Abstract
DNA molecules in mitochondria, just like those in the nucleus of eukaryotic cells, are constantly damaged by noxious agents. Eukaryotic cells have developed efficient mechanisms to deal with this assault. The process of DNA repair in mitochondria, initially believed nonexistent, has now evolved into a mature area of research. In recent years, it has become increasingly appreciated that mitochondria possess many of the same DNA repair pathways that the nucleus does. Moreover, a unique pathway that is enabled by high redundancy of the mitochondrial DNA and allows for the disposal of damaged DNA molecules operates in this organelle. In this review, we attempt to present a unified view of our current understanding of the process of DNA repair in mitochondria with an emphasis on issues that appear controversial.
Mitochondria possess many of the same DNA repair pathways as the nucleus. Moreover, since a cell can lose mtDNA molecules without detrimental effects, mitochondria employ a unique pathway to degrade damaged mtDNA.
In mammalian cells, genetic information is stored in two locations: in the nucleus and in mitochondria. Nuclear DNA (nDNA) is organized into chromosomes of which two sets are present per cell: one paternal and one maternal. In contrast, mitochondrial DNA (mtDNA) inheritance is (with a few exceptions) exclusively maternal, and this DNA species is highly redundant, typically a few hundred to a few thousand copies per cell. In many (but not all) (Noll et al. 1990) cell types, the bulk of ATP is produced by oxidative phosphorylation (OXPHOS) in mitochondria. Because mtDNA encodes components of four out of five mitochondrial respiratory complexes, it is not surprising that mutations in mtDNA may result in (pathological) alterations in mitochondrial function and mitochondrial diseases (Holt et al. 1988; Lestienne and Ponsot 1988; Wallace et al. 1988). Apart from mitochondrial diseases, mutations in mtDNA are linked to a spectrum of other pathologies including cancer, diabetes, cardiovascular diseases, and neurodegenerative disorders, as well as the normal process of aging (Wallace 2005). Moreover, it has been established that not only mtDNA mutations but also a reduction in the mtDNA copy number can be pathogenic (Clay Montier et al. 2009; Rotig and Poulton 2009). Understanding cellular mechanisms for the maintenance of mtDNA integrity and copy number is, therefore, of utmost importance because it can provide targets for clinical interventions aimed at prevention and treatment of human diseases.
ORGANIZATION OF THE MITOCHONDRIAL GENOME
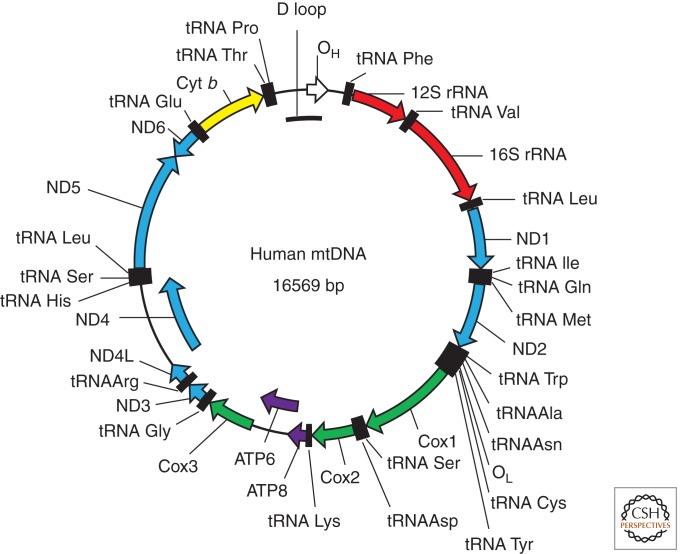
Human mtDNA (Fig. 1) is ∼16.6 kb long and encodes two rRNAs, 22 tRNAs, and 13 polypeptides of which seven are subunits of complex I (NADH dehydrogenase), three are subunits of complex IV (cytochrome c oxidase), and two are subunits of complex V (ATP synthase) and cytochrome b (a subunit of complex III). The density of genetic information in mtDNA is relatively high, with very short intergenic regions. To increase this density, some genes overlap, and some others lack complete termination codons, which are created by polyadenylation of the corresponding mRNAs (Ojala et al. 1981). A short noncoding regulatory region in mtDNA harbors an origin of replication plus two promoters, one on each of the two complementary strands (Fig. 1). These promoters generate polycistronic transcripts that are processed to produce mature rRNAs, tRNAs, and mRNAs and also are involved in the generation of the primer for replication of one of the strands.
Figure 1.
The map of human mitochondrial DNA. (OH and OL) Origins of heavy- and light-strand replication, respectively; (ND1–ND6) subunits of NADH dehydrogenase (ETC complex I) subunits 1–6; (COX1–COX3) subunits of cytochrome oxidase subunits 1–3 (ETC complex IV); (ATP6 and ATP8) subunits 6 and 8 of mitochondrial ATPase (complex V); (Cyt b) cytochrome b (complex III).
In the nucleus, genetic material is represented by nucleoprotein complexes consisting of DNA wrapped around a core octamer of histones forming “beads on a string.” This nucleosomal chromatin is further organized to form chromosomes. In contrast, the mitochondrial genome lacks histones, which has led to the belief that the observed high rate of mtDNA mutagenesis (∼10-fold greater than in nDNA) (Brown et al. 1979; Ballard and Whitlock 2004; Tatarenkov and Avise 2007) is explained by the lack of “protective” histones. This belief lacks direct experimental support and remains controversial because it contradicts some experimental evidence, which suggests that histones, at least under some conditions, may enhance, rather than reduce DNA damage (Liang et al. 1999; Liang and Dedon 2001), and that mtDNA-associated proteins may be at least as protective against mutagenic insults as histones under other conditions (Guliaeva et al. 2006; Alexeyev 2009). Moreover, mtDNA may be physically covered with TFAM, an HMG-like protein involved in mtDNA transcription and replication. It is present in mitochondria in quantities of one molecule per 10–20 bp of mtDNA (Alam et al. 2003; Ekstrand et al. 2004; Pellegrini et al. 2009). Limited accessibility of mtDNA to methyltransferases (Rebelo et al. 2009) is in agreement with the notion that TFAM coating may impede access to other proteins.
In view of the endosymbiotic theory of mitochondrial origin from an ancient prokaryote, it is perhaps not surprising that recent studies revealed similarities in packaging of mtDNA and bacterial chromosomes. Thus, it has been established that mtDNA is organized in nucleoids. However, the precise structure of these nucleoids is still debated. Initial studies revealed that nucleoids are ∼70 nm in diameter, and each of them carries six to 10 copies of mtDNA (Iborra et al. 2004). This organization ensures similar DNA densities in mitochondrial and Escherichia coli nucleoids, ∼35 mg/mL (Iborra et al. 2004). Subsequent studies refined this model, leading to the view that nucleoids are layered structures consisting of a core, where replication and transcription of mtDNA occur, and peripheral regions, where translation of mitochondrial transcripts and assembly of newly synthesized polypeptides into respiratory complexes occur (Bogenhagen et al. 2008). Further refinement has been achieved using recently developed super-resolution imaging techniques. Thus, it has been established, with the help of stimulated emission depletion (STED) microscopy, that each nucleoid may contain only one or two (on average, 1.4) molecules of mtDNA (Kukat et al. 2011). However, studies that used another super-resolution technique, photoactivated localization microscopy (PALM and iPALM), produced a conflicting finding that, on average, the predominantly elliptical nucleoids in 3T3 cells contain three mtDNA genomes (Brown et al. 2011; Kopek et al. 2012). Interestingly, these latter studies also suggest that nucleoid size varies with mtDNA content and that mtDNA in nucleoids is extremely condensed, requiring an access mechanism for DNA repair and replication proteins (Brown et al. 2011).
Nucleoids may conserve their mtDNA, or they may dynamically exchange mtDNA molecules. Recently, these alternatives were tested directly by fusing two cell lines, each homoplasmic for a partially deleted mtDNA in which the deletions were nonoverlapping and each deficient in mitochondrial protein synthesis. Upon fusion, restoration of mitochondrial protein synthesis was observed, but mutant nucleoids remained spatially separated, and mixing of two mutant mtDNA species in a single nucleoid was not observed. These results indicate that mitochondrial nucleoids tightly regulate their genetic content and do not freely exchange mtDNA (Gilkerson et al. 2008).
RELEVANT SOURCES OF mtDNA LESIONS AND TYPES OF DAMAGE
mtDNA is constantly exposed to external and internal noxious agents. There are five main types of mtDNA damage:
Alkylation damage. This type of damage comes from both exogenous (e.g., chemotherapeutic agents) and endogenous (e.g., S-adenosylmethionine, which can methylate DNA nonenzymatically) (Rydberg and Lindahl 1982) sources (De Bont and van Larebeke 2004). Importantly, mitochondria house ∼30% of the cellular pool of S-adenosylmethionine (Horne et al. 1997). Endogenous alkylation damage has been shown experimentally (Xiao and Samson 1993).
Hydrolytic damage. This damage comes in two types: formation of abasic sites (as a result of hydrolysis of the glycosidic bond) and hydrolytic deamination of bases, predominantly cytosine (Lindahl 1993; De Bont and van Larebeke 2004).
Formation of adducts (De Bont and van Larebeke 2004; Blair 2008). This type of damage can come from both endogenous (e.g., estrogens) (Liehr 2000; Thomas and Roy 2001) and exogenous (e.g., tobacco smoke and chemical exposure) sources. In some instances, upon exposure to these agents, mtDNA is damaged preferentially (Wunderlich et al. 1970; Miyaki et al. 1977; Neubert et al. 1981; Niranjan et al. 1982; Tomasi et al. 1987; Rossi et al. 1988; Balansky et al. 1996; Bartsch 1996; Thomas and Roy 2001; Nair et al. 2005; Yang et al. 2006; Jung et al. 2009).
Mismatched bases (due to replication errors or incorporation of nucleotides containing modified, e.g., oxidized, bases during replication) (Kamiya 2003; Boesch et al. 2011).
DNA strand breaks. These come in the form of both single-strand breaks (SSBs) (el-Khamisy and Caldecott 2007) and double-strand breaks (DSBs) (Kasparek and Humphrey 2011). Both types of strand breaks can be induced by noxious stimuli both directly and indirectly (e.g., in the process of the repair of other lesions).
Oxidative damage. This type of damage is by far the best studied and the most prevalent, primarily because mitochondria are a major cellular source of reactive oxygen species (ROS). Therefore, we consider it here in greater detail.
