Abstract
The compound eye of Drosophila melanogaster is configured by a differentiating wave, the morphogenetic furrow, that sweeps across the eye imaginal disc and transforms thousands of undifferentiated cells into a precisely ordered repetitive array of 800 ommatidia. The initiation of the furrow at the posterior margin of the epithelium and its subsequent movement across the eye field is controlled by the activity of the Hedgehog (Hh) signaling pathway. Differentiating photoreceptors that lie behind the furrow produce and secrete the Hh morphogen, which is captured by cells within the furrow itself. This leads to the stabilization of the full-length form of the zinc-finger transcription factor Cubitus interruptus (Ci155), the main effector of Hh signaling. Ci155 functions as a transcriptional activator of a number of downstream targets, including decapentaplegic (dpp), a TGFβ homolog. In this report, we describe a mechanism that is in place within the fly retina to limit Hh pathway activity within and ahead of the furrow. We demonstrate that the helix-loop-helix (HLH) protein Extramacrochaetae (Emc) regulates Ci155 levels. Loss of emc leads to an increase in Ci155 levels, nuclear migration, apical cell constriction and an acceleration of the furrow. We find that these roles are distinct from the bHLH protein Hairy (H), which we show restricts atonal (ato) expression ahead of the furrow. Secondary furrow initiation along the dorsal and ventral margins is blocked by the activity of the Wingless (Wg) pathway. We also show that Emc regulates and cooperates with Wg signaling to inhibit lateral furrow initiation.
Keywords: Extramacrochaetae, Hairy, Hedgehog, Suppressor of fused, Wingless, Morphogenetic furrow, Eye
INTRODUCTION
Overt patterning of the Drosophila retina begins during the third and final larval instar when a wave of morphogenesis initiates at the posterior margin of the eye imaginal disc and proceeds across the epithelium towards the eye-antenna border. The anterior-most edge of this differentiating wave can be visualized by a dorso-ventral groove in the epithelium known as the morphogenetic furrow (Ready et al., 1976). As the furrow traverses the retinal epithelium, the field of non-patterned and undifferentiated cells is transformed into columns of periodically spaced unit eyes called ommatidia. Because the retina continues to grow while it is patterned, the movement of the furrow across the eye disc must be synchronized with the rate of cell proliferation. The phenotypes of several furrow-stop mutants such as hhbar3 and Drop1 demonstrate that if the two are uncoupled the resulting adult will contain too few ommatidia (Ives, 1950; Heberlein et al., 1993; Ma et al., 1993; Mozer, 2001). In addition, correct patterning is achieved when only a single furrow initiates and moves across the eye disc. The initiation of multiple furrows from the margins results in a small, disorganized eye (Ma and Moses, 1995; Chanut and Heberlein, 1997b; Pignoni and Zipursky, 1997).
Furrow initiation is restricted to the intersection of the posterior margin and the midline (called the posterior center) by the JAK/STAT, Hedgehog (Hh) and Wingless (Wg) signaling pathways. Both hh and unpaired (upd), the ligand for the JAK/STAT pathway, are expressed at the posterior center (Domínguez and Hafen, 1997; Borod and Heberlein, 1998; Ekas et al., 2006; Tsai et al., 2007). Removal of either pathway from the posterior center prevents furrow initiation, while ectopic expression along the margins or within the anterior regions of the disc results in ectopic furrow initiation (Heberlein et al., 1995; Ma and Moses, 1995; Pan and Rubin, 1995; Strutt et al., 1995; Wehrli and Tomlinson, 1995; Wiersdorff et al., 1996; Domínguez and Hafen, 1997; Borod and Heberlein, 1998). An additional input into furrow initiation is the EGF receptor pathway, as its removal also prevents the furrow from initiating (Kumar and Moses, 2001). By contrast, wg is absent from the posterior center and is instead expressed along the lateral margins (Baker, 1988; Ma and Moses, 1995; Treisman and Rubin, 1995). Inhibition of Wg signaling leads to ectopic furrow initiation at the margins, whereas overexpression of wg within the eye field proper is sufficient to block furrow initiation and progression (Ma and Moses, 1995; Treisman and Rubin, 1995; Ekas et al., 2006; Tsai et al., 2007).
The progression of the furrow across the eye field is under the control of Hh signaling. The Hh signal is produced and secreted in differentiating photoreceptors and received by cells that reside within the furrow. Reception of the Hh signal prevents the cleavage of the full-length form of Cubitus Interruptus (Ci155 or CiACT) into its shorter repressor form (Ci75 or CiR) (Pappu et al., 2003). Hh signals through CiACT to activate transcription of decapentaplegic (dpp), which encodes a TGFβ homolog, within the morphogenetic furrow (Heberlein et al., 1993; Borod and Heberlein, 1998; Greenwood and Struhl, 1999). Reducing hh expression within developing photoreceptors leads to the loss of dpp expression and a furrow-stop phenotype (Ives, 1950; Mohler, 1988; Ma et al., 1993; Heberlein et al., 1993). It appears, however, that dpp plays only a minor role in furrow progression as clones lacking downstream components of the Dpp pathway only slow the furrow (Burke and Basler, 1996; Wiersdorff et al., 1996; Greenwood and Struhl, 1999; Curtiss and Mlodzik, 2000). Similarly, although dpp is necessary for furrow initiation and is sufficient to induce ectopic furrows, it appears to promote furrow initiation by a feedback loop that activates hh expression (Chanut and Heberblein, 1997a; Pignoni and Zipursky, 1997; Borod and Heberlein, 1998). Thus, it appears that Hh signaling is the main regulator of furrow initiation and progression.
Here, we focus on the role that extramacrochaetae (emc) plays in refining the activity of the Hh signaling gradient during furrow progression. Emc, a helix-loop-helix (HLH) protein, modulates transcription by sequestering basic helix-loop-helix (bHLH) proteins away from target DNA enhancers (Ellis et al., 1990; Garrell and Modolell, 1990; Van Doren et al., 1991). The furrow accelerates through emc-null mutant clones suggesting that during normal development Emc functions to regulate the rate of pattern formation (Bhattacharya and Baker, 2009). However, a clear mechanism by which this is achieved has remained elusive. Here, we show that Emc adjusts the rate at which the furrow moves across the eye field by regulating the levels of the active form of Ci (CiACT). This is accomplished in part through the regulation of Suppressor of fused [Su(fu)], a member of the Hh cascade. Its role in the pathway is poorly understood but here we show that Su(fu) is required to stabilize CiACT levels. We go on to demonstrate that increasing the level of CiACT alone is sufficient to accelerate the pace at which the furrow progresses. We find that emc is also required for regulating two cellular features of the furrow: nuclear migration and apical surface constrictions. These roles are different from that of the bHLH protein Hairy (H), which has also been implicated in regulating furrow progression. We demonstrate that H is required for suppressing precocious neurogenesis ahead of the furrow through repression of atonal (ato). Finally, we demonstrate that emc functions upstream of wg to prevent furrow initiation along the margins. Overall, our results indicate that the role of Emc in eye development is to impose order on the retina by regulating the activity of the Hh and Wg signaling pathways.
MATERIALS AND METHODS
Fly strains and mosaic clones
The following fly stocks were used: (1) w1118; (2) emc-GFPYB0040; (3) emc-GFPYB0067; (4) emc-GFPYB0217; (5) emc-GFPCB03369; (6) P(PZ) emc04322 ry506/TM3; (7) y w hs-flp[22]; FRT80B emcAP6/TM6B; (8) y w hs-flp[22]; FRT80B M(3)i55 Ubi-GFP; (9) y w eyflp; (10) y w hs-flp[22]; (11) Act5C >y+ >GAL4, UAS-GFP; (12) UAS-emc4M; (13) UAS-emc5M; (14) FRT80B h22/TM3; (15) FRT80B h22 emcAP6/TM3; (16) UAS-Su(fu); (17) UAS-Su(fu) RNAi; (18) UAS-ci; (19) UAS-ci RNAi; (20) UAS-h; (21) UAS-arm RNAi; (22) UAS-pan; (23) dpp-lacZ; (24) slimb-lacZ; (25) ci-lacZ; (26) 5′F-ato-lacZ; (27) wg-lacZ; (28) wg1-12/CyO; (29) wgGla/CyO; (30) 54C-GAL4; (31) ato-GAL4; (32) ey-GAL4; and (33) dpp-GAL4. emc-null clones were generated with the following genotypes: y w ey-flp; FRT80B emcAP6/ FRT80B M(3)i55 Ubi-GFP and/or y w hs-flp[22}; FRT80B emcAP6/ FRT80B M(3)i55 Ubi-GFP. Overexpression clones were generated with the following genotypes: (1) y w hs-flp[22]; Act5C >y+ >GAL4, UAS-GFP/UAS-Su(fu); (2) y w hs-flp[22]; Act5C >y+ >GAL4, UAS-GFP/UAS-Su(fu) RNAi; (3) y w hs-flp[22]; Act5C >y+ >GAL4, UAS-GFP/UAS-ci; (4) y w hs-flp[22]; Act5C >y+ >GAL4, UAS-GFP/UAS-ci RNAi; and (5) y w hs-flp[22]; Act5C >y+ >GAL4, UAS-GFP/UAS-arm RNAi.
