Abstract
Genetic variation plays a major role in drug response variability. CsA (cyclosporin A), a widely used immunosuppressive agent, is a specific antagonist for FPR1 (formyl peptide receptor 1), which is an important G-protein-coupled chemoattractant receptor in the innate immune system. In order to study the variable responses of cyclosporins to different FPR1 mutants, we investigated the distribution of human FPR1 haplotypes among 209 healthy Han Chinese subjects. The haplotype pattern in Han Chinese were characterized on the basis of five SNPs (single nucleotide polymorphisms), including rs5030878 (p.T11I), rs2070745 (p.V101L), rs5030880 (p.R190W), rs1042229 (p.N192K) and rs867228 (p.A346E). Receptor binding affinity of cyclosporins to FPR1 haplotypes was assessed using N-formyl-Nle-Leu-Phe-Nle-Tyr-Lys–FITC in CHO-Gα16 cells stably transfected with cDNAs encoding the top 12 FPR1 haplotypes in the Han Chinese. Variants of FPR1 carrying a single amino acid substitution of leucine for valine at position 101 (p.Leu101) displayed significantly higher pKi values for CsA and CsH (cyclosporin H), indicative of an improved receptor affinity. The polymorphism of FPR1 p.Leu101 also enhanced the inhibitory effects of cyclosporins on fMLF (N-formyl-methionyl-leucyl-phenylalanine)-induced activities, including calcium mobilization, cell chemotaxis and MAPK (mitogen-activated protein kinase) phosphorylation. These results point to a possible complication for clinical use of CsA in patients carrying the p.Leu101 allele of FPR1.
Keywords: cyclosporin, formyl peptide receptor 1 (FPR1), haplotype, pharmacogenomics, receptor affinity, single nucleotide polymorphism
Abbreviations: CHO, Chinese-hamster ovary; CsA, cyclosporin A; CsH, cyclosporin H; ERK, extracellular-signal-regulated kinase; Fluo-4/AM, Fluo-4 acetoxymethyl ester; fMLF, N-formyl-methionyl-leucyl-phenylalanine; fNLFNYK, N-formyl-Nle-Leu-Phe-Nle-Tyr-Lys; FPR, formyl peptide receptor; HBSS, Hanks balanced saline solution; LD, linkage disequilibrium; MAF, minor allele frequency; MAPK, mitogen-activated protein kinase; ORF, open reading frame; SNP, single nucleotide polymorphism
INTRODUCTION
Pharmacogenomics is an emerging research field that focuses on the study of how genes modulate drug responses among individuals [1,2]. An abundant volume of literature in this area indicates that genetic variation plays a major role in drug response variability, which is involved in pharmacodynamics and pharmacokinetics of a drug. Such variations, including SNPs (single nucleotide polymorphisms), base insertions or deletions, copy-number variations and variable numbers of tandem repeats, can influence, and hence potentially predict, both the efficacy and toxicity of a drug. As generally used markers throughout the genome, SNPs may exist in individual genes in coding regions, introns and surrounding regions. The haplotypes of several SNPs provide greater statistical power to detect genes that are associated with disease traits and drug responses. Genetically distinct populations may differ in both the extent of the LD (linkage disequilibrium) of these SNPs and their haplotype frequencies, which reflect the population structure and human evolution [3].
For an individual, their genetic make-up exerts influences in how well a medication works and what side effects are likely to occur within the body. Small differences in DNA sequence between different population groups or families within a population group, building up over generations, can lead to different reactivity to therapeutic agents. Following the completion of the Human Genome Project (http://www.ornl.gov/sci/techresources/Human_Genome/home.shtml), the International HapMap Consortium (2003, 2005) and the 1000 Genomes Project Consortium (2010), breakthroughs in genomics have boosted the development and application of pharmacogenomics, which has a potential clinical value when deciding between multiple treatment options to maximize therapeutic benefits and to limit the risk of side effects for individuals or special populations [4]. The number of pharmacogenetic associations has steadily increased over the years. PharmGKB (http://www.pharmgkb.org), a pharmacogenomics knowledge resource that encompasses clinical information, including dosing guidelines, drug labels, potentially clinically actionable gene–drug associations and genotype–phenotype relationships, annotated over 2000 genes involved in drug responses. Approximately 10% of the labels for FDA (Food and Drug Administration)-approved drugs contain pharmacogenomic information (http://www.fda.gov/drugs/scienceresearch/researchareas/pharmacogenetics/ucm083378.htm) [2,5]. It is the ultimate goal of pharmacogenomics to use an understanding of drug–gene associations to devise personalized treatment strategies for any given medication.
Cyclosporins are cyclic undecapeptides produced by fungi. Among them, CsA (cyclosporin A) was initially identified as an immunosuppressant and has been widely used clinically to prevent organ rejection following transplantation in humans [6]. CsA is lipophilic, readily penetrates the plasma membrane and exerts its immunosuppressive action through binding to the cytosolic protein cyclophilin, which causes inhibition of the Ca2+ and calmodulin-dependent phosphatase calcineurin, thereby disrupting transcriptional induction of interleukin-2 and T-cell activation [7]. A variety of structural analogues of CsA are produced either by fungi or through chemical synthesis [8–11]. Among them, CsH (cyclosporin H) is an optical isomer of CsA that contains the amino acid residue N-methyl-D-valine at position 11 instead of N-methyl-L-valine seen in CsA. CsH lacks immunosuppressive activity and only weakly interferes with the multidrug-resistance protein P-glycoprotein (MDR1). Several lines of evidence have indicated that cyclosporins are antagonists for human FPR (formyl peptide receptor) 1 [12].
The human FPR1 gene is located in chromosome 19q13.3-19q13.4, clustered with the genes of another two receptors in the same receptor family, FPR2/ALX and FPR3 [13]. It contains three exons, but the ORF (open reading frame), which encodes a protein of 350 amino acids, locates exclusively in the third exon. At least ten SNP loci, four synonymous and six non-synonymous, with the least abundant allele showing a frequency of 0.1 or more, have been identified in the human FPR1 coding region. It is known that SNPs occur throughout the genome at a frequency of approximately 1 in 300–1000 bp [14,15]. It is quite unique for the human FPR1 gene to have such a high frequency of polymorphism. When nucleotide diversity, which is defined as the number of nucleotide differences per site between two randomly chosen sequences from a population, was used to quantify genetic variability of human FPR1, the value of FPR1 was approximately 3-fold higher compared with the mean value reported for other genes [16]. Furthermore, the number of FPR1 variants varies markedly in different mammalian species, indicating differential gene expansion or extinction [13]. Among different human racial groups, the FPR1 SNPs are also detected at variable levels, which may suggest that the FPR1 gene has been subjected to strong natural selection throughout evolution [16].
The protein encoded by the human FPR1 gene is a seven-membrane-span G-protein-coupled receptor in the rhodopsin family. When stimulated by chemoattractants, such as the bacterial peptide fMLF (N-formyl-methionyl-leucyl-phenylalanine), formyl peptides released by mitochondria of ruptured cells [17,18], neutrophil granule protein cathepsin G [19], virus-derived peptides HIV-T20, T21 and HCV (hepatitis C virus) nonstructural peptide 5A [20–22], FPR1 activates heterotrimeric G-proteins and downstream signalling molecules, leading to neutrophil chemotaxis, degranulation and superoxide production [13]. These activities are essential to the elimination of invading pathogens and damaged tissues, suggesting that FPR1 plays an important role in the innate immune system. Experimental data indicate that mice lacking the Fpr1 gene are more susceptible to Listeria monocytogenes infection with an increased mortality rate [23]. The pharmacogenomic survey of human FPR1 polymorphism provides some hints about the function of this gene. It was first reported in 1977 by Clark et al. [24] that defective chemotactic responses were observed in neutrophils from seven out of nine juvenile periodontitis patients. Since then, an association between aggressive periodontitis and defects in neutrophil responses to formyl peptides was documented from time to time, involving SNPs including p.F110S, p.C126W, p.R190W, p.N192K, c.−12915C>T, c.301G>C, c.546C>A and c.348T>C [25–28]. There are also some inconsistent reports about the association of FPR1 c.32C>T with C-reactive protein. Recently, El Shamieh et al. [29] reported that FPR1 c.32C>T interacted with age, and was associated with high blood pressure in healthy individuals under 45 years of age. Owing to the different allele and genotype frequencies in various populations, studies on FPR1 haplotypes in different racial groups were also carried out. To date, more than 20 haplotypes have been reported [16,30], and the investigation by Gripentrog et al. [30] found that some of the FPR1 haplotypes are functionally distinct when activated by formyl peptides (e.g. fMFEDAVAWF) from other bacterial strains such as Mycobacterium avium rather than the Escherichia coli-derived fMLF. The different SNPs and haplotypes of FPR1 may respond differently to some special modulators of the receptor. However, no drug-response association with the FPR1 gene polymorphism has been described so far.