Hierarchy of the ETC-Generated ROS with Respect to Their Reactivity toward mtDNA
Mitochondria possess at least nine enzyme systems capable of producing ROS under favorable conditions (Andreyev et al. 2005). However, only those produced by the ETC are usually considered when mtDNA damage is concerned. Early studies established that 1%–2% of oxygen consumed by isolated mitochondria at atmospheric oxygen tension can be converted to H2O2 under conditions of so-called reverse electron transport from complex II to complex I (Boveris et al. 1972; Andreyev et al. 2005; Murphy 2009). Although it is not likely that conditions favoring reverse electron transport can be encountered under physiological conditions (Murphy 2009), it is these numbers that get frequently quoted. Electron “leak” from the electron transport chain (ETC) results in the production of superoxide (O2−), which is charged, comparatively unstable, and has low reactivity toward DNA (Lesko et al. 1980; Brawn and Fridovich 1981; Rowley and Halliwell 1983; Blakely et al. 1990; Halliwell and Aruoma 1991). The negative charge has been proposed to render O2− impermeable to membranes (Muller et al. 2004), and this hypothesis has found experimental support (Takahashi and Asada 1983; Mao and Poznansky 1992; Frimer et al. 1996).
Importantly, O2− production by mitochondria can be self-limiting because of inactivation of mitochondrial aconitase by O2−. This inactivation can reduce NADH output by the citric acid cycle and, consequently, electron flow through ETC. The net effect would be a lowered steady-state reduction of complexes I and III, which would diminish O2− production (Gardner and Fridovich 1991; Gardner 1997).
The O2− generated by the ETC is quickly and efficiently converted to H2O2, which is the principal cellular mediator of oxidative stress because of its relative stability and membrane permeability. Still, H2O2 fails to efficiently attack mtDNA directly (Lesko et al. 1980; Brawn and Fridovich 1981; Rowley and Halliwell 1983; Blakely et al. 1990; Halliwell and Aruoma 1991). However, in the presence of redox-active metal ions like Fe2+, H2O2 can undergo Fenton chemistry, generating extremely reactive hydroxyl radical that efficiently damages DNA (Henle et al. 1996a,b; Alexeyev 2009).
Relative Prevalence of Various Oxidative mtDNA Lesions
In human fibroblasts, mtDNA is particularly vulnerable to oxidative stress and accumulates more polymerase-blocking lesions (e.g., strand breaks, abasic sites and oxidative DNA adducts) (Blair 2008) than nDNA upon treatment with either a H2O2-generating system (glucose oxidase plus glucose) (Salazar and Van Houten 1997) or by pure H2O2 (Yakes and Van Houten 1997). The same pattern was observed in endothelial (Ballinger et al. 2000) and in retinal pigmented epithelial cells (Ballinger et al. 1999). Similarly, using an independent technique, we observed a higher frequency of H2O2-induced lesions (strand breaks and possibly abasic sites, which are readily converted to strand breaks under the assay conditions) in mtDNA as compared with nDNA (Shokolenko et al. 2009). Importantly, H2O2-induced strand breaks predominated over mtDNA mutations by at least 10-fold, implicating damage to the sugar–phosphate backbone as the predominant lesion induced in mtDNA by oxidative stress (Shokolenko et al. 2009).
Unlike damage to the sugar–phosphate backbone, oxidative base damage to mtDNA compared with nDNA has been the subject of controversy. Initial studies indicated that oxidative base damage in mtDNA may be higher than in nDNA (Richter et al. 1988). However, subsequent refinement in analytical techniques reversed this notion (Anson et al. 2000; Lim et al. 2005) and revealed that some oxidative lesions such as 8-hydroxyguanine, Fapy-adenine, 8-hydroxyadenine, 5,6-dihydroxyuracil, 5-hydroxyuracil, 5-hydroxycytosine, and 5-hydroxymethyluracil are less prevalent in liver mtDNA of aged animals as compared with nDNA (Anson et al. 2000; Alexeyev 2009). The wide spectrum of oxidative base lesions and methodological difficulties prevent precise elucidation of their frequencies (ESCODD 2003; De Bont and van Larebeke 2004; Gedik and Collins 2005). The main pyrimidine product of oxidative DNA base damage is believed to be a thymine glycol (Wang et al. 1998), and the main purine product is probably 7,8-dihydro-8-oxo-2′-deoxyguanosine (8-oxodG) (Bohr 2002; Christmann et al. 2003; De Bont and van Larebeke 2004). The former has low mutagenicity but is a block to polymerase, whereas the latter is less inhibitory toward the actions of polymerase, but can cause characteristic G:T transversions in 10% of the cases in vitro (Hanes et al. 2006). The mutagenic potential of some oxidative lesions has been established (for review, see Kamiya 2003).
mtDNA REPAIR PATHWAYS
The discovery that mitochondria are unable to repair ultraviolet (UV)–induced pyrimidine dimers (Clayton et al. 1974, 1975) and some types of alkylation damage (Miyaki et al. 1977) showed that they contain a reduced complement of DNA repair pathways. Over the years this view has evolved, and now we have documented some evidence for the existence in mitochondria of almost all major DNA repair pathways (for review, see Gredilla 2010; Liu and Demple 2010; Boesch et al. 2011). Inferences about the presence of novel DNA repair pathways in mitochondria are often made based on the assignment of these components to the mitochondrial compartment. Typically, this assignment is made with the help of subcellular fractionation, which remains a gold standard. Other techniques such as subcellular localization of fluorescent fusion proteins, immunogold labeling combined with electron microscopy (EM), and in silico detection of putative matrix targeting sequences are often used as accessory techniques to unambiguously localize a protein to the matrix compartment. However, it should be noted here that these accessory techniques have their inherent limitations and often provide inconclusive results. Fluorescence microscopy has relatively low spatial resolution to discriminate between four mitochondrial compartments, immunogold EM requires highly specific antibodies, and the existence of poorly understood internal matrix targeting sequences limits utility of the in silico predictions. Perhaps the most dramatic example of the latter limitation is in vivo import into the matrix of mammalian mitochondria of heterologously expressed chlorella virus ligase and Escherichia coli LigA proteins accompanied by the rescue of the lethality of Lig3 knockout (Simsek et al. 2011).
Direct Reversal
Direct reversal is conceptually the simplest pathway exemplified by photoreactivation of cyclobutane pyrimidine dimers by photolyase in E. coli. Photolyase activity has been reported in yeast (Yasui et al. 1992) and plant mitochondria (Takahashi et al. 2011). Despite the existence of a report suggesting the presence of photolyase activity in Xenopus mitochondria (Ryoji et al. 1996), it is generally believed that mitochondria in higher eukaryotes do not possess photolyase activity. Instead, in mammalian cells, homologs of photolyase, transcriptional repressors Cryptochromes 1 and 2 (CRY1 and CRY2), are involved in the regulation of circadian rhythm (Christmann et al. 2003; Khan et al. 2012).
In the nucleus of mammalian cells, the major direct repair enzyme is O6-methylguanine-DNA methyltransferase (MGMT), which is involved in the repair of O6-alkylguanines. Mitochondria are able to repair O6-methyl-2′-deoxyguanosine and O6-ethyl-2′-deoxyguanosine (Myers et al. 1988; Satoh et al. 1988), and a protein with the molecular weight of MGMT was shown to localize to the mitochondrial matrix (Myers et al. 1988). This report of mitochondrial localization of MGMT, however, has not been corroborated, and in an independent study, the investigators were unable to detect MGMT in mitochondria using Western blotting technique (Cai et al. 2005). In addition, this protein appears to lack typical amino-terminal cleavable mitochondrial targeting sequence (however, see above) (Tano et al. 1990).
Recently, another direct reversal pathway mediated by ABH2 dioxygenase has been identified in mammalian cells (Ringvoll et al. 2008). This protein can directly repair 1,N6-ethenoadenine adducts in nDNA in an iron, oxoglutarate-dependent manner. However, mitochondrial localization of this protein has not been reported yet.
Nucleotide Excision Repair
There appears to exist a consensus in the literature that substrates of the nucleotide excision repair pathway are not repaired in mtDNA, unless they can be processed by an alternative pathway (Clayton et al. 1974, 1975).
Mismatch Repair and Sanitation of the dNTP Pool
DNA mismatch repair (MMR) is a conserved pathway that corrects misincorporation and slippage errors introduced by DNA polymerase during DNA replication as well as base mismatches caused by spontaneous and induced base deamination, oxidation, and alkylation. In the eukaryotic nucleus, MMR begins with the binding of MSH (MutS homolog) proteins (Msh2–Msh6 = MutSα or Msh2–Msh3 = MutSβ) to base–base mismatches and loop mismatches (Kunkel and Erie 2005). This event is followed by the recruitment of MLH (MutL homolog) proteins (primarily Mlh1–Pms1 in Saccharomyces cerevisiae) followed by the initiation of downstream excision and resynthesis steps that maintain template strand information (Modrich 2006; Mendillo et al. 2009). Although mismatch repair activity is present in mammalian mitochondria, it appears to be independent of MSH2, MSH3, MSH6, or MLH1 (Mason et al. 2003; de Souza-Pinto et al. 2009). Instead, mismatch recognition in mitochondria appears to be dependent on the Y-box binding protein 1 (YB-1) (de Souza-Pinto et al. 2009). In a recent study, MLH1 was reported in mitochondria, but to date, no MSH2 was detected (Martin et al. 2010).
Damaged (predominantly, oxidized) deoxyribonucleotide triphosphates (dNTPs) are an important source of mismatch errors during DNA synthesis. In vitro, replication fidelity of mitochondrial DNA polymerase γ (Pol γ) was reduced when the fraction of oxidized dGTP (8-oxodGTP) constituted only 0.06%–0.6% of the total dGTP pool, whereas the mitochondrial fraction of this species was as high as 10% of total dGTP and on par with dTTP, with which it competes for incorporation opposite A in mtDNA (Pursell et al. 2008). To counter this threat, mitochondria possess MTH1 (Kang et al. 1995; Nakabeppu 2001), which can hydrolyze 8-oxo-2′-deoxyguanosine triphosphate as well as 8-oxo-2′-deoxyadenosine triphosphate and 2-hydroxy-2′-deoxtadenosine triphosphate to corresponding monophosphates, which are not substrates for DNA polymerase. Although sanitation of the mitochondrial dNTP pool is not a bona fide DNA repair mechanism, it preserves mtDNA integrity through the reduction of mismatches in mtDNA and therefore reduces mutagenesis.