Antibodies and microscopy
The following primary antibodies were used: (1) rat anti-ELAV (1:20, DSHB); (2) mouse anti-Hairy (1:100, a gift from Nadean Brown, UC Davis, CA, USA); (3) rabbit anti-PKA RII (1:500, a gift from Dan Kalderon, Columbia University, New York, USA); (4) mouse anti-Fu (1:200, DSHB); (5) rabbit anti-CK1 (1:500, a gift from Claudio Sunkel, Institute for Molecular and Cell Biology, Singapore); (6) mouse anti-Arm (1:20, DSHB); (7) mouse anti-β-Gal (1:100, Promega); (8) mouse anti-GFP (1:100, Santa Cruz Biotechnology); (9) rat anti-CiACT (1:50, a gift from Robert Holmgren, Northwestern University, Evanston, IL, USA); (10) mouse anti-GSK (1:50, DSHB); (11) rabbit anti-Cad86C (1:10,000, a gift from Christian Dahmann, Max Planck Institute, Dresden, Germany); (12) mouse anti-Costal-2 (1:20, DSHB); (13) mouse anti-Ptc (1:100, DSHB); (14) mouse anti-Smo (1:100, DSHB); (15) mouse anti-Su[fu] (1:200, DSHB); (16) mouse anti-HA (1:1000, Santa Cruz Biotechnology); and (17) mouse anti-Myc (1:1000, Santa Cruz Biotechnology). Fluorophore-conjugated secondary antibodies and phalloidin-fluorophore conjugate were obtained from Jackson Laboratories and Molecular Probes. Imaginal discs and adult heads were prepared as described by Anderson et al. (Anderson et al., 2012).
Number of ommatidial rows and furrow velocity
w1118 flies were allowed to lay eggs for 2 hours at 25°C. Individual eggs were placed in tubes containing ∼500 μl of media and aged for varying numbers of hours at 25°C. Eye-antennal discs were dissected and stained with rat anti-ELAV antibodies and phalloidin. The number of ommatidial rows was determined by counting the number of rows of ELAV-positive clusters. At least 19 discs were analyzed for each time point. Standard deviations were calculated in Excel. Furrow velocity was calculated by dividing the difference between the average number of ommatidial rows at two consecutive time points by 180 minutes (the time interval between each time point).
Calculation of degree of furrow advancement through emc and hairy clones
Images of imaginal discs containing emc-, h- or emc h-null mutant clones were imported into ImageJ. Three lines were drawn on each eye-antennal disc: (1) a vertical line marking the position of the most posteriorly located ommatidial row; (2) a vertical line marking the position of the normal furrow; and (3) a vertical line marking the position of the furrow that has advanced through mutant clone. The distance between 1 and 2 was measured three times and averaged (NF), as was the distance between 1 and 3 (AF). This process was repeated for 30 clones. The contribution made by emc and h to the distance travelled by the furrow was calculated by dividing the values for the AF by those for the NF. We then averaged the set of AF/NF ratios and multiplied by 100 to obtain a final percentage. Standard deviations were calculated in Excel. An F-test was used to determine equal or unequal variance. If the F-value was more than 0.05, then P-values were calculated using a Student’s t-test of equal variance. If the F-value was less than 0.05, then a Student’s t-test of unequal variance was used.
emc-GFP expression pattern analysis
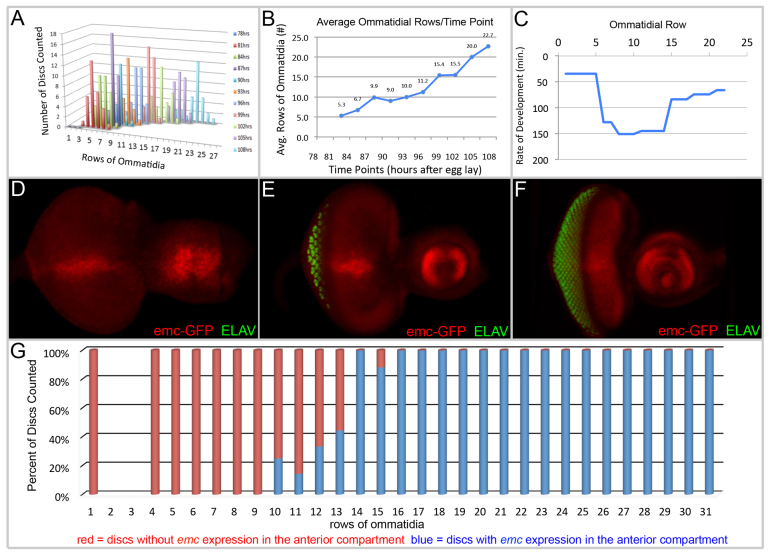
More than 400 eye-antennal discs from four emc-GFP enhancer trap lines were dissected and stained with Phalloidin and anti-ELAV antibodies. The presence or absence of emc transcription in the anterior compartment was registered against the number of ommatidial rows.
Immunoprecipitation assays
Kc167 cells were transfected with combinations of the following plasmids: (1) mt-GAL4, (2) UAS-Emc-HA, (3) UAS-Emc-MYC, (4) UAS-H-HA, (5) UAS-H-MYC. Immunoprecipitations were carried out as described by Anderson et al. (Anderson et al., 2012). Each experiment was carried out in both directions (immunoprecipitation with Emc-MYC, immunoblotting with H-HA and immunoprecipitation with H-HA, immunoblotting with Emc-MYC).
Transcriptional activation assay
A transcriptional reporter containing five copies of the consensus Ci binding site was built (IDT) and cloned into a plasmid containing the hsp70 promoter and the firefly luciferase gene. This plasmid was simultaneously transfected (Qiagen Effectene Transfection Reagent) into Kc167 cells with plasmids containing the pMT-GAL4 driver and a UAS-Renilla luciferase reporter. These cells were also transfected with combinations of the following plasmids: UAS-GFP, UAS-Su(fu) and UAS-CiU. Deleting amino acids 612-760 from the full-length Ci protein generated the CiU protein that was used in this assay. This modified protein cannot be cleaved into the smaller repressor isoform and functions as a transcriptional activator of Hedgehog target genes (Wang and Holmgren, 1999). Equal amounts of each construct were transfected into Kc167 cells, which were then incubated for 12 hours, followed by protein induction and an incubation period. The Promega Dual-Luciferase Reporter Assay System and a Glomax 20/20 Luminometer were used to detect luminescence signals from both firefly and Renilla luciferases, which were then divided to generate luminescence ratios. For each experiment, three independent biological and technical replicates were conducted, thus the luminescence ratio for each genotype in Fig. 4M represents the average of nine experimental ratios.
Fig. 4.

Su(fu) regulates CiACT protein but does not promote transcriptional activation. (A) Distribution of Su(fu) protein in wild-type retinas. (B-D) Su(fu) levels appear elevated in emc clones (hs-flp[22]; FRT80B emcAP6/FRT80B M(3)i55 Ubi-GFP). (E-H) Reductions in Su(fu) levels (hs-flp[22]; Act5C >y+ >GAL4, UAS-GFP; UAS-Su(fu) RNAi) reduce CiACT levels but do not inhibit furrow progression. (I-L) Elevation of Su(fu) levels (hs-flp[22]; Act5C >y+ >GAL4, UAS-GFP; UAS-Su(fu)) is not sufficient to increase CiACT levels. Anterior is towards the right. All markers and abbreviated genotypes are listed in each panel. (M) Expression of Su(fu) leads to a reduction in the activation of a luciferase reporter by CiU that is under the control of a synthetic Ci-responsive element (compare blue and pink bars).
RESULTS
Emc regulates furrow velocity during eye development
An accurate measure of furrow velocity across the retinal epithelium is a prerequisite for fully appreciating the role that emc plays in regulating the pace of pattern formation. Two prior studies of furrow speed provide conflicting estimates of its velocity. One measurement indicates that the furrow moves at a constant rate with one ommatidial row being laid down every 120 minutes (Basler and Hafen, 1989). A second study determined that an ommatidial row was generated every 100 minutes in the posterior half of the eye field. The pace quickens in the anterior half where a row is produced every 60 minutes (Campos-Ortega and Hofbauer, 1977). These conflicting measurements prompted us to measure directly the velocity of the furrow by determining the number of ommatidial rows that had been produced in eye discs of individually staged larvae at defined time points (Fig. 1A,B; supplementary material Table S1). We calculated the velocity of the furrow by dividing the difference in the number of rows between time points by the 180-minute interval that we had incorporated into the experiment (Fig. 1C). We observe a dynamic range to the pace at which the furrow progresses across the eye field. The generation time for a single ommatidial row ranges from 35 to 150 minutes (Fig. 1C). Thus, in contrast to prior studies, we demonstrate that the furrow moves dynamically across the eye field with alternating periods of accelerations and decelerations.