We have previously demonstrated that both CsA and CsH are selective antagonists for human FPR1 [12]. They inhibited the uptake of fNLFNYK (N-formyl-Nle-Leu-Phe-Nle-Tyr-Lys)–fluorescein and [3H]fMLF binding to FPR1. In functional assays, both cyclosporins inhibited fMLF-stimulated degranulation, chemotaxis, calcium mobilization and phosphorylation of MAPKs [mitogen-activated protein kinases; ERK1/2 (extracellular-signal-regulated kinase)] and serine/threonine protein kinase Akt. In 2002, Loor et al. [9] established the structure–activity relationships of 59 cyclosporins for FPR1 inhibition in vitro using intact live cells. However, no data could be found relative to investigations on pharmacogenomics of cyclosporins at different FPR1 haplotypes, especially in special racial groups. In the present study, we studied the distribution of human FPR1 haplotypes in the Han Chinese population. The variability of biological responses elicited by cyclosporins to these FPR1 haplotypes was also evaluated.
EXPERIMENTAL
Subjects
Blood samples were collected by ADICON Clinical Laboratories (Shanghai, China) from 209 healthy Han ethnic volunteers (ages ranged from 18 to 88 years, including 78 males and 131 females) in accordance with the procedure approved by the institutional review board. The participants were all aware of and agreed upon the tests to be performed through execution of individual informed consent forms. The samples were studied and analysed for FPR1 gene polymorphisms anonymously and the present study was conducted according to the principles expressed in the Declaration of Helsinki.
Reagents
Cyclosporins were produced at the Fujian Institute of Microbiology (Fuzhou, China). Fluo-4/AM (Fluo-4 acetoxymethyl ester) and fNLFNYK–FITC were obtained from Molecular Probes. Probenecid and fMLF were the products of Sigma–Aldrich. The anti-phospho-ERK1/2 and anti-phospho-Akt (recognizing Ser473) antibodies were from Cell Signaling Technologies. All restriction enzymes, DNA polymerase and DNA ligation kits were purchased from TaKaRa Biotechnology. DNA purification after electrophoresis was done with the TIANgel Mini Purification kit (Tiangen Biotech). PCR products were purified with the AxyPrep PCR Clean-up kit (Axyegen). Site-directed mutagenesis kits were bought from Stratagene.
Vectors and cell lines
The expression vector pGEN-IRES Neo was provided by AstraZeneca. The cDNA encoding human FPR1 (NCBI accession number NM_002029.3) was obtained from a human HL-60 granulocyte library [31]. The plasmid pUC18 was obtained from Tiangen Biotech.
The cell line CHO-Gα16 (RD-HGA16) was purchased from Molecular Devices. It consists of a CHO (Chinese-hamster ovary) cell host and the stably transfected plasmid for the Gα16 protein. Cells were routinely maintained in Ham's F-12 medium supplemented with 10% (v/v) fetal bovine serum (Invitrogen), 2 mM L-glutamine, 100 μg/ml streptomycin, 100 units/ml penicillin and 100 μg/ml hygromycin.
Genotype and haplotype determination
Genomic DNA was isolated from the cellular component of 1 ml of whole blood with the TIANamp Blood DNA kit according to the manufacturer's instructions (Tiangen Biotech). The ORF of the FPR1 gene was amplified with PCR using primers located upstream and downstream of exon 3. The forward primer was 5′-TTGCCCAACAGGTACAATAA-3′ and the reverse primer was 5′-ATTTCAGGCAAACTAGGATG-3′. The products were sequenced with the primers 5′-AGAACCACCGCACCGTGA-3′ and 5′-GGATGTTCCGGCTGTTGT-3′. Gene polymorphisms were determined with sequencing by Shanghai DNA BioTechnologies. Five polymorphisms in the FPR1 gene, including rs5030878 (p.T11I), rs2070745 (p.V101L), rs5030880 (p.R190W), rs1042229 (p.N192K) and rs867228 (p.A346E), were further studied. The construction and population frequencies of haplotypes were calculated with the SHEsis software using its haplotype construction element, the algorithm of which is designed based on an improved expectation maximization method [32]. Analysis of Hardy–Weinberg equilibria was performed using the Pearson χ-squared test.
Site-directed mutagenesis
Site-directed mutagenesis was carried out according to the manufacturer's instructions (Stratagene). The FPR1 cDNA was amplified with PCR using the forward primer 5′-TCCGAATTCATGGAGACAAATTCCTCTCTC-3′ and the reverse primer 5′-GCTTCTAGATCACTTTGCCTGTAACGCCAC-3′. The fragment was subcloned into the pUC18 vector between the EcoRI and XbaI restriction sites to produce pUC18-FPR1, which is the template for mutation analysis. PCRs were performed as follows: 95°C for 1 min, then 95°C for 1 min, 55°C for 1 min and 65°C for 8 min for 30 cycles. Primers used for the site-directed mutagenesis are listed in Supplementary Table S1 (at http://www.biochemj.org/bj/451/bj4510245add.htm). The resultant FPR1 cDNA inserts encoding different haplotypes were verified with sequencing and ligated into the mammalian expression vector pGEN-IRES Neo between EcoRI and XbaI restriction sites.
Transfection of FPR1 haplotype cDNAs into CHO-Gα16 cells was carried out with the Gene Pulser Xcell Electroporation System from Bio-Rad Laboratories. Briefly, approximately 4×106 cells were harvested and resuspended in 400 μl of RPMI 1640 medium containing 10 mM glucose and 0.1 mM dithiothreitol. Vectors containing the ORFs of the human FPR1 gene (8 μg in a volume of 8 μl) were added to the cells. Following a 280 V and 20 ms pulse (squarewave), the cells were immediately returned to 10 ml of culture medium and incubated for 24 h. Stable clones expressing FPR1 haplotypes were selected after 10 days of culture under the selection of 600 μg/ml G418 and identified by FACScan analysis after binding of 20 nM of the fluorescent ligand fNLFNYK–FITC to the CHO cell transfectants for 45 min at 4°C. Transfectants with similar expression levels of different FPR1 haplotypes were further confirmed by FACScan analysis after labelling with FITC-conjugated anti-FPR1 antibodies according to the manufacturer's instructions (BD Biosciences).
FACScan analysis
CHO cells were added to 3 mM KCl, 100 mM NaCl, 10 mM sodium phosphate, 1 mM Mg2+ and 1 mM Ca2+, pH 7.4, containing 5% fetal bovine serum and incubated at 4°C for 1 h with various concentrations of fNLFNYK–FITC. The mean fluorescence of the cells was determined after subtracting the readout of cells without fNLFNYK–FITC. The results were analysed by non-linear least squares analysis to determine Kd and Bmax values. For assessment of compound binding affinities to FPR1 haplotypes, cells were incubated with the indicated concentrations of cyclosporins together with 1 nM fNLFNYK–FITC. Ki values of different FPR1 variants were determined by non-linear least squares analysis using the known Kd for fNLFNYK–FITC and the observed IC50 numbers of cyclosporins in each case. Data from at least two different experiments in duplicate were analysed.
Calcium mobilization
A calcium mobilization assay was performed as described previously with minor modifications [33]. Briefly, CHO-Gα16-FPR1 cells were plated on to 96-well plates at a density of 15000 cells in 100 μl of medium per well and incubated for 24 h. Cells were loaded with 5 μM Fluo-4/AM in HBSS (Hanks balanced saline solution) supplemented with 2.5 mM probenecid for 45 min and then washed twice with HBSS. After incubation with or without cyclosporins for 15 min, 1 nM fMLF was added followed by analysis of calcium mobilization using a FlexStation (Molecular Devices) with the excitation wavelength at 485 nm and the emission wavelength at 525 nm.
Chemotaxis
The chemotaxis assay was performed as described previously [12] in a 48-well microchemotaxis chamber (NeuroProbe). Briefly, fMLF (10 nM, 30 μl) was placed in the lower chamber and CHO-Gα16-FPR1 cells (50 μl at 1×106 cells/ml) were pre-incubated with or without cyclosporins for 15 min and then loaded on to the upper chamber, which was separated from the lower chamber by a polycarbonate filter (pore size of 8 μm). Chemotaxis was allowed to proceed for 4 h at 37°C. After that, cells on the upper face of the filter were removed. The cells that adhered to the underside of the filter were fixed and stained with 0.1% Crystal Violet [in 20% (v/v) methanol] for 30 min. The integrated absorbance of stained cells from three random fields of each well (triplicates for each concentration) was quantified using an image analyser (Image-Pro Plus). The chemotaxis of CHO-Gα16-FPR1 cells induced with 1 nM fMLF was set as 100%.
