Abstract
Nonhomologous end joining (NHEJ) refers to a set of genome maintenance pathways in which two DNA double-strand break (DSB) ends are (re)joined by apposition, processing, and ligation without the use of extended homology to guide repair. Canonical NHEJ (c-NHEJ) is a well-defined pathway with clear roles in protecting the integrity of chromosomes when DSBs arise. Recent advances have revealed much about the identity, structure, and function of c-NHEJ proteins, but many questions exist regarding their concerted action in the context of chromatin. Alternative NHEJ (alt-NHEJ) refers to more recently described mechanism(s) that repair DSBs in less-efficient backup reactions. There is great interest in defining alt-NHEJ more precisely, including its regulation relative to c-NHEJ, in light of evidence that alt-NHEJ can execute chromosome rearrangements. Progress toward these goals is reviewed.
Canonical nonhomologous end joining (NHEJ) uses Ku, DNA ligase IV, and associated factors to fix certain double-strand breaks in DNA. Alternative NHEJ can repair double-strand breaks in less-efficient backup reactions.
DNA double-strand breaks (DSBs) are serious lesions that threaten a loss of chromosomal content. Repair of DSBs is particularly challenging because, unlike all other lesions, the DNA substrate is inherently bimolecular. Bringing two DNA molecules together is also dangerous because local mutations and chromosome rearrangements can arise if ends are inappropriately coupled.
The cell has two general strategies for repairing DSBs. Homologous recombination (HR) refers to mechanisms in which an intact homologous donor duplex is used to guide DNA synthesis across the DSB gap. HR is considered in Rothstein (2013) and is discussed here only in the context of influences on the relative utilization of different DSB repair pathways.
Nonhomologous end joining (NHEJ) is defined as repair in which two DSB ends are joined by direct ligation. The resulting joints are characterized by little (less than ∼10 bp) or no homology between the joined ends that could have been used to guide repair, which, when it exists, is referred to as “microhomology.” NHEJ is thus recognized as having a high potential for error. The term “NHEJ” was for a long time used to refer to a specific DNA repair pathway characterized by its dependence on Ku, DNA ligase IV, and associated factors (Table 1), referred to here as “canonical NHEJ” (c-NHEJ). However, some homology-independent repair occurs in the absence of these proteins, variably referred to as “backup NHEJ,” “microhomology-mediated end joining” (MMEJ), or “alternative NHEJ” (alt-NHEJ; the main terminology used here). This article describes these various NHEJ pathways with an emphasis on new findings in eukaryotic systems since 2005.
Table 1.
Major eukaryotic NHEJ proteins
| Category | Protein | c-NHEJ | alt-NHEJ | Description |
|---|---|---|---|---|
| DNA-PK | Ku70 | + | − | DSB recognition, dRP lyase |
| Ku80 | + | − | Ku70 partner protein | |
| DNA-PKcs | + | − | DSB-dependent protein kinase | |
| Ligase | Lig4 | + | − | Ligase catalytic subunit |
| XRCC4 | + | − | Lig4 structural scaffold | |
| XLF | + | − | Lig4 structural scaffold | |
| Lig1 | − | +/− | Ligase catalytic subunit | |
| Lig3 | − | + | Ligase catalytic subunit | |
| XRCC1 | − | +/− | Lig3 structural scaffold | |
| MRN | Mre11 | + | + | Dimerization, nuclease |
| Rad50 | + | + | Regulatory ATPase | |
| Nbs1 | + | + | Protein recruitment | |
| PARP | PARP-1 | +/− | + | Poly-ADP ribose polymerase |
| PARP-3 | + | Poly-ADP ribose polymerase | ||
| Polymerase | Pol μ | + | Gap filling | |
| Pol λ | + | Gap filling, dRP lyase | ||
| Nuclease | Artemis | + | Endo/5′ exonuclease | |
| Tdp1 | + | 3′ phosphoesterase | ||
| APLF | + | Endo/3′ exonuclease | ||
| CtIP | + | Supports 5′ resection | ||
| Other | PNKP | + | 5′ kinase, 3′ phosphatase | |
| Aprataxin | + | 5′-AMP intermediate removal |
A summary of NHEJ proteins discussed in the text, with a focus on structural and enzymatic components. This list is not exhaustive because other proteins have been suggested to influence NHEJ. (+) Described role in c-NHEJ or alt-NHEJ; (−) not involved by definitions of c-NHEJ and alt-NHEJ; (+/−) conflicting reports or uncertain role.