DNA Strand Break Repair
Single-Strand Break Repair (SSBR)
Single-strand breaks (SSBs) are discontinuities in one strand of the DNA double helix and are usually accompanied by loss of a single nucleotide and by damaged 5′ and/or 3′ termini at the site of the break. SSBs can arise either directly (e.g., from attack of deoxyribose by reactive oxygen species) or indirectly (e.g., via enzymatic cleavage of the phosphodiester backbone during DNA base excision repair [BER]) (el-Khamisy and Caldecott 2007). Upon exposure to H2O2, ∼1000-fold more SSBs are induced in nDNA as compared with DSBs (Caldecott 2008). SSBs can also arise as a result of erroneous or abortive activity of DNA topoisomerase 1 (Hudson et al. 2012), which is present in mitochondria, where it participates in transcription and replication (Zhang et al. 2001). 5′-AMP–SSBs, in which AMP is covalently linked to a 5′-phosphate through a pyrophosphate bond, arise from abortive DNA-ligase activity at existing SSBs and are processed by aprataxin (Ahel et al. 2006), another mitochondrial protein (Sykora et al. 2011). Overall, SSBR is very similar to BER (see below) and shares with it two of the three functional steps: gap tailoring and DNA synthesis/ligation. Therefore, it is often considered to be a subpathway of the joint BER/SSBR (Hegde et al. 2012). This subpathway involves distinct gap-tailoring enzymes such as aprataxin (Ahel et al. 2006) and TDP1 (Das et al. 2010) in addition to APE and does not involve DNA glycosylases (Hegde et al. 2012).
Double-Strand Break Repair
Double-strand breaks (DSBs) can be repaired by either homologous recombination (HR) or nonhomologous end joining (NHEJ). Although Drosophila melanogaster cells can efficiently repair bleomycin-induced DSBs (Morel et al. 2008), a similarly efficient mechanism has not been described yet in the mitochondria of mammalian cells.
Homologous Recombination
Homologous recombination (HR) in yeast and Chlamydomonas reinhardtii mitochondria is well established and is used as a tool for genetic manipulation of mtDNA in these organisms (Zhou et al. 2010; Mileshina et al. 2011). Recently, homologous recombination also was shown in plant mitochondria (Manchekar et al. 2006; Davila et al. 2011; Mileshina et al. 2011). In mammalian mitochondria, mtDNA recombination appears to be an infrequent event. When FC cells containing two species of mtDNA each containing a unique deletion were grown in ketogenic media, no recombinant mtDNA species were detected by Southern blot (Gilkerson et al. 2008). Thus, even a harsh selective environment did not induce or select for recombination of the two parental mtDNAs. Although D’Aurelio et al. (2004) did observe intermolecular recombination at low levels, in that case it was necessary to first deplete mtDNA with ethidium bromide to facilitate detection of recombinants. Similar low-frequency recombination was detectable in cultured cells and in livers of mice after introduction of mitochondrially targeted restriction endonucleases (Bacman et al. 2009). In these experiments, DSB repair was accompanied by the formation of mtDNA deletions, some of which had breakpoints flanked by direct repeats, thus implicating homologous recombination in the repair (Srivastava and Moraes 2005; Fukui and Moraes 2008). Recent studies revealed that mtDNA in adult human heart is organized in multimeric networks joined by abundant recombination junctions and catenation (Kajander et al. 2001). Together with an absence of θ replication in this tissue, this suggests that mammalian mtDNA in some cases uses recombination-dependent initiation for replication (Pohjoismaki et al. 2009). Curiously, extensive recombination junctions appear to be absent in human infant mtDNA and in the mtDNA of rodents (Pohjoismaki et al. 2009, 2010).
Nonhomologous End Joining
Currently, our understanding of the mitochondrial NHEJ pathway is incomplete. Mammalian mitochondria possess DNA end-binding activity, which is preserved in Ku-deficient cells (Coffey et al. 1999). Mitochondrial protein extracts can join blunt-ended DNA fragments, although with low efficiency and precision as compared with “sticky” ends, and results in deletions reminiscent of those observed in human disease (Lakshmipathy and Campbell 1999). Mitochondrial targeting of restriction endonucleases typically results in large deletions that have preferential end points, therefore interrupting the structural integrity of mtDNA and suggesting a role for topological organization in mtDNA recombination (Srivastava and Moraes 2005; Bacman et al. 2009). Moreover, in this model, DSBs are accompanied by degradation of the bulk of the damaged mtDNA, which has allowed for the use of mitochondrially targeted restriction endonucleases to both shift heteroplasmy in favor of mtDNA species that do not contain recognition sites for the restriction endonuclease (Alexeyev et al. 2008), and to produce cells totally devoid of mtDNA (Kukat et al. 2008).
Collectively, the available evidence for HR and NHEJ suggests that recombination is a viable, if minor, pathway for mtDNA repair in mammalian mitochondria.
Base Excision Repair
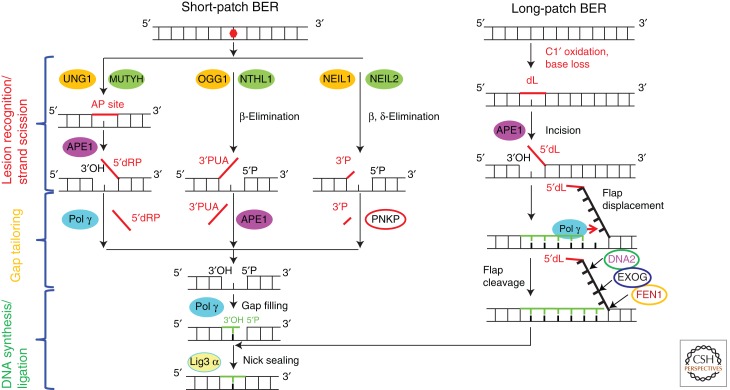
Base excision repair (BER) is the predominant and best understood DNA repair pathway in mitochondria (Fig. 2). The existence of this pathway in mitochondria was first suggested by the discovery of mitochondrial uracil–DNA glycosylase (Anderson and Friedberg 1980). Subsequently, repair of diverse BER substrates in mitochondria has been shown directly (Pettepher et al. 1991; LeDoux et al. 1992; Driggers et al. 1993). BER is the pathway of choice for the repair of most common oxidative DNA lesions (Svilar et al. 2011), and mitochondrial proficiency in this pathway is particularly important in the context of mitochondria being a major cellular source of reactive oxygen species (Murphy 2009). In fact, 8-oxodG, the most prominent oxidative DNA base lesion, is repaired more efficiently in mitochondria than in the nucleus (Thorslund et al. 2002).
Figure 2.
Mitochondrial BER in mammalian cells. Three different subpathways of SP-BER converge at the gap-filling step. SP-BER (left) and LP-BER (right) converge at the nick-sealing step. In addition to FEN1, DNA2 is required for efficient flap processing during LP-BER in mitochondria.
Nuclear BER consists of two subpathways (short-patch BER [SP-BER] and long-patch BER [LP-BER]), and each subpathway includes three functional steps: lesion recognition/strand scission, gap tailoring, and DNA synthesis/ligation (Svilar et al. 2011). Although the cast of specific players in nuclear BER appears to be more diverse than that in its mitochondrial counterpart, no differences in basic functional steps have been reported for the two compartments so far. Therefore, here we present a view of the mitochondrial BER based on what is known about nuclear BER and assuming, in the absence of evidence to the contrary, that identical proteins perform similar or identical functions.
Short-Patch BER
Lesion Recognition/Strand Scission
The initial step in the BER pathway is recognition of a damaged base by DNA glycosylase. Two types of DNA glycosylases are recognized: monofunctional and bifunctional glycosylases (Fig. 2).
Monofunctional DNA Glycosylases. Upon recognition of a damaged base, monofunctional DNA glycosylase hydrolyzes the N-glycosidic bond, leaving in DNA an apurinic/apyrimidinic (abasic or AP) site. Two monofunctional DNA glycosylases have been described in mitochondria: uracil-DNA glycosylase 1 (UDG1; or uracil-N-glycosylase1 [UNG1]) (Anderson and Friedberg 1980) and a homolog of the Escherichia coli MutY glycosylase, MUTYH (MYH) (Ohtsubo et al. 2000). Substrates for UNG1 and MUTYH have been reviewed recently (Svilar et al. 2011). Although we (LeDoux et al. 1998) and others (Pirsel and Bohr 1993) have shown that mitochondria are capable of repairing alkylation lesions typically processed by another monofunctional glycosylase, MPG (Chakravarti et al. 1991), the presence of this enzyme in mitochondria has not been shown yet. Following the removal of a damaged DNA base by a monofunctional DNA glycosylase, the resulting AP site is incised 5′ to the lesion by AP endonuclease (APE1). A special mitochondrial isoform of this enzyme is generated by proteolytic amino-terminal truncation of 33 residues that contain the nuclear localization sequence (Chattopadhyay et al. 2006). The product of the concerted action of a monofunctional DNA glycosylase and APE1 is an SSB with a 5′-blocking deoxyribosophosphate (dRP) group and 3′-OH (Fig. 2). The 5′-dRP group is nonligatable and is removed during the gap-tailoring step (see below).
Bifunctional DNA Glycosylases. Unlike monofunctional DNA glycosylases, bifunctional DNA glycosylases possess an intrinsic AP-lyase activity and not only cleave the N-glycosidic bond but also incise the resulting AP site via either a β-elimination or β,δ-elimination reaction (Dodson and Lloyd 2002). Incision via the β-elimination reaction results in a strand break at the AP site with 3′ phospho-αβ-unsaturated aldehyde (3′PUA) and 5′P, whereas β,δ-elimination results in a single nucleotide gap with 3′P and 5′P termini. In both cases, the ends are nonligatable and have to be further processed. Four bifunctional DNA glycosylases have been described in the mitochondria: 8-oxoguanine DNA glycosylase (OGG1) (Takao et al. 1998; de Souza-Pinto et al. 2001) and nth (E. coli endonuclease III)-like 1 (NTHL1, or NTH1) (Karahalil et al. 2003), as well as Nei (E. coli endonuclease VIII)-like-1 and -2 (NEIL1 and NEIL2) (Hu et al. 2005; Mandal et al. 2012). Two of them (OGG1 and NTHL1) possess a β-lyase activity, whereas the remaining two (NEIL1 and NEIL2) are β,δ-lyases (Fig. 2). Importantly, bifunctional DNA glycosylases are predominant in the repair of oxidative base lesions.
All known DNA glycosylases appear to use the same mechanism to excise damaged base: The base is “flipped out” of the stack into the active-site pocket of the enzyme, where hydrolysis of the glycosidic bond takes place (Friedman and Stivers 2010).
Gap Tailoring
The product of the first step of the BER is an SSB, whose ends need further processing (gap tailoring) to remove blocking groups at either the 5′ or 3′ end before insertion of the correct base in place of a damaged one so that restoration of the DNA continuity (ligation) may take place. Enzymatic activities recruited during tailoring depend on the chemical nature of blocking groups, and therefore are determined by the upstream glycosylase (Fig. 2).