ERK1/2 activation
Activation of the p44/p42 MAPKs (ERK1/2) was determined as described previously [12] with modest modifications. Briefly, cells were cultured in 24-well plates at 5×105 cells/well for 24 h and serum starved for 2 h. They were pretreated with the indicated amounts of cyclosporins for 5 min before fMLF (10 nM) stimulation in HBSS/BSA. After 5 min of stimulation, the reaction was terminated by adding 50 μl of ice-cold SDS/PAGE loading buffer [15% (v/v) glycerol, 125 mM Tris/HCl, pH 6.8, 5 mM EDTA, 2% (w/v) SDS, 0.1% Bromophenol Blue and 1% (v/v) 2-mercaptoethanol]. Samples were transferred to microcentrifuge tubes and sonicated twice for 5 s each to disperse DNA contents. After boiling, samples were analysed by SDS/PAGE and Western blotting using anti-ERK1/2 and anti-phospho-ERK1/2 antibodies at a 1:1000 dilution. Horseradish peroxidase-conjugated anti-rabbit antibody (1:3000 dilution) was used as the secondary antibody. The resulting immunocomplex was visualized using the SuperSignal West Pico chemiluminescence kit (Pierce Biotechnology) according to the manufacturer's instructions. The relative intensities of the bands from the PVDF membrane (Millipore) were quantified using the ChemDoc MP System (Bio-Rad) and Image-Pro Plus software (MediaCybernetics).
Data analysis
The distribution of genotypes for each polymorphism was assessed for deviation from the Hardy–Weinberg equilibrium using w2 tests (StatView). Data on CHO cell transfectants were analysed using GraphPad Prism software. Non-linear regression analyses were performed to generate dose–response curves to calculate IC50 values.
RESULTS
FPR1 polymorphisms in the Han Chinese
Using genomic DNA isolated from whole blood cells derived from 209 healthy Han Chinese subjects, we analysed the distribution of allele and genotype of the human FPR1 gene in this population. We selected all non-synonymous SNPs that have a MAF (minor allele frequency) higher than 0.1 for human FPR1 from the dbSNP database (http://www.ncbi.nlm.nih.gov/SNP/snp_ref.cgi?locusId=2357). Five SNPs met this requirement, namely, rs5030878 (p.T11I), rs2070745 (p.V101L), rs5030880 (p.R190W), rs1042229 (p.N192K) and rs867228 (p.A346E). The SNP p.T11I is located at the N-terminus of the FPR1 protein, whereas p.V101L resides near the transmembrane–extracellular interface [13]. SNPs p.R190W and p.N192K are located in the second extracellular loop of FPR1. p.A346E is near the intracellular C-terminus of the receptor.
Next, we examined the prevalence of genetic variants containing these five FPR1 SNPs in all subjects, including allele and genotype frequencies (Tables 1 and 2). The MAFs for p.T11I (c.32T), p.V101L (c.301C), p.R190W (c.568T) and p.A346E (c.1037A) were 0.029, 0.45, 0.172 and 0.297 respectively. The MAF value for p.V101L (0.45) found in the present study is consistent with that (0.477) derived from the HapMap resource (NCBI assay ID ss24686736), but it is approximately 1.5-fold higher than that found in other two ethnic groups (0.292 in European and 0.258 in Yoruba African populations respectively; Supplementary Figure S1 at http://www.biochemj.org/bj/451/bj4510245add.htm).The frequency of homozygous p.Ile11 was 0.005, closely resembling the value in Chinese from metropolitan Denver and differing from that in Chinese from Beijing. Although the variant for p.N192K was generally believed to be c.576T>G, our results displayed three alleles for p.N192K, with a frequency of 0.514 for c.576T, 0.304 for c.576G and 0.182 for c.576C respectively. Alleles C and T at this position are synonymous, and both are translated into p.Asn192, resulting in a much lower genotype frequency of p.Lys192 than those documented in the HapMap (0.30 compared with the reported values of 0.48 for Han Chinese in Beijing and 0.52 for Chinese in metropolitan Denver; Table 1). The LD pattern of these FPR1 SNPs was analysed with pair-wise Lewontin's D’ and correlation coefficient r2 (Supplementary Figure S2 at http://www.biochemj.org/bj/451/bj4510245add.htm). The two adjacent SNPs, p.R190W and p.N192K, exhibited relative strong LD compared with other pairs of SNPs [34].
Table 1. Allele frequencies in the Han Chinese studied.
Data are derived from direct sequencing of genomic DNA from 209 Han Chinese subjects.
| Reference ID | SNP | Allele | Phenotype | Number | Frequency |
|---|---|---|---|---|---|
| rs5030878 | T11I | c.32C | Thr11 | 406 | 0.971 |
| c.32T | Ile11 | 12 | 0.029 | ||
| rs2070745 | V101L | c.301G | Val101 | 230 | 0.550 |
| c.301C | Leu101 | 188 | 0.450 | ||
| rs5030880 | R190W | c.568A | Arg190 | 346 | 0.828 |
| c.568T | Trp190 | 72 | 0.172 | ||
| rs1042229 | N192K | c.576T | Asn192 | 215 | 0.514 |
| c.576G | Lys192 | 127 | 0.304 | ||
| c.576C | Asn192 | 76 | 0.182 | ||
| rs867228 | A346E | c.1037C | Ala346 | 294 | 0.703 |
| c.1037A | Glu346 | 124 | 0.297 |
Table 2. Genotype frequencies in the Han Chinese studied.
Data are derived from direct sequencing of genomic DNA from 209 Han Chinese subjects. Amino acids are in the single-letter code.
| Reference ID | SNP | Genotype | Number | Frequency |
|---|---|---|---|---|
| rs5030878 | T11I | T/T | 198 | 0.947 |
| T/I | 10 | 0.048 | ||
| I/I | 1 | 0.005 | ||
| rs2070745 | V101L | V/V | 62 | 0.297 |
| V/L | 106 | 0.507 | ||
| L/L | 41 | 0.196 | ||
| rs5030880 | R190W | R/R | 144 | 0.689 |
| R/W | 58 | 0.278 | ||
| W/W | 7 | 0.033 | ||
| rs1042229 | N192K | N/N | 98 | 0.469 |
| N/K | 95 | 0.455 | ||
| K/K | 16 | 0.077 | ||
| rs867228 | A346E | A/A | 106 | 0.507 |
| A/E | 82 | 0.392 | ||
| E/E | 21 | 0.100 |
Haplotype frequencies in the Han Chinese were inferred from the genotype information with SHEsis, which is based on the PLCSEM (Partition-Ligation Combination-Subdivision Expectation Maximization) algorithm and is robust for efficient estimation of haplotypes constructed from large numbers of biallelic or multiallelic loci in diploid individuals [32]. As depicted in Table 3, nine FPR1 haplotypes with a frequency higher than 0.03 were constructed and eight more were inferred in the Han Chinese. Haplotypes CH1–CH13 have all been reported previously [16,30]. However, the distribution of these haplotypes differs markedly from that reported in other races. As described in the literature [16,30], the top two haplotypes in the Caucasian population were CH3 (p.Lys192, H-1) and CH6 (p.Trp190, H-2), whereas the third one carrying p.Ile11/Lys192/Glu346 in the Caucasian population could not be detected in Han Chinese. CH14 (Ile11/Leu101/Glu346) is a newly discovered haplotype never reported before. These results demonstrate that the pattern and distribution of FPR1 haplotypes differ between Caucasian and Han Chinese populations, thus representing the different structure and evolution pathway of these two populations.
Table 3. Haplotype frequencies in the Han Chinese studied.
Amino acids are in the single-letter code.
| SNP | |||||||
|---|---|---|---|---|---|---|---|
| Haplotype* | Original nomenclature† | I11T | V101L | R190W | N192K | A346E | Frequency (%) |
| CH1 | H-6A | T | L | R | N-t‡ | A | 15.9 |
| CH2 | H-11 | T | V | R | N-t | A | 13.2 |
| CH3 | H-1 | T | V | R | K | A | 12.1 |
| CH4 | H-12 | T | V | R | N-t | E | 11.5 |
| CH5 | H-5 | T | L | R | K | A | 10.2 |
| CH6 | H-2 | T | V | W | N-c | A | 8.8 |
| CH7 | H-16 | T | V | R | K | E | 8.1 |
| CH8 | H-19 | T | L | R | N-t | E | 7.9 |
| CH9 | H-17 | T | L | W | N-c | A | 6.5 |
| CH10 | H-20 | T | L | W | N-c | E | 2.0 |
| CH11 | H-8 | I | V | R | N-t | A | 1.3 |
| CH12 | H-4 | I | L | R | N-t | A | 1.3 |
| CH13 | H-6B | T | L | R | N-c | A | 1.0 |
| CH14 | -§ | I | L | R | N-t | E | 0.2 |
| CH15 | H-25A | T | L | R | K | E | 0 |
| CH16 | H-21 | T | V | W | N-c | E | 0 |
| CH17 | H-18 | I | V | W | N-c | A | 0 |
*Each haplotype is defined by the shown linkage of nucleotides in the corresponding row, and each corresponds to the SNPs listed in the top row. Haplotypes were determined with the SHEsis software based on the genotype information from sequencing.