Fig. 1.
emc expression and the progression of the furrow is dynamic during development. (A,B) Graphs depicting the average number of ommatidial rows that are found at various times after egg deposition. (C) The furrow progresses across the eye field at a dynamic rate. (D,E) In young discs, emc is elevated at the midline. (F) In more mature discs, emc expression is enriched in the anterior compartment. Anterior is towards the right. All markers are listed in each panel. (G) Graph demonstrating that onset of emc expression ahead of the furrow coincides with the formation of the tenth row of ommatidia.
We next set out to analyze whether the expression pattern of emc correlates with the changing velocity of the furrow. Several GFP enhancer trap lines, including YB0067, which was used throughout this study, faithfully recapitulate the normal expression pattern of emc (Fig. 1D-F). In young eye discs, emc is expressed along the D/V midline (Fig. 1D,E). Several genes, such as four-jointed, that are involved in the establishment and maintenance of dorsal-ventral compartment boundaries are expressed along the midline in a similar pattern to emc (Brodsky and Steller, 1996). This might hint at an earlier role for emc in compartment boundary formation. In older third instar eye discs, emc is transcribed throughout the entire retinal field with low expression levels within the furrow and significant enrichment ahead of the advancing furrow and at the equator (Fig. 1F) (Brown et al., 1995). Eye discs were dissected from larvae of various ages from these enhancer trap lines and the expression pattern of emc was compared with the number of ommatidial rows. We find that although emc is expressed at low levels throughout the eye disc, enriched expression in the anterior compartment does not appear until ∼10 rows of ommatidia have been generated (Fig. 1G). We note that the furrow has already begun to decelerate prior to the onset of this enrichment and that the furrow continues to accelerate later in development despite the continued elevated expression of emc throughout the anterior compartment (Fig. 1C,G). Coupled with our findings that emc is required to slow the furrow across the entire disc (see below) we conclude that the lower basal level of Emc protein is sufficient to serve as a brake on the furrow and that mechanistically Emc is not the rate-limiting reagent.
Despite the recognition that emc plays a role in regulating the speed of the furrow, the mechanisms that underlie this activity remain poorly understood. A recent report using a null emc allele concluded that emc could regulate furrow velocity on its own but only in the ventral half of the retina (Bhattacharya and Baker, 2009). However, emc was removed throughout the entire disc, resulting in a small, disorganized eye. This complicates the interpretation of the emc loss-of-function phenotypes (Bhattacharya and Baker, 2009). We set out to determine unambiguously whether the loss of emc alone is sufficient to slow the furrow throughout the whole eye field during the patterning process. Similar to prior reports, discs that are lacking emc throughout the entire primordium are small and extremely disorganized. These animals die as pharate adults and the eye fields contain very few ommatidia and are instead populated with head cuticle and extra macrochaetae type bristles (Fig. 2A-D).
Fig. 2.

The morphogenetic furrow accelerates through emc-null clones. (A,B) Scanning electron microscopy and confocal images of wild-type retinas. (C,D) Removal of emc from the entire eye disc (ey-flp; FRT80B emcAP6/FRT80B M(3)i55 Ubi-GFP) leads to severe patterning and proliferation defects. (E-O) The furrow (assayed by F-actin, ELAV and dpp-lacZ) accelerates through emc clones (hs-flp[22]; FRT80B emcAP6/FRT80B M(3)i55 Ubi-GFP). Anterior is towards the right. All markers and abbreviated genotypes are listed in each panel. NF, normal furrow; AF, advanced furrow.
We then made patches of emc-null mutant tissue and observed the morphogenetic furrow accelerating through the clone, irrespective of compartmental location (Fig. 2E-K). Using physical distance as a measure of how far the furrow has advanced through wild-type and emc-null mutant tissue, we calculated the contribution that emc plays in regulating furrow velocity (Fig. 2I-K). We find that the furrow moves an average of 30% further through the emc-null clones than wild-type tissue. This is unaffected by the size, width or location of the clone. We confirmed that the furrow was in fact accelerating through emc mutant tissue by analyzing the dpp-lacZ transcriptional reporter, which, during the late third instar, is expressed solely within the advancing furrow (Blackman et al., 1991; Heberlein et al., 1993; Chanut and Heberlein, 1997a; Chanut and Heberlein, 1997b). As the furrow passes through emc-null clones dpp appears to be expressed in a broader swathe of cells and at higher levels (Fig. 2L-O, arrows). From these results, we conclude that emc is able to regulate the furrow throughout the entire eye disc.
Emc controls the pace of furrow movement through regulation of Hh signaling
Progression of the furrow and dpp activation are both dependent upon Hh signaling (Heberlein et al., 1993; Ma et al., 1993; Heberlein et al., 1995; Domínguez and Hafen, 1997; Borod and Heberlein, 1998). We attempted to ascertain whether furrow acceleration and increased dpp transcription within emc-null clones are due to elevated Hh signaling. We first examined the protein levels of Cubitus interruptus (Ci), a transcription factor that resides at the bottom of the Hh pathway (Aza-Blanc and Kornberg, 1999). Two different versions of Ci are found in developing tissues: a full-length form that functions as a transcriptional activator (CiACT) (Alexandre et al., 1996; Hepker et al., 1997) and a smaller form that serves as a repressor (CiR) (Aza-Blanc et al., 1997). CiACT is present in cells responding to Hh signaling (Méthot and Basler, 1999; Price and Kalderon, 1999; Wang and Holmgren, 1999). Within wild-type eye discs the highest levels of CiACT are found within the furrow (Fig. 3A) (Eaton and Kornberg, 1990). Removal of emc within the furrow results in a significant increase in the level and number of cells expressing CiACT, suggesting that emc regulates the progression of the furrow by controlling the transcriptional output of Hh signaling (Fig. 3B-D).
Fig. 3.
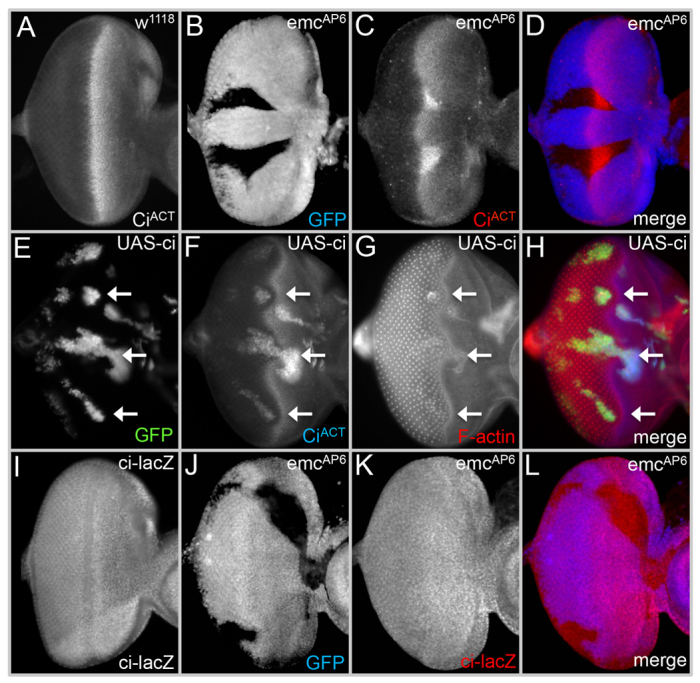
Emc regulates the active form of the Ci transcription factor (Ci155). (A,I) Wild-type expression patterns of CiACT and ci-lacZ in third instar discs. (B-D,J-L) Elevated levels of CiACT seen in emc clones (hs-flp[22]; FRT80B emcAP6/FRT80B M(3)i55 Ubi-GFP) are not due to increased ci transcription. (E-H) Overexpression of full-length Ci (hs-flp[22]; Act5C >y+ >GAL4, UAS-GFP; UAS-Ci155) results in the acceleration of the furrow. Arrows in E-H indicate accelerated regions of the furrow. Anterior is towards the right. All markers and abbreviated genotypes are listed in each panel.
Several reports have suggested that the Hh pathway is bifurcated and that the Hh signal can be transduced independently of Ci (Krishnan et al., 1997; Lessing and Nusse, 1998; Lewis et al., 1999; Suzuki and Saigo, 2000; Gallet et al., 2000). The existence of this distal branch point has been disputed (Méthot and Basler, 2001). We wanted to determine whether Hh-mediated control of the furrow is through Ci or not. We observed that the furrow accelerates through clones in which full-length Ci (Ci155) is overexpressed, thereby suggesting that increased CiACT is sufficient to advance the furrow and is responsible for the emc mutant phenotype (Fig. 3E-H, arrows).