Processing of the Ends Generated by a Combination of Monofunctional Glycosylase Plus APE1. The 5′-dRP group at the nicks generated in mtDNA by the action of UNG1 plus APE1 or MUTYH1 plus APE1 on damaged bases is removed by dRP lyase activity of mitochondrial Pol γ. This polymerase is involved in both replication and repair of mtDNA and possesses DNA polymerase, 5′–3′ exonuclease and dRP-lyase activities (Longley et al. 1998). The product of this step is a single-stranded gap with 5′P and 3′-OH groups (Fig. 2).
Processing of the Ends Generated by a Bifunctional Glycosylase/β-Lyase. SSBs generated in mtDNA by OGG1 and NTHL1 have 5′P and 3′-phospho-α,β-unsaturated aldehyde (4-hydroxypentenal phosphate) (Altieri et al. 2008), which is removed by the phosphodiesterase activity of APE1 (Svilar et al. 2011). The resulting product is a single-stranded gap with 5′P and 3′-OH groups (Fig. 2).
Processing of the Ends Generated by a Bifunctional Glycosylase/β, δ-Lyase. NEIL1 and NEIL2 removal of damaged bases from mtDNA results in a single-strand one-nucleotide gap flanked by 5′P and 3′P. 3′P is then processed to generate 3′-OH by phosphatase activity of the polynucleotide kinase 3′-phosphatase (PNKP) (Mandal et al. 2012; Tahbaz et al. 2012). Although APE1 also possesses 3′-phosphatase activity, it is weak (Xu et al. 2003), and its contribution appears to be minor, if present. The resulting product is, again, a single-stranded gap with 5′P and 3′-OH groups (Fig. 2).
DNA Synthesis/Ligation
As we saw above, the product of the gap-tailoring step in the SP-BER is invariably a single-nucleotide gap with 5′P and 3′-OH groups. This gap is filled by activity of the Pol γ, and the remaining nick is sealed by DNA ligase IIIα (Lig3) (Fig. 2). Lig3 is an essential gene, and its loss is associated with early lethality both at the organismal (Puebla-Osorio et al. 2006; Gao et al. 2011) and the cellular (Puebla-Osorio et al. 2006; Simsek et al. 2011; Arakawa et al. 2012) levels. In cultured cells, viability can be restored by expressing either eukaryotic or prokaryotic DNA ligase targeted to mitochondria, but not to the nucleus, indicating an essential mitochondrial function of Lig3 (Simsek et al. 2011; Arakawa et al. 2012). Antisense-mediated down-regulation (Lakshmipathy and Campbell 2001) and knockout (Gao et al. 2011) of Lig3 have been reported to reduce mtDNA copy number. Paradoxically in this context, levels of Lig 3 transcripts do not change when mtDNA depletion is induced through mitochondrial targeting of restriction endonuclease, and are actually decreased during the recovery of mtDNA copy number back to original levels (Bacman et al. 2009).
Long-Patch BER
Certain 5′ blocking groups, such as 5′ deoxyribonolactone (5′-dL, oxidized 5′-dRP group) can not be efficiently removed by dRP-lyase activity of Pol γ. Yet, the yield of this lesion may be as high as 72% of total oxidative sugar damage in DNA upon irradiation accompanied by a base loss (Roginskaya et al. 2005). Formation of the dL lesion is not associated with glycosylase activity because the base is lost as a result of hydrogen abstraction and oxidation of the C1′ of deoxyribose. The removal of this lesion in nDNA is performed by LP-BER. Similarly, mitochondrial LP-BER has been described recently (Akbari et al. 2008; Copeland and Longley 2008; Liu et al. 2008; Szczesny et al. 2008). During LP-BER, the 5′-blocking group is displaced by DNA polymerase extending from the 3′-OH end and forming a so called “flap.” This flap in mitochondria has a length of up to 6–9 nucleotides (Szczesny et al. 2008) and is subsequently removed by the concerted activity of two nucleases, FEN1 and DNA2 (Copeland and Longley 2008; Zheng et al. 2008; Duxin et al. 2009; Kalifa et al. 2009) or by EXOG (Tann et al. 2011), resulting in the nicked DNA duplex, which can be sealed by LIG3 (Fig. 2). Although it is currently unclear whether lesions other than dL are repaired in mitochondria by LP-BER, it has been suggested that given the weak dRP lyase activity of Pol γ and the strong strand-displacement activity of Pol γ in vitro, LP-BER might be the predominant mode for repairing damaged bases in vivo.
mtDNA Degradation
Unlike the nuclear genome, the mitochondrial genome is redundant, consisting of hundreds to thousands of copies per cell. Therefore, a “repair or die” constraint is not imposed on mtDNA. Conceivably, a substantial fraction of damaged mtDNA can be lost without detrimental effects, provided that this loss can be compensated for by replication of the remaining genomes. In fact, the loss and resynthesis of mtDNA was observed more than 40 years ago by Gross and Rabinowitz, who described mtDNA turnover (Gross and Rabinowitz 1969). Many cell lines are fairly tolerant to the loss of mtDNA and can survive both a gradual loss of mtDNA through chronic treatment with ethidium bromide (King and Attardi 1989) and acute destruction of a fraction (Alexeyev et al. 2008) or even all of their mtDNA (Kukat et al. 2008) by mitochondrially targeted restriction endonucleases. This is in a stark contrast to nDNA, in which persistent DSB can activate apoptosis. However, the hypothesis that turnover (degradation) of damaged mtDNA can be a mechanism used by mitochondria to deal with either excessive damage or damage that cannot be repaired did not take hold in part because of the lack of direct experimental evidence supporting it and in part because of discovery of mitochondrial BER (Pettepher et al. 1991), which has shifted attention from unrepairable lesions to those that can be repaired. However, recent evidence reignited interest in mtDNA degradation.
Ethanol has been reported to induce the loss of mtDNA in yeast (Ibeas and Jimenez 1997). In mice, intragastric administration of ethanol induced oxidative stress and was accompanied by a reversible loss of mtDNA (Mansouri et al. 1999). The loss of mtDNA was ∼50% in all organs studied. It could be partially prevented by the antioxidants melatonin, vitamin E, and coenzyme Q, and was followed by adaptive mtDNA resynthesis (Mansouri et al. 2001). Lipopolysaccharide, a known inducer of in vivo oxidative stress, also induced mtDNA depletion (Suliman et al. 2003). Angiotensin II induced mitochondrial ROS production and decreased skeletal muscle mtDNA content in mice (Mitsuishi et al. 2008). Degradation of mtDNA was observed in a rat model of cerebral ischemia/reperfusion (Chen et al. 2001). Similar to mtDNA depletion induced by intragastric ethanol administration, mtDNA levels returned to normal within 24 h of cerebral ischemia/reperfusion (Chen et al. 2001). Finally, H2O2-induced oxidative stress in hamster fibroblasts was accompanied by Ca2+-dependent degradation of mtDNA (Crawford et al. 1998). Taken together, these findings strongly suggested a link between oxidative stress (which may result in oxidative mtDNA damage) and mtDNA degradation, yet they stopped short of invoking degradation as a protective mechanism. In an unrelated study, it was observed that mtDNA is resistant to mutagenesis induced by alkylating agents, and the investigators suggested degradation of damaged mtDNA as one of the potential mechanisms to explain this phenomenon (Mita et al. 1988). However, mtDNA degradation was not shown in that study (Mita et al. 1988).
Recently, we attempted to study the relationship between experimentally induced oxidative stress and mtDNA mutagenesis. In initial experiments, superoxide radicals were generated on the matrix side of the mitochondrial inner membrane by treating cells with sublethal concentrations of the complex I inhibitor rotenone (St-Pierre et al. 2002; Muller et al. 2004). However, exposing human colon carcinoma cells or mouse embryonic fibroblasts to rotenone for 30 d did not result in a significant increase in the rate of mtDNA mutagenesis (Shokolenko et al. 2009). Similarly, repeated treatment of HCT116 colon cancer cells with H2O2 failed to induce significant mtDNA mutagenesis. Instead, DNA lesions that manifest themselves as strand breaks under denaturing conditions (single-strand breaks, SSBs; DSBs, abasic sites; etc.) prevailed over premutagenic base modifications by a factor of 10. Consistent with the hypothesis that unrepairable mtDNA molecules are degraded, when BER was inhibited by methoxyamine, which blocks one of the initial steps of BER, processing of the abasic sites by apurinic, apyrymidinic endonuclease (APE1), mtDNA degradation was enhanced in response to both oxidative and alkylation damage (Shokolenko et al. 2009). The elimination of damaged mtDNA was preceded by the accumulation of linear mtDNA molecules, which may represent degradation intermediates, because, unlike undamaged circular molecules, they are susceptible to exonucleolytic degradation.
The high rate of SSBs and other polymerase-blocking lesions induced in mtDNA by reactive oxygen species (ROS) suggests a mechanism by which mitochondria may maintain the integrity of their genetic information. In this model, in mtDNA, oxidative stress induces strand breaks with a much higher (by the factor of 10) (Shokolenko et al. 2009) frequency than mutagenic lesions. These lesions represent a block to transcription and replication of mtDNA, and when accumulated above a threshold level, they induce degradation of mtDNA molecules. Therefore, degradation of the mtDNA molecule is triggered before it accumulates mutagenic lesions. This model provides a mechanistic explanation for the observations made by Suter and Richter (1999), who found that the 8-oxodG content of circular mtDNA is low and does not increase in response to oxidative insult. However, fragmented mtDNA had a very high 8-oxodG content, which increased further after oxidative stress. It incorporates the previously suggested notion of the possible contribution of APE1 to mtDNA degradation (Tomkinson et al. 1988, 1990). The model is consistent with the observations of Yakes and Van Houten (1997), who found that oxidative stress promoted a higher incidence of polymerase-blocking strand breaks and abasic sites in mtDNA than in nDNA. Recent studies using qPCR for the analysis of mtDNA provide further support for the notion of mtDNA degradation in response to oxidative stress (Rothfuss et al. 2010; Furda et al. 2012). Therefore, degradation of severely damaged mtDNA emerges as a unique, mitochondria-specific mechanism for the maintenance of DNA integrity.
Degradation of damaged organellar DNA appears not to be restricted to mammalian cells. Known examples of rapid organellar DNA turnover in plants and protists in response to ROS were recently reviewed by Bendich (2010).