†Original nomenclature follows that of [7,21], and the number represents the ranking of haplotypes in the Caucasian population.
‡SNP that does not change the amino acid.
§Newly discovered haplotype that has not been reported previously.
Binding affinity of cyclosporins to FPR1 mutants
CsA is an immunosuppressant widely used in the clinic for patients receiving organ transplantation. In order to study whether the polymorphism of the human FPR1 gene affects the pharmacology of cyclosporins, especially in the Han Chinese, we constructed a batch of FPR1 cDNAs encoding the first 12 FPR1 haplotypes in this population. Vectors including FPR1 ORFs were transfected into CHO-Gα16 cells and stable cell lines expressing similar levels of individual FPR1 haplotypes were selected. The expression of the FPR1 haplotype in each clone was determined by FACScan analysis using the peptide fNLFNYK–FITC as a probe, which was subsequently confirmed with FITC-conjugated anti-FPR1 antibodies. As shown in Table 4, the Bmax values of CHO-Gα16-FPR1 clones containing different FPR1 haplotypes were in the range of 33860 in CH4 (p.Glu346)-expressing cells (CH4) to 65059 in CH12 (p.Ile11/Leu101)-expressing cells (CH12). The receptor affinity of fNLFNYK–FITC to different FPR1 haplotypes varied moderately. Compared with CH1 (p.Leu101), haplotypes CH5 (p.Leu101/Lys192) and CH12 (p.Ile11/Leu101) had the highest affinity (Kd=2.8±0.1 and 3.2±0.3 nM respectively), whereas haplotypes CH4 (p.Glu346) and CH8 (p.Leu101/Glu346) showed the lowest affinity (Kd=1.3±0.2 and 1.3±0.1 nM respectively).
Table 4. Receptor-binding affinity of cyclosporins to FPR1 haplotypes.
| fNLFNYK–FITC-binding parameters* | pKi for cyclosporins† | |||
|---|---|---|---|---|
| Haplotype | Kd (nM) | Bmax | pKi for CsA | pKi for CsH |
| CH1 | 2.0±0.2 | 48336±4553 | 7.15±0.05 | 8.16±0.16 |
| CH2 | 1.7±0.1 | 47657±9098 | 5.26±0.02‡ | 6.34±0.04‡ |
| CH3 | 1.8±0.2 | 59135±4185 | 5.62±0.09‡ | 6.48±0.04‡ |
| CH4 | 1.3±0.2‡ | 33860±4977 | 5.99±0.12‡ | 6.16±0.08‡ |
| CH5 | 2.8±0.1‡ | 42575±4902 | 6.86±0.08 | 7.75±0.09 |
| CH6 | 1.8±0.4 | 34093±693 | 5.63±0.02‡ | 6.46±0.05‡ |
| CH7 | 1.7±0.3 | 60688±10388 | 5.85±0.02‡ | 6.47±0.04‡ |
| CH8 | 1.3±0.1‡ | 36362±353 | 6.96±0.11 | 7.90±0.08 |
| CH9 | 1.9±0.5 | 39009±6672 | 7.05±0.09 | 7.91±0.11 |
| CH10 | 2.4±0.3 | 41751±615 | 7.13±0.12 | 7.94±0.06 |
| CH11 | 2.2±0.7 | 37713±5841 | 5.59±0.01‡ | 6.27±0.02‡ |
| CH12 | 3.2±0.3‡ | 65059±3570 | 7.26±0.02 | 7.91±0.01 |
*CHO-Gα16 cells stably transfected with individual variants of FPR1 (CH1–CH12) were incubated with various concentrations of fNLFNYK–FITC. The data were analysed by non-linear least-squares analysis to determine Kd and Bmax values.
†Cells were incubated with various concentrations of cyclosporins together with 1 nM fNLFNYK–FITC. Ki was determined by non-linear least-squares analysis using the known Kd for fNLFNYK–FITC and the observed IC50 of cyclosporins for different FPR1 variants.
‡P<0.05 compared with CH1.
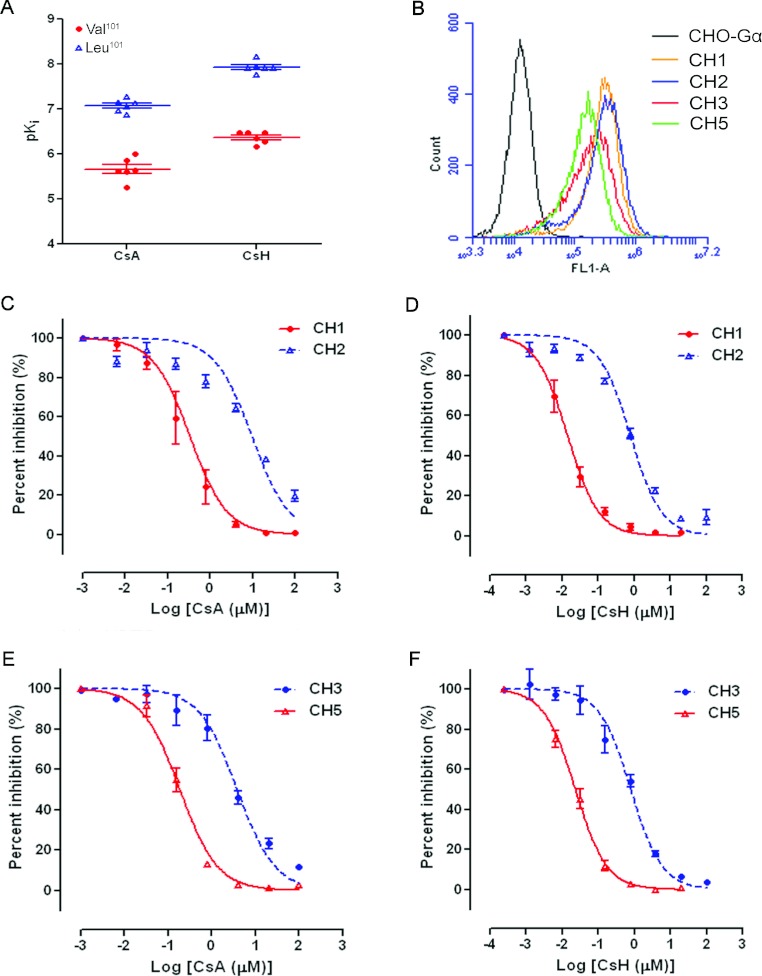
The binding affinity of CsA and CsH to mutant FPR1 receptors was also assessed in a competitive binding assay using 1 nM fNLFNYK–FITC (Table 4). We observed that the binding affinity of cyclosporins to different FPR1 mutants varied significantly. Haplotypes CH1, CH5, CH8, CH9, CH10 and CH12 all showed decreased Ki values for both CsA and CsH. These variants share a common allele, p.Leu101 (Figure 1A), indicating that p.Leu101 is a major factor that contributes variations to ligand binding of the receptor. The average Ki values of FPR1 for CsA and CsH increased in Leu101-expressing cells by 25.9- and 36.8-fold respectively. No other SNP demonstrated significant and consistent variation between these haplotypes. In order to determine the influence of amino acid variation of p.V101L on FPR1 affinity for cyclosporins, pairs of haplotypes containing only one variation at position 101, including CH1 (p.Leu101) compared with CH2 and CH3 (p.Lys192) compared with CH5 (p.Leu101/Lys192), were selected for further analysis. When assessed with anti-FPR1 antibodies, the surface expression profile of FPR1 on the cells expressing these haplotypes was comparable between each of the pairs tested (Figure 1B), but their affinity for CsA and CsH varied. The pKi value of CsA increased from 5.26±0.02 in CH2-expressing cells to 7.15±0.05 in CH1-expressing cells, whereas those of CsH improved from 6.34±0.04 in CH2 to 8.16±0.16 in CH1 (Table 4, Figures 1C and 1D). The results for the haplotype pair CH3 and CH5 displayed a similar tendency: the pKi value of CsA increased from 5.62±0.09 in CH3-expressing cells to 6.86±0.08 in CH5-expressing cells (Figure 1E). In this pair, the binding affinity of CsH also improved significantly (pKi=6.48±0.04 in CH3 compared with pKi=7.75±0.09 in CH5; Figure 1F).
