Abstract
Aneuploidies are a major cause of perinatal morbidity and mortality. Therefore, it is the most common indication for invasive prenatal diagnosis. Initially, screening for aneuploidies started with maternal age risk estimation. Later on, serum testing for biochemical markers and ultrasound markers were added. Women detected to be at high-risk for aneuploidies were offered invasive testing. New research is now focusing on non-invasive prenatal testing using cell-free fetal DNA in maternal circulation. The advantage of this technique is the ability to reduce the risk of miscarriage associated with invasive diagnostic procedures. However, this new technique has its own set of technical limitations and ethical issues at present and careful consideration is required before broad implementation
Keywords: Biochemical screening, Cell-free fetal DNA, Non-invasive prenatal testing, Prenatal diagnosis, Trisomy
Introduction
Screening is the process of surveying a population, using a specific marker or markers and defined screening cut-off levels, to identify the individuals in the population at higher risk for a particular disorder. Screening is applicable to a population; diagnosis is applied at the individual patient level.[1]
A couple may approach the doctor seeking preconception or early pregnancy genetic advice for a variety of reasons, including:
A possibly heritable condition in one (or both) of the couple
A history of infertility
History of recurrent pregnancy loss
A family history of one or more possibly heritable conditions
The couple is from a population group with a high frequency of certain genetic diseases
The couple is blood relatives (a consanguineous marriage)
Advanced age
The couple is anxious about reproductive risks, even though there is no specific indication that they are at increased risk.
Aneuploidies are major causes of perinatal death and childhood handicap. Consequently, the detection of chromosomal disorders constitutes the most frequent indication for invasive prenatal diagnosis. Multiple marker screening uses a combination of maternal age and two or more biochemical tests, with or without an ultrasound examination, to produce a single result for risk of Down syndrome, trisomy 18, and open neural tube defects (ONTDs), which is used to offer options for clinical management. A screen is positive when the risk of one or more of the screened disorders falls above a designated risk cut-off. Counseling and further testing options are offered when a screen is positive.
Information was obtained through a literature search via PubMed and Google using key words like “aneuploidy screening”, “non-invasive prenatal diagnosis” and “cell-free fetal DNA (cffDNA)”. The internet search was accompanied by a detailed search of our library database. The articles were then reviewed and summarized in a comprehensive manner.
Criteria and Indication for Prenatal Diagnosis
When prenatal diagnosis is being considered in genetic counseling, several basic factors must be examined, but the most important is whether the couple concerned actively wish for prenatal diagnosis; all too often it is suggested simply because it may be technically feasible and without adequate information. Because most prenatal diagnostic procedures involve a large amount of worry to the parents, and a significant morbidity and mortality to the fetus (with 100% mortality if the test proves abnormal and termination is requested), prenatal diagnosis should normally be carried out only if the general criteria summarized in [Table 1] are fulfilled. These are self-evident, but as in most clinical situations, cases of real doubt may occur.
Table 1.
Criteria for prenatal diagnosis

Severity of disorder
This is beyond doubt in most of the disorders for which prenatal diagnosis is employed, including Down syndrome and other autosomal trisomies, ONTD and the rare neurodegenerative metabolic disorders. Other conditions may be more questionable, especially those where physical abnormalities (e.g., limb defects, cleft lip and palate) are likely to be accompanied by normal intellect and life expectancy. Albinism, which has few general health implications in northern climates, may, because of the likelihood of skin cancer, be a fatal disorder in tropical countries. Such variable categories are increasing, particularly as molecular analysis increasingly recognizes specific mutations with relatively mild clinical effects, and this may present difficult decisions, the outcome of which will vary from family to family, and between different societies.
Treatment availability
Treatment may be clear-cut and satisfactory in some disorders that might otherwise be considered for prenatal diagnosis. Thus, in phenylketonuria, now detectable prenatally by molecular analysis, most children treated from birth have near normal health and intelligence, at least in countries where dietary treatment is available. In contrast, in galactosaemia, liver damage is occasionally present at birth and the long-term outlook for the infant is less clear. Whether prenatal diagnosis is undertaken here will probably depend on the attitudes and previous experience of the parents. In congenital adrenal hyperplasia the outlook with treatment for a second child is much better than for the first-born, in whom delayed diagnosis commonly results in death or serious morbidity, while treatment in utero is also a possibility.
Acceptability of termination
The acceptability of termination of pregnancy to a couple must be determined before any prenatal procedures are contemplated. In some cases, it is unacceptable on religious grounds or because of the prevailing attitude of the community; in others, it is a more personal ethical view. Acceptability may be a relative phenomenon. Thus, in the past many couples found fetal sexing by amniocentesis–with late termination of a male pregnancy which might be normal–unacceptable, whereas these same individuals may accept first-trimester termination, following chorionic villus sampling (CVS), of a definitely affected male pregnancy. Similarly, in some religious traditions, early termination may be allowable, while late termination is forbidden. It is essential to know the attitude of a couple before pregnancy occurs because this may well affect their decision whether or not to have further children. Unacceptability of termination should not be considered as automatically ruling out prenatal diagnosis. In rare instances, parents may feel that they will gain by being able to prepare for an affected child, although this is exceptional, especially when the risk of prenatal procedures is pointed out. Serious potential ethical problems arise if prenatal diagnosis is undertaken for a late-onset disorder and the pregnancy continues.