CANONICAL NHEJ CORE PROTEINS
Ku and DNA-PKcs
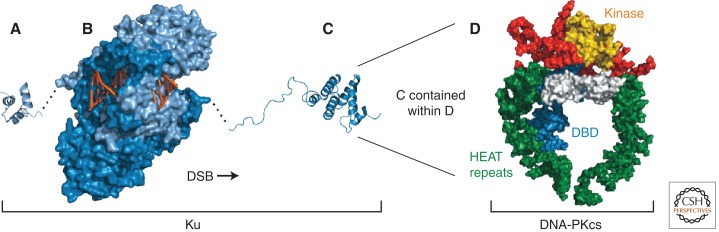
The prototypical c-NHEJ protein is Ku, a heterodimer of two related proteins, Ku70 and Ku80. Ku is a DSB-specific end-binding protein by virtue of the oriented threading of a DNA end into a hole in the protein dimer that allows its further translocation onto the DNA duplex (Fig. 1) (Walker et al. 2001). This configuration means that degradation of Ku is required for its removal from DNA following, and possibly during, repair, which is promoted by the ubiquitylation of Ku80 by the E3 ligase RNF8 (Postow et al. 2008; Feng and Chen 2012).
Figure 1.
Ku and DNA-PKcs. (A) Structural representation of the CTD of human Ku70 (PDB 1JJR) (Zhang et al. 2001). (B) The human Ku70–Ku80 heterodimer bound to DNA (PDB 1JEY) (Walker et al. 2001). (C) The CTD of human Ku80 (PDB 1RW2) (Zhang et al. 2004). (Dashed black lines) Connections between structures. (Light blue) Ku70; (blue) Ku80; (orange) DNA. (D) Low-resolution structure of human DNA-PKcs plus the Ku80 CTD (PDB 3KGV) (Sibanda et al. 2010). (Yellow) Kinase domain, (green) HEAT repeats, (white) “brow,” (blue) putative DBD, and (red) remainder.
Ku is one part of the DNA-dependent protein kinase (DNA-PK). The DNA-PK catalytic subunit (DNA-PKcs), related to the Ataxia-Telangiectasia Mutated (ATM) checkpoint kinase (Lempiainen and Halazonetis 2009), is also a core c-NHEJ protein, although there are situations in which the requirement for DNA-PKcs is less stringent than for Ku (Gu et al. 2000; Gapud and Sleckman 2011). For example, DNA-PKcs deficiency does not retard growth as in Ku-deficient mice (Gu et al. 1997). When activated, DNA-PKcs extensively phosphorylates itself, other c-NHEJ proteins, and other targets (Wang et al. 2004; Douglas et al. 2005; Yu et al. 2008).
Ku interacts with DNA-PKcs mainly via the Ku80 carboxy-terminal domain (CTD) (Falck et al. 2005; Hammel et al. 2010b), attached to the Ku DNA-binding domain (DBD) by a flexible tether (Fig. 1C). Low-resolution crystallographic analysis of DNA-PKcs plus the Ku80 CTD revealed a structure mainly composed of Huntingtin-Elongation-A-TOR (HEAT) repeats with an overall clamp-like fold (Fig. 1D) (Sibanda et al. 2010) generally consistent with globular shapes revealed by electron microscopy (EM) and small-angle X-ray scattering (SAXS) (Hammel et al. 2010b; Morris et al. 2011). The precise meaning of this fold is unknown, but the regulated entry and exit of duplex DNA into the clamp might be important.
Ku is the main DNA-binding subunit of DNA-PK, but DNA-PKcs itself also interacts directly with DNA ends (Yaneva et al. 1997; Hammarsten et al. 2000). Many factors influence this binding and subsequent kinase activation, including DSB base content and overhang polarity (Pawelczak et al. 2005). Unwound DSB structures can also stimulate DNA-PKcs activity in vitro, suggesting that DNA-PKcs might splay ends during binding (Hammarsten et al. 2000; Pawelczak and Turchi 2008). Ku bound to DNA is nonetheless required for DNA-PK activation in vivo. Ku undergoes conformational changes on DNA that alter the accessibility of the Ku80 CTD (Lehman et al. 2008) so that the CTD must be tethered to the Ku DBD to fully support DNA-PKcs activation (Bennett et al. 2012).
DNA Ligase IV
Lig4 is the DNA ligase required for, and specific to, c-NHEJ. It catalyzes the same ATP-dependent transfer of phosphate bonds that results in strand ligation in all eukaryotic DNA repair (Ellenberger and Tomkinson 2008). However, unusual biochemical properties of Lig4 modify this core reaction in ways important to c-NHEJ. Lig4 was the only ligase with the mechanistic flexibility to ligate one strand independently of another (Ma et al. 2004) or incompatible DSB ends as well as gaps of several nucleotides (Gu et al. 2007a), properties consistent with the joining of the wide variety of DSB structures relevant to c-NHEJ in vivo.