We next investigated the molecular mechanism by which emc regulates CiACT levels by generating emc-null clones and examining the expression levels of known Hh signaling components. Using a ci-lacZ transcriptional reporter (Fig. 3I), we demonstrate that the increased levels of CiACT seen within emc-null clones are not due to an increase in ci transcription (Fig. 3J-L). From this, we conclude that CiACT levels are elevated in emc clones via regulation of at least one other member of the Hh signaling network. A systematic screen of Hh signaling components in emc clones revealed that nearly all Hh signaling components, including hh, patched (ptc), smoothened (smo), fused (fu), costal-2 (cos-2; cos - FlyBase), slimb (slmb - FlyBase), protein kinase A (PKA), glycogen synthase kinase-3 (GSK3) and casein kinase-1 (CK1) are not regulated by emc (supplementary material Fig. 1A-P) (Østerlund and Kogerman, 2006). The lone exception is Su(fu), which encodes a novel PEST domain-containing protein. In emc-null clones, Su(fu) protein levels appear elevated (Fig. 4A-D, arrows). This apparent increase in Su(fu) does not appear to be a response to elevated CiACT levels as forced increases or decreases in CiACT levels are insufficient to alter the amount of Su(fu) protein (supplementary material Fig. 2A-H).
Su(fu) appears to be required for maintaining the levels of CiACT as reductions in Su(fu) levels are sufficient to drastically reduce levels of CiACT (Fig. 4E-H; supplementary material Fig. S1Q,R) (Ohlmeyer and Kalderon, 1998). The increase in CiACT levels is not a result of increased transcription of ci as the ci-lacZ reporter is unaffected in emc clones despite elevated Su(fu) protein levels (Fig. 3J-L; Fig. 4B-D). Instead, the increased CiACT levels may result from post-translational stabilization by Su(fu), which physically associates with Ci and its mammalian homologs, the Gli proteins (Monnier et al., 1998; Pearse et al., 1999; Stegman et al., 2000; Monnier et al., 2002; Paces-Fessy et al., 2004). Su(fu) does not, however, appear to be sufficient to prevent the full-length activator form of Ci from being cleaved into the smaller repressor form as overexpression clones of Su(fu) do not lead to increased levels of CiACT (Fig. 4I-L).
The role that Su(fu) plays in regulating Ci activity is not well understood. Several reports suggest that Su(fu) antagonizes Ci/Gli activity by blocking its entry into the nucleus and/or by repressing target genes within cells that have received the Hh signal (Kogerman et al., 1999; Lefers et al., 2001; Méthot and Basler, 2000; Wang and Holmgren, 2000; Wang et al., 2000; Taylor et al., 2002; Ho et al., 2005; Dussillol-Godar et al., 2006). Consistent with these findings, we observe that Su(fu) is also capable of inhibiting the ability of CiACT to activate the transcription of a luciferase reporter that has been fused to an artificial enhancer containing five perfect Ci-binding sites (Fig. 4M). These results suggest that in the developing eye Su(fu) is unlikely to cooperate with CiACT in the activation of Hh pathway target genes that are needed for furrow progression. Our results are also consistent with the fact that Su(fu)-null mutant animals have no obvious eye defects and movement of the furrow is not retarded as it passes through clones that lack Su(fu) and/or CiACT (Fig. 4E-H) (Préat, 1992; Pappu et al., 2003). These results suggest that emc-dependent regulation of CiACT relies on the activity of a yet to be identified Hh signaling component.
Emc functions as an inhibitor of apical surface constriction and nuclear migration
In addition to changes in gene expression profiles, two cellular features distinguish cells within the furrow from all others cells within the eye disc. The first is the constriction of apical cellular surfaces and the second is the basal migration of nuclei. These two cellular activities result in a dorsoventral indentation in the disc epithelium, hence the description of this zone as a furrow (Ready et al., 1976). The constriction of the apical surfaces of cells within the furrow is dependent upon Hh signaling. A decrease in F-actin accumulation and Armadillo (Arm) levels (two apical profile markers) are observed in clones that are mutant for smoothened (smo), which encodes a transducer of the Hh signal (Corrigall et al., 2007). The cell shape changes that are induced by Hh signaling are crucial for limiting the spread of the Hh signal itself. Cells within clones mutant for the Drosophila homolog of cyclase-associated protein homolog act up (acu; capt - FlyBase) fail to constrict their apical profiles, and both Hh and CiACT proteins accumulate more anteriorly than in surrounding wild-type cells (Benlali et al., 2000). As our data indicate that emc regulates Hh signaling, we turned our attention to the cellular dynamics of cells that lack emc. As expected, Arm and Cad86C, two proteins that localize to adherens junctions and are enriched within the furrow (Fig. 5A,E) (McCrea et al., 1991; Schlichting and Dahmann, 2008), are elevated within emc-null clones (Fig. 5B-D,F-H). Cad86C is a transcriptional target of Hh signaling (Schlichting and Dahmann, 2008); therefore, the increase in Cad86C is consistent with the increases that we see in CiACT levels (Fig. 3B-D). Arm does not appear to play a role in stabilizing CiACT as expression of an arm RNAi construct has no effect on CiACT (supplementary material Fig. S3A-D). Sagittal sections of discs containing emc mutant clones reveal accumulations of F-actin and cell shape changes within the mutant tissue (Fig. 5I,K,L). These cellular changes result in depressions at the apical surface (Fig. 5I, yellow asterisk) that mimic cells within the endogenous furrow (Fig. 5I, white asterisk).
Fig. 5.

Emc regulates Hedgehog-dependent constriction of apical cell surfaces. (A,E) Arm and Cad86C proteins are enriched within the furrow. (B-D,F-H) Both Arm and Cad86C appear elevated in emc clones (hs-flp[22]; FRT80B emcAP6/FRT80B M(3)i55 Ubi-GFP). (I) F-actin accumulation (yellow asterisk) and nuclei migration (yellow arrow) within an emc clone. The normal furrow is marked by a white asterisk, whereas a white arrow marks nuclear migration. The yellow and orange lines indicate regions of the disc that are viewed in the side sagittal sections. (J-L) High-resolution images demonstrating apical surface constriction and nuclear migration (white asterisk indicates the wild-type furrow). Anterior is towards the right. All markers and abbreviated genotypes are listed in each panel.
Nuclear migrations are a second distinguishing cellular feature of the morphogenetic furrow (Ready et al., 1976; Fischer-Vize and Mosley, 1994). The basal migration of nuclei within the furrow and their subsequent apical rise in post-mitotic cells is dependent upon microtubule motors such as Dynein and associated proteins (Fan and Ready, 1997; Mosley-Bishop et al., 1999; Swan et al., 1999; Houalla et al., 2005). We have used this feature to further analyze the cellular behavior of cells lacking emc. In emc mutant tissue, nuclei appear to migrate basally (Fig. 5I, yellow arrowhead) as they do within the normal furrow (Fig. 5I, white arrowhead). The strongest effect is seen within and ahead of the furrow (Fig. 5J), suggesting that cells lacking emc behave as if they are in a ‘furrow-like’ state. From these observations, we conclude that emc is required to prevent changes in cell shape and basal migration of nuclei, two distinct characteristics of the cells within the furrow that receive high levels of Hh signaling.
Emc and H cooperate to regulate pattern formation through different targets
Although it has been demonstrated that the furrow will accelerate through a clone that is null for emc (Fig. 2E-H) (Bhattacharya and Baker, 2009) we wanted to determine what role, if any, does h play in regulating the furrow velocity. h is normally expressed in a stripe of cells ahead of the morphogenetic furrow (Fig. 6A) (Brown et al., 1991). Consistent with prior reports, we find that the furrow does not accelerate through h-null mutant clones (supplementary material Fig. S4A-G) (Brown et al., 1995). By contrast, the distance traveled by the furrow through emc h double-null mutant clones is significantly greater than the distance traveled through just emc mutant tissue (Fig. 6B-F). A question that arises from these results concerns the molecular and genetic relationship between emc and h. As both genes are co-expressed in a subset of cells ahead of the furrow, we set out to determine whether emc regulates h expression. In very large emc-null mutant clones, h expression is reduced but not eliminated from the entire clone (Fig. 6G-I, arrow). In small- to medium-sized clones, the level of h expression appears unchanged (Fig. 6J-L). These results suggest that h expression in emc clones can be restored non-autonomously from surrounding wild-type tissue. A potential candidate is the Dpp pathway, which activates h expression ahead of the furrow (Greenwood and Struhl, 1999).
Fig. 6.