Feasibility of prenatal diagnosis
The feasibility of diagnosis is something that continues to change rapidly with scientific advances, so it cannot be too strongly stressed that the person giving genetic counseling must obtain accurate information on this point before suggesting the possibility to a couple, and must be satisfied that the technique is reliably applicable as a service rather than just as a research procedure. Failure to do this is as reprehensible as submitting a patient to some new surgical procedure without enquiring as to its benefit and mortality. This is especially relevant when using new molecular advances, where the boundary between research discovery and established techniques can be hard to define, especially for very rare disorders, or those where the gene has been recently isolated.
Current Screening Technologies
Nuchal translucency combined with biochemical markers (first trimester)
Nuchal translucency refers to the subcutaneous layer of fluid behind the fetal neck and lower cranium, which can be visualized on ultrasound. Increased nuchal translucency in the fetus is associated with increased risk of chromosomal abnormality and other diseases.[2] Sonographic measurement of the thickness of the nuchal fold between 11 + 0 weeks and 13 + 6 weeks of pregnancy, together with maternal age and biochemical markers allows an individualized risk of aneuploidies such as trisomy 21, 13, and 18 to be calculated.
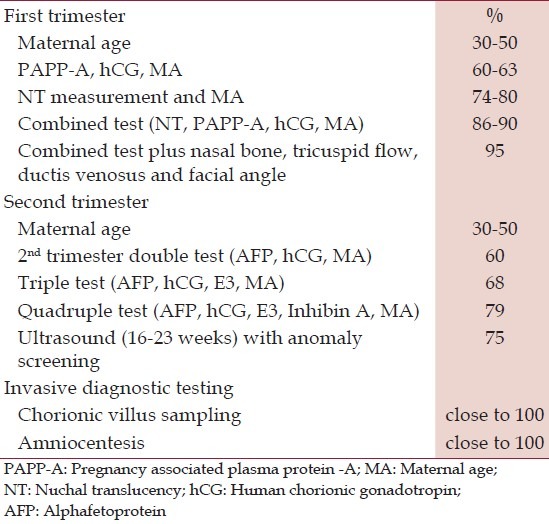
Prerequisites for an interpretable nuchal thickness measurement include operator qualification, choice of appropriate duration of investigation, and technical considerations. The Down syndrome detection rate in major studies ranges from 79% to 90% with a 5% false positive rate.[3] Table 2 showing inclusion of additional parameters such as measurement of the nasal bone, doppler assessment of the tricuspid valve, and the ductus venosus, and the facial angle allows individualized detection rates for trisomy 21 to be increased to up to 95%.[4,5] Two first trimester maternal serum biochemical markers emerged at the same time as NT was being investigated. These are PAPP-A and human chorionic gonadotropin (hCG) (free beta or total). PAPP-A is lower in Down syndrome pregnancies and hCG is higher.[6,7]
Table 2.
Detection rate of trisomy ten plotted against the type of screening parameters and tests used
Second trimester biochemical screening
Traditionally at 16-20 weeks the concentration of maternal serum alphafetoprotein (MSAFP), unconjugated estriol, hCG in the “triple screen” and additionally inhibin A in the “quadruple screen” are measured and the composite risk for neural tube defect (NTD), trisomy 21and trisomy 18 is estimated. For a 5% false positivity rate the sensitivity of triple test is 70% and that of quadruple test is 75% [Table 2]. In foetuses with Down's syndrome, the MSAFP and estriol levels are decreased and hCG and inhibin levels are higher. In trisomy 18, levels of all the three markers are decreased.
Screening for NTD
Elevated MSAFP can identify 75-90% ONTDs, 95% anencephaly and 85% of ventral wall defects with a false positive rate of 5%. Raised MSAFP levels should be offered genetic counselling and targeted ultrasound for further evaluation.[8]
Integrated prenatal screening
In an effort to further improve performance, the first and second trimester screening tests have been combined into a process called IPS. IPS was based on the use of PAPP-A and NT in the first trimester and the quad screen in the second trimester, with results released when all the testing was completed.[9] This approach has been controversial, with some authors suggesting women had the right to know their results early and that it was unethical to withhold the first trimester results.[10] However, when IPS utilizes a quad screen in the second trimester, studies have shown a detection rate of 85% to 87% with an FPR of 0.8% to 1.5%.[6,11] The benefit of IPS over first trimester screening (FTS) is the achievement of a lower FPR and reduction of the number of invasive diagnostic procedures needed.
When NT is not available, IPS still can be offered, using PAPP-A in the first trimester and triple or quad screening in the second trimester. This approach has an 83% DR for a 4% FPR.[11] Given that timing is critical for serum analysis, accurate dating of the pregnancy is very important. Ultrasound dating should be performed if menstrual or conception dating is unreliable. For any abnormal serum screen (serum IPS, quad) calculated using menstrual dating, an ultrasound should be done to confirm gestational age.
Contingent screening
The concept of contingent screening has been suggested by Wright et al.[12] as an alternative to IPS. In contingent screening, the majority of women receive their result after FTS. Women at high risk (risk > 1/50) are offered invasive testing, and women at low risk (risk < 1/1500) require no further testing. A proportion of women with a risk between two cut-offs (1/50 and 1/1500) will go on to have second trimester screening and will receive a combined result.
