators of stomatal development, we first performed a bioinformatic analysis and surveyed cis-regulatory elements within the promoters of genes regulating stomatal development. Among the 18 genes surveyed, a significant (P=0.003) enrichment of L1-box motifs in their promoter regions was revealed (supplementary material Table S4). Indeed, the P value for significance of enrichment for the L1 box was the lowest among the 47 motifs analyzed (supplementary material Table S4).
To demonstrate that HDG2 activates transcription of genes regulating stomatal development, we generated reporter luciferase constructs containing native promoters of TMM and MUTE, which possess one and two L1 boxes, respectively, within their proximal regions (Fig. 7A). These reporter and effector constructs were co-expressed in N. benthamiana. High luciferase activity was observed in a combination of HDG2 and TMMpro::LUC, and even higher activity with MUTEpro::LUC, consistent with the number of L1 boxes present (Fig. 7B). Therefore, HDG2 is capable of directly transactivating TMM and MUTE in planta, supporting the possibility that HDG2 promotes stomatal differentiation via binding to the promoters and activating the expression of genes regulating stomatal development.
Fig. 7.
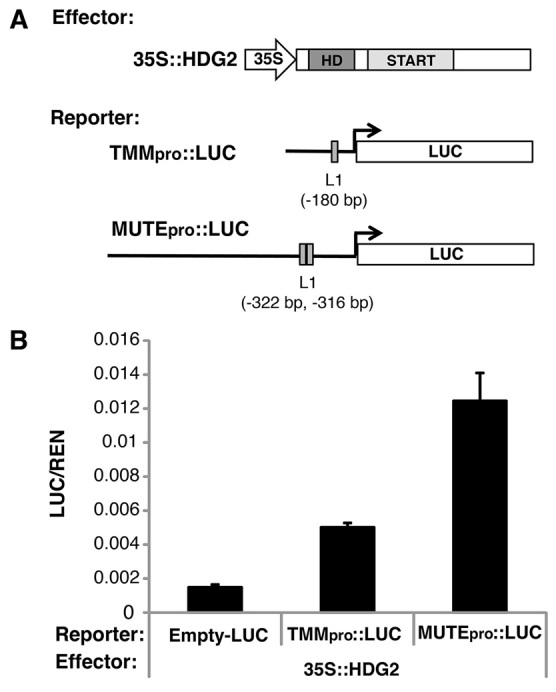
HDG2 transactivates TMM and MUTE promoters in planta. (A) Effector and reporter constructs used. The exact locations of L1 boxes are indicated. (B) Relative luciferase activity in N. benthamiana transiently transformed with 35S::HDG2 and the indicated reporters. Luciferase activities were measured 7 days after infiltration. Relative luciferase activities are shown by firefly luciferase (LUC) activities normalized to renilla luciferase (REN) activities. Bars indicate mean; error bars indicate s.e.m. (n=4).
DISCUSSION
Our work reveals the unique role of the AtHD-ZIP IV protein HDG2 as a transcriptional activator that promotes stomatal differentiation. HDG2 is necessary for progression of stomatal cell lineages and, most strikingly, is sufficient to drive stomatal differentiation in internal mesophyll tissues. The internal stomata in HDG2-OX are normal in appearance, and thus different from FAMA-OX, which confers abnormally shaped mesophyll cells expressing mature GC markers (Ohashi-Ito and Bergmann, 2006). Our data support the idea that HDG2-OX upregulates whole sets of gene regulatory cascades, which leads to stomatal differentiation. As HDG2 accumulates at high levels in actively proliferating stomatal cell lineages, the stomatal bHLH proteins may promote HDG2, which in turn promotes stomatal bHLHs for reinforcing stomatal differentiation. The transactivation of TMM and MUTE promoters in planta by HDG2 (Fig. 7) strongly supports this idea.