Extramacrochaetae and Hairy regulate different aspects of pattern formation. (A) During normal retinal development, h is expressed in a stripe ahead of the furrow. (B-E) The furrow accelerates through emc h double mutant clones (hs-flp22]; FRT80B emcAP6 h22/FRT80B M(3)i55 Ubi-GFP). (F) Comparison of measured furrow acceleration through emc single and emc h double mutant clones. The y-axis refers the percentage advancement of the furrow through clones when compared with wild-type tissue. (G-L) h expression in emc clones (hs-flp[22]; FRT80B emcAP6/FRT80B M(3)i55 Ubi-GFP). Arrow in I indicates the non-autonomous rescue of h expression by the surrounding wild-type tissue. (M) Co-immunoprecipitation from Kc167 cells demonstrating that Emc and H do not form a biochemical complex. (N-R) Simultaneous expression of h and emc by ato-GAL4 results in a synergistic reduction of eye development. (S-U) Expression of ato within the furrow is reduced in emc clones (arrows). (V-X) Loss of h (hs-flp[22]; FRT80B h22/FRT80B M(3)i55 Ubi-GFP) results in ectopic ato expression (arrows). Anterior is towards the right. All markers and abbreviated genotypes are listed in each panel.
We then attempted to ascertain whether Emc and H cooperate to regulate the furrow by forming a biochemical complex. We performed immunoprecipitation assays on Drosophila Kc167 cell extracts that contain epitope-tagged versions of both proteins and were unable to detect any physical interactions between Emc and H (Fig. 6M). The immunoprecipitations were performed in both directions and in both instances we failed to detect an Emc-H interaction. As Emc and H cooperate to regulate the furrow, the lack of an Emc-H complex suggests that the two proteins function in different pathways. Forced expression of emc within the furrow using an atonal-GAL4 driver has no effect on the structure of the retina (Fig. 6N), whereas expression of h leads to defects in photoreceptor specification and a mild roughening of the adult eye (Fig. 6O,P) (Brown et al., 1991). Simultaneous expression of both factors has a synergistic effect, with neuronal specification being nearly completely blocked and an adult eye almost devoid of ommatidia (Fig. 6Q,R). This synergism, which was first reported by Brown et al. (Brown et al., 1995), supports a model in which emc and h are functioning separately rather than being bound together as a biochemical complex.
As the developmental and adult retinal phenotypes of atonal (ato) mutants are nearly identical to those observed when emc and h are overexpressed (Fig. 6Q,R) (Jarman et al., 1994; Jarman et al., 1995), we examined the effects that loss of either emc or h has on ato. An enhancer element that lies 5′ to the ato-coding sequence can direct expression to the intermediate groups, the R8 equivalence group and within the R8 cell itself (Sun et al., 1998). Expression of the ato reporter is reduced in emc-null clones (Fig. 6S-U, arrows), whereas the reporter is ectopically activated in h-null clones (Fig. 6V-X, arrows). Ectopic ato expression in h clones is not accompanied by neuronal development (marked by ELAV), indicating that ato activation, by itself, is insufficient to induce neurogenesis. We conclude that emc and h regulate different aspects of the furrow: h is primarily tasked with repressing ato expression ahead of the furrow, whereas emc functions to inhibit Hh signaling and the cellular dynamics of the furrow.
Emc both regulates and cooperates with Wg signaling to prevent ectopic furrow formation
As emc appears to slow the progression of the normal furrow through regulation of Hh signaling, we next asked the following two questions: (1) would elevated levels of emc at the posterior margin be sufficient to inhibit the initiation of the furrow and (2) does emc play a role in preventing ectopic furrows from initiating at the dorsal and ventral margins of the eye disc? The impetus for asking the first question lies in the fact that Hh signaling is also required for furrow initiation (Ma et al., 1993; Heberlein et al., 1993; Chanut and Heberlein, 1995; Heberlein et al., 1995; Ma and Moses, 1995; Pan and Rubin, 1995; Strutt et al., 1995; Wehrli and Tomlinson, 1995). However, expression of emc under the control of either dpp or ey enhancers, both of which drive expression at the posterior margin of the disc (Blackman et al., 1991; Hauck et al., 1999), failed to have any observable effect on furrow initiation (supplementary material Fig. S5A,B).
An earlier study has shown that emc is required for Wingless (Wg)-dependent repression of bristle formation in the retina (Cadigan et al., 2002). Wg signaling also plays an important role in preventing ectopic furrow initiation at the margins of the eye disc (Ma and Moses, 1995; Treisman and Rubin, 1995), so it seemed possible that emc may function with wg to prevent ectopic furrow initiation. emc-null clones that contact the margins result in ectopic furrow initiation as assayed by dpp-lacZ (Fig. 7A-D, arrow) and ato-lacZ (supplementary material Fig. S5C-F) reporters. These ectopic furrows can be seen originating from both the dorsal and ventral margins. Our next effort was to determine whether emc lies genetically upstream or downstream of the Wg signaling cascade. Removal of Wg activity via a temperature-sensitive allele (wg1-12) (Baker, 1988) has no visible effect on emc transcription (Fig. 7E-H). Similarly, activation of the Wg pathway through expression of pangolin (pan) in clones failed to influence emc expression (Fig. 7I-L). And finally, neither removal nor overexpression of emc was sufficient to significantly modify the adult eye phenotype of wgGla (supplementary material Fig. S6A-D), a gain-of-function allele of wg (Brunner et al., 1999). These results suggest that emc does not function downstream of Wg and are consistent with an earlier observation that elevating wg expression does not induce either emc or h transcription in the eye disc (Cadigan et al., 2002). We then used a wg-lacZ reporter, which recapitulates wg transcription in the eye (Fig. 7M) (Ma and Moses, 1995; Treisman and Rubin, 1995), to monitor wg transcription in emc-null mutant clones. Under these circumstances, the wg reporter is silenced along the ventral margin (Fig. 7N-S, arrows). This indicates that emc lies upstream of wg at the ventral margin but cooperates with wg to block inappropriate pattern formation at the dorsal margin.
Fig. 7.

Activation of Wingless signaling by Emc prevents ectopic furrow initiation. (A-D) Ectopic furrows initiate from the margins (arrow) in the absence of emc (hs-flp[22]; FRT80B emcAP6/FRT80B M(3)i55 Ubi-GFP). (E-H) emc expression is not altered when wg activity is removed via a temperature-sensitive mutant wg1-12. (I-L) Activation of Wg signaling (hs-flp[22]; Act5C >y+ >GAL4, UAS-GFP; UAS-pan) fails to alter emc expression. (M-S) wg expression at the ventral (arrows), but not dorsal, margin is lost in emc clones. Clones in M-P were generated with hs-flp[22]; FRT80B emcAP6/FRT80B M(3)i55 Ubi-GFP, whereas clones in Q-S were generated with ey-flp; FRT80B emcAP6/FRT80B M(3)i55 Ubi-GFP. Anterior is towards the right. All markers and abbreviated genotypes are listed in each panel.
DISCUSSION
Here, we describe a molecular mechanism by which the HLH protein Emc refines the activity of the Hh signaling gradient and thereby ensures that the morphogenetic furrow patterns the retina evenly and at an appropriate set of velocities (Fig. 8A,B). As the furrow progresses across the eye field, Hh is secreted by differentiating photoreceptors and received by cells within the furrow (Heberlein et al., 1993; Ma et al., 1993). As a result, the active version of the transcription factor Ci (CiACT) is stabilized, which in turn leads to the transcriptional activation of dpp, a long-range morphogen (Heberlein et al., 1993; Borod and Heberlein, 1998; Fu and Baker, 2003; Pappu et al., 2003). Dpp is then used to prepare more anteriorly positioned cells for entry into the furrow (Greenwald and Struhl, 1999). In addition to activating dpp expression, Hh signaling within the furrow is required for apical surface constriction and the transcriptional repression of h, which encodes a bHLH transcription factor that, at elevated levels, can induce numerous structural defects (Brown et al., 1991; Fu and Baker, 2003; Corrigall et al., 2007; Schlichting and Dahmann, 2008). Together with H, Emc is thought to regulate the rate by which the furrow traverses the retinal epithelium (Brown et al., 1995; Bhattacharya and Baker, 2009). However, a molecular link between H/Emc and the signaling pathways controlling furrow velocity has remained elusive.
Fig. 8.

Models for the involvement of Emc in furrow initiation and progression. (A,B) During furrow progression, Emc counterbalances the activity of Hh signaling via regulation of CiACT levels. This puts the rates of pattern formation and proliferation into equilibrium. (C) A summary of the genetic interactions by which Emc regulates furrow initiation and progression.
In this report, we have demonstrated that Emc regulates the levels of CiACT and this in turn has a direct impact on the speed at which the furrow travels across the eye field. The furrow travels through emc loss-of-function clones, which show elevated levels of CiACT, ∼30% faster on average than it does through wild-type tissue. As a result, the eye does not have sufficient time to proliferate and the adult retina is small and disorganized (Fig. 8A,B). The elevated CiACT levels are solely responsible for the accelerated furrow speed, as overexpression clones of CiACT also show an advanced furrow. But how does Emc control CiACT levels within and ahead of the furrow? Our experimental data suggest that higher CiACT levels are not achieved through direct transcriptional regulation but rather through post-translational stabilization via Su(fu). Su(fu) is the only other Hh signaling component that was observed to be elevated in emc loss-of-function tissue, and reductions in Su(fu) lead to the near elimination of CiACT. These results suggest that Emc regulation of Su(fu) is essential for the maintenance of appropriate CiACT levels.