Sequential screening
Sequential screening selects women for second trimester testing on the basis of their FTS results. Women found to be at high-risk on the basis of the FTS (e.g. risk ≥ 1 in 50) are offered invasive testing. Those with a risk lower than the cut-off are offered additional serum screening in the second trimester. The removal of screen positive affected cases in the first trimester decreases the prevalence of Down syndrome in the second trimester and consequently lowers the PPV of second trimester serum screening.[13]
A substantial proportion of rural women with at-risk pregnancies go through their pregnancy period without significant modern antenatal care.[14] The current screening programs for aneuploidy require women to have multiple visits especially for contingent, integrated and sequential screening. This is inconvenient for the women as well as causes delay in diagnosis. Furthermore, expertise for NT is not available in all centers, thereby, limiting the detection of aneuploidies in FTS.
The use of ultrasound in screening for chromosomal anomalies
At 18-20 weeks’ gestation, all pregnant women should be offered a detailed ultrasound that meets previously established minimum standards.[15] Most major fetal anatomic abnormalities should be detected by this screen. In particular, the majority of ONTDs should be detected by this ultrasound.[16] In addition, ultrasound can detect “soft markers,” which are features that increase the a priori risk of fetal aneuploidy but can also be variations of normal. When used alone, second trimester ultrasound soft markers do not effectively discriminate between unaffected fetuses and fetuses with Down syndrome, because of the high positive rate from the large number of potential markers.[17]
Ultrasound soft markers and anomalies identified in the 18-20 week ultrasound can be used to modify any a priori risk established by age or prior screening. In the absence of soft markers and anomalies, a reduction of risk can be applied.
Screening in twin pregnancies
The biochemical markers in twin pregnancies are on average twice that in singleton pregnancies. A “Pseudo risk” is calculated whereby the measured result (in Multiple of Median [MOM]) is divided by corresponding median MOM value. The risk is evaluated as for singleton pregnancy. This does decrease the sensitivity of the screening test compared to singleton pregnancy, however, remains a useful approach for evaluation.
Following points to be noted while ordering a screening test which have a significant impact on the screening performance–correct date of birth, gestation by USG, maternal weight, number of foetuses, chorionicity, natural/in vitro fertilization (IVF) conception, if ART date of embryo transfer/age of egg donor, maternal age, insulin dependent diabetes, family history of Down's syndrome.
CVS or Amniocentesis
An alternative to screening is invasive prenatal diagnosis by CVS or amniocentesis which directly assesses the chromosome constitution of the fetus through cells from the pregnancy. The advantage is the diagnostic certainty of detecting trisomy 21, 18, and 13. In addition, testing fetal cells and the amniotic fluid may allow for the detection of other chromosome abnormities, genetic conditions, or ONTDs [Table 2]. Although, this approach to the fetal testing is gold standard and gives definitive diagnosis, the chances of miscarriage (around 1%) and invasiveness makes it inconvenient to pregnant women.[18] Thus, the need for the non-invasive methods of detection of fetal cells led to detection of these fetal cells in the cervical mucus[19] and in maternal blood.
Non-invasive prenatal testing
The presence of cffDNA in the blood of pregnant women and its potential use in NIPT was first described in the 1990s.[20,21] Fetal DNA can be detected from the 4th week of gestation, though only reliably from 7 weeks, and the concentration increases with gestational age-from the 16 fetal genomes per ml of maternal blood in the first trimester to 80 fetal genomes per ml in the third trimester, with a sharp peak during the last 8 weeks of pregnancy. Fetal DNA is believed to originate from trophoblast cells, and comprises around less than 10% of the total cell-free DNA in maternal circulation during pregnancy.[22] Unlike cellular DNA, circulating cffDNA consists predominantly of short DNA fragments rather than whole chromosomes, of which 80% are <193 base-pairs in length. In contrast to fetal cells, cffDNA is rapidly cleared from the maternal circulation with a half-life of 16 min and is undetectable after 2 h of delivery.[23]
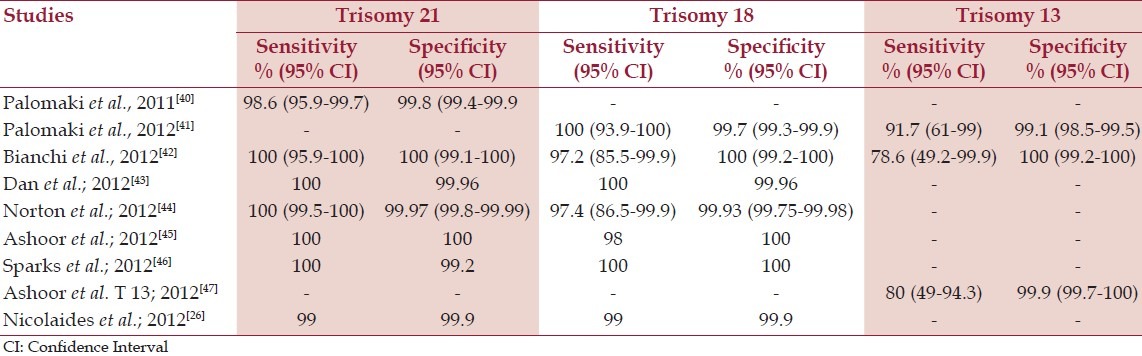
Different published clinical trials validated cell free DNA analysis to detect common aneuploidies with a high sensitivity and specificity [Table 3]. This led to the clinical availability of NIPT in high-risk pregnancies in the United States, beginning in late 2011.