In addition, HDG2-OX confers multiple epidermal layers, indicating that it also specifies protodermal identity. In vitro HD-binding site selection experiments have revealed 5′-GCATTATTTTGC-3′ as consensus sequence for four HD-ZIP IVs: HDG7, HDG9, AtML1 and PDF2 (Nakamura et al., 2006). The well-described L1 and HAHR1 boxes overlap with this consensus sequence (Abe et al., 2001; Tron et al., 2001). Although this experiment has not been performed for HDG2, it is most likely that HDG2 and AtML1, when overexpressed, bind to common in vivo target sequences for the following reasons. First, both HDG2 and AtML1 bind to L1 and HAHR1 boxes to similar extents in yeast (Fig. 6E,F). Second, four amino acid residues predicted to serve as a DNA sequence-specific contact surface (F47, Q50, N51 and T54) within the HD domain are absolutely conserved in HDG2 and AtML1 (Gehring et al., 1994; Nakamura et al., 2006). Third, AtML1-OX confers ectopic internal stomatal differentiation, phenocopying HDG2-OX (Fig. 4B). Future genome-wide identification and comparison of their target genes will address the extent of unique versus shared components of gene regulatory networks specified by HDG2 and AtML1.
Autoregulation of transcription factors is crucial for robust specification of cell fate. AtML1 auto-activates by directly binding to the L1 box in its own promoter to reinforce protodermal identity (Abe et al., 2003; Takada and Jürgens, 2007). That HDG2-OX triggers ectopic AtML1 expression (Fig. 3B) is consistent with the presence of such an auto-activation loop, in this case initially triggered by the ectopic HDG2. The HDG2 promoter also possesses L1 box (Nakamura et al., 2006), and its auto-activation may contribute to robust progression of stomatal development. Specification of the protoderm (L1 layer) in Arabidopsis involves cell-cell signaling mediated by receptor-like kinases ARABIDOPSIS CRINKLY4 (ACR4) and ABNORMAL LEAF SHAPE2 (ALE2), a putative cysteine peptidase DEFECTIVE KERNEL1 (AtDEK1) and a putative subtilase ALE1 (Becraft et al., 1996; Cao et al., 2005; Johnson et al., 2005; Tanaka et al., 2001; Tanaka et al., 2007). The signaling pathways appear to act upstream of AtML1 and PDF2, which in turn may promote expression of AtDEK1 and ALE2 as part of a positive-feedback loop (Javelle et al., 2011b). Although these signaling components are required for normal epidermal differentiation, none of them has been shown to drive supernumerary epidermal differentiation. Our work additionally establishes AtML1 as a master regulatory transcription factor that is both necessary and sufficient for the protodermal identity. It is surprising that an AtML1 overexpression phenotype has not been reported since its identification in 1996 (Lu et al., 1996). PDF2-OX confers a later-flowering phenotype, likely mimicking the effects of a distantly related AtHD-ZIP IV, FWA (Abe et al., 2003); thus, HDG2 may share more common targets with AtML1 than PDF2.
Although our ectopic overexpression study revealed the equivalence of HDG2 and AtML1, expression patterns and loss-of-function phenotypes distinguish their endogenous functions and provide evidence for a major role of HDG2 in stomatal development. We further uncovered an unexpected redundant role for AtML1 in promoting stomatal differentiation, which was not shared by PDF2. Thus, these HD-ZIP IVs have unique yet partially overlapping functions in specifying multiple aspects of epidermal development. Bioinformatic co-expression network ATTED II (Obayashi et al., 2009) places HDG2 at the ‘hub’ connecting stomatal and protodermal regulators: HDG2 clusters with SPCH, TMM and SCRM/ICE1, with a direct connection to SCRM/ICE1 (supplementary material Fig. S6). At the same time, HDG2 is directly connected to AtML1 and clusters together with PDF2, HDG1 and ERECTA, a receptor kinase that inhibits stomatal-lineage initiation (Shpak et al., 2005) (supplementary material Fig. S6).