Figure 1. Effects of FPR1 gene polymorphisms on the receptor-binding affinity for CsA and CsH.
(A) Distribution of pKi values for CsA and CsH in CHO-Gα16-FPR1 haplotype-expressing cells. (B) Flow cytometric analysis of cell-surface expression of FPR1 in control CHO-Gα16 cells and haplotype CH1, CH2, CH3 or CH5-expressing CHO-Gα16-FPR1 cells. (C–F) Concentration–response data of CH1, CH2, CH3 and CH5 cells to CsA and CsH were analysed. fNLFNYK–FITC (1 nM) was used as an agonist, and the value of agonist alone was set as 100%. Data are presented as means±S.E.M. derived from two to three independent experiments.
Enhanced antagonism of cyclosporins on calcium mobilization in FPR1 p.Leu101 mutants
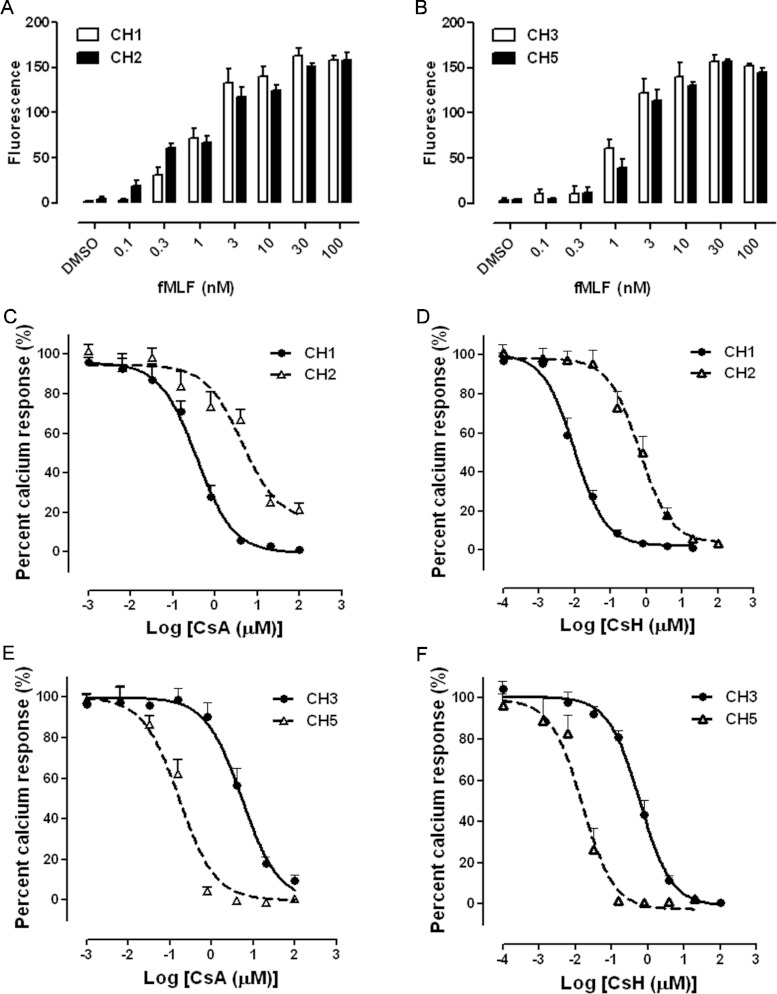
Our previous study elucidated that both CsA and CsH affect the signal transduction and cellular function of FPR1, including calcium mobilization, degranulation and chemotaxis [12]. In order to investigate whether the substitution of leucine for valine at position 101 of FPR1 would affect the pharmacological property of cyclosporins at the receptor, calcium mobilization induced by fMLF in haplotype-expressing CHO-Gα16-FPR1 cells was carried out. The peptide fMLF elicited calcium mobilization dose-dependently in the four cell lines tested (Figures 2A and 2B). When CsA was added 15 min before stimulation with 1 nM fMLF, its IC50 decreased greatly from 4.35 μM in CH2 to 0.36 μM in CH1 and from 5.29 μM in CH3 to 0.17 μM in CH5 (Figures 2C and 2E). Upon the addition of CsH, IC50 values in FPR1 p.Leu101-expressing cells were also markedly lower than those in FPR1 p.Val101-expressing cells (9.9 nM in CH1 compared with 647 nM in CH2, and 15.5 nM in CH5 compared with 605 nM in CH3) (Figures 2D and 2F).
Figure 2. Inhibitory effects of cyclosporins on fMLF-mediated calcium mobilization in CHO-Gα16-FPR1 haplotype-expressing cells.
Different concentrations of fMLF-induced calcium mobilization in CH1 and CH2 cells (A) and in CH3 and CH5 cells (B) are shown. (C–F) Cells were cultured overnight, loaded with 5 μM Fluo-4/AM and incubated with the indicated concentrations of CsA or CsH for 15 min before stimulation with 1 nM fMLF. Calcium mobilization was recorded on a FlexStationIII. The fluorescence signal stimulated with fMLF alone was arbitrarily set as 100%. Data are presented as means±S.E.M. derived from at least three independent experiments.
Enhanced antagonism of cyclosporins on chemotaxis in FPR1 p.Leu101 mutants
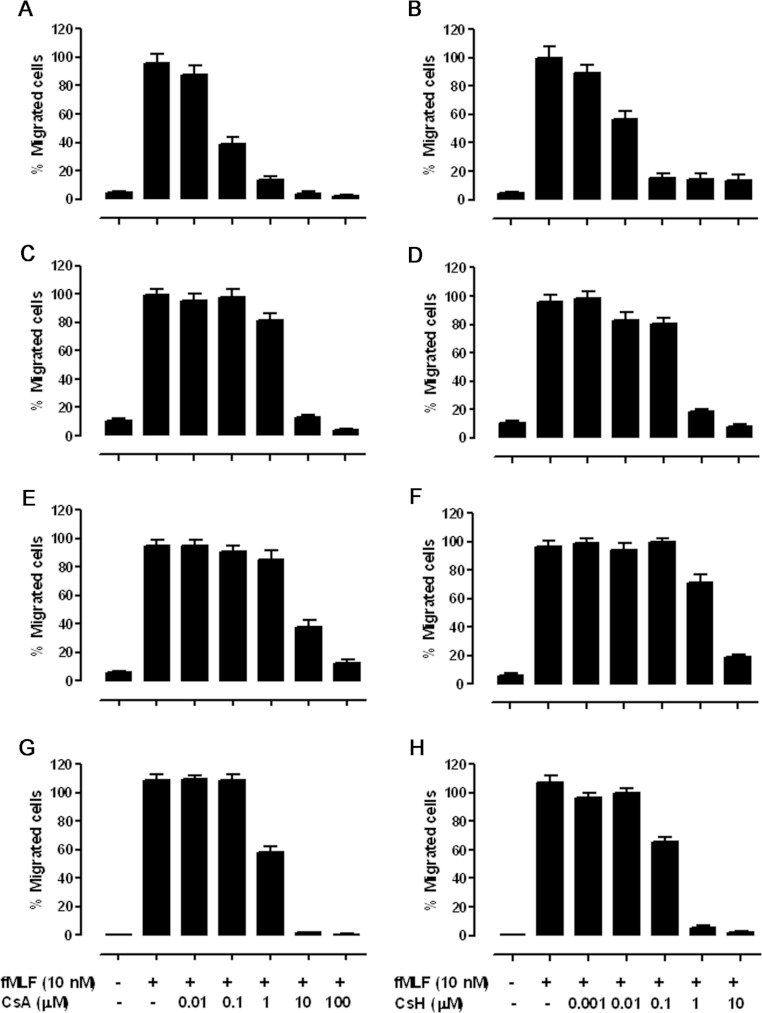
Chemotaxis towards bacterial formylated peptides is one of the primary functions of FPR1. Besides the influence of FPR1 gene polymorphism on fMLF-mediated calcium mobilization, we also monitored the inhibitory effects of CsA and CsH on cell chemotaxis induced by fMLF in CHO-Gα16-FPR1 cells. The CHO transfectants with different FPR1 haplotypes were allowed to migrate for 4 h across a polycarbonate filter with 8 μm pores towards a lower well containing 10 nM fMLF. The inhibition on chemotaxis was greatly enhanced when p.Val101 of FPR1 was changed to p.Leu101, as depicted in Figure 3. In CH2 cells, the IC50 of CsA was 3.07 μM, whereas that of CsH was 320 nM. In CH1 cells, the inhibitory effects on chemotaxis were evident at CsA concentrations above 10 nM (IC50=61.8 nM) and at CsH concentrations above 1 nM (IC50=9.7 nM). Likewise, CsA inhibited fMLF-induced chemotaxis with a potency approximately 5-fold lower in CH5 (IC50=1.16 μM) than in CH3 (IC50=5.86 μM) cells, whereas the antagonism exerted by CsH was more significant between them (with IC50 values of 2.87 μM for CH3 compared with 165.1 nM for CH5 cells).