Table 3.
Clinical trials validated cffDNA analysis for detection of fetal aneuploidies
Methods of detecting cffDNA
The basic principle in extracting the cffDNA is to take initially maternal plasma, separate cellular matter by centrifugation, followed by isolation and purification of all cell-free DNA, followed by exploiting the small differences between the fetal and maternal DNA sequences in order to make a specific fetal diagnosis.[23] The most common technique currently used for detection and identification of specific cffDNA sequence is polymerase chain reaction (PCR) with its different variants (nested PCR, real time PCR, digital PCR) and followed by DNA sequencing.
NIPT for fetal aneuploidy
One of the applications of NIPT that appears to be close to clinical implementation is a test for fetal-chromosome abnormalities, notably Down syndrome. This testing is envisaged as being available to all women in the first trimester of pregnancy and would potentially replace current screening and diagnostic methods. Recently, NIPT by analysis of cffDNA in maternal blood has shown promise for highly accurate detection of common fetal autosomal trisomies.[24] Analysis of cffDNA has been validated in several clinical studies utilizing next generation DNA sequencing technology.[25] Clinical studies have primarily included women identified by prior screening, with maternal age and biochemical and/or sonographic testing in the first or second trimester of pregnancy, to be at high-risk for aneuploidies.
In a recently published article of NIPT of fetal trisomies in a routinely screened first trimester population showed that NIPT with a chromosome-selective sequencing approach is highly accurate for fetal aneuploidy detection with very low FPR. The estimated trisomy risk score was >99% in all cases of trisomy 21 and trisomy 18 and <1% in 99.9% of the euploid cases.[26]
There is less confidence in NIPT as a screen for trisomy 13 due to technical issues and the infrequency of the condition. Detection rates between 79% and 92% have been reported, meaning between 8 and 21 out of 100 pregnancies with affected fetuses will be missed. The false-positive rate may be about 1%, so 1 out of 100 unaffected pregnancies may be positive for trisomy 13, so confirmatory testing is recommended.
Positive result
NIPT is highly sensitive and specific for trisomies 21 and 18; positive results are “near diagnostic”. However, false positives have been reported so at this time it is recommended that positive results be followed with confirmatory testing by CVS or amniocentesis.[27] Confirmatory testing can also provide important information about the cause of the trisomy; specifically, CVS or amniocentesis will identify cases of Down syndrome that are due to a 21 chromosome translocation as opposed to the more common trisomy 21. This has important recurrence risk implications for the parents and other family members.
Fetal anatomic ultrasound can also be a helpful tool for pregnancies that test positive on NIPT, looking for additional ultrasound findings that support the diagnosis.
Negative result
Even though NIPT is highly sensitive and specific, it is important to remember that it is not 100%. There are false-negative results, so a negative result cannot absolutely rule out an affected fetus. A laboratory may provide a risk score, allowing the clinician to quantify risk for trisomy.
“Unreportable” or “no-call” result
Depending on the laboratory, 0.5-7% of women who undergo NIPT will not get a result, often because there is an insufficient amount of fetal DNA in the sample (low fetal fraction) due to various clinical reasons which may include high maternal weight or early gestational age.[27] A laboratory may decline to report results near the cut-off. In any case, the clinician must determine, in conjunction with the NIPT laboratory and the patient, whether to draw another sample later in the pregnancy, revert to conventional serum or ultrasound screening, move on to invasive testing, or decline any further testing.
Other clinical uses of NIPT
Sex determination was the first application of NIPT. Because the Y chromosome is absent in the genome of the pregnant woman, detection and measurement of fetal-derived paternally inherited DNA was the first focus of researchers in the prenatal screening field. While, detection and identification of fetal sex-linked or sex limited conditions is considered a legitimate medical reason for NIPT testing of sex, the more common use of pre-conception and prenatal technology for sex-determination is based on preference.[28] It is mainly helpful for sex-linked disease, such as haemophilia, Duchenne muscular dystrophy, X-linked mental retardation, adrenoleukodystrophy, Alport's syndrome, X-linked severe immunodeficiency, retinitis pigmentosa.[23]
NIPT allows for faster determinations of the Rh factor status of the mother and fetus.[29] It lessens misdiagnosis (where both the mother and fetus are Rh D negative), and the unnecessary exhaustion of medical resources.
Technical difficulties
There are a number of technical and clinical obstacles to achieving high diagnostic accuracy:[23]
It is important to emphasize that complete fetal genotyping is not conceivable using cffDNA in the maternal circulation and that the genetic information derived from cffDNA is entirely restricted to the specific DNA sequence (or chromosome) detected.
False negatives can be the result of failure to extract or detect sufficient material, due to individual variability in the amount of total cell-free DNA and the small proportion of fetal versus maternal cell-free DNA.
False positives can be the result of either technical issues, such as contamination, or clinical abnormalities such as the presence of a non-identical vanishing twin.
Ethical issues
Widespread clinical implementation of NIPT is likely to have significant societal consequences. One issue will be the degree of equity as to which groups will have access.