CONCLUDING REMARKS
Our understanding of DNA repair processes in mitochondria has advanced dramatically in recent years. Discoveries of mitochondrial mismatch repair, LP-BER, and mtDNA degradation in response to damage establish that not only do mitochondria possess most DNA repair pathways available in the nucleus, they also have evolved a specific pathway, which is enabled by the intrinsic redundancy of mtDNA and allows these organelles to dispose of damaged DNA molecules and replicate new ones. At the same time, we saw an expansion of the repertoire of DNA repair enzymes with proven mitochondrial localization. However, despite these advances, our understanding of the details of mitochondrial DNA repair remains limited and is often extrapolated from our knowledge of nDNA repair. In addition, physiological levels of DNA damaging species (most importantly, ROS) in mitochondria remain to be determined. Finally, the enzymatic apparatus behind mtDNA degradation remains obscure. We expect that in the future a greater level of detail will become available on these and other critical issues.
ACKNOWLEDGMENTS
The authors are supported by the National Institutes of Health grants ES03456, PO1 HL66299, and RR031286/OD010944.
Footnotes
Editors: Errol C. Friedberg, Stephen J. Elledge, Alan R. Lehmann, Tomas Lindahl, and Marco Muzi-Falconi
Additional Perspectives on DNA Repair, Mutagenesis, and Other Responses to DNA Damage available at www.cshperspectives.org
REFERENCES
- Ahel I, Rass U, El-Khamisy SF, Katyal S, Clements PM, McKinnon PJ, Caldecott KW, West SC 2006. The neurodegenerative disease protein aprataxin resolves abortive DNA ligation intermediates. Nature 443: 713–716 [DOI] [PubMed] [Google Scholar]
- Akbari M, Visnes T, Krokan HE, Otterlei M 2008. Mitochondrial base excision repair of uracil and AP sites takes place by single-nucleotide insertion and long-patch DNA synthesis. DNA Repair 7: 605–616 [DOI] [PubMed] [Google Scholar]
- Alam TI, Kanki T, Muta T, Ukaji K, Abe Y, Nakayama H, Takio K, Hamasaki N, Kang D 2003. Human mitochondrial DNA is packaged with TFAM. Nucleic Acids Res 31: 1640–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexeyev MF 2009. Is there more to aging than mitochondrial DNA and reactive oxygen species? FEBS J 276: 5768–5787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexeyev MF, Venediktova N, Pastukh V, Shokolenko I, Bonilla G, Wilson GL 2008. Selective elimination of mutant mitochondrial genomes as therapeutic strategy for the treatment of NARP and MILS syndromes. Gene Ther 15: 516–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altieri F, Grillo C, Maceroni M, Chichiarelli S 2008. DNA damage and repair: From molecular mechanisms to health implications. Antioxid Redox Signal 10: 891–937 [DOI] [PubMed] [Google Scholar]
- Anderson CT, Friedberg EC 1980. The presence of nuclear and mitochondrial uracil-DNA glycosylase in extracts of human KB cells. Nucleic Acids Res 8: 875–888 [PMC free article] [PubMed] [Google Scholar]
- Andreyev AY, Kushnareva YE, Starkov AA 2005. Mitochondrial metabolism of reactive oxygen species. Biochemistry (Mosc) 70: 200–214 [DOI] [PubMed] [Google Scholar]
- Anson RM, Hudson E, Bohr VA 2000. Mitochondrial endogenous oxidative damage has been overestimated. FASEB J 14: 355–360 [DOI] [PubMed] [Google Scholar]
- Arakawa H, Bednar T, Wang M, Paul K, Mladenov E, Bencsik-Theilen AA, Iliakis G 2012. Functional redundancy between DNA ligases I and III in DNA replication in vertebrate cells. Nucleic Acids Res 40: 2599–2610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacman SR, Williams SL, Moraes CT 2009. Intra- and inter-molecular recombination of mitochondrial DNA after in vivo induction of multiple double-strand breaks. Nucleic Acids Res 37: 4218–4226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balansky R, Izzotti A, Scatolini L, D’Agostini F, De Flora S 1996. Induction by carcinogens and chemoprevention by N-acetylcysteine of adducts to mitochondrial DNA in rat organs. Cancer Res 56: 1642–1647 [PubMed] [Google Scholar]
- Ballard JW, Whitlock MC 2004. The incomplete natural history of mitochondria. Mol Ecol 13: 729–744 [DOI] [PubMed] [Google Scholar]
- Ballinger SW, Van Houten B, Jin GF, Conklin CA, Godley BF 1999. Hydrogen peroxide causes significant mitochondrial DNA damage in human RPE cells. Exp Eye Res 68: 765–772 [DOI] [PubMed] [Google Scholar]
- Ballinger SW, Patterson C, Yan CN, Doan R, Burow DL, Young CG, Yakes FM, Van Houten B, Ballinger CA, Freeman BA, et al. 2000. Hydrogen peroxide- and peroxynitrite-induced mitochondrial DNA damage and dysfunction in vascular endothelial and smooth muscle cells. Circ Res 86: 960–966 [DOI] [PubMed] [Google Scholar]
- Bartsch H 1996. DNA adducts in human carcinogenesis: Etiological relevance and structure–activity relationship. Mutat Res 340: 67–79 [DOI] [PubMed] [Google Scholar]
- Bendich AJ 2010. Mitochondrial DNA, chloroplast DNA and the origins of development in eukaryotic organisms. Biol Direct 5: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair IA 2008. DNA adducts with lipid peroxidation products. J Biol Chem 283: 15545–15549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakely WF, Fuciarelli AF, Wegher BJ, Dizdaroglu M 1990. Hydrogen peroxide-induced base damage in deoxyribonucleic acid. Radiat Res 121: 338–343 [PubMed] [Google Scholar]
- Boesch P, Weber-Lotfi F, Ibrahim N, Tarasenko V, Cosset A, Paulus F, Lightowlers RN, Dietrich A 2011. DNA repair in organelles: Pathways, organization, regulation, relevance in disease and aging. Biochim Biophys Acta 1813: 186–200 [DOI] [PubMed] [Google Scholar]
- Bogenhagen DF, Rousseau D, Burke S 2008. The layered structure of human mitochondrial DNA nucleoids. J Biol Chem 283: 3665–3675 [DOI] [PubMed] [Google Scholar]
- Bohr VA 2002. Repair of oxidative DNA damage in nuclear and mitochondrial DNA, and some changes with aging in mammalian cells. Free Radic Biol Med 32: 804–812 [DOI] [PubMed] [Google Scholar]
- Boveris A, Oshino N, Chance B 1972. The cellular production of hydrogen peroxide. Biochem J 128: 617–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brawn K, Fridovich I 1981. DNA strand scission by enzymically generated oxygen radicals. Arch Biochem Biophys 206: 414–419 [DOI] [PubMed] [Google Scholar]
- Brown WM, George M Jr, Wilson AC 1979. Rapid evolution of animal mitochondrial DNA. Proc Natl Acad Sci 76: 1967–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown TA, Tkachuk AN, Shtengel G, Kopek BG, Bogenhagen DF, Hess HF, Clayton DA 2011. Superresolution fluorescence imaging of mitochondrial nucleoids reveals their spatial range, limits, and membrane interaction. Mol Cell Biol 31: 4994–5010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai S, Xu Y, Cooper RJ, Ferkowicz MJ, Hartwell JR, Pollok KE, Kelley MR 2005. Mitochondrial targeting of human O6-methylguanine DNA methyltransferase protects against cell killing by chemotherapeutic alkylating agents. Cancer Res 65: 3319–3327 [DOI] [PubMed] [Google Scholar]
- Caldecott KW 2008. Single-strand break repair and genetic disease. Nat Rev Genet 9: 619–631 [DOI] [PubMed] [Google Scholar]
- Chakravarti D, Ibeanu GC, Tano K, Mitra S 1991. Cloning and expression in Escherichia coli of a human cDNA encoding the DNA repair protein N-methylpurine-DNA glycosylase. J Biol Chem 266: 15710–15715 [PubMed] [Google Scholar]
- Chattopadhyay R, Wiederhold L, Szczesny B, Boldogh I, Hazra TK, Izumi T, Mitra S 2006. Identification and characterization of mitochondrial abasic (AP)-endonuclease in mammalian cells. Nucleic Acids Res 34: 2067–2076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Hu CJ, He YY, Yang DI, Xu J, Hsu CY 2001. Reduction and restoration of mitochondrial DNA content after focal cerebral ischemia/reperfusion. Stroke 32: 2382–2387 [DOI] [PubMed] [Google Scholar]
- Christmann M, Tomicic MT, Roos WP, Kaina B 2003. Mechanisms of human DNA repair: An update. Toxicology 193: 3–34 [DOI] [PubMed] [Google Scholar]
- Clay Montier LL, Deng JJ, Bai Y 2009. Number matters: Control of mammalian mitochondrial DNA copy number. J Genet Genomics 36: 125–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton DA, Doda JN, Friedberg EC 1974. The absence of a pyrimidine dimer repair mechanism in mammalian mitochondria. Proc Natl Acad Sci 71: 2777–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton DA, Doda JN, Friedberg EC 1975. Absence of a pyrimidine dimer repair mechanism for mitochondrial DNA in mouse and human cells. Basic Life Sci 5B: 589–591 [DOI] [PubMed] [Google Scholar]
- Coffey G, Lakshmipathy U, Campbell C 1999. Mammalian mitochondrial extracts possess DNA end-binding activity. Nucleic Acids Res 27: 3348–3354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland WC, Longley MJ 2008. DNA2 resolves expanding flap in mitochondrial base excision repair. Mol Cell 32: 457–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford DR, Abramova NE, Davies KJ 1998. Oxidative stress causes a general, calcium-dependent degradation of mitochondrial polynucleotides. Free Radic Biol Med 25: 1106–1111 [DOI] [PubMed] [Google Scholar]
- Das BB, Dexheimer TS, Maddali K, Pommier Y 2010. Role of tyrosyl-DNA phosphodiesterase (TDP1) in mitochondria. Proc Natl Acad Sci 107: 19790–19795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Aurelio M, Gajewski CD, Lin MT, Mauck WM, Shao LZ, Lenaz G, Moraes CT, Manfredi G 2004. Heterologous mitochondrial DNA recombination in human cells. Hum Mol Genet 13: 3171–3179 [DOI] [PubMed] [Google Scholar]