The meristemoid-to-GMC transition represents a key step from proliferation to differentiation within the stomatal lineages, and is specified by MUTE and SCRM. Loss of HDG2 may delay the onset of MUTE or SCRM expression or deregulate their functions in a small fraction of meristemoids. The observation that MUTE can be directly activated by HDG2 (Fig. 7) supports this hypothesis. scrm loss-of-function mutant produces aberrant stomatal complexes at low penetrance (Kanaoka et al., 2008), a phenotype shared by hdg2 (Fig. 5L-N). The very low penetrance of hdg2 stomatal phenotype may be due to functional redundancy with the remaining members of the AtHD-ZIP IV family or, alternatively, HDG2 could indirectly influences stomatal development through promoting cell differentiation.
Proper stomatal differentiation requires an intricate balance of cell proliferation and differentiation through coordinated activities of cell-cycle regulators and transcription factors. Among plant cell-cycle regulators, A-type cyclin CycA2;3 and its partner CDKB1;1 are required for GMC divisions, and their losses of function lead to singular GC phenotype (Boudolf et al., 2004; Vanneste et al., 2011). Myb transcription factors FOUR LIPS (FLP) and Myb88 directly bind to the promoters of CycA2;3 and CDKB1;1 and repress their expression (Vanneste et al., 2011; Xie et al., 2010). Owing to the very low-penetrant hdg2 stomatal phenotype, it is difficult to draw clear connections between HDG2 and these regulators. Nevertheless, it is tempting to speculate on a potential role of HDG2 in cell proliferation and differentiation. Interestingly, in addition to meristemoids, HDG2-GFP is highly expressed in developing trichomes, where active endoreduplication occurs (supplementary material Fig. S7) (Hülskamp, 2004). hdg2 trichomes do not have growth/morphological phenotypes, but exhibit altered cuticle textures (Marks et al., 2009). Further studies, such as cell type-specific manipulation of HDG2 activity and identification of its targets, may provide a direct link between HDG2-mediated epidermal identity, cell proliferation and stomatal differentiation.
Supplementary Material
Acknowledgments
We thank Lynn Pillitteri, Hongya Gu, Nam-Hai Chua, Tsuyoshi Nakagawa, John Schiefelbein, Xing-Wang Deng, Adrienne Roeder and Naoyuki Uchida for kindly providing us with plasmids and reporter lines; Valerie Soza for molecular phylogenetic analysis; Ya-Chen Lin for plant care; Hatsumi Fukada for assisting with cloning; Janneke Hille Ris Lambers for assistance in statistical analysis; Wim Lewis for assistance with cell counting; Shinobu Takada for communicating results; and Lynn Pillitteri and Jin Suk Lee for comments.
Footnotes
Funding
The work was initially supported by the University of Washington Royalty Research Fund and subsequently by the Japan Science and Technology Agency-Precursory Research for Embryonic Science and Technology award and National Science Foundation [MCB-0855659] to K.U.T.; and by Japan Society for the Promotion of Science KAKENHI [23012021, 24113510 and 4570045 to M.M.K. and 23012014 to Y.S.]. K.M.P. was an NSF Graduate Research Fellow and K.U.T. is an Howard Hughes Medical Institute-Gordon and Betty Moore Foundation investigator. Deposited in PMC for release after 6 months.
Competing interests statement
The authors declare no competing financial interests.