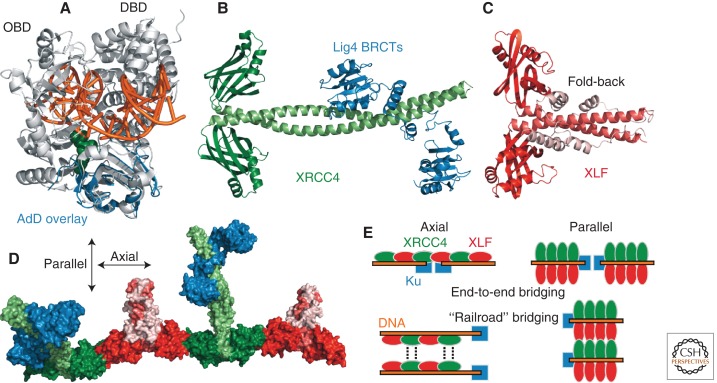
Structurally, little is known about Lig4, including how mechanistic flexibility might be realized at the protein level. One possibility is that the Lig4 catalytic domain is structurally as well as functionally flexible (Perry et al. 2010), although SAXS experiments suggest it may, in fact, have an extended shape with limited flexibility (Ochi et al. 2012). Lig4 shares a catalytic domain structure with DNA ligase I (Lig1) that allows the latter protein to completely encircle DNA during catalysis (Fig. 2A) (Pascal et al. 2004). The simplest assumption is that this architecture will be recapitulated in Lig4, an idea supported by the similar folding of the isolated Lig4 adenylation domain compared with Lig1 (Fig. 2A) (Ochi et al. 2012). However, the Lig4 DBD is a weakly conserved subdomain, so it is possible that an extended shape is maintained throughout the c-NHEJ reaction.
Figure 2.
DNA ligase IV assembly. (A) The adenylation domain (AdD) of Lig4 (blue, PDB 3VNN) (Ochi et al. 2012) is superimposed on a structural representation of Lig1 bound to a DNA nick (light gray, PDB 1X9N) (Pascal et al. 2004) as a surrogate model of how Lig4 might bind DNA. (OBD) Oligonucleotide/oligosaccharide binding domain; (green) 5′ AMP; (orange) DNA. (B) The human XRCC4 homodimer bound to the Lig4 tandem BRCT repeat region (PDB 3II6) (Wu et al. 2009). (C) Human XLF homodimer (PDB 2QM4) (Li et al. 2008b). (D) Surface representation of the XRCC4–XLF axial filament with a bound Lig4 BRCT region, created by superimposing PDB 3II6 onto PDB 3RWR (Andres and Junop 2011). (Blue) Lig4; (shades of green) XRCC4; (shades of red) XLF. (E) Idealized models of DNA engagement and end bridging by XRCC4–XLF multimers, colored the same as in D. “Axial” and “parallel” refer to the orientation of XRCC4–XLF interactions that drive the assembly.
XRCC4 and XLF
XRCC4 is a nonenzymatic Lig4 partner protein, with the two copurifying as a complex. XRCC4 has a homodimeric structure with paired globular head domains, an elongated coiled-coil (Fig. 2B), and a structurally ill-defined and less conserved CTD that nonetheless has strong influences on c-NHEJ in vivo (Koch et al. 2004; Palmbos et al. 2005). The Lig4–XRCC4 interaction is mediated by tandem BRCA1 carboxy-terminal (BRCT) repeats at the Lig4 carboxyl terminus, and especially the inter-BRCT linker, that intimately wrap around the XRCC4 coiled-coil in a clamp-like fashion (Fig. 2B) (Dore et al. 2006; Wu et al. 2009). The catalytic and XRCC4-binding domains account for nearly all of Lig4 except for a small region between them that carries a basic patch implicated in DNA binding (Hammel et al. 2011).
An XRCC4-like factor (XLF, also called Cernunnos) proved to be the unidentified c-NHEJ factor inferred from the study of biochemical extracts and human patients (Ahnesorg et al. 2006; Buck et al. 2006). XLF shares a similar homodimeric architecture with XRCC4, although in XLF the coiled-coil region is disrupted in a manner that supports a fold-back of the XLF carboxyl terminus toward its amino-terminal globular head (Fig. 2C) (Andres et al. 2007; Li et al. 2008b). In part because of this difference, XLF alone does not support stable Lig4 binding (Deshpande and Wilson 2007; Lu et al. 2007). Lig4–XRCC4 and XLF interact via their head domains (Ahnesorg et al. 2006; Deshpande and Wilson 2007), but this is less stable than the Lig4–XRCC4 interaction such that XLF is typically purified separately. Like XRCC4, XLF has no enzymatic activity but does stimulate Lig4 activity and readenylation (Riballo et al. 2009), especially at incompatible DSBs that might require XLF to align the DNA (Gu et al. 2007b; Tsai et al. 2007).