- Davila JI, Arrieta-Montiel MP, Wamboldt Y, Cao J, Hagmann J, Shedge V, Xu YZ, Weigel D, Mackenzie SA 2011. Double-strand break repair processes drive evolution of the mitochondrial genome in Arabidopsis. BMC Biol 9: 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bont R, van Larebeke N 2004. Endogenous DNA damage in humans: A review of quantitative data. Mutagenesis 19: 169–185 [DOI] [PubMed] [Google Scholar]
- de Souza-Pinto NC, Eide L, Hogue BA, Thybo T, Stevnsner T, Seeberg E, Klungland A, Bohr VA 2001. Repair of 8-oxodeoxyguanosine lesions in mitochondrial DNA depends on the oxoguanine DNA glycosylase (OGG1) gene and 8-oxoguanine accumulates in the mitochondrial DNA of OGG1-defective mice. Cancer Res 61: 5378–5381 [PubMed] [Google Scholar]
- de Souza-Pinto NC, Mason PA, Hashiguchi K, Weissman L, Tian J, Guay D, Lebel M, Stevnsner TV, Rasmussen LJ, Bohr VA 2009. Novel DNA mismatch-repair activity involving YB-1 in human mitochondria. DNA Repair (Amst) 8: 704–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodson ML, Lloyd RS 2002. Mechanistic comparisons among base excision repair glycosylases. Free Radic Biol Med 32: 678–682 [DOI] [PubMed] [Google Scholar]
- Driggers WJ, LeDoux SP, Wilson GL 1993. Repair of oxidative damage within the mitochondrial DNA of RINr 38 cells. J Biol Chem 268: 22042–22045 [PubMed] [Google Scholar]
- Duxin JP, Dao B, Martinsson P, Rajala N, Guittat L, Campbell JL, Spelbrink JN, Stewart SA 2009. Human Dna2 is a nuclear and mitochondrial DNA maintenance protein. Mol Cell Biol 29: 4274–4282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekstrand MI, Falkenberg M, Rantanen A, Park CB, Gaspari M, Hultenby K, Rustin P, Gustafsson CM, Larsson NG 2004. Mitochondrial transcription factor A regulates mtDNA copy number in mammals. Hum Mol Genet 13: 935–944 [DOI] [PubMed] [Google Scholar]
- el-Khamisy SF, Caldecott KW 2007. DNA single-strand break repair and spinocerebellar ataxia with axonal neuropathy-1. Neuroscience 145: 1260–1266 [DOI] [PubMed] [Google Scholar]
- European Standards Committee on Oxidative DNA Damage (ESCODD) 2003. Measurement of DNA oxidation in human cells by chromatographic and enzymic methods. Free Radic Biol Med 34: 1089–1099 [DOI] [PubMed] [Google Scholar]
- Friedman JI, Stivers JT 2010. Detection of damaged DNA bases by DNA glycosylase enzymes. Biochemistry 49: 4957–4967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frimer AA, Strul G, Buch J, Gottlieb HE 1996. Can superoxide organic chemistry be observed within the liposomal bilayer? Free Radic Biol Med 20: 843–852 [DOI] [PubMed] [Google Scholar]
- Fukui H, Moraes CT 2008. Mechanisms of formation and accumulation of mitochondrial DNA deletions in aging neurons. Hum Mol Genet 18: 1028–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furda AM, Marrangoni AM, Lokshin A, Van Houten B 2012. Oxidants and not alkylating agents induce rapid mtDNA loss and mitochondrial dysfunction. DNA Repair 11: 684–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Katyal S, Lee Y, Zhao J, Rehg JE, Russell HR, McKinnon PJ 2011. DNA ligase III is critical for mtDNA integrity but not Xrcc1-mediated nuclear DNA repair. Nature 471: 240–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner PR 1997. Superoxide-driven aconitase FE-S center cycling. Biosci Rep 17: 33–42 [DOI] [PubMed] [Google Scholar]
- Gardner PR, Fridovich I 1991. Superoxide sensitivity of the Escherichia coli aconitase. J Biol Chem 266: 19328–19333 [PubMed] [Google Scholar]
- Gedik CM, Collins A 2005. Establishing the background level of base oxidation in human lymphocyte DNA: Results of an interlaboratory validation study. FASEB J 19: 82–84 [DOI] [PubMed] [Google Scholar]
- Gilkerson RW, Schon EA, Hernandez E, Davidson MM 2008. Mitochondrial nucleoids maintain genetic autonomy but allow for functional complementation. J Cell Biol 181: 1117–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gredilla R 2010. DNA damage and base excision repair in mitochondria and their role in aging. J Aging Res 2011: 257093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross NJ, Rabinowitz M 1969. Synthesis of new strands of mitochondrial and nuclear deoxyribonucleic acid by semiconservative replication. J Biol Chem 244: 1563–1566 [PubMed] [Google Scholar]
- Guliaeva NA, Kuznetsova EA, Gaziev AI 2006. Proteins associated with mitochondrial DNA protect it against the action of X-rays and hydrogen peroxide. Biofizika 51: 692–697 [PubMed] [Google Scholar]
- Halliwell B, Aruoma OI 1991. DNA damage by oxygen-derived species. Its mechanism and measurement in mammalian systems. FEBS Lett 281: 9–19 [DOI] [PubMed] [Google Scholar]
- Hanes JW, Thal DM, Johnson KA 2006. Incorporation and replication of 8-oxo-deoxyguanosine by the human mitochondrial DNA polymerase. J Biol Chem 281: 36241–36248 [DOI] [PubMed] [Google Scholar]
- Hegde ML, Izumi T, Mitra S 2012. Oxidized base damage and single-strand break repair in mammalian genomes: Role of disordered regions and posttranslational modifications in early enzymes. Prog Mol Biol Transl Sci 110: 123–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henle ES, Luo Y, Gassmann W, Linn S 1996a. Oxidative damage to DNA constituents by iron-mediated Fenton reactions. The deoxyguanosine family. J Biol Chem 271: 21177–21186 [PubMed] [Google Scholar]
- Henle ES, Luo Y, Linn S 1996b. Fe2+, Fe3+, and oxygen react with DNA-derived radicals formed during iron-mediated Fenton reactions. Biochemistry 35: 12212–12219 [DOI] [PubMed] [Google Scholar]
- Holt IJ, Harding AE, Morgan-Hughes JA 1988. Deletions of muscle mitochondrial DNA in patients with mitochondrial myopathies. Nature 331: 717–719 [DOI] [PubMed] [Google Scholar]
- Horne DW, Holloway RS, Wagner C 1997. Transport of S-adenosylmethionine in isolated rat liver mitochondria. Arch Biochem Biophys 343: 201–206 [DOI] [PubMed] [Google Scholar]
- Hu J, de Souza-Pinto NC, Haraguchi K, Hogue BA, Jaruga P, Greenberg MM, Dizdaroglu M, Bohr VA 2005. Repair of formamidopyrimidines in DNA involves different glycosylases: Role of the OGG1, NTH1, and NEIL1 enzymes. J Biol Chem 280: 40544–40551 [DOI] [PubMed] [Google Scholar]
- Hudson JJ, Chiang SC, Wells OS, Rookyard C, El-Khamisy SF 2012. SUMO modification of the neuroprotective protein TDP1 facilitates chromosomal single-strand break repair. Nat Commun 3: 733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibeas JI, Jimenez J 1997. Mitochondrial DNA loss caused by ethanol in Saccharomyces flor yeasts. Appl Environ Microbiol 63: 7–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iborra FJ, Kimura H, Cook PR 2004. The functional organization of mitochondrial genomes in human cells. BMC Biol 2: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung D, Cho Y, Collins LB, Swenberg JA, Di Giulio RT 2009. Effects of benzo[a]pyrene on mitochondrial and nuclear DNA damage in Atlantic killifish (Fundulus heteroclitus) from a creosote-contaminated and reference site. Aquat Toxicol 95: 44–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajander OA, Karhunen PJ, Holt IJ, Jacobs HT 2001. Prominent mitochondrial DNA recombination intermediates in human heart muscle. EMBO Rep 2: 1007–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalifa L, Beutner G, Phadnis N, Sheu SS, Sia EA 2009. Evidence for a role of FEN1 in maintaining mitochondrial DNA integrity. DNA Repair 8: 1242–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya H 2003. Mutagenic potentials of damaged nucleic acids produced by reactive oxygen/nitrogen species: Approaches using synthetic oligonucleotides and nucleotides: Survey and summary. Nucleic Acids Res 31: 517–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang D, Nishida J, Iyama A, Nakabeppu Y, Furuichi M, Fujiwara T, Sekiguchi M, Takeshige K 1995. Intracellular localization of 8-oxo-dGTPase in human cells, with special reference to the role of the enzyme in mitochondria. J Biol Chem 270: 14659–14665 [DOI] [PubMed] [Google Scholar]
- Karahalil B, de Souza-Pinto NC, Parsons JL, Elder RH, Bohr VA 2003. Compromised incision of oxidized pyrimidines in liver mitochondria of mice deficient in NTH1 and OGG1 glycosylases. J Biol Chem 278: 33701–33707 [DOI] [PubMed] [Google Scholar]
- Kasparek TR, Humphrey TC 2011. DNA double-strand break repair pathways, chromosomal rearrangements and cancer. Semin Cell Dev Biol 22: 886–897 [DOI] [PubMed] [Google Scholar]
- Khan SK, Xu H, Ukai-Tadenuma M, Burton B, Wang Y, Ueda HR, Liu AC 2012. Identification of a novel cryptochrome differentiating domain required for feedback repression in circadian clock function. J Biol Chem 287: 25917–25926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King MP, Attardi G 1989. Human cells lacking mtDNA: Repopulation with exogenous mitochondria by complementation. Science 246: 500–503 [DOI] [PubMed] [Google Scholar]
- Kopek BG, Shtengel G, Xu CS, Clayton DA, Hess HF 2012. Correlative 3D superresolution fluorescence and electron microscopy reveal the relationship of mitochondrial nucleoids to membranes. Proc Natl Acad Sci 109: 6136–6141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukat A, Kukat C, Brocher J, Schafer I, Krohne G, Trounce IA, Villani G, Seibel P 2008. Generation of ρ0 cells utilizing a mitochondrially targeted restriction endonuclease and comparative analyses. Nucleic Acids Res 36: e44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukat C, Wurm CA, Spahr H, Falkenberg M, Larsson NG, Jakobs S 2011. Super-resolution microscopy reveals that mammalian mitochondrial nucleoids have a uniform size and frequently contain a single copy of mtDNA. Proc Natl Acad Sci 108: 13534–13539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel TA, Erie DA 2005. DNA mismatch repair. Annu Rev Biochem 74: 681–710 [DOI] [PubMed] [Google Scholar]