Author contributions
K.M.P. and K.U.T. conceived and designed the research; K.M.P., C.S., C.A.B., R.J.H. and K.U.T. performed the research apart from the in situ hybridization (carried out by (M.M.K., M.O. and Y.S.); K.M.P., C.S., C.A.B., M.M.K. and K.U.T. generated research materials and reagents; K.M.P., R.J.H. and K.U.T. analyzed the data; K.U.T. wrote the manuscript.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.090209/-/DC1
References
- Abascal F., Zardoya R., Posada D. (2005). ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21, 2104–2105 [DOI] [PubMed] [Google Scholar]
- Abe M., Takahashi T., Komeda Y. (2001). Identification of a cis-regulatory element for L1 layer-specific gene expression, which is targeted by an L1-specific homeodomain protein. Plant J. 26, 487–494 [DOI] [PubMed] [Google Scholar]
- Abe M., Katsumata H., Komeda Y., Takahashi T. (2003). Regulation of shoot epidermal cell differentiation by a pair of homeodomain proteins in Arabidopsis. Development 130, 635–643 [DOI] [PubMed] [Google Scholar]
- Akaike H. (1974). A new look at the statistical model identification. IEEE Trans. Automatic Control AC 19, 716–723 [Google Scholar]
- Becraft P. W., Stinard P. S., McCarty D. R. (1996). CRINKLY4: A TNFR-like receptor kinase involved in maize epidermal differentiation. Science 273, 1406–1409 [DOI] [PubMed] [Google Scholar]
- Boudolf V., Barrôco R., Engler J. A., Verkest A., Beeckman T., Naudts M., Inzé D., De Veylder L. (2004). B1-type cyclin-dependent kinases are essential for the formation of stomatal complexes in Arabidopsis thaliana. Plant Cell 16, 945–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X., Li K., Suh S. G., Guo T., Becraft P. W. (2005). Molecular analysis of the CRINKLY4 gene family in Arabidopsis thaliana. Planta 220, 645–657 [DOI] [PubMed] [Google Scholar]
- Chinnusamy V., Ohta M., Kanrar S., Lee B. H., Hong X., Agarwal M., Zhu J. K. (2003). ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 17, 1043–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depège-Fargeix N., Javelle M., Chambrier P., Frangne N., Gerentes D., Perez P., Rogowsky P. M., Vernoud V. (2011). Functional characterization of the HD-ZIP IV transcription factor OCL1 from maize. J. Exp. Bot. 62, 293–305 [DOI] [PubMed] [Google Scholar]
- Dong J., Bergmann D. C. (2010). Stomatal patterning and development. Curr. Top. Dev. Biol. 91, 267–297 [DOI] [PubMed] [Google Scholar]
- Gehring W. J., Qian Y. Q., Billeter M., Furukubo-Tokunaga K., Schier A. F., Resendez-Perez D., Affolter M., Otting G., Wüthrich K. (1994). Homeodomain-DNA recognition. Cell 78, 211–223 [DOI] [PubMed] [Google Scholar]
- Guseman J. M., Lee J. S., Bogenschutz N. L., Peterson K. M., Virata R. E., Xie B., Kanaoka M. M., Hong Z., Torii K. U. (2010). Dysregulation of cell-to-cell connectivity and stomatal patterning by loss-of-function mutation in Arabidopsis chorus (glucan synthase-like 8). Development 137, 1731–1741 [DOI] [PubMed] [Google Scholar]
- Hara K., Yokoo T., Kajita R., Onishi T., Yahata S., Peterson K. M., Torii K. U., Kakimoto T. (2009). Epidermal cell density is autoregulated via a secretory peptide, EPIDERMAL PATTERNING FACTOR 2 in Arabidopsis leaves. Plant Cell Physiol. 50, 1019–1031 [DOI] [PubMed] [Google Scholar]
- Hellens R. P., Allan A. C., Friel E. N., Bolitho K., Grafton K., Templeton M. D., Karunairetnam S., Gleave A. P., Laing W. A. (2005). Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1, 13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratsu K., Matsui K., Koyama T., Ohme-Takagi M. (2003). Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 34, 733–739 [DOI] [PubMed] [Google Scholar]
- Huelsenbeck J. P., Ronquist F. (2001). MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17, 754–755 [DOI] [PubMed] [Google Scholar]