In both Drosophila and mammals, Su(fu) binds to and negatively regulates the activity of Ci/Gli proteins (Monnier et al., 1998; Kogerman et al., 1999; Pearse et al., 1999; Stone et al., 1999; Methot and Basler, 2000; Stegman et al., 2000; Wang and Holmgren, 2000; Wang et al., 2000; Lefers et al., 2001; Monnier et al., 2002; Taylor et al., 2002; Paces-Fessy et al., 2004; Ho et al., 2005; Dussillol-Godar et al., 2006; Sisson et al., 2006). Our own results support these findings, as expression of Su(fu) reduces the ability of CiACT to activate a Ci-dependent luciferase reporter. Within the mouse eye, loss of Su(fu) leads to an increase in Hh signaling which in turn results in retinal hyperplasia, late onset premature exit from the cell cycle, and an increase in the number of horizontal and amacrine cells at the expense of other cell types (Cwinn et al., 2011). By contrast, Su(fu) does not appear to be a rate-limiting component of the Hh pathway in Drosophila and, as such, null mutants are completely viable and have normally constructed eyes (Préat, 1992; Shi et al., 2011; Zhou and Kalderon, 2011). Despite the observed increases in Su(fu) within emc-null clones, a role for Su(fu) in furrow progression remains unclear, as reduced or elevated levels do not lead to furrow acceleration.
Our data also indicate that Emc plays a role in preventing ectopic furrow initiation at the dorsal and ventral margins of the eye field. We observed that new furrows initiate from both dorsal and ventral margins in the absence of emc. Until now, the only other molecule that is known to block ectopic furrows is Wg/Wnt signaling (Ma and Moses, 1995; Treisman and Rubin, 1995). We demonstrate that at the dorsal margin Emc and the Wg pathway probably function in parallel to block pattern formation while at the ventral margin this genetic hierarchy is altered and Emc regulates wg transcription (Fig. 8C). The underlying reasons for these different genetic relationships are not known. One possible explanation lies with the earlier differences in the developmental history of these two compartments. The eye initially comprises only ventral tissue, with Wg signaling becoming activated later within the dorsal region of the eye field to specify dorsal fate (Maurel-Zaffran and Treisman, 2000; Singh and Choi, 2003). The genetic separation of emc and wg in the dorsal half of the retina may stem from the need to activate Wg signaling independently of emc.
Finally, the original description of emc and furrow regulation proposed that Emc and H cooperate to regulate furrow velocity (Brown et al., 1995). A subsequent study has suggested that Emc on its own is solely responsible for the acceleration of the furrow (Bhattacharya and Baker, 2009). Our data indicate that although the furrow can accelerate through tissue that is lacking only emc, h does in fact contribute to the pace of furrow movement. We measured the rate of the furrow in emc, h double mutant clones and demonstrate that the furrow proceeds faster through these clones than it does through clones that just lack emc. It appears that Emc and H regulate different aspects of furrow progression with Emc regulating the constriction of apical surfaces, the basal migration of nuclei and the Hh signaling cascade whereas H, on the other hand, appears to restrict ato expression in cells that have not yet entered the furrow.
Supplementary Material
Acknowledgments
We thank Brandon Weasner for technical support with co-immunoprecipitation and luciferase assays; Abby Anderson, Bonnie Weasner and Brandon Weasner for comments on the manuscript; Nick Baker, Antonio Baonza, Konrad Basler, Nadean Brown, Lynn Cooley, Michael Buszczak, Christian Dahmann, Barry Dickson, Wei Du, Robert Holmgren, Daniel Kalderon, Monn Myat, Ilaria Rebay, Alan Spradling, Claudio Sunkel, Jessica Treisman, Janice Fischer, Georg Halder, the DSHB, the DGRC and the BDSC for antibodies and fly stocks; and the Indiana University Light Microscopy Imaging Center and Olivier Brun for technical support with Leica SP5 confocal microscope.
Footnotes
Funding
This work is supported by a stipend from the National Institutes of Health (NIH) GMCS Training Grant [T32-GM007757] to C.M.S. and a grant from the National Eye Institute [2R01 EY014863] to J.P.K. Deposited in PMC for release after 12 months.
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.088963/-/DC1
References
- Alexandre C., Jacinto A., Ingham P. W. (1996). Transcriptional activation of hedgehog target genes in Drosophila is mediated directly by the cubitus interruptus protein, a member of the GLI family of zinc finger DNA-binding proteins. Genes Dev. 10, 2003–2013 [DOI] [PubMed] [Google Scholar]
- Anderson A. M., Weasner B. M., Weasner B. P., Kumar J. P. (2012). Dual transcriptional activities of SIX proteins define their roles in normal and ectopic eye development. Development 139, 991–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aza-Blanc P., Kornberg T. B. (1999). Ci: a complex transducer of the hedgehog signal. Trends Genet. 15, 458–462 [DOI] [PubMed] [Google Scholar]
- Aza-Blanc P., Ramírez-Weber F. A., Laget M. P., Schwartz C., Kornberg T. B. (1997). Proteolysis that is inhibited by hedgehog targets Cubitus interruptus protein to the nucleus and converts it to a repressor. Cell 89, 1043–1053 [DOI] [PubMed] [Google Scholar]
- Baker N. E. (1988). Embryonic and imaginal requirements for wingless, a segment-polarity gene in Drosophila. Dev. Biol. 125, 96–108 [DOI] [PubMed] [Google Scholar]
- Basler K., Hafen E. (1989). Dynamics of Drosophila eye development and temporal requirements of sevenless expression. Development 107, 723–731 [DOI] [PubMed] [Google Scholar]
- Benlali A., Draskovic I., Hazelett D. J., Treisman J. E. (2000). act up controls actin polymerization to alter cell shape and restrict Hedgehog signaling in the Drosophila eye disc. Cell 101, 271–281 [DOI] [PubMed] [Google Scholar]
- Bhattacharya A., Baker N. E. (2009). The HLH protein Extramacrochaetae is required for R7 cell and cone cell fates in the Drosophila eye. Dev. Biol. 327, 288–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackman R. K., Sanicola M., Raftery L. A., Gillevet T., Gelbart W. M. (1991). An extensive 3′ cis-regulatory region directs the imaginal disk expression of decapentaplegic, a member of the TGF-beta family in Drosophila. Development 111, 657–666 [DOI] [PubMed] [Google Scholar]
- Borod E. R., Heberlein U. (1998). Mutual regulation of decapentaplegic and hedgehog during the initiation of differentiation in the Drosophila retina. Dev. Biol. 197, 187–197 [DOI] [PubMed] [Google Scholar]
- Brodsky M. H., Steller H. (1996). Positional information along the dorsal-ventral axis of the Drosophila eye: graded expression of the four-jointed gene. Dev. Biol. 173, 428–446 [DOI] [PubMed] [Google Scholar]
- Brown N. L., Sattler C. A., Markey D. R., Carroll S. B. (1991). hairy gene function in the Drosophila eye: normal expression is dispensable but ectopic expression alters cell fates. Development 113, 1245–1256 [DOI] [PubMed] [Google Scholar]
- Brown N. L., Sattler C. A., Paddock S. W., Carroll S. B. (1995). Hairy and emc negatively regulate morphogenetic furrow progression in the Drosophila eye. Cell 80, 879–887 [DOI] [PubMed] [Google Scholar]
- Brunner E., Brunner D., Fu W., Hafen E., Basler K. (1999). The dominant mutation Glazed is a gain-of-function allele of wingless that, similar to loss of APC, interferes with normal eye development. Dev. Biol. 206, 178–188 [DOI] [PubMed] [Google Scholar]
- Burke R., Basler K. (1996). Hedgehog-dependent patterning in the Drosophila eye can occur in the absence of Dpp signaling. Dev. Biol. 179, 360–368 [DOI] [PubMed] [Google Scholar]
- Cadigan K. M., Jou A. D., Nusse R. (2002). Wingless blocks bristle formation and morphogenetic furrow progression in the eye through repression of Daughterless. Development 129, 3393–3402 [DOI] [PubMed] [Google Scholar]
- Campos-Ortega J. A., Hofbauer A. (1977). Cell clones and pattern formation on the lineage of photoreceptor cells in the compound eye of Drosophila. Roux’s Arch. Dev. Biol. 181, 227–245 [DOI] [PubMed] [Google Scholar]