XRCC4 and XLF together form long superhelical filaments (Fig. 2D) (Hammel et al. 2010a; Andres and Junop 2011; Ropars et al. 2011; Wu et al. 2011). Further contacts support interactions between parallel filaments (Fig. 2E) (Hammel et al. 2011). Binding of the Lig4 BRCT region likely influences the nature and extent of these various modes of XRCC4–XLF multimerization, in part by preventing the XRCC4 carboxyl terminus (not present in Fig. 2D) from interacting with the XRCC4–XLF interface (Hammel et al. 2010a; Ochi et al. 2012). The importance of XRCC4–XLF higher-order structures appears to be DNA binding, which might include collinear protein and DNA filaments as well as a channel whose base is the XRCC4/XLF head domains and sides are the coiled-coil stalks, with DNA binding perpendicularly to the XRCC4–XLF filament (Fig. 2E) (Hammel et al. 2011; Andres et al. 2012).
The assembly models in Figure 2E require substantial validation. No XRCC4–XLF-DNA cocrystal has yet been described to confirm proposed modes of DNA engagement. More importantly, a major gap in current knowledge is the stoichiometry and geometry of Lig4–XRCC4–XLF at DSBs in vivo. This information is needed to support models of ligase assembly and to give insight into whether c-NHEJ likely entails coordinated or sequential ligation of two strands. Another challenge is that XLF deficiency confers a less severe phenotype than XRCC4 (Li et al. 2008a), including a functional redundancy of XLF with the structurally unrelated proteins ATM and 53BP1 (Zha et al. 2011; Oksenych et al. 2012). This difference is not easily rationalized by models in which ligation strictly depends on a precise equimolar XRCC4–XLF structural coassembly. XRCC4, unlike XLF, is required for stability and therefore normal levels of Lig4 protein (Grawunder et al. 1997; Riballo et al. 2009) as an alternative explanation for their differential phenotypes.
Mre11–Rad50–Nbs1
Unlike Ku and Lig4, the MRN complex of proteins formed by Mre11, Rad50, and Nbs1 is involved in most aspects of DSB repair, including ATM-dependent checkpoint signaling. MRN was first identified as a c-NHEJ factor in budding yeast, where it is as required as Ku or Lig4 (Milne et al. 1996). In contrast, MRN is not required for NHEJ in fission yeast (Manolis et al. 2001), and early efforts gave conflicting but often nonsupportive observations regarding a role for MRN in vertebrate NHEJ (Yamaguchi-Iwai et al. 1999; Huang and Dynan 2002; Di Virgilio and Gautier 2005). However, a series of more recent studies using refined genetic tools has established a less penetrant but consistent contribution of MRN to some mammalian NHEJ (Deriano et al. 2009; Dinkelmann et al. 2009; Helmink et al. 2009; Rass et al. 2009; Xie et al. 2009). Conditional Mre11 loss in mouse B lymphocytes caused NHEJ deficiencies during immunoglobulin class switch recombination that could not be explained by impairment of ATM activation (Dinkelmann et al. 2009). In a distinct approach, siRNA-mediated depletion of Mre11 reduced end joining in a reporter assay in both XRCC4+/+ and XRCC4−/− cells, suggesting MRN roles in both c-NHEJ and alt-NHEJ (Xie et al. 2009). Such studies support a checkpoint-independent NHEJ role of MRN that might include structural stabilization of DSBs and/or end processing.
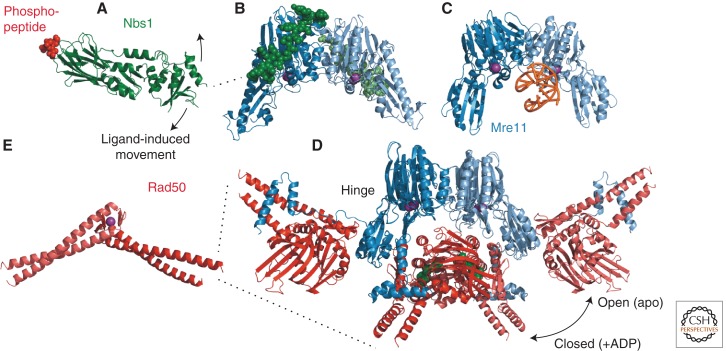
Much effort has been applied to the intricate structural biology of MRN (Fig. 3). Mre11 can be seen as the center of the complex, forming a homodimer capable of binding DNA (Fig. 3C) (Williams et al. 2008). Once bound, Mre11 is an Mn2+-dependent nuclease that supports both endonucleolytic and 3′–5′ exonucleolytic cleavages (Trujillo et al. 1998).