- Hülskamp M. (2004). Plant trichomes: a model for cell differentiation. Nat. Rev. Mol. Cell Biol. 5, 471–480 [DOI] [PubMed] [Google Scholar]
- Ingram G. C., Boisnard-Lorig C., Dumas C., Rogowsky P. M. (2000). Expression patterns of genes encoding HD-ZipIV homeo domain proteins define specific domains in maize embryos and meristems. Plant J. 22, 401–414 [DOI] [PubMed] [Google Scholar]
- Javelle M., Vernoud V., Depège-Fargeix N., Arnould C., Oursel D., Domergue F., Sarda X., Rogowsky P. M. (2010). Overexpression of the epidermis-specific homeodomain-leucine zipper IV transcription factor Outer Cell Layer1 in maize identifies target genes involved in lipid metabolism and cuticle biosynthesis. Plant Physiol. 154, 273–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javelle M., Klein-Cosson C., Vernoud V., Boltz V., Maher C., Timmermans M., Depège-Fargeix N., Rogowsky P. M. (2011a). Genome-wide characterization of the HD-ZIP IV transcription factor family in maize: preferential expression in the epidermis. Plant Physiol. 157, 790–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javelle M., Vernoud V., Rogowsky P. M., Ingram G. C. (2011b). Epidermis: the formation and functions of a fundamental plant tissue. New Phytol. 189, 17–39 [DOI] [PubMed] [Google Scholar]
- Johnson K. L., Degnan K. A., Ross Walker J., Ingram G. C. (2005). AtDEK1 is essential for specification of embryonic epidermal cell fate. Plant J. 44, 114–127 [DOI] [PubMed] [Google Scholar]
- Kanaoka M. M., Pillitteri L. J., Fujii H., Yoshida Y., Bogenschutz N. L., Takabayashi J., Zhu J. K., Torii K. U. (2008). SCREAM/ICE1 and SCREAM2 specify three cell-state transitional steps leading to arabidopsis stomatal differentiation. Plant Cell 20, 1775–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouchi H., Hata S. (1993). Isolation and characterization of novel nodulin cDNAs representing genes expressed at early stages of soybean nodule development. Mol. Gen. Genet. 238, 106–119 [DOI] [PubMed] [Google Scholar]
- Lee J. S., Kuroha T., Hnilova M., Khatayevich D., Kanaoka M. M., McAbee J. M., Sarikaya M., Tamerler C., Torii K. U. (2012). Direct interaction of ligand-receptor pairs specifying stomatal patterning. Genes Dev. 26, 126–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Li G., Gao S., Martinez C., He G., Zhou Z., Huang X., Lee J.-H., Zhang H., Shen Y., et al. (2010). Arabidopsis transcription factor ELONGATED HYPOCOTYL5 plays a role in the feedback regulation of phytochrome A signaling. Plant Cell 22, 3634–3649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- Lu P., Porat R., Nadeau J. A., O’Neill S. D. (1996). Identification of a meristem L1 layer-specific gene in Arabidopsis that is expressed during embryonic pattern formation and defines a new class of homeobox genes. Plant Cell 8, 2155–2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAlister C. A., Ohashi-Ito K., Bergmann D. C. (2007). Transcription factor control of asymmetric cell divisions that establish the stomatal lineage. Nature 445, 537–540 [DOI] [PubMed] [Google Scholar]
- Marks M. D., Wenger J. P., Gilding E., Jilk R., Dixon R. A. (2009). Transcriptome analysis of Arabidopsis wild-type and gl3-sst sim trichomes identifies four additional genes required for trichome development. Mol. Plant 2, 803–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masucci J. D., Rerie W. G., Foreman D. R., Zhang M., Galway M. E., Marks M. D., Schiefelbein J. W. (1996). The homeobox gene GLABRA2 is required for position-dependent cell differentiation in the root epidermis of Arabidopsis thaliana. Development 122, 1253–1260 [DOI] [PubMed] [Google Scholar]