- Chanut F., Heberlein U. (1995). Role of the morphogenetic furrow in establishing polarity in the Drosophila eye. Development 121, 4085–4094 [DOI] [PubMed] [Google Scholar]
- Chanut F., Heberlein U. (1997a). Retinal morphogenesis in Drosophila: hints from an eye-specific decapentaplegic allele. Dev. Genet. 20, 197–207 [DOI] [PubMed] [Google Scholar]
- Chanut F., Heberlein U. (1997b). Role of decapentaplegic in initiation and progression of the morphogenetic furrow in the developing Drosophila retina. Development 124, 559–567 [DOI] [PubMed] [Google Scholar]
- Corrigall D., Walther R. F., Rodriguez L., Fichelson P., Pichaud F. (2007). Hedgehog signaling is a principal inducer of Myosin-II-driven cell ingression in Drosophila epithelia. Dev. Cell 13, 730–742 [DOI] [PubMed] [Google Scholar]
- Curtiss J., Mlodzik M. (2000). Morphogenetic furrow initiation and progression during eye development in Drosophila: the roles of decapentaplegic, hedgehog and eyes absent. Development 127, 1325–1336 [DOI] [PubMed] [Google Scholar]
- Cwinn M. A., Mazerolle C., McNeill B., Ringuette R., Thurig S., Hui C. C., Wallace V. A. (2011). Suppressor of fused is required to maintain the multipotency of neural progenitor cells in the retina. J. Neurosci. 31, 5169–5180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domínguez M., Hafen E. (1997). Hedgehog directly controls initiation and propagation of retinal differentiation in the Drosophila eye. Genes Dev. 11, 3254–3264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dussillol-Godar F., Brissard-Zahraoui J., Limbourg-Bouchon B., Boucher D., Fouix S., Lamour-Isnard C., Plessis A., Busson D. (2006). Modulation of the Suppressor of fused protein regulates the Hedgehog signaling pathway in Drosophila embryo and imaginal discs. Dev. Biol. 291, 53–66 [DOI] [PubMed] [Google Scholar]
- Eaton S., Kornberg T. B. (1990). Repression of ci-D in posterior compartments of Drosophila by engrailed. Genes Dev. 4, 1068–1077 [DOI] [PubMed] [Google Scholar]
- Ekas L. A., Baeg G. H., Flaherty M. S., Ayala-Camargo A., Bach E. A. (2006). JAK/STAT signaling promotes regional specification by negatively regulating wingless expression in Drosophila. Development 133, 4721–4729 [DOI] [PubMed] [Google Scholar]
- Ellis H. M., Spann D. R., Posakony J. W. (1990). extramacrochaetae, a negative regulator of sensory organ development in Drosophila, defines a new class of helix-loop-helix proteins. Cell 61, 27–38 [DOI] [PubMed] [Google Scholar]
- Fan S. S., Ready D. F. (1997). Glued participates in distinct microtubule-based activities in Drosophila eye development. Development 124, 1497–1507 [DOI] [PubMed] [Google Scholar]
- Fischer-Vize J. A., Mosley K. L. (1994). Marbles mutants: uncoupling cell determination and nuclear migration in the developing Drosophila eye. Development 120, 2609–2618 [DOI] [PubMed] [Google Scholar]
- Fu W., Baker N. E. (2003). Deciphering synergistic and redundant roles of Hedgehog, Decapentaplegic and Delta that drive the wave of differentiation in Drosophila eye development. Development 130, 5229–5239 [DOI] [PubMed] [Google Scholar]
- Gallet A., Angelats C., Kerridge S., Thérond P. P. (2000). Cubitus interruptus-independent transduction of the Hedgehog signal in Drosophila. Development 127, 5509–5522 [DOI] [PubMed] [Google Scholar]
- Garrell J., Modolell J. (1990). The Drosophila extramacrochaetae locus, an antagonist of proneural genes that, like these genes, encodes a helix-loop-helix protein. Cell 61, 39–48 [DOI] [PubMed] [Google Scholar]
- Greenwood S., Struhl G. (1999). Progression of the morphogenetic furrow in the Drosophila eye: the roles of Hedgehog, Decapentaplegic and the Raf pathway. Development 126, 5795–5808 [DOI] [PubMed] [Google Scholar]
- Hauck B., Gehring W. J., Walldorf U. (1999). Functional analysis of an eye specific enhancer of the eyeless gene in Drosophila. Proc. Natl. Acad. Sci. USA 96, 564–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heberlein U., Wolff T., Rubin G. M. (1993). The TGF beta homolog dpp and the segment polarity gene hedgehog are required for propagation of a morphogenetic wave in the Drosophila retina. Cell 75, 913–926 [DOI] [PubMed] [Google Scholar]
- Heberlein U., Singh C. M., Luk A. Y., Donohoe T. J. (1995). Growth and differentiation in the Drosophila eye coordinated by hedgehog. Nature 373, 709–711 [DOI] [PubMed] [Google Scholar]
- Hepker J., Wang Q. T., Motzny C. K., Holmgren R., Orenic T. V. (1997). Drosophila cubitus interruptus forms a negative feedback loop with patched and regulates expression of Hedgehog target genes. Development 124, 549–558 [DOI] [PubMed] [Google Scholar]
- Ho K. S., Suyama K., Fish M., Scott M. P. (2005). Differential regulation of Hedgehog target gene transcription by Costal2 and Suppressor of Fused. Development 132, 1401–1412 [DOI] [PubMed] [Google Scholar]
- Houalla T., Hien Vuong D., Ruan W., Suter B., Rao Y. (2005). The Ste20-like kinase misshapen functions together with Bicaudal-D and dynein in driving nuclear migration in the developing drosophila eye. Mech. Dev. 122, 97–108 [DOI] [PubMed] [Google Scholar]
- Ives P. (1950). New mutant report: bar-3. Drosoph. Inf. Serv. 24, 58 [Google Scholar]
- Jarman A. P., Grell E. H., Ackerman L., Jan L. Y., Jan Y. N. (1994). Atonal is the proneural gene for Drosophila photoreceptors. Nature 369, 398–400 [DOI] [PubMed] [Google Scholar]
- Jarman A. P., Sun Y., Jan L. Y., Jan Y. N. (1995). Role of the proneural gene, atonal, in formation of Drosophila chordotonal organs and photoreceptors. Development 121, 2019–2030 [DOI] [PubMed] [Google Scholar]
- Kogerman P., Grimm T., Kogerman L., Krause D., Undén A. B., Sandstedt B., Toftgård R., Zaphiropoulos P. G. (1999). Mammalian suppressor-of-fused modulates nuclear-cytoplasmic shuttling of Gli-1. Nat. Cell Biol. 1, 312–319 [DOI] [PubMed] [Google Scholar]
- Krishnan V., Pereira F. A., Qiu Y., Chen C. H., Beachy P. A., Tsai S. Y., Tsai M. J. (1997). Mediation of Sonic hedgehog-induced expression of COUP-TFII by a protein phosphatase. Science 278, 1947–1950 [DOI] [PubMed] [Google Scholar]
- Kumar J. P., Moses K. (2001). The EGF receptor and notch signaling pathways control the initiation of the morphogenetic furrow during Drosophila eye development. Development 128, 2689–2697 [DOI] [PubMed] [Google Scholar]
- Lefers M. A., Wang Q. T., Holmgren R. A. (2001). Genetic dissection of the Drosophila Cubitus interruptus signaling complex. Dev. Biol. 236, 411–420 [DOI] [PubMed] [Google Scholar]
- Lessing D., Nusse R. (1998). Expression of wingless in the Drosophila embryo: a conserved cis-acting element lacking conserved Ci-binding sites is required for patched-mediated repression. Development 125, 1469–1476 [DOI] [PubMed] [Google Scholar]
- Lewis K. E., Drossopoulou G., Paton I. R., Morrice D. R., Robertson K. E., Burt D. W., Ingham P. W., Tickle C. (1999). Expression of ptc and gli genes in talpid3 suggests bifurcation in Shh pathway. Development 126, 2397–2407 [DOI] [PubMed] [Google Scholar]
- Ma C., Moses K. (1995). Wingless and patched are negative regulators of the morphogenetic furrow and can affect tissue polarity in the developing Drosophila compound eye. Development 121, 2279–2289 [DOI] [PubMed] [Google Scholar]
- Ma C., Zhou Y., Beachy P. A., Moses K. (1993). The segment polarity gene hedgehog is required for progression of the morphogenetic furrow in the developing Drosophila eye. Cell 75, 927–938 [DOI] [PubMed] [Google Scholar]
- Maurel-Zaffran C., Treisman J. E. (2000). pannier acts upstream of wingless to direct dorsal eye disc development in Drosophila. Development 127, 1007–1016 [DOI] [PubMed] [Google Scholar]
- McCrea P. D., Turck C. W., Gumbiner B. (1991). A homolog of the armadillo protein in Drosophila (plakoglobin) associated with E-cadherin. Science 254, 1359–1361 [DOI] [PubMed] [Google Scholar]
- Méthot N., Basler K. (1999). Hedgehog controls limb development by regulating the activities of distinct transcriptional activator and repressor forms of Cubitus interruptus. Cell 96, 819–831 [DOI] [PubMed] [Google Scholar]