NIPT may be costly and only covered by some types of medical insurance policies. If so, it could be yet another technology disproportionately available to the affluent. Indeed, a consequence of these inequalities could be the perception that NIPT constitutes a contemporary form of eugenics with the affluent, educated, or other selected groups having a greater ability to determine the genetic characteristics of their children.[30]
The ethical implications of sex-selection are well documented. Sex selective breeding and sex selective abortion are most commonly associated with the nations of India and China, whose overpopulations concerns have led to growth control policies, generally targeting girl children.[31]
Professional society statements
Professional societies are beginning to make statements about the use of NIPT. American College of Obstetricians and Gynecologists (ACOG) recommends offering aneuploidy screening or invasive testing to all women, regardless of age. The ACOG and Soceity of Maternal Fetal Medicine (SMFM) both say that cffDNA testing can be offered to pregnant women at increased risk for trisomy 13, 18, or 21. Women age 35 and older, women with a history of a child with trisomy, and women carrying a fetus that shows abnormalities on an ultrasound are at increased risk. The cffDNA test should not be offered to low-risk women or women carrying multiple fetuses because it has not been sufficiently tested in these groups.
A patient with positive result should be referred for genetic counselling and offered invasive prenatal testing for confirmation of test results. cffDNA does not replace the accuracy and diagnostic precision of prenatal diagnosis with CVS or amniocentesis, which remain an option for women.[32]
International society of prenatal diagnosis
ISPD recognizes that NIPT can be helpful as a screening test for women who are at high-risk for Trisomy 21 with suitable genetic counseling. A positive test should be confirmed through invasive testing.
National society of genetic counselors
NSGC recognizes NIPT as an option for aneuploidy assessment in pregnancy: Peer-reviewed data currently supports NIPT only as a screening tool for select populations. While abnormal NIPT results have a high positive predictive value, NIPT results should not be considered diagnostic at this time, and any abnormal results should be confirmed through a conventional prenatal diagnostic procedure, such as CVS or amniocentesis.[33]
Rapid aneuploidy detection
Newer molecular cytogenetic techniques have been introduced which provide rapid results. They allow a rapid diagnosis of the common aneuploidies
-
1.
Quantitative fluorescence polymerase chain reaction (QF-PCR): Highly polymorphic short tandem repeats on chromosomes 13, 18, 21, X and Y are identified using fluorescently labeled primers and PCR. After amplification, electrophoresis and automated analysis of fluorescence intensity of the alleles in a genetic analyzer analysis is performed.
QF-PCR has the advantage that it is less expensive and allows automation and simultaneous processing of larger number of samples than FISH. The sensitivity and specificity of QF-PCR for targeted aneuploidies is 95.65% and 99.97%.[34]
There is a residual risk of a chromosome aberration when interphase FISH or QF-PCR shows a normal result. This risk was estimated to be overall 0.9% for all indications for invasive prenatal diagnosis in a meta-analysis of 12 studies, and in 0.4% of all invasive tests the chromosome aberration was deemed to have clinical significance.[35]
-
2.
Multiplex ligation dependent probe amplification (MLPA): It is a PCR based technique that utilizes the amplification and quantification of the probes instead of nucleic acids. MLPA is a new PCR-based technology that discriminates between copy numbers of specific sequences of DNA. MLPA uses two-part probes of unique length that, when hybridized to adjacent target sequences on genomic DNA, can be joined together by the enzyme DNA ligase. This then allows all target sites to be amplified using a single primer pair that is complementary to the two free ends common to all probes. The products are run on a capillary electrophoresis system and separated by size. This method is not being used routinely and is still under investigation for its potential role.
Though they provide rapid results, RAD techniques are unable to detect structural chromosomal aberrations apart from aneuploidies. They appear suitable for prenatal diagnosis in women undergoing invasive testing for aneuploidies alone. For women with risk factors such as structural malformations on ultrasound or family history of chromosomal translocations, a full cytogenetic karyotype analysis is required.
Preimplantation genetic screening
PGS is performed on oocytes or a blastomere obtained from a preimplantation embryo. The goal is to identify de-novo aneuploidy in embryo (s) of couples known or presumed to be chromosomally normal to allow selection of only those embryos with normal karyotypes for transfer. A high percentage of preimplantation embryos are aneuploid, indicating a potential etiology for the relatively low implantation efficiency of both natural conceptions and IVF. Theoretically, avoiding transfer of aneuploid embryos will reduce the risk of pregnancy failure and improve the probability of conceiving a viable pregnancy. The primary populations targeted for this approach are women with the highest risk of having aneuploid embryos, such as women over age 35 or who have had multiple IVF failures or recurrent pregnancy loss. However, PGS has also been promoted to support embryo selection in patients with low probability of having aneuploid embryos.