Figure 3.
Mre11–Rad50–Nbs1 (MRN) complex. (A) Fission yeast Nbs1 FHA domain bound to a Ctp1 phosphopeptide (PDB 3HUF) (Williams et al. 2009). (B) Fission yeast Mre11 globular domain bound to an Nbs1 internal peptide (PDB 4FBW) (Schiller et al. 2012). (C) Pyrococcus furiosus Mre11 globular domain bound to DNA (PDB 3DSD) (Williams et al. 2008). (D) Superimposed structures of the Thermotoga maritima Mre11 globular domain in the open (PDB 3QG5) (Lammens et al. 2011) and closed (PDB 3THO) (Mockel et al. 2012) conformations showing the large Rad50 domain movement induced by adenosine nucleotide binding. (E) P. furiosus Rad50 Zn hook motif (PDB 1L8D) (Hopfner et al. 2002). (Red) Ctp1 peptide; (green) Nbs1; (shades of blue) Mre11; (shades of red) Rad50; (green) ADP; (purple) Zn and Mn ions; (orange) DNA. (Dashed black lines) Connections between structures.
Rad50 is an ABC-family ATPase with a catalytic domain composed of four subdomains, two from each end of two different Rad50 molecules (Fig. 3D) (Hopfner et al. 2000). The intervening region is a long 50-nm coiled-coil, capped at its fold-back by a Zn-hook dimerization motif (Fig. 3E) (Hopfner et al. 2002). The Zn hook can tether distant DNA molecules bound by different MRN complexes (Moreno-Herrero et al. 2005), apparently essential for its stable binding to chromosomal DSBs (He et al. 2012).
Together, Mre11 and Rad50 form a unit with probably many dynamic states and functions. Mre11 binds to the Rad50 coiled-coil near the ATPase head via a carboxy-terminal helix–loop–helix motif on a flexible tether (Fig. 3D) (Lammens et al. 2011). Binding of ATP by Rad50 induces a large conformational change at this hinge from an open to a closed configuration with respect to the channel by which DNA gains access to Mre11 (Fig. 3D) (Mockel et al. 2012). This changes the disposition of the Rad50 coiled-coils and influences Mre11 nuclease activity, with the open and closed configurations promoting exo- and endonuclease activities, respectively (Majka et al. 2012).
Nbs1 binds to Mre11 via a small internal domain, extending the MRN globular core (Fig. 3B) (Schiller et al. 2012). At its amino terminus, Nbs1 has fused Forkhead Associated (FHA) and BRCT domains. A conformational change is propagated through this coupling when the FHA domain binds to threonine phosphopeptides in target proteins (Fig. 3A) (Williams et al. 2009). This change might propagate to the MRN globular head, or perhaps Nbs1 simply acts as a flexible tether for recruiting proteins (see below).
The most precise studies of the MRN c-NHEJ structure–function relationship come from budding yeast, where Rad50 ATPase activity is required for efficient NHEJ (Chen et al. 2005), as is the presence of a long Rad50 coiled-coil (Hohl et al. 2011). Zn-hook alterations gave a complex pattern of results where hook deletion had a more severe effect on c-NHEJ than cleaving away the hook in vivo (Hohl et al. 2011). All such manipulations are confounded by the potential for influences of one portion of MRN on another, including conformational transitions propagated along coiled-coils; thus it is not yet fully clear how MRN supports c-NHEJ.
END-PROCESSING ENZYMES
DSBs bear distinct structures such as varying overhang polarities and lengths as well as chemical modifications of nearby nucleotides. Although simple religation is possible with fully compatible overhangs, more highly damaged DSBs delay repair kinetics and challenge or eliminate the possibility of accurate rejoining. Excision and resynthesis of damaged nucleotides must occur as in all DNA repair (Table 1) but is complicated by the separation of the DSB termini.