- Lakshmipathy U, Campbell C 1999. Double strand break rejoining by mammalian mitochondrial extracts. Nucleic Acids Res 27: 1198–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakshmipathy U, Campbell C 2001. Antisense-mediated decrease in DNA ligase III expression results in reduced mitochondrial DNA integrity. Nucleic Acids Res 29: 668–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeDoux SP, Wilson GL, Beecham EJ, Stevnsner T, Wassermann K, Bohr VA 1992. Repair of mitochondrial DNA after various types of DNA damage in Chinese hamster ovary cells. Carcinogenesis 13: 1967–1973 [DOI] [PubMed] [Google Scholar]
- LeDoux SP, Shen CC, Grishko VI, Fields PA, Gard AL, Wilson GL 1998. Glial cell-specific differences in response to alkylation damage. Glia 24: 304–312 [PubMed] [Google Scholar]
- Lesko SA, Lorentzen RJ, Ts’o PO 1980. Role of superoxide in deoxyribonucleic acid strand scission. Biochemistry 19: 3023–3028 [DOI] [PubMed] [Google Scholar]
- Lestienne P, Ponsot G 1988. Kearns-Sayre syndrome with muscle mitochondrial DNA deletion. Lancet 1: 885. [DOI] [PubMed] [Google Scholar]
- Liang Q, Dedon PC 2001. Cu(II)/H2O2-induced DNA damage is enhanced by packaging of DNA as a nucleosome. Chem Res Toxicol 14: 416–422 [DOI] [PubMed] [Google Scholar]
- Liang R, Senturker S, Shi X, Bal W, Dizdaroglu M, Kasprzak KS 1999. Effects of Ni(II) and Cu(II) on DNA interaction with the N-terminal sequence of human protamine P2: Enhancement of binding and mediation of oxidative DNA strand scission and base damage. Carcinogenesis 20: 893–898 [DOI] [PubMed] [Google Scholar]
- Liehr JG 2000. Is estradiol a genotoxic mutagenic carcinogen? Endocr Rev 21: 40–54 [DOI] [PubMed] [Google Scholar]
- Lim KS, Jeyaseelan K, Whiteman M, Jenner A, Halliwell B 2005. Oxidative damage in mitochondrial DNA is not extensive. Ann NY Acad Sci 1042: 210–220 [DOI] [PubMed] [Google Scholar]
- Lindahl T 1993. Instability and decay of the primary structure of DNA. Nature 362: 709–715 [DOI] [PubMed] [Google Scholar]
- Liu P, Demple B 2010. DNA repair in mammalian mitochondria: Much more than we thought? Environ Mol Mutagen 51: 417–426 [DOI] [PubMed] [Google Scholar]
- Liu P, Qian L, Sung JS, de Souza-Pinto NC, Zheng L, Bogenhagen DF, Bohr VA, Wilson DM 3rd, Shen B, Demple B 2008. Removal of oxidative DNA damage via FEN1-dependent long-patch base excision repair in human cell mitochondria. Mol Cell Biol 28: 4975–4987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longley MJ, Prasad R, Srivastava DK, Wilson SH, Copeland WC 1998. Identification of 5′-deoxyribose phosphate lyase activity in human DNA polymerase gamma and its role in mitochondrial base excision repair in vitro. Proc Natl Acad Sci 95: 12244–12248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manchekar M, Scissum-Gunn K, Song D, Khazi F, McLean SL, Nielsen BL 2006. DNA recombination activity in soybean mitochondria. J Mol Biol 356: 288–299 [DOI] [PubMed] [Google Scholar]
- Mandal SM, Hegde ML, Chatterjee A, Hegde PM, Szczesny B, Banerjee D, Boldogh I, Gao R, Falkenberg M, Gustafsson CM, et al. 2012. Role of human DNA glycosylase Nei-like 2 (NEIL2) and single strand break repair protein polynucleotide kinase 3′-phosphatase in maintenance of mitochondrial genome. J Biol Chem 287: 2819–2829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansouri A, Gaou I, De Kerguenec C, Amsellem S, Haouzi D, Berson A, Moreau A, Feldmann G, Letteron P, Pessayre D, et al. 1999. An alcoholic binge causes massive degradation of hepatic mitochondrial DNA in mice. Gastroenterology 117: 181–190 [DOI] [PubMed] [Google Scholar]
- Mansouri A, Demeilliers C, Amsellem S, Pessayre D, Fromenty B 2001. Acute ethanol administration oxidatively damages and depletes mitochondrial DNA in mouse liver, brain, heart, and skeletal muscles: Protective effects of antioxidants. J Pharmacol Exp Ther 298: 737–743 [PubMed] [Google Scholar]
- Mao GD, Poznansky MJ 1992. Electron spin resonance study on the permeability of superoxide radicals in lipid bilayers and biological membranes. FEBS Lett 305: 233–236 [DOI] [PubMed] [Google Scholar]
- Martin SA, McCabe N, Mullarkey M, Cummins R, Burgess DJ, Nakabeppu Y, Oka S, Kay E, Lord CJ, Ashworth A 2010. DNA polymerases as potential therapeutic targets for cancers deficient in the DNA mismatch repair proteins MSH2 or MLH1. Cancer Cell 17: 235–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason PA, Matheson EC, Hall AG, Lightowlers RN 2003. Mismatch repair activity in mammalian mitochondria. Nucleic Acids Res 31: 1052–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendillo ML, Hargreaves VV, Jamison JW, Mo AO, Li S, Putnam CD, Woods VL Jr, Kolodner RD 2009. A conserved MutS homolog connector domain interface interacts with MutL homologs. Proc Natl Acad Sci 106: 22223–22228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mileshina D, Koulintchenko M, Konstantinov Y, Dietrich A 2011. Transfection of plant mitochondria and in organello gene integration. Nucleic Acids Res 39: e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mita S, Monnat RJ Jr, Loeb LA 1988. Resistance of HeLa cell mitochondrial DNA to mutagenesis by chemical carcinogens. Cancer Res 48: 4578–4583 [PubMed] [Google Scholar]
- Mitsuishi M, Miyashita K, Muraki A, Itoh H 2008. Angiotensin II reduces mitochondrial content in skeletal muscle and affects glycemic control. Diabetes 58: 710–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyaki M, Yatagai K, Ono T 1977. Strand breaks of mammalian mitochondrial DNA induced by carcinogens. Chem Biol Interact 17: 321–329 [DOI] [PubMed] [Google Scholar]
- Modrich P 2006. Mechanisms in eukaryotic mismatch repair. J Biol Chem 281: 30305–30309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel F, Renoux M, Lachaume P, Alziari S 2008. Bleomycin-induced double-strand breaks in mitochondrial DNA of Drosophila cells are repaired. Mutat Res 637: 111–117 [DOI] [PubMed] [Google Scholar]
- Muller FL, Liu Y, Van Remmen H 2004. Complex III releases superoxide to both sides of the inner mitochondrial membrane. J Biol Chem 279: 49064–49073 [DOI] [PubMed] [Google Scholar]
- Murphy MP 2009. How mitochondria produce reactive oxygen species. Biochem J 417: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers KA, Saffhill R, O’Connor PJ 1988. Repair of alkylated purines in the hepatic DNA of mitochondria and nuclei in the rat. Carcinogenesis 9: 285–292 [DOI] [PubMed] [Google Scholar]
- Nair J, Strand S, Frank N, Knauft J, Wesch H, Galle PR, Bartsch H 2005. Apoptosis and age-dependant induction of nuclear and mitochondrial etheno-DNA adducts in Long-Evans Cinnamon (LEC) rats: Enhanced DNA damage by dietary curcumin upon copper accumulation. Carcinogenesis 26: 1307–1315 [DOI] [PubMed] [Google Scholar]
- Nakabeppu Y 2001. Molecular genetics and structural biology of human MutT homolog, MTH1. Mutat Res 477: 59–70 [DOI] [PubMed] [Google Scholar]
- Neubert D, Hopfenmuller W, Fuchs G 1981. Manifestation of carcinogenesis as a stochastic process on the basis of an altered mitochondrial genome. Arch Toxicol 48: 89–125 [DOI] [PubMed] [Google Scholar]
- Niranjan BG, Bhat NK, Avadhani NG 1982. Preferential attack of mitochondrial DNA by aflatoxin B1 during hepatocarcinogenesis. Science 215: 73–75 [DOI] [PubMed] [Google Scholar]
- Noll T, Wissemann P, Mertens S, Krutzfeldt A, Spahr R, Piper HM 1990. Hypoxia tolerance of coronary endothelial cells. Adv Exp Med Biol 277: 467–476 [DOI] [PubMed] [Google Scholar]
- Ohtsubo T, Nishioka K, Imaiso Y, Iwai S, Shimokawa H, Oda H, Fujiwara T, Nakabeppu Y 2000. Identification of human MutY homolog (hMYH) as a repair enzyme for 2-hydroxyadenine in DNA and detection of multiple forms of hMYH located in nuclei and mitochondria. Nucleic Acids Res 28: 1355–1364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojala D, Montoya J, Attardi G 1981. tRNA punctuation model of RNA processing in human mitochondria. Nature 290: 470–474 [DOI] [PubMed] [Google Scholar]
- Pellegrini M, Asin-Cayuela J, Erdjument-Bromage H, Tempst P, Larsson NG, Gustafsson CM 2009. MTERF2 is a nucleoid component in mammalian mitochondria. Biochim Biophys Acta 1787: 296–302 [DOI] [PubMed] [Google Scholar]
- Pettepher CC, LeDoux SP, Bohr VA, Wilson GL 1991. Repair of alkali-labile sites within the mitochondrial DNA of RINr 38 cells after exposure to the nitrosourea streptozotocin. J Biol Chem 266: 3113–3117 [PubMed] [Google Scholar]
- Pirsel M, Bohr VA 1993. Methyl methanesulfonate adduct formation and repair in the DHFR gene and in mitochondrial DNA in hamster cells. Carcinogenesis 14: 2105–2108 [DOI] [PubMed] [Google Scholar]
- Pohjoismaki JL, Goffart S, Tyynismaa H, Willcox S, Ide T, Kang D, Suomalainen A, Karhunen PJ, Griffith JD, Holt IJ, et al. 2009. Human heart mitochondrial DNA is organized in complex catenated networks containing abundant four-way junctions and replication forks. J Biol Chem 284: 21446–21457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohjoismaki JL, Goffart S, Taylor RW, Turnbull DM, Suomalainen A, Jacobs HT, Karhunen PJ 2010. Developmental and pathological changes in the human cardiac muscle mitochondrial DNA organization, replication and copy number. PLoS ONE 5: e10426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puebla-Osorio N, Lacey DB, Alt FW, Zhu C 2006. Early embryonic lethality due to targeted inactivation of DNA ligase III. Mol Cell Biol 26: 3935–3941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pursell ZF, McDonald JT, Mathews CK, Kunkel TA 2008. Trace amounts of 8-oxo-dGTP in mitochondrial dNTP pools reduce DNA polymerase γ replication fidelity. Nucleic Acids Res 36: 2174–2181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebelo AP, Williams SL, Moraes CT 2009. In vivo methylation of mtDNA reveals the dynamics of protein-mtDNA interactions. Nucleic Acids Res 37: 6701–6715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter C, Park JW, Ames BN 1988. Normal oxidative damage to mitochondrial and nuclear DNA is extensive. Proc Natl Acad Sci 85: 6465–6467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringvoll J, Moen MN, Nordstrand LM, Meira LB, Pang B, Bekkelund A, Dedon PC, Bjelland S, Samson LD, Falnes PO, et al. 2008. AlkB homologue 2-mediated repair of ethenoadenine lesions in mammalian DNA. Cancer Res 68: 4142–4149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roginskaya M, Bernhard WA, Marion RT, Razskazovskiy Y 2005. The release of 5-methylene-2-furanone from irradiated DNA catalyzed by cationic polyamines and divalent metal cations. Radiat Res 163: 85–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi SC, Gorman N, Wetterhahn KE 1988. Mitochondrial reduction of the carcinogen chromate: Formation of chromium(V). Chem Res Toxicol 1: 101–107 [DOI] [PubMed] [Google Scholar]