Early PGS protocols examined chromosomes 13, 18, 21, X, and Y, (which are sometimes compatible with viable pregnancies), thus, screening for aneuploid syndromes (i.e. trisomy 13, 18, 21, Turner [45, X], and Klinefelter [47, XXY]). This probe combination led to a reduction in the incidence of aneuploid syndromes but did not result in any statistically significant improvement of implantation rates. Current protocols involve the combination of up to five probes in a single experiment and also investigate up to 15 chromosomes in two sequential FISH rounds.[36] Preliminary data obtained with such protocols have demonstrated a doubling in implantation rates and a significant increase in pregnancies per retrieval for women of advanced maternal age and/or couples who have had recurrent miscarriage.[37]
While it is generally accepted that PGS succeeds in reducing miscarriage rates and the incidence of aneuploid syndromes, there are conflicting data concerning the efficacy of PGS in raising implantation and birth rates. Recently, Mastenbroek et al. reported the results of a large, multicentre, randomized, double-blind trial demonstrating that a planned one-cell biopsy with FISH for nine chromosomes is not an effective means of improving pregnancy outcomes for women 35-41 years of age. Given these findings, PGD for aneuploidy screening should not be performed solely because of advanced maternal age. Adequately powered randomized trials are needed to assess whether the same is true when this procedure is used for recurrent unexplained miscarriage and recurrent implantation failure; its use for these conditions should be restricted to research studies pending evidence of effectiveness.[38]
Future
Prenatal screening and diagnosis are routinely offered in antenatal care, and are considered to be important in managing pregnancy and allowing women to make informed choices about the continuation of pregnancies affected by developmental abnormalities.[39] The feasibility of prenatal diagnosis is something that continues to change rapidly with scientific advances. Validation studies of NIPT are underway in “low risk” women and results should be available within a few years. It is expected that labs will continue to explore the number of conditions that can be detected using circulating cffDNA. As the sensitivity and specificity in the general population are better established, it is likely that NIPT will become a diagnostic test for fetal chromosomal aneuploidy for routine use in all pregnancies.
Footnotes
Source of Support: Nil.
Conflict of Interest: None declared.
References
- 1.Cuckle HS, Wald N. Tests using single markers. In: Wald N, Leck I, editors. Antenatal and Neonatal Screening. Oxford: Oxford University Press; 2000. pp. 3–19. [Google Scholar]
- 2.Wald NJ, Morris JK, Walker K, Simpson JM. Prenatal screening for serious congenital heart defects using nuchal translucency: A meta-analysis. Prenat Diagn. 2008;28:1094–104. doi: 10.1002/pd.2124. [DOI] [PubMed] [Google Scholar]
- 3.Driscoll DA, Gross SJ. First trimester diagnosis and screening for fetal aneuploidy. Genet Med. 2008;10:73–5. doi: 10.1097/GIM.0b013e31815efde8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bethune M. Literature review and suggested protocol for managing ultrasound soft markers for Down syndrome: Thickened nuchal fold, echogenic bowel, shortened femur, shortened humerus, pyelectasis and absent or hypoplastic nasal bone. Australas Radiol. 2007;51:218–25. doi: 10.1111/j.1440-1673.2007.01713.x. [DOI] [PubMed] [Google Scholar]
- 5.Nicolaides KH. Some thoughts on the true value of ultrasound. Ultrasound Obstet Gynecol. 2007;30:671–4. doi: 10.1002/uog.5156. [DOI] [PubMed] [Google Scholar]
- 6.Macri JN, Spencer K, Aitken D, Garver K, Buchanan PD, Muller F, et al. First-trimester free beta (hCG) screening for Down syndrome. Prenat Diagn. 1993;13:557–62. doi: 10.1002/pd.1970130704. [DOI] [PubMed] [Google Scholar]
- 7.Orlandi F, Damiani G, Hallahan TW, Krantz DA, Macri JN. First-trimester screening for fetal aneuploidy: Biochemistry and nuchal translucency. Ultrasound Obstet Gynecol. 1997;10:381–6. doi: 10.1046/j.1469-0705.1997.10060381.x. [DOI] [PubMed] [Google Scholar]
- 8.Milunsky A. Genetic Disorders and the Fetus: Diagnosis, Prevention and Treatment. 4th ed. Baltimore, MD: Johns Hopkin's University Press; 1998. Maternal serum screening for neural tube and other defects; pp. 635–701. [Google Scholar]
- 9.Wald NJ, Watt HC, Hackshaw AK. Integrated screening for Down's syndrome on the basis of tests performed during the first and second trimesters. N Engl J Med. 1999;341:461–7. doi: 10.1056/NEJM199908123410701. [DOI] [PubMed] [Google Scholar]