DNA Polymerases
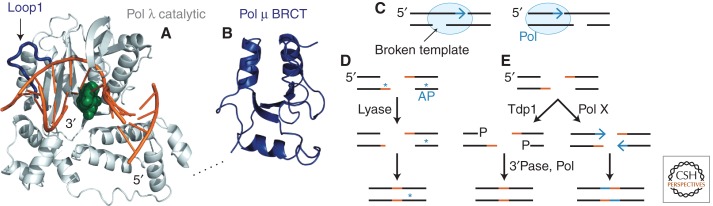
The yeast Pol X family DNA polymerase, Pol 4, is absolutely required for c-NHEJ events in which gaps must be filled on both strands of 3′-overhanging DSBs (Daley et al. 2005). This pattern shows the critical importance of DSB configuration, because the template for extending a 3′ overhang is the other side of the DSB, requiring a polymerase that can engage a disrupted template (Fig. 4C). Structural studies of mammalian Pol λ and Pol μ, themselves also c-NHEJ enzymes, provide insight into this function (Garcia-Diaz et al. 2005; Nick McElhinny et al. 2005; Moon et al. 2007; Ramsden and Asagoshi 2012). The catalytic domains of these Pol X polymerases are bipartite with binding pockets for the 5′ and 3′ termini of the broken strand whose gap is being filled (Fig. 4A). Loop 1 of the enzymes can be seen positioned near the point on the template strand that might be broken in a DSB (Fig. 4A), the same loop that replaces the template strand in terminal deoxynucleotidyl transferase (TdT) (Delarue et al. 2002). Biochemical analyses support the idea that Loop 1 and other Pol X residues promote catalysis by intrinsically stabilizing the weak association of primer terminus to a disrupted template (Juarez et al. 2006; Moon et al. 2007; Bebenek et al. 2010; Martin et al. 2012).
Figure 4.
Pol X polymerases. (A) Human Pol λ catalytic domain bound to a 1-base gap and incoming nucleotide (light gray; PDB 1XSN) (Garcia-Diaz et al. 2005). (Dark blue) Loop 1; (green) ddTTP; (orange) DNA. (B) Human Pol μ BRCT domain (PDB 2DUN, RIKEN Structural Genomics/Proteomics Initiative, similar to PDB 2HTF) (DeRose et al. 2007). (Dashed black line) Connection between structures. (C) Line diagrams depicting the different requirements imposed on a DNA polymerase (blue) filling a DSB gap (blue arrow) at 3′ (left panel) versus 5′ (right panel) overhangs with respect to placement of the template strand break. (D) dRP lyase activity as an example of end processing, illustrating different requirements for handling terminal versus internal base damage. AP, Abasic site. (E) The Tdp1 fidelity control mechanism that prevents Pol X-dependent insertional mutagenesis by transiently cleaving and blocking termini with a 3′ phosphate.
5′-Overhanging DSBs in yeast do not require Pol 4 (Daley et al. 2005). Pol λ and Pol μ are similarly required for only subsets of c-NHEJ in vivo, implying that other polymerases must act (Capp et al. 2006, 2007). Yeast replicative polymerases Pol 2 and Pol 3 have been suggested to participate (Chan et al. 2008; Tseng et al. 2008), and c-NHEJ translesion synthesis has been observed (Covo et al. 2009), although the role of bypass polymerases is not well explored.
Nucleases and Related Activities
The Mre11 nuclease does not have a clear role in trimming overhangs in yeast where the most precise correlation of DSB and joint structures can be performed (TE Wilson, unpubl.). However, it has been suggested to support microhomology pairing (Zhang and Paull 2005; Williams et al. 2008), and the mammalian Mre11 nuclease does promote DSB end processing that leads to nucleotide deletions (Fig. 5) (Xie et al. 2009). Mre11 nuclease deficiency confers a milder NHEJ defect than loss of MRN during class switch recombination, although the exact mode of end processing in the nuclease-dependent events is not known (Dinkelmann et al. 2009).
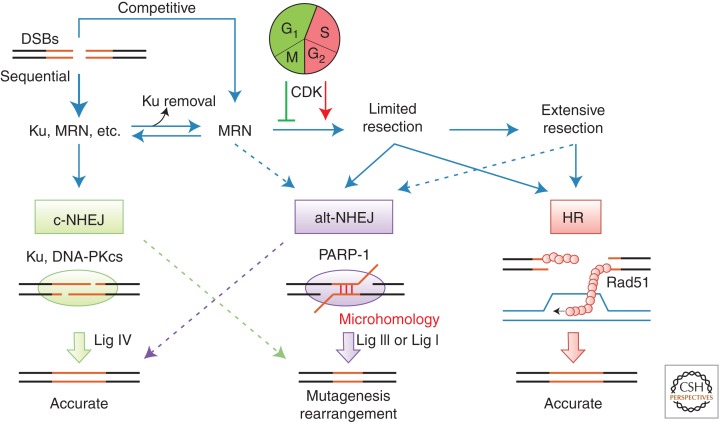
Figure 5.
Disposition of DSBs between repair pathways. Diagram illustrating the relationships between c-NHEJ, alt-NHEJ, and HR and the factors that influence the disposition of DSBs between these repair pathways. See text for further discussion.