Figure 3. Effects of cyclosporins on fMLF-stimulated chemotaxis in FPR1 haplotype-expressing CHO-Gα16 cells.
(A and B) CH1, (C and D) CH2, (E and F) CH3 and (G and H) CH5. Cells were pre-incubated with different concentrations of cyclosporins for 15 min before loading into the upper wells of a 48-well chemotaxis chamber with 10 nM fMLF placed in the lower wells. The chemotaxis assay was conducted at 37°C for 4 h, and the numbers of migrated cells were determined by analysis with the Image-Pro Plus software, i.e. counting the integrated absorbance of cells after staining with 0.01% Crystal Violet in 20% methanol. Three fields were photographed for each well. The results presented are means±S.E.M. from three independent experiments, each in triplicate, showing the percentage of maximal chemotaxis induced with fMLF alone.
Improved inhibition of cyclosporins on ERK1/2 activation in FPR1 p.Leu101 mutants
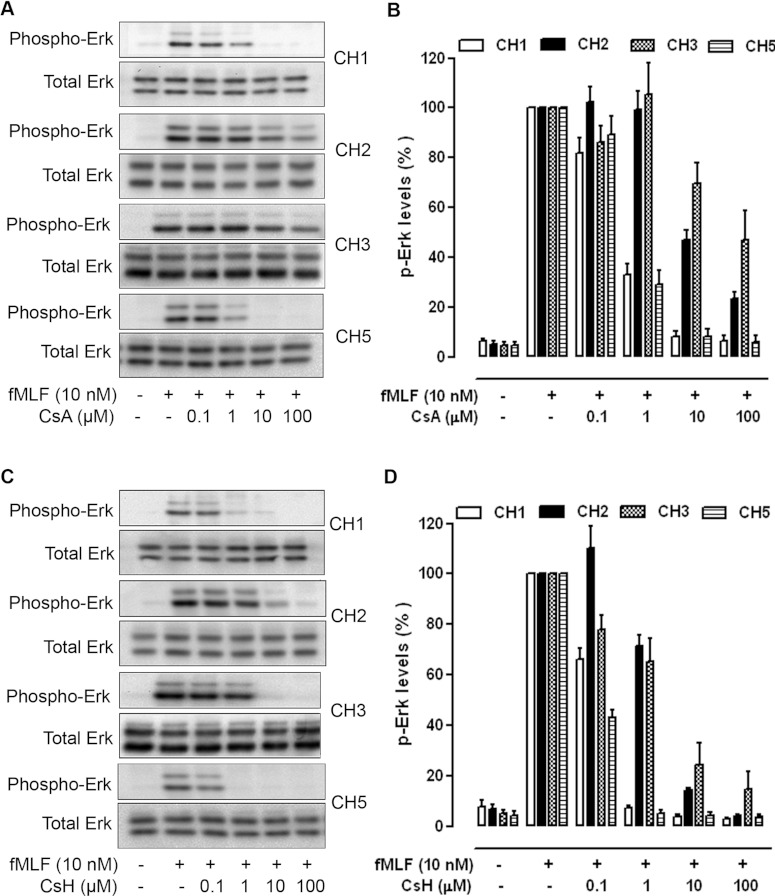
fMLF-induced FPR1 activation signals through heterotrimeric G-proteins and stimulates downstream kinases such as ERK1/2 and Akt. Since phosphorylation of ERK1/2 and Akt are early signalling events triggered by chemoattractants in leucocytes, we performed experiments to evaluate whether variation in the FPR1 gene polymorphism p.V101L affects ERK1/2 phosphorylation. In CHO-Gα16-FPR1 cells, fMLF (10 nM) elicited a significant increase in the phosphorylation of ERK1/2 (Figure 4). Treatment of the cells with CsA or CsH prior to fMLF stimulation concentration-dependently inhibited phosphorylation of the kinases. In this assay, CsH is approximately 10-fold more potent than CsA on the basis of the level of protein phosphorylation. The variants carrying p.Leu101 displayed a marked inhibition of fMLF-stimulated ERK1/2 phosphorylation at CsA and CsH concentrations above 1 μM and 100 nM respectively. In cells expressing FPR1 p.Val101, the potency of CsA and CsH in suppressing fMLF-induced ERK1/2 phosphorylation was approximately 10-fold lower than those in FPR1 p.Leu101-expressing cells.
Figure 4. Inhibition of ERK1/2 phosphorylation by CsA and CsH.
CHO-Gα16-FPR1 cells were treated with either CsA (A and B) or CsH (C and D) for 15 min as indicated and stimulated with fMLF (10 nM) for 5 min. Cell lysates were prepared, separated by SDS/PAGE and blotted with specific anti-phospho-ERK1/2 antibodies as described in the Experimental section. The total protein kinase levels in cell lysates were determined by blotting with antibodies against the non-phosphorylated ERK1/2, as shown at the bottom of each panel. Images displayed in (A) and (C) are representative of at least three independent experiments with similar results. The results in (B) and (D) are the means±S.E.M. of three independent experiments, with the ERK1/2 phosphorylation induced by fMLF set as 100%.
DISCUSSION
In the present study, we investigated the distribution of human FPR1 haplotypes in Han Chinese subjects using five SNPs, p.T11I, p.V101L, p.R190W, p.N192K and p.A346E. Stable cell lines expressing the top 12 FPR1 haplotypes in the Han Chinese were developed in CHO-Gα16 cells and binding affinities of CsA and CsH to these FPR1 variants were evaluated. The results of the present study indicate that FPR1 haplotypes carrying a single amino acid substitution of leucine for valine at position 101 possess significantly higher affinities for CsA and CsH (Figure 1A and Table 4), thereby inhibiting fMLF-induced calcium mobilization, chemotaxis and MAPK phosphorylation in a more potent manner. The present study is among the first to investigate variable responses of FPR1 haplotypes to cyclosporins and to characterize the effects of FPR1 gene polymorphisms such as that in the Han Chinese population.
SNPs are the simplest form of DNA variation among individuals, but covers approximately 80% of genome variations. Most SNPs in modern humans probably arose by single base-modifying events that took place a long time ago, which at the instance of creation would have been surrounded by a series of alleles at polymorphic loci and transmitted together. A unique grouping of alleles was established to be a haplotype. Thus haplotype is a more discriminative state of a chromosomal region and the basis for linkage analysis, which is very valuable for studying the genetics behind common diseases, genetic demography and chromosomal evolution [14]. For this purpose, haplotype has been investigated extensively in the human species by the International HapMap Project. In the present study we characterized the FPR1 haplotype pattern in the Han Chinese using five SNPs, p.T11I [29,35], p.V101L [25,27], p.R190W [25], p.N192K [25,28] and p.A346E, most of which were previously reported to be associated with the activation, downstream signal transduction and disease association of FPR1. When comparing with the HapMap data, we found that all five polymorphisms were common in the three racial groups, but the allele frequencies varied markedly among different racial groups, especially for alleles p.Ile11 and p.Leu101 (Supplementary Figure S1). In the Han Chinese, the frequency of p.Ile11 was less than a half of the frequencies seen in Caucasian and Yoruba populations. The frequency of p.Leu101 in the present study is 0.45 in the Han Chinese, approximately 1.5-fold higher than that reported in the other two ethnic groups. We also identified a third allele for p.N192K, and the resultant minor allele frequency of p.Lys192 was much lower than that documented in the HapMap database. On the basis of this information, we constructed nine FPR1 haplotypes with frequencies higher than 3% and inferred eight more in the Han Chinese (Table 3). Except for the newly identified haplotype 14, which carries FPR1 p.Ile11/Leu101/Glu346, all other haplotypes exist in both Han Chinese and Caucasian populations with considerable frequency variations.
The regional distribution of haplotypes will reflect not only the biological processes such as mutation and recombination, but also population-specific demographic history, such as bottlenecks, admixture, inbreeding, migration, immigration and assortative mating. Due to its high and variable levels of polymorphism in different racial groups, human FPR1 is a good case to study the evolution of the ethnic population. Sahagun-Ruiz et al. [16] once compared the differences among primate haplotypes at sites that are polymorphic in human FPR1, and deduced that the presence of these polymorphisms in all racial groups tested argues against the possibility of a population bottleneck, but in favour of natural selection. FPR1 is known to be important in the chemotaxis of neutrophils and monocytes in response to bacterium-derived N-formyl peptides. Thus it is conceivable that infection of epidemic bacterial plagues, such as cholera, salmonella, bubonic plague, tuberculosis and pertussis might have created selective pressure on the evolution of the FPR1 gene.