- 10.Copel JA, Bahado-Singh RO. Prenatal screening for Down's syndrome – A search for the family's values. N Engl J Med. 1999;341:521–2. doi: 10.1056/NEJM199908123410709. [DOI] [PubMed] [Google Scholar]
- 11.Wald NJ, Rodeck C, Hackshaw AK, Walters J, Chitty L, Mackinson AM. First and second trimester antenatal screening for Down's syndrome: The results of the Serum, Urine and Ultrasound Screening Study (SURUSS) J Med Screen. 2003;10:56–104. doi: 10.1258/096914103321824133. [DOI] [PubMed] [Google Scholar]
- 12.Wright D, Bradbury I, Benn P, Cuckle H, Ritchie K. Contingent screening for Down syndrome is an efficient alternative to non-disclosure sequential screening. Prenat Diagn. 2004;24:762–6. doi: 10.1002/pd.974. [DOI] [PubMed] [Google Scholar]
- 13.Platt LD, Greene N, Johnson A, Zachary J, Thom E, Krantz D, et al. Sequential pathways of testing after first-trimester screening for trisomy 21. Obstet Gynecol. 2004;104:661–6. doi: 10.1097/01.AOG.0000139832.79658.b9. [DOI] [PubMed] [Google Scholar]
- 14.Oyibo PG, Ebeigbe PN, Nwonwu EU. Assessment of the risk status of pregnant women presenting for antenatal care in a rural health facility in Ebonyi State, South Eastern Nigeria. N Am J Med Sci. 2011;3:424–7. doi: 10.4297/najms.2011.3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van den Hof MC, Demianczuk N. SOGC diagnostic imaging committee. Content of a complete obstetrical report. SOGC committee opinion no. 103, May 2001. J Soc Obstet Gynaecol Can. 2001;23:427–8. [Google Scholar]
- 16.Van den Hof MC, Nicolaides KH, Campbell J, Campbell S. Evaluation of the lemon and banana signs in one hundred thirty fetuses with open spina bifida. Am J Obstet Gynecol. 1990;162:322–7. doi: 10.1016/0002-9378(90)90378-k. [DOI] [PubMed] [Google Scholar]
- 17.Smith-Bindman R, Hosmer W, Feldstein VA, Deeks JJ, Goldberg JD. Second-trimester ultrasound to detect fetuses with Down syndrome: A meta-analysis. JAMA. 2001;285:1044–55. doi: 10.1001/jama.285.8.1044. [DOI] [PubMed] [Google Scholar]
- 18.Mujezinovic F, Alfirevic Z. Procedure-related complications of amniocentesis and chorionic villous sampling: A systematic review. Obstet Gynecol. 2007;110:687–94. doi: 10.1097/01.AOG.0000278820.54029.e3. [DOI] [PubMed] [Google Scholar]
- 19.Mantzaris D, Cram D, Healy C, Howlett D, Kovacs G. Preliminary report: Correct diagnosis of sex in fetal cells isolated from cervical mucus during early pregnancy. Aust N Z J Obstet Gynaecol. 2005;45:529–32. doi: 10.1111/j.1479-828X.2005.00492.x. [DOI] [PubMed] [Google Scholar]
- 20.Kazakov VI, Bozhkov VM, Linde VA, Repina MA, Mikhaĭlov VM. Extracellular DNA in the blood of pregnant women. Tsitologiia. 1995;37:232–6. [PubMed] [Google Scholar]
- 21.Lo YM, Corbetta N, Chamberlain PF, Rai V, Sargent IL, Redman CW, et al. Presence of fetal DNA in maternal plasma and serum. Lancet. 1997;350:485–7. doi: 10.1016/S0140-6736(97)02174-0. [DOI] [PubMed] [Google Scholar]
- 22.Barrett AN, Zimmermann BG, Wang D, Holloway A, Chitty LS. Implementing prenatal diagnosis based on cell-free fetal DNA: Accurate identification of factors affecting fetal DNA yield. PLoS One. 2011;6:e25202. doi: 10.1371/journal.pone.0025202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nigam A, Saxena P, Prakash A, Acharya AS. Detection of fetal nucleic acid in maternal plasma: A novel noninvasive prenatal diagnostic technique. JIMSA. 2012;25:119–200. [Google Scholar]
- 24.Chitty LS, Hill M, White H, Wright D, Morris S. Noninvasive prenatal testing for aneuploidy-ready for prime time? Am J Obstet Gynecol. 2012;206:269–75. doi: 10.1016/j.ajog.2012.02.021. [DOI] [PubMed] [Google Scholar]
- 25.Chiu RW, Akolekar R, Zheng YW, Leung TY, Sun H, Chan KC, et al. Non-invasive prenatal assessment of trisomy 21 by multiplexed maternal plasma DNA sequencing: Large scale validity study. BMJ. 2011;342:c7401. doi: 10.1136/bmj.c7401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nicolaides KH, Syngelaki A, Ashoor G, Birdir C, Touzet G. Noninvasive prenatal testing for fetal trisomies in a routinely screened first-trimester population. Am J Obstet Gynecol. 2012;207:374.e1–6. doi: 10.1016/j.ajog.2012.08.033. [DOI] [PubMed] [Google Scholar]
- 27.Noninvasive prenatal testing (NIPT) factsheet. NCHPEG+NSGC August 2012.National coalition for health professional education group and National soceity of genetic counselors. 2012 Aug 08; [Google Scholar]
- 28.Mukherjee T. Unexpected gender bias found in IVF cycles for sex selection. Fertil Steril. 2007;106:S134. [Google Scholar]
- 29.Bianchi DW, Avent ND, Costa JM, van der Schoot CE. Noninvasive prenatal diagnosis of fetal Rhesus D: Ready for Prime (r) Time. Obstet Gynecol. 2005;106:841–4. doi: 10.1097/01.AOG.0000179477.59385.93. [DOI] [PubMed] [Google Scholar]