- Miller M. A., Pfeiffer W., Schwartz T. (2010). Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Proceedings of the Gateway Computing Environments Workshop (GCE), pp. 1–8 New Orleans, LA: IEEE; [Google Scholar]
- Mukherjee K., Brocchieri L., Bürglin T. R. (2009). A comprehensive classification and evolutionary analysis of plant homeobox genes. Mol. Biol. Evol. 26, 2775–2794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau J. A., Sack F. D. (2002). Control of stomatal distribution on the Arabidopsis leaf surface. Science 296, 1697–1700 [DOI] [PubMed] [Google Scholar]
- Nakagawa T., Suzuki T., Murata S., Nakamura S., Hino T., Maeo K., Tabata R., Kawai T., Tanaka K., Niwa Y., et al. (2007). Improved Gateway binary vectors: high-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci. Biotechnol. Biochem. 71, 2095–2100 [DOI] [PubMed] [Google Scholar]
- Nakagawa T., Nakamura S., Tanaka K., Kawamukai M., Suzuki T., Nakamura K., Kimura T., Ishiguro S. (2008). Development of R4 gateway binary vectors (R4pGWB) enabling high-throughput promoter swapping for plant research. Biosci. Biotechnol. Biochem. 72, 624–629 [DOI] [PubMed] [Google Scholar]
- Nakamura M., Katsumata H., Abe M., Yabe N., Komeda Y., Yamamoto K. T., Takahashi T. (2006). Characterization of the class IV homeodomain-Leucine Zipper gene family in Arabidopsis. Plant Physiol. 141, 1363–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor T. R., Dyreson C., Wyrick J. J. (2005). Athena: a resource for rapid visualization and systematic analysis of Arabidopsis promoter sequences. Bioinformatics 21, 4411–4413 [DOI] [PubMed] [Google Scholar]
- Obayashi T., Hayashi S., Saeki M., Ohta H., Kinoshita K. (2009). ATTED-II provides coexpressed gene networks for Arabidopsis. Nucleic Acids Res. 37, D987–D991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi-Ito K., Bergmann D. C. (2006). Arabidopsis FAMA controls the final proliferation/differentiation switch during stomatal development. Plant Cell 18, 2493–2505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillitteri L. J., Torii K. U. (2012). Mechanisms of stomatal development. Annu. Rev. Plant Biol. 63, 591–614 [DOI] [PubMed] [Google Scholar]
- Pillitteri L. J., Sloan D. B., Bogenschutz N. L., Torii K. U. (2007). Termination of asymmetric cell division and differentiation of stomata. Nature 445, 501–505 [DOI] [PubMed] [Google Scholar]
- Pillitteri L. J., Bogenschutz N. L., Torii K. U. (2008). The bHLH protein, MUTE, controls differentiation of stomata and the hydathode pore in Arabidopsis. Plant Cell Physiol. 49, 934–943 [DOI] [PubMed] [Google Scholar]
- Pillitteri L. J., Peterson K. M., Horst R. J., Torii K. U. (2011). Molecular profiling of stomatal meristemoids reveals new component of asymmetric cell division and commonalities among stem cell populations in Arabidopsis. Plant Cell 23, 3260–3275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt R. E., Vielle-Calzada J.-P., Ploense S. E., Grossniklaus U., Lolle S. J. (2000). FIDDLEHEAD, a gene required to suppress epidermal cell interactions in Arabidopsis, encodes a putative lipid biosynthetic enzyme. Proc. Natl. Acad. Sci. USA 97, 1311–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rerie W. G., Feldmann K. A., Marks M. D. (1994). The GLABRA2 gene encodes a homeo domain protein required for normal trichome development in Arabidopsis. Genes Dev. 8, 1388–1399 [DOI] [PubMed] [Google Scholar]
- Roeder A. H. K., Cunha A., Ohno C. K., Meyerowitz E. M. (2012). Cell cycle regulates cell type in the Arabidopsis sepal. Development 139, 4416–4427 [DOI] [PubMed] [Google Scholar]
- Ronquist F., Huelsenbeck J. P. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574 [DOI] [PubMed] [Google Scholar]
- Serna L., Martin C. (2006). Trichomes: different regulatory networks lead to convergent structures. Trends Plant Sci. 11, 274–280 [DOI] [PubMed] [Google Scholar]