- Rothfuss O, Gasser T, Patenge N 2010. Analysis of differential DNA damage in the mitochondrial genome employing a semi-long run real-time PCR approach. Nucleic Acids Res 38: e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotig A, Poulton J 2009. Genetic causes of mitochondrial DNA depletion in humans. Biochim Biophys Acta 1792: 1103–1108 [DOI] [PubMed] [Google Scholar]
- Rowley DA, Halliwell B 1983. DNA damage by superoxide-generating systems in relation to the mechanism of action of the anti-tumour antibiotic adriamycin. Biochim Biophys Acta 761: 86–93 [DOI] [PubMed] [Google Scholar]
- Rydberg B, Lindahl T 1982. Nonenzymatic methylation of DNA by the intracellular methyl group donor S-adenosyl-l-methionine is a potentially mutagenic reaction. EMBO J 1: 211–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryoji M, Katayama H, Fusamae H, Matsuda A, Sakai F, Utano H 1996. Repair of DNA damage in a mitochondrial lysate of Xenopus laevis oocytes. Nucleic Acids Res 24: 4057–4062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar JJ, Van Houten B 1997. Preferential mitochondrial DNA injury caused by glucose oxidase as a steady generator of hydrogen peroxide in human fibroblasts. Mutat Res 385: 139–149 [DOI] [PubMed] [Google Scholar]
- Satoh MS, Huh N, Rajewsky MF, Kuroki T 1988. Enzymatic removal of O6-ethylguanine from mitochondrial DNA in rat tissues exposed to N-ethyl-N-nitrosourea in vivo. J Biol Chem 263: 6854–6856 [PubMed] [Google Scholar]
- Shokolenko I, Venediktova N, Bochkareva A, Wilson GL, Alexeyev MF 2009. Oxidative stress induces degradation of mitochondrial DNA. Nucleic Acids Res 37: 2539–2548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simsek D, Furda A, Gao Y, Artus J, Brunet E, Hadjantonakis AK, Van Houten B, Shuman S, McKinnon PJ, Jasin M 2011. Crucial role for DNA ligase III in mitochondria but not in Xrcc1-dependent repair. Nature 471: 245–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava S, Moraes CT 2005. Double-strand breaks of mouse muscle mtDNA promote large deletions similar to multiple mtDNA deletions in humans. Hum Mol Genet 14: 893–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Pierre J, Buckingham JA, Roebuck SJ, Brand MD 2002. Topology of superoxide production from different sites in the mitochondrial electron transport chain. J Biol Chem 277: 44784–44790 [DOI] [PubMed] [Google Scholar]
- Suliman HB, Carraway MS, Piantadosi CA 2003. Postlipopolysaccharide oxidative damage of mitochondrial DNA. Am J Respir Crit Care Med 167: 570–579 [DOI] [PubMed] [Google Scholar]
- Suter M, Richter C 1999. Fragmented mitochondrial DNA is the predominant carrier of oxidized DNA bases. Biochemistry 38: 459–464 [DOI] [PubMed] [Google Scholar]
- Svilar D, Goellner EM, Almeida KH, Sobol RW 2011. Base excision repair and lesion-dependent subpathways for repair of oxidative DNA damage. Antioxid Redox Signal 14: 2491–2507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykora P, Croteau DL, Bohr VA, Wilson DM 3rd 2011. Aprataxin localizes to mitochondria and preserves mitochondrial function. Proc Natl Acad Sci 108: 7437–7442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczesny B, Tann AW, Longley MJ, Copeland WC, Mitra S 2008. Long patch base excision repair in mammalian mitochondrial genomes. J Biol Chem 283: 26349–26356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahbaz N, Subedi S, Weinfeld M 2012. Role of polynucleotide kinase/phosphatase in mitochondrial DNA repair. Nucleic Acids Res 40: 3484–3495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi MA, Asada K 1983. Superoxide anion permeability of phospholipid membranes and chloroplast thylakoids. Arch Biochem Biophys 226: 558–566 [DOI] [PubMed] [Google Scholar]
- Takahashi M, Teranishi M, Ishida H, Kawasaki J, Takeuchi A, Yamaya T, Watanabe M, Makino A, Hidema J 2011. Cyclobutane pyrimidine dimer (CPD) photolyase repairs ultraviolet-B-induced CPDs in rice chloroplast and mitochondrial DNA. Plant J 66: 433–442 [DOI] [PubMed] [Google Scholar]
- Takao M, Aburatani H, Kobayashi K, Yasui A 1998. Mitochondrial targeting of human DNA glycosylases for repair of oxidative DNA damage. Nucleic Acids Res 26: 2917–2922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tann AW, Boldogh I, Meiss G, Qian W, Van Houten B, Mitra S, Szczesny B 2011. Apoptosis induced by persistent single-strand breaks in the mitochondrial genome: Critical role of EXOG (5′ EXO/endonuclease) in their repair. J Biol Chem 286: 31975–31983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tano K, Shiota S, Collier J, Foote RS, Mitra S 1990. Isolation and structural characterization of a cDNA clone encoding the human DNA repair protein for O6-alkylguanine. Proc Natl Acad Sci 87: 686–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatarenkov A, Avise JC 2007. Rapid concerted evolution in animal mitochondrial DNA. Proc Biol Sci 274: 1795–1798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas RD, Roy D 2001. Stilbene estrogen produces higher levels of mitochondrial DNA adducts than nuclear DNA adducts in the target organ of cancer (liver) of male Sprague Dawley rats. Oncol Rep 8: 1035–1038 [DOI] [PubMed] [Google Scholar]
- Thorslund T, Sunesen M, Bohr VA, Stevnsner T 2002. Repair of 8-oxoG is slower in endogenous nuclear genes than in mitochondrial DNA and is without strand bias. DNA Repair (Amst) 1: 261–273 [DOI] [PubMed] [Google Scholar]
- Tomasi A, Albano E, Banni S, Botti B, Corongiu F, Dessi MA, Iannone A, Vannini V, Dianzani MU 1987. Free-radical metabolism of carbon tetrachloride in rat liver mitochondria. A study of the mechanism of activation. Biochem J 246: 313–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomkinson AE, Bonk RT, Linn S 1988. Mitochondrial endonuclease activities specific for apurinic/apyrimidinic sites in DNA from mouse cells. J Biol Chem 263: 12532–12537 [PubMed] [Google Scholar]
- Tomkinson AE, Bonk RT, Kim J, Bartfeld N, Linn S 1990. Mammalian mitochondrial endonuclease activities specific for ultraviolet-irradiated DNA. Nucleic Acids Res 18: 929–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC 2005. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: A dawn for evolutionary medicine. Annu Rev Genet 39: 359–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC, Singh G, Lott MT, Hodge JA, Schurr TG, Lezza AM, Elsas LJ 2nd, Nikoskelainen EK 1988. Mitochondrial DNA mutation associated with Leber’s hereditary optic neuropathy. Science 242: 1427–1430 [DOI] [PubMed] [Google Scholar]
- Wang D, Kreutzer DA, Essigmann JM 1998. Mutagenicity and repair of oxidative DNA damage: Insights from studies using defined lesions. Mutat Res 400: 99–115 [DOI] [PubMed] [Google Scholar]
- Wunderlich V, Schutt M, Bottger M, Graffi A 1970. Preferential alkylation of mitochondrial deoxyribonucleic acid by N-methyl-N-nitrosourea. Biochem J 118: 99–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W, Samson L 1993. In vivo evidence for endogenous DNA alkylation damage as a source of spontaneous mutation in eukaryotic cells. Proc Natl Acad Sci 90: 2117–2121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu YJ, DeMott MS, Hwang JT, Greenberg MM, Demple B 2003. Action of human apurinic endonuclease (Ape1) on C1′-oxidized deoxyribose damage in DNA. DNA Repair (Amst) 2: 175–185 [DOI] [PubMed] [Google Scholar]
- Yakes FM, Van Houten B 1997. Mitochondrial DNA damage is more extensive and persists longer than nuclear DNA damage in human cells following oxidative stress. Proc Natl Acad Sci 94: 514–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Schumaker LM, Egorin MJ, Zuhowski EG, Guo Z, Cullen KJ 2006. Cisplatin preferentially binds mitochondrial DNA and voltage-dependent anion channel protein in the mitochondrial membrane of head and neck squamous cell carcinoma: Possible role in apoptosis. Clin Cancer Res 12: 5817–5825 [DOI] [PubMed] [Google Scholar]
- Yasui A, Yajima H, Kobayashi T, Eker AP, Oikawa A 1992. Mitochondrial DNA repair by photolyase. Mutat Res 273: 231–236 [DOI] [PubMed] [Google Scholar]
- Zhang H, Barcelo JM, Lee B, Kohlhagen G, Zimonjic DB, Popescu NC, Pommier Y 2001. Human mitochondrial topoisomerase I. Proc Natl Acad Sci 98: 10608–10613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Zhou M, Guo Z, Lu H, Qian L, Dai H, Qiu J, Yakubovskaya E, Bogenhagen DF, Demple B, et al. 2008. Human DNA2 is a mitochondrial nuclease/helicase for efficient processing of DNA replication and repair intermediates. Mol Cell 32: 325–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Liu L, Chen J 2010. Mitochondrial DNA heteroplasmy in Candida glabrata after mitochondrial transformation. Eukaryot Cell 9: 806–814 [DOI] [PMC free article] [PubMed] [Google Scholar]