The nuclease with the clearest requirement in c-NHEJ is Artemis, a vertebrate SNM1-family enzyme that has endonuclease as well as 3′-phosphoglycolate processing activities (Povirk et al. 2007; Kurosawa and Adachi 2010; Yan et al. 2010). A 5′-exonuclease activity has also been reported (Ma et al. 2002; Yannone et al. 2008), but exonuclease-free Artemis can be purified (Pawelczak and Turchi 2010). Artemis activity is tightly regulated by DNA-PKcs-mediated phosphorylation and remodeling (Goodarzi et al. 2006; Morris et al. 2011). Artemis was first identified as the enzyme that opens hairpin DSB termini during V(D)J recombination (Moshous et al. 2001; Ma et al. 2002). It is further critical for repairing the subset of DSBs induced by ionizing radiation (IR) that are both the slowest to repair and the most associated with cell survival (Woodbine et al. 2011), presumably by cleaving challenging but incompletely characterized damage configurations.
Three mammalian proteins form a trio of processing enzymes that use a common mechanism to engage the c-NHEJ complex (see below). Polynucleotide kinase 3′ phosphatase (PNKP) acts to generate ligatable 5′-phosphate/3′-hydroxyl termini (Coquelle et al. 2011; Garces et al. 2011). Aprataxin removes AMP groups from the 5′ termini of abortive ligation intermediates (Gong et al. 2011; Tumbale et al. 2011) and also has 3′-phosphoesterase activity (Takahashi et al. 2007). The aprataxin-PNKP-like factor (APLF; also called Xip1 and PALF) has endonuclease and 3′-exonuclease activity (Li et al. 2011). The importance of PNKP’s catalytic function to c-NHEJ ligation in vivo is clear (Chappell et al. 2002; Karimi-Busheri et al. 2007), although it is less certain how often processing of abortive ligation intermediates is required at DSBs (Daley et al. 2010).
A specific function in strand break repair is removal of 5′-deoxyribose phosphates (dRPs), a breakdown product of abasic sites. Pol X polymerases sometimes have the required dRP lyase activity, but the yeast Pol 4 lyase was not seen to be required for c-NHEJ, perhaps because of redundancy (Daley and Wilson 2008). Surprisingly, Ku itself was discovered to possess lyase activity via many different lysines capable of forming the catalytic Schiff’s base intermediate (Roberts et al. 2010). The Ku lyase also cleaves internal abasic sites in DSB termini but is restricted by an unclear mechanism to overhang positions that would interfere with ligation, given that some internal abasic sites can be incorporated into ligation products and resolved subsequently (Fig. 4D) (Strande et al. 2012).
Another surprising development is that some cleavage of DSB termini might be executed to promote joining fidelity rather than the necessity of ligation. The tyrosyl-DNA phosphodiesterase (Tdp1) is a general 3′ phosphoesterase capable of removing lesions such as 3′ phosphoglycolates (Zhou et al. 2009). Tdp1 leaves a terminal 3′ phosphate that must be removed for repair to continue. An interesting model developed in yeast proposes that Tdp1 removes intact bases from otherwise undamaged 5′-overhanging DSBs such that the new 3′-phosphate lesion prevents undesirable filling of the overhang and insertional mutagenesis (Fig. 4E) (Bahmed et al. 2010).
c-NHEJ ASSEMBLY AND EXECUTION
Local Histone Remodeling
DSBs in vivo arise in chromatin that might block c-NHEJ access to the DNA ends. In addition to a series of chromatin modifications spanning many kilobases from a damage site, discussed in Sirbu and Cortez (2013), there are two ways that histones at the DSB site itself might be managed during c-NHEJ. First, damage signaling might be required to remove local histones. This phenomenon has been documented in yeast, where the Ino80 and RSC chromatin remodeling complexes accumulate at DSBs and facilitate the removal and/or movement of nucleosomes and recruitment of Ku and MRN to DSBs (Tsukuda et al. 2005; Shim et al. 2007). However, the situation is likely more complicated than simple competition because Ku reciprocally promotes RSC binding to DSBs (Shim et al. 2005).
Alternatively, c-NHEJ proteins themselves could interact with histones in a competitive or cooperative fashion. Ku can bind in vitro to nucleosome-associated DNA ends by peeling up to 50 bp away from the histone octamer, in addition to displacing the linker histone H1 (Roberts and Ramsden 2007). The role of the highly abundant H1 protein is again likely more complicated, however, because DNA-PK can phosphorylate histone H1 to promote local release (Kysela et al. 2005), but H1 can paradoxically stimulate Lig4 activity (Rosidi et al. 2008). A further role of H1 in promoting c-NHEJ is mediated through its poly-ADP ribosylation (PAR) by the PAR polymerase (PARP) PARP-3 in coordination with APLF. In addition to its enzymatic activity, APLF has a zinc finger domain that binds PAR (Rulten et al. 2008). It further interacts with XRCC4 and Ku80 (Iles et al. 2007; Macrae et al. 2008). PARP-3-mediated PARylation of histone H1 at DSB sites thus recruits APLF, which then promotes the binding and retention of the Lig4 complex (Rulten et al. 2011).