Whether different patterns of haplotypes affect FPR1 expression or function in primary cells remains unknown. The high frequency of FPR1 polymorphic alleles implies that they may be suitable to disease association and drug response studies. Obviously, the highly divergent haplotype patterns for human FPR1 point to the potential of developing therapeutic strategies to target different ethnic populations. Our previous work has established that both CsA and CsH interact with FPR1 through cognitive binding to the receptor [12]. Inhibition by CsA and CsH on fMLF-induced FPR1 activities affects downstream events such as calcium mobilization, chemotaxis, degranulation, MAPK and Akt activation. However, it is still unclear how CsA and CsH interact with FPR1 and what structural basis dictates the different potency between CsA and CsH in FPR1 antagonism. Previous site-directed mutagenesis of the FPR1 gene and structure–activity relationship studies on cyclosporins revealed some key sites that may be important for the formation of a binding pocket of FPR1. Loor et al. [9] reported that cyclosporins might bind to a single pharmacophore on FPR1. The latter normally maintains itself in an inactive conformation by an ion pair between Lys85 in the second transmembrane α-helix and Asp284 in the seventh transmembrane α-helix near the transmembrane–extracellular interface. It is believed that binding of agonists activates the receptor by disrupting such pairing or recruiting FPR1 conformers with a disrupted ion pair. Cyclosporins could compete with the binding of agonists to FPR1, and their interaction with FPR1 might also locate at the same region. Our experimental data indicate that Val101 of FPR1 may be a part of the binding site for cyclosporins. Val101 locates at the interface between the second transmembrane domain and the first extracellular loop, proximal to the predicted disulfide bond between Cys98 and Cys176 [13]. Compared with p.Val101, variants of FPR1 carrying p.Leu101 displayed significantly improved receptor affinity, thereby enhancing the inhibition of cyclosporins on FPR1-mediated functions, including calcium mobilization, chemotaxis and MAPK phosphorylation. Both CsA and CsH demonstrated the same trend, but questions remain as to why Leu101 could enhance the antagonism of cyclosporins on FPR1 so dramatically. Clearly, it would be of interest to learn whether cyclosporins could differentially inhibit fMLF-mediated superoxide generation and degranulation among different FPR1 variants. Functional FPR1 was initially identified and is mainly detectable in phagocytic leucocytes. Although cells of non-haemopoietic origin (e.g. epithelial cells and hepatocytes) also express this receptor, the associated biological significance has yet to be defined [13,36]. Using cells that express endogenous FPR1 (e.g. neutrophils) may be helpful in the conduction of functional assays for the variants.
A primary aim of pharmacogenomics is to identify novel human genetic variants responsible for phenotypic differences in drug efficacies. Biomarkers applicable to pharmacogenomics are becoming a major player in personalized medicine [5]. The success of this approach will depend upon having accurate diagnostic tests that identify patients who can benefit from targeted therapies while minimizing the risk of adverse effects. In the clinic, most drugs are often administered at maximally tolerated doses, which are typically based on population averages, resulting in unnecessary toxicity in some population and poor or no response in the other. As a widely used immunosuppressant, concerns remain relative to the therapeutic window of CsA. Although CsA achieves its therapeutic effects through signalling pathways unrelated to FPR1, over-suppression of FPR1 function by CsA may result in side effects that are not fully recognized. The most important effect of CsA is to lower the activity of T-cells and their immune responses through binding to the cytosolic protein cyclophilin (immunophilin). It also inhibits lymphokine production and interleukin release, leading to reduced functions of effector T-cells. This implies that in patients undergoing CsA therapy, immune responses mediated by T-cells are weakened. Therefore it is not unreasonable to deduce that CsA recipients who carry p.Leu101 variants of FPR1 may be more susceptible to pathogen infection than those displaying the p.Val101 phenotype, and this is especially noteworthy in a population with a relatively high p.Leu101 allele frequency such as the Han Chinese.
In conclusion, the results of the present study suggest that haplotypic variation in FPR1, especially the SNP p.V101L, alters the receptor's response to cyclosporins, which may have implications in guiding the clinical application of CsA. In order to translate this discovery into bedside practices, further work is necessary to replicate the findings in subjects carrying this variant in a larger population scale.
Online data
AUTHOR CONTRIBUTION
Jinglun Xue, Richard Ye and Ming-Wei Wang conceived and designed the experiments. Caihong Zhou, Yan Zhou, Jia Wang, Yang Feng, Haonan Wang and Yani Chen performed the experiments. Caihong Zhou, Yan Zhou, Jia Wang and Ming-Wei Wang analysed the data. Richard Ye and Yani Chen contributed reagents/materials/analysis tools. Caihong Zhou, Richard Ye and Ming-Wei Wang wrote the paper.
ACKNOWLEDGEMENTS
We are indebted to Ms Linyun Zhu, Dr Weiwei Gao, Dr Meng Zhang, Dr Zhiyun Zhang and Dr Min He for valuable discussions and technical assistance, and to Dr Dale E. Mais for a critical review of this paper prior to submission.
FUNDING
This work was partly supported by the Ministry of Science and Technology of China [grant number 2009ZX09302-001], the Ministry of Health [grant numbers 2012ZX09304-011 and 2013ZX09507002], the Chinese Academy of Sciences [grant number SIMM1105KF-03], the Shanghai Science and Technology Development Fund [grant numbers 10ZR1406900 and 11DZ2292200], an AstraZeneca Grant-In-Aid and the CAS-Novo Nordisk Research Fund. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the paper.
References
- 1.Salari K., Watkins H., Ashley E. A. Personalized medicine: hope or hype? Eur. Heart J. 2012;33:1564–1570. doi: 10.1093/eurheartj/ehs112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Madian A. G., Wheeler H. E., Jones R. B., Dolan M. E. Relating human genetic variation to variation in drug responses. Trends Genet. 2012;28:487–495. doi: 10.1016/j.tig.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amatruda T. T., 3rd, Dragas-Graonic S., Holmes R., Perez H. D. Signal transduction by the formyl peptide receptor. Studies using chimeric receptors and site-directed mutagenesis define a novel domain for interaction with G-proteins. J. Biol. Chem. 1995;270:28010–28013. doi: 10.1074/jbc.270.47.28010. [DOI] [PubMed] [Google Scholar]
- 4.di Iulio J., Rotger M. Pharmacogenomics: what is next? Front. Pharmacol. 2011;2:86. doi: 10.3389/fphar.2011.00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hamburg M. A., Collins F. S. The path to personalized medicine. New Engl. J. Med. 2010;363:301–304. doi: 10.1056/NEJMp1006304. [DOI] [PubMed] [Google Scholar]
- 6.Ruegger A., Kuhn M., Lichti H., Loosli H. R., Huguenin R., Quiquerez C., von Wartburg A. Cyclosporin A, a peptide metabolite from Trichoderma polysporum (Link ex Pers.) rifai, with a remarkable immunosuppressive activity. Helv. Chim. Acta. 1976;59:1075–1092. doi: 10.1002/hlca.19760590412. [DOI] [PubMed] [Google Scholar]
- 7.Liu J., Farmer J. D., Jr, Lane W. S., Friedman J., Weissman I., Schreiber S. L. Calcineurin is a common target of cyclophilin-cyclosporin A and FKBP-FK506 complexes. Cell. 1991;66:807–815. doi: 10.1016/0092-8674(91)90124-h. [DOI] [PubMed] [Google Scholar]
- 8.Wenger R. M., France J., Bovermann G., Walliser L., Widmer A., Widmer H. The 3D structure of a cyclosporin analogue in water is nearly identical to the cyclophilin-bound cyclosporin conformation. FEBS Lett. 1994;340:255–259. doi: 10.1016/0014-5793(94)80149-5. [DOI] [PubMed] [Google Scholar]