- Méthot N., Basler K. (2000). Suppressor of fused opposes hedgehog signal transduction by impeding nuclear accumulation of the activator form of Cubitus interruptus. Development 127, 4001–4010 [DOI] [PubMed] [Google Scholar]
- Méthot N., Basler K. (2001). An absolute requirement for Cubitus interruptus in Hedgehog signaling. Development 128, 733–742 [DOI] [PubMed] [Google Scholar]
- Mohler J. (1988). Requirements for hedgehog, a segmental polarity gene, in patterning larval and adult cuticle of Drosophila. Genetics 120, 1061–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monnier V., Dussillol F., Alves G., Lamour-Isnard C., Plessis A. (1998). Suppressor of fused links fused and Cubitus interruptus on the hedgehog signalling pathway. Curr. Biol. 8, 583–586 [DOI] [PubMed] [Google Scholar]
- Monnier V., Ho K. S., Sanial M., Scott M. P., Plessis A. (2002). Hedgehog signal transduction proteins: contacts of the Fused kinase and Ci transcription factor with the kinesin-related protein Costal2. BMC Dev. Biol. 2, 4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosley-Bishop K. L., Li Q., Patterson L., Fischer J. A. (1999). Molecular analysis of the klarsicht gene and its role in nuclear migration within differentiating cells of the Drosophila eye. Curr. Biol. 9, 1211–1220 [DOI] [PubMed] [Google Scholar]
- Mozer B. A. (2001). Dominant Drop mutants are gain-of-function alleles of the muscle segment homeobox gene (msh) whose overexpression leads to the arrest of eye development. Dev. Biol. 233, 380–393 [DOI] [PubMed] [Google Scholar]
- Ohlmeyer J. T., Kalderon D. (1998). Hedgehog stimulates maturation of Cubitus interruptus into a labile transcriptional activator. Nature 396, 749–753 [DOI] [PubMed] [Google Scholar]
- Østerlund T., Kogerman P. (2006). Hedgehog signalling: how to get from Smo to Ci and Gli. Trends Cell Biol. 16, 176–180 [DOI] [PubMed] [Google Scholar]
- Paces-Fessy M., Boucher D., Petit E., Paute-Briand S., Blanchet-Tournier M. F. (2004). The negative regulator of Gli, Suppressor of fused (Sufu), interacts with SAP18, Galectin3 and other nuclear proteins. Biochem. J. 378, 353–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D., Rubin G. M. (1995). cAMP-dependent protein kinase and hedgehog act antagonistically in regulating decapentaplegic transcription in Drosophila imaginal discs. Cell 80, 543–552 [DOI] [PubMed] [Google Scholar]
- Pappu K. S., Chen R., Middlebrooks B. W., Woo C., Heberlein U., Mardon G. (2003). Mechanism of hedgehog signaling during Drosophila eye development. Development 130, 3053–3062 [DOI] [PubMed] [Google Scholar]
- Pearse R. V., 2nd, Collier L. S., Scott M. P., Tabin C. J. (1999). Vertebrate homologs of Drosophila suppressor of fused interact with the gli family of transcriptional regulators. Dev. Biol. 212, 323–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pignoni F., Zipursky S. L. (1997). Induction of Drosophila eye development by decapentaplegic. Development 124, 271–278 [DOI] [PubMed] [Google Scholar]
- Préat T. (1992). Characterization of Suppressor of fused, a complete suppressor of the fused segment polarity gene of Drosophila melanogaster. Genetics 132, 725–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price M. A., Kalderon D. (1999). Proteolysis of cubitus interruptus in Drosophila requires phosphorylation by protein kinase A. Development 126, 4331–4339 [DOI] [PubMed] [Google Scholar]
- Ready D. F., Hanson T. E., Benzer S. (1976). Development of the Drosophila retina, a neurocrystalline lattice. Dev. Biol. 53, 217–240 [DOI] [PubMed] [Google Scholar]
- Schlichting K., Dahmann C. (2008). Hedgehog and Dpp signaling induce cadherin Cad86C expression in the morphogenetic furrow during Drosophila eye development. Mech. Dev. 125, 712–728 [DOI] [PubMed] [Google Scholar]
- Shi Q., Li S., Jia J., Jiang J. (2011). The Hedgehog-induced Smoothened conformational switch assembles a signaling complex that activates Fused by promoting its dimerization and phosphorylation. Development 138, 4219–4231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A., Choi K. W. (2003). Initial state of the Drosophila eye before dorsoventral specification is equivalent to ventral. Development 130, 6351–6360 [DOI] [PubMed] [Google Scholar]
- Sisson B. E., Ziegenhorn S. L., Holmgren R. A. (2006). Regulation of Ci and Su(fu) nuclear import in Drosophila. Dev. Biol. 294, 258–270 [DOI] [PubMed] [Google Scholar]
- Stegman M. A., Vallance J. E., Elangovan G., Sosinski J., Cheng Y., Robbins D. J. (2000). Identification of a tetrameric hedgehog signaling complex. J. Biol. Chem. 275, 21809–21812 [DOI] [PubMed] [Google Scholar]
- Stone D. M., Murone M., Luoh S., Ye W., Armanini M. P., Gurney A., Phillips H., Brush J., Goddard A., de Sauvage F. J., et al. (1999). Characterization of the human suppressor of fused, a negative regulator of the zinc-finger transcription factor Gli. J. Cell Sci. 112, 4437–4448 [DOI] [PubMed] [Google Scholar]
- Strutt D. I., Wiersdorff V., Mlodzik M. (1995). Regulation of furrow progression in the Drosophila eye by cAMP-dependent protein kinase A. Nature 373, 705–709 [DOI] [PubMed] [Google Scholar]
- Sun Y., Jan L. Y., Jan Y. N. (1998). Transcriptional regulation of atonal during development of the Drosophila peripheral nervous system. Development 125, 3731–3740 [DOI] [PubMed] [Google Scholar]
- Suzuki T., Saigo K. (2000). Transcriptional regulation of atonal required for Drosophila larval eye development by concerted action of eyes absent, sine oculis and hedgehog signaling independent of fused kinase and cubitus interruptus. Development 127, 1531–1540 [DOI] [PubMed] [Google Scholar]
- Swan A., Nguyen T., Suter B. (1999). Drosophila Lissencephaly-1 functions with Bic-D and dynein in oocyte determination and nuclear positioning. Nat. Cell Biol. 1, 444–449 [DOI] [PubMed] [Google Scholar]
- Taylor M. D., Liu L., Raffel C., Hui C. C., Mainprize T. G., Zhang X., Agatep R., Chiappa S., Gao L., Lowrance A., et al. (2002). Mutations in SUFU predispose to medulloblastoma. Nat. Genet. 31, 306–310 [DOI] [PubMed] [Google Scholar]
- Treisman J. E., Rubin G. M. (1995). wingless inhibits morphogenetic furrow movement in the Drosophila eye disc. Development 121, 3519–3527 [DOI] [PubMed] [Google Scholar]
- Tsai Y. C., Yao J. G., Chen P. H., Posakony J. W., Barolo S., Kim J., Sun Y. H. (2007). Upd/Jak/STAT signaling represses wg transcription to allow initiation of morphogenetic furrow in Drosophila eye development. Dev. Biol. 306, 760–771 [DOI] [PubMed] [Google Scholar]
- Van Doren M., Ellis H. M., Posakony J. W. (1991). The Drosophila extramacrochaetae protein antagonizes sequence-specific DNA binding by daughterless/achaete-scute protein complexes. Development 113, 245–255 [DOI] [PubMed] [Google Scholar]
- Wang Q. T., Holmgren R. A. (1999). The subcellular localization and activity of Drosophila cubitus interruptus are regulated at multiple levels. Development 126, 5097–5106 [DOI] [PubMed] [Google Scholar]
- Wang Q. T., Holmgren R. A. (2000). Nuclear import of cubitus interruptus is regulated by hedgehog via a mechanism distinct from Ci stabilization and Ci activation. Development 127, 3131–3139 [DOI] [PubMed] [Google Scholar]
- Wang G., Amanai K., Wang B., Jiang J. (2000). Interactions with Costal2 and suppressor of fused regulate nuclear translocation and activity of cubitus interruptus. Genes Dev. 14, 2893–2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehrli M., Tomlinson A. (1995). Epithelial planar polarity in the developing Drosophila eye. Development 121, 2451–2459 [DOI] [PubMed] [Google Scholar]
- Wiersdorff V., Lecuit T., Cohen S. M., Mlodzik M. (1996). Mad acts downstream of Dpp receptors, revealing a differential requirement for dpp signaling in initiation and propagation of morphogenesis in the Drosophila eye. Development 122, 2153–2162 [DOI] [PubMed] [Google Scholar]
- Zhou Q., Kalderon D. (2011). Hedgehog activates fused through phosphorylation to elicit a full spectrum of pathway responses. Dev. Cell 20, 802–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.