End Synapsis
Not only do the core c-NHEJ complexes all bind DNA, but also all additionally support the apposition, or synapsis, of DSB ends. DNA-PKcs is suggested to mediate synapsis by facilitating DNA contacts across the break (DeFazio et al. 2002; Morris et al. 2011). Synapsis can also occur when two DSB ends bind two Mre11 monomers in an MRN globular head (Fig. 3C) (Williams et al. 2008). XRCC4–XLF filaments finally also support associations between DNA molecules (Fig. 2E) (Andres et al. 2012). In addition to these structural modes of end bridging, Lig4 and Pol X act enzymatically on a locally synapsed DSB in a manner that appears exclusive of other factors (Figs. 2 and 4). c-NHEJ must therefore be a dynamic assembly that allows DSB access to different proteins at different times.
Transient Protein Interactions
A series of weaker solution interactions between the c-NHEJ core complexes is important for creating a higher-order repair assembly at DSBs. Interactions in budding yeast include contacts between the Ku80 CTD and Lig4, where yeast lack DNA-PKcs that might otherwise occupy this Ku80 domain, and between the Nbs1 FHA and threonine phosphorylated XRCC4 CTD (Palmbos et al. 2005; Matsuzaki et al. 2008). Together, these interactions are necessary for accumulation of Lig4 at a chromosomal DSB (Palmbos et al. 2008). In mammals, similar types of interactions exist between Ku and XRCC4 (Mari et al. 2006), Ku and DNA ligase IV via its tandem BRCT domain (Costantini et al. 2007), and Ku and the CTD of XLF (Yano et al. 2011).
Other contacts recruit end-processing factors. c-NHEJ specificity of Pol X polymerases is supported by an amino-terminal BRCT domain that interacts with the core c-NHEJ enzymes (Fig. 4B) (DeRose et al. 2007; Mueller et al. 2008). Artemis interacts closely with DNA-PKcs (Kurosawa and Adachi 2010; Yan et al. 2010) as well as Lig4 (Malu et al. 2012). PNKP, aprataxin, and APLF all have amino-terminal FHA domains that contact constitutively phosphorylated threonine residues in the XRCC4 CTD (Clements et al. 2004; Koch et al. 2004; Iles et al. 2007), in a manner similar to yeast Nbs1 and XRCC4. The consequences of at least PNKP–XRCC4 interaction are more complicated than simple recruitment, however, because it actually represses enzyme activity (Mani et al. 2010), which might help orchestrate a stepwise reaction (Weinfeld et al. 2011).
c-NHEJ Reaction Progression
In yeast, chromatin immunoprecipitation (ChIP) at site-specific DSBs has probed c-NHEJ assembly and progression in vivo. Ku and MRN binding to DSBs occurs in the same time course as DSB formation, suggesting rapid binding, whereas Lig4 accumulates with an ∼10-min delay (Wu et al. 2008). Ku and MRN binding is largely independent of each other (Wu et al. 2008), although decreased MRN binding in the absence of Ku in one study suggests that Ku stabilizes MRN (Zhang et al. 2007). Binding of Lig4 and its cofactors requires Ku, whereas Ku binds largely independently of Lig4 (Wu et al. 2008), although again a stabilizing effect of especially XLF on Ku binding has been noted (Chen and Tomkinson 2011). Together, a picture emerges that yeast Ku and MRN bring the ligase to breaks but with an interdependence such that ligase components play structural roles before ligation.
In mammalian cells, the precise time resolution of laser microirradiation followed by live-cell imaging of fluorescently labeled proteins has revealed that Ku (Mari et al. 2006), DNA-PKcs (Uematsu et al. 2007), and XLF all appear at DSBs within seconds (Yano et al. 2008). XLF again acts in the early phases of assembly of the NHEJ machinery through interaction with Ku on DNA (Yano et al. 2008). An important observation from associated photobleaching experiments is that c-NHEJ proteins exchange rapidly at DSBs. Protein interactions refine exchange rates such that, for example, XRCC4 is not required for XLF recruitment to a DSB but does stabilize its binding (Yano and Chen 2008). In contrast to a sequential view of c-NHEJ assembly, these and related data suggest that c-NHEJ proteins bind DSBs independently, with protein interactions and exchanges dynamically modulating the assembly as repair proceeds, such that