- Sessions A., Weigel D., Yanofsky M. F. (1999). The Arabidopsis thaliana MERISTEM LAYER 1 promoter specifies epidermal expression in meristems and young primordia. Plant J. 20, 259–263 [DOI] [PubMed] [Google Scholar]
- Shpak E. D., Lakeman M. B., Torii K. U. (2003). Dominant-negative receptor uncovers redundancy in the Arabidopsis ERECTA Leucine-rich repeat receptor-like kinase signaling pathway that regulates organ shape. Plant Cell 15, 1095–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpak E. D., McAbee J. M., Pillitteri L. J., Torii K. U. (2005). Stomatal patterning and differentiation by synergistic interactions of receptor kinases. Science 309, 290–293 [DOI] [PubMed] [Google Scholar]
- Takada S., Jürgens G. (2007). Transcriptional regulation of epidermal cell fate in the Arabidopsis embryo. Development 134, 1141–1150 [DOI] [PubMed] [Google Scholar]
- Tanaka H., Onouchi H., Kondo M., Hara-Nishimura I., Nishimura M., Machida C., Machida Y. (2001). A subtilisin-like serine protease is required for epidermal surface formation in Arabidopsis embryos and juvenile plants. Development 128, 4681–4689 [DOI] [PubMed] [Google Scholar]
- Tanaka H., Watanabe M., Sasabe M., Hiroe T., Tanaka T., Tsukaya H., Ikezaki M., Machida C., Machida Y. (2007). Novel receptor-like kinase ALE2 controls shoot development by specifying epidermis in Arabidopsis. Development 134, 1643–1652 [DOI] [PubMed] [Google Scholar]
- Tron A. E., Bertoncini C. W., Palena C. M., Chan R. L., Gonzalez D. H. (2001). Combinatorial interactions of two amino acids with a single base pair define target site specificity in plant dimeric homeodomain proteins. Nucleic Acids Res. 29, 4866–4872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truernit E., Bauby H., Dubreucq B., Grandjean O., Runions J., Barthélémy J., Palauqui J. C. (2008). High-resolution whole-mount imaging of three-dimensional tissue organization and gene expression enables the study of Phloem development and structure in Arabidopsis. Plant Cell 20, 1494–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida N., Lee J. S., Horst R. J., Lai H.-H., Kajita R., Kakimoto T., Tasaka M., Torii K. U. (2012). Regulation of inflorescence architecture by intertissue layer ligand-receptor communication between endodermis and phloem. Proc. Natl. Acad. Sci. USA 109, 6337–6342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste S., Coppens F., Lee E., Donner T. J., Xie Z., Van Isterdael G., Dhondt S., De Winter F., De Rybel B., Vuylsteke M., et al. (2011). Developmental regulation of CYCA2s contributes to tissue-specific proliferation in Arabidopsis. EMBO J. 30, 3430–3441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R., Li S., He S., Wassmann F., Yu C., Qin G., Schreiber L., Qu L. J., Gu H. (2011). CFL1, a WW domain protein, regulates cuticle development by modulating the function of HDG1, a class IV homeodomain transcription factor, in rice and Arabidopsis. Plant Cell 23, 3392–3411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Lee E., Lucas J. R., Morohashi K., Li D., Murray J. A., Sack F. D., Grotewold E. (2010). Regulation of cell proliferation in the stomatal lineage by the Arabidopsis MYB FOUR LIPS via direct targeting of core cell cycle genes. Plant Cell 22, 2306–2321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. H., Rannala B. (1997) Bayesian phylogenetic inference using DNA sequences: a Markov Chain Monte Carlo method. Mol. Biol. Evol. 14, 717–724 [DOI] [PubMed] [Google Scholar]
- Zuo J., Niu Q. W., Chua N. H. (2000). Technical advance: an estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J. 24, 265–273 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.