- 30.Benn PA, Chapman AR. Ethical challenges in providing noninvasive prenatal diagnosis. Curr Opin Obstet Gynecol. 2010;22:128–34. doi: 10.1097/GCO.0b013e3283372352. [DOI] [PubMed] [Google Scholar]
- 31.Ethics Committee of the American Society for Reproductive Medicine. Preconception gender selection for nonmedical reasons. Fertil Steril. 2004;82:S232–5. doi: 10.1016/j.fertnstert.2004.05.013. [DOI] [PubMed] [Google Scholar]
- 32.American College of Obstetricians and Gynecologists Committee on Genetics. Committee Opinion No. 545: Noninvasive prenatal testing for fetal aneuploidy. 2012;120:1532–4. doi: 10.1097/01.AOG.0000423819.85283.f4. [DOI] [PubMed] [Google Scholar]
- 33.Devers PL, Cronister A, Ormond KE, Facio F, Brasington CK, Flodman P. Noninvasive prenatal testing/noninvasive prenatal diagnosis: The position of the national society of genetic counselors. J Genet Couns. 2013 doi: 10.1007/s10897-012-9564-0. [In Press] [DOI] [PubMed] [Google Scholar]
- 34.Grimshaw GM, Szczepura A, Hultén M, MacDonald F, Nevin NC, Sutton F, et al. Evaluation of molecular tests for prenatal diagnosis of chromosome abnormalities. Health Technol Assess. 2003;7:1–146. doi: 10.3310/hta7100. [DOI] [PubMed] [Google Scholar]
- 35.Leung WC, Lao TT. Rapid aneuploidy testing, traditional karyotyping, or both? Lancet. 2005;366:97–8. doi: 10.1016/S0140-6736(05)66791-8. [DOI] [PubMed] [Google Scholar]
- 36.Baart EB, van den Berg I, Martini E, Eussen HJ, Fauser BC, van Opstal D. FISH analysis of 15 chromosomes in human day 4 and 5 preimplantation embryos: The added value of extended aneuploidy detection. Prenat Diagn. 2007;27:55–63. doi: 10.1002/pd.1623. [DOI] [PubMed] [Google Scholar]
- 37.Munné S, Chen S, Fischer J, Colls P, Zheng X, Stevens J, et al. Preimplantation genetic diagnosis reduces pregnancy loss in women aged 35 years and older with a history of recurrent miscarriages. Fertil Steril. 2005;84:331–5. doi: 10.1016/j.fertnstert.2005.02.027. [DOI] [PubMed] [Google Scholar]
- 38.Mastenbroek S, Twisk M, van Echten-Arends J, Sikkema-Raddatz B, Korevaar JC, Verhoeve HR, et al. In vitro fertilization with preimplantation genetic screening. N Engl J Med. 2007;357:9–17. doi: 10.1056/NEJMoa067744. [DOI] [PubMed] [Google Scholar]
- 39.Wright C. Cell-free fetal nucleic acids for noninvasive prenatal diagnosis, 2009. [Accessed February 16, 2009];PHG Foundation. at http://www.phgfoundation.org . [Google Scholar]
- 40.Palomaki GE, Kloza EM, Lambert-Messerlian GM, Haddow JE, Neveux LM, Ehrich M, et al. DNA sequencing of maternal plasma to detect Down syndrome: An international clinical validation study. Genet Med. 2011;13:913–20. doi: 10.1097/GIM.0b013e3182368a0e. [DOI] [PubMed] [Google Scholar]
- 41.Palomaki GE, Deciu C, Kloza EM, Lambert-Messerlian GM, Haddow JE, Neveux LM, et al. DNA sequencing of maternal plasma reliably identifies trisomy 18 and trisomy 13 as well as Down syndrome: An international collaborative study. Genet Med. 2012;14:296–305. doi: 10.1038/gim.2011.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bianchi DW, Platt LD, Goldberg JD, Abuhamad AZ, Sehnert AJ, Rava RP, et al. Genome-wide fetal aneuploidy detection by maternal plasma DNA sequencing. Obstet Gynecol. 2012;119:890–901. doi: 10.1097/AOG.0b013e31824fb482. [DOI] [PubMed] [Google Scholar]
- 43.Dan S, Wang W, Ren J, Li Y, Hu H, Xu Z, et al. Clinical application of massively parallel sequencing-based prenatal noninvasive fetal trisomy test for trisomies 21 and 18 in 11,105 pregnancies with mixed risk factors. Prenat Diagn. 2012;32:1225–32. doi: 10.1002/pd.4002. [DOI] [PubMed] [Google Scholar]
- 44.Norton ME, Brar H, Weiss J, Karimi A, Laurent LC, Caughey AB, et al. Non-Invasive Chromosomal Evaluation (NICE) Study: Results of a multicenter prospective cohort study for detection of fetal trisomy 21 and trisomy 18. Am J Obstet Gynecol. 2012;207:137.e1–8. doi: 10.1016/j.ajog.2012.05.021. [DOI] [PubMed] [Google Scholar]
- 45.Ashoor G, Syngelaki A, Wagner M, Birdir C, Nicolaides KH. Chromosome-selective sequencing of maternal plasma cell-free DNA for first-trimester detection of trisomy 21 and trisomy 18. Am J Obstet Gynecol. 2012;206:322.e1–5. doi: 10.1016/j.ajog.2012.01.029. [DOI] [PubMed] [Google Scholar]
- 46.Sparks AB, Struble CA, Wang ET, Song K, Oliphant A. Noninvasive prenatal detection and selective analysis of cell-free DNA obtained from maternal blood: Evaluation for trisomy 21 and trisomy. Am J Obstet Gynecol. 2012;206:319.e1–9. doi: 10.1016/j.ajog.2012.01.030. [DOI] [PubMed] [Google Scholar]
- 47.Ashoor G, Syngelaki A, Wang E, Struble C, Oliphant A, Song K, et al. Trisomy 13 detection in the first trimester of pregnancy using a chromosome-selective cell-free DNA analysis method. Ultrasound Obstet Gynecol. 2013;41:21–5. doi: 10.1002/uog.12299. [DOI] [PubMed] [Google Scholar]