- 9.Loor F., Tiberghien F., Wenandy T., Didier A., Traber R. Cyclosporins: structure-activity relationships for the inhibition of the human FPR1 formylpeptide receptor. J. Med. Chem. 2002;45:4613–4628. doi: 10.1021/jm010987v. [DOI] [PubMed] [Google Scholar]
- 10.Giaccone G., Linn S. C., Welink J., Catimel G., Stieltjes H., van der Vijgh W. J., Eeltink C., Vermorken J. B., Pinedo H. M. A dose-finding and pharmacokinetic study of reversal of multidrug resistance with SDZ PSC 833 in combination with doxorubicin in patients with solid tumors. Clin. Cancer Res. 1997;3:2005–2015. [PubMed] [Google Scholar]
- 11.Gonzalez O., Colombo T., De Fusco M., Imperatori L., Zucchetti M., D’Incalci M. Changes in doxorubicin distribution and toxicity in mice pretreated with the cyclosporin analogue SDZ PSC 833. Cancer Chemother. Pharmacol. 1995;36:335–340. doi: 10.1007/BF00689051. [DOI] [PubMed] [Google Scholar]
- 12.Yan P., Nanamori M., Sun M., Zhou C., Cheng N., Li N., Zheng W., Xiao L., Xie X., Ye R. D., Wang M. W. The immunosuppressant cyclosporin A antagonizes human formyl peptide receptor through inhibition of cognate ligand binding. J. Immunol. 2006;177:7050–7058. doi: 10.4049/jimmunol.177.10.7050. [DOI] [PubMed] [Google Scholar]
- 13.Ye R. D., Boulay F., Wang J. M., Dahlgren C., Gerard C., Parmentier M., Serhan C. N., Murphy P. M. International Union of Basic and Clinical Pharmacology. LXXIII. Nomenclature for the formyl peptide receptor (FPR) family. Pharmacol. Rev. 2009;61:119–161. doi: 10.1124/pr.109.001578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brookes A. J. The essence of SNPs. Gene. 1999;234:177–186. doi: 10.1016/s0378-1119(99)00219-x. [DOI] [PubMed] [Google Scholar]
- 15.Cargill M., Altshuler D., Ireland J., Sklar P., Ardlie K., Patil N., Shaw N., Lane C. R., Lim E. P., Kalyanaraman N., et al. Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nat. Genet. 1999;22:231–238. doi: 10.1038/10290. [DOI] [PubMed] [Google Scholar]
- 16.Sahagun-Ruiz A., Colla J. S., Juhn J., Gao J. L., Murphy P. M., McDermott D. H. Contrasting evolution of the human leukocyte N-formylpeptide receptor subtypes FPR and FPRL1R. Genes Immun. 2001;2:335–342. doi: 10.1038/sj.gene.6363787. [DOI] [PubMed] [Google Scholar]
- 17.Rabiet M. J., Huet E., Boulay F. Human mitochondria-derived N-formylated peptides are novel agonists equally active on FPR and FPRL1, while Listeria monocytogenes-derived peptides preferentially activate FPR. Eur. J. Immunol. 2005;35:2486–2495. doi: 10.1002/eji.200526338. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Q., Raoof M., Chen Y., Sumi Y., Sursal T., Junger W., Brohi K., Itagaki K., Hauser C. J. Circulating mitochondrial DAMPs cause inflammatory responses to injury. Nature. 2010;464:104–107. doi: 10.1038/nature08780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun R., Iribarren P., Zhang N., Zhou Y., Gong W., Cho E. H., Lockett S., Chertov O., Bednar F., Rogers T. J., et al. Identification of neutrophil granule protein cathepsin G as a novel chemotactic agonist for the G protein-coupled formyl peptide receptor. J. Immunol. 2004;173:428–436. doi: 10.4049/jimmunol.173.1.428. [DOI] [PubMed] [Google Scholar]
- 20.Lin Q., Fang D., Hou X., Le Y., Fang J., Wen F., Gong W., Chen K., Wang J. M., Su S. B. HCV peptide (C5A), an amphipathic α-helical peptide of hepatitis virus C, is an activator of N-formyl peptide receptor in human phagocytes. J. Immunol. 2011;186:2087–2094. doi: 10.4049/jimmunol.1002340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Su S. B., Gong W. H., Gao J. L., Shen W. P., Grimm M. C., Deng X., Murphy P. M., Oppenheim J. J., Wang J. M. T20/DP178, an ectodomain peptide of human immunodeficiency virus type 1 gp41, is an activator of human phagocyte N-formyl peptide receptor. Blood. 1999;93:3885–3892. [PubMed] [Google Scholar]
- 22.Su S. B., Gao J., Gong W., Dunlop N. M., Murphy P. M., Oppenheim J. J., Wang J. M. T21/DP107, a synthetic leucine zipper-like domain of the HIV-1 envelope gp41, attracts and activates human phagocytes by using G-protein-coupled formyl peptide receptors. J. Immunol. 1999;162:5924–5930. [PubMed] [Google Scholar]
- 23.Gao J. L., Lee E. J., Murphy P. M. Impaired antibacterial host defense in mice lacking the N-formylpeptide receptor. J. Exp. Med. 1999;189:657–662. doi: 10.1084/jem.189.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clark R. A., Page R. C., Wilde G. Defective neutrophil chemotaxis in juvenile periodontitis. Infect. Immun. 1977;18:694–700. doi: 10.1128/iai.18.3.694-700.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y., Syed R., Uygar C., Pallos D., Gorry M. C., Firatli E., Cortelli J. R., VanDyke T. E., Hart P. S., Feingold E., Hart T. C. Evaluation of human leukocyte N-formylpeptide receptor (FPR1) SNPs in aggressive periodontitis patients. Genes Immun. 2003;4:22–29. doi: 10.1038/sj.gene.6363900. [DOI] [PubMed] [Google Scholar]
- 26.Gwinn M. R., Sharma A., De Nardin E. Single nucleotide polymorphisms of the N-formyl peptide receptor in localized juvenile periodontitis. J. Periodontol. 1999;70:1194–1201. doi: 10.1902/jop.1999.70.10.1194. [DOI] [PubMed] [Google Scholar]
- 27.Gunji T., Onouchi Y., Nagasawa T., Katagiri S., Watanabe H., Kobayashi H., Arakawa S., Noguchi K., Hata A., Izumi Y., Ishikawa I. Functional polymorphisms of the FPR1 gene and aggressive periodontitis in Japanese. Biochem. Biophys. Res. Commun. 2007;364:7–13. doi: 10.1016/j.bbrc.2007.09.105. [DOI] [PubMed] [Google Scholar]
- 28.Maney P., Emecen P., Mills J. S., Walters J. D. Neutrophil formylpeptide receptor single nucleotide polymorphism 348T>C in aggressive periodontitis. J. Periodontol. 2009;80:492–498. doi: 10.1902/jop.2009.080225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.El Shamieh S., Herbeth B., Azimi-Nezhad M., Benachour H., Masson C., Visvikis-Siest S. Human formyl peptide receptor 1 C32T SNP interacts with age and is associated with blood pressure levels. Clin. Chim. Acta. 2012;413:34–38. doi: 10.1016/j.cca.2010.11.038. [DOI] [PubMed] [Google Scholar]
- 30.Gripentrog J. M., Mills J. S., Saari G. J., Miettinen H. M. Variable responses of formyl peptide receptor haplotypes toward bacterial peptides. Immunogenetics. 2008;60:83–93. doi: 10.1007/s00251-008-0277-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He R., Tan L., Browning D. D., Wang J. M., Ye R. D. The synthetic peptide Trp-Lys-Tyr-Met-Val-D-Met is a potent chemotactic agonist for mouse formyl peptide receptor. J. Immunol. 2000;165:4598–4605. doi: 10.4049/jimmunol.165.8.4598. [DOI] [PubMed] [Google Scholar]
- 32.Li Z., Zhang Z., He Z., Tang W., Li T., Zeng Z., He L., Shi Y. A partition-ligation-combination-subdivision EM algorithm for haplotype inference with multiallelic markers: update of the SHEsis ( http://analysis.bio-x.cn) Cell Res. 2009;19:519–523. doi: 10.1038/cr.2009.33. [DOI] [PubMed] [Google Scholar]
- 33.Zhou C., Zhang S., Nanamori M., Zhang Y., Liu Q., Li N., Sun M., Tian J., Ye P. P., Cheng N., et al. Pharmacological characterization of a novel nonpeptide antagonist for formyl peptide receptor-like 1. Mol. Pharmacol. 2007;72:976–983. doi: 10.1124/mol.107.037564. [DOI] [PubMed] [Google Scholar]
- 34.Gabriel S. B., Schaffner S. F., Nguyen H., Moore J. M., Roy J., Blumenstiel B., Higgins J., DeFelice M., Lochner A., Faggart M., et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 35.Benachour H., Zaiou M., Herbeth B., Lambert D., Lamont J. V., Pfister M., Siest G., Tiret L., Blankenberg S., Fitzgerald P. S., Visvikis-Siest S. Human formyl peptide receptor 1 (FPR1) c.32C>T SNP is associated with decreased soluble E-selectin levels. Pharmacogenomics. 2009;10:951–959. doi: 10.2217/pgs.09.29. [DOI] [PubMed] [Google Scholar]
- 36.Le Y., Murphy P. M., Wang J. M. Formyl-peptide receptors revisited. Trends Immunol. 2002;23:541–548. doi: 10.1016/s1471-4906(02)02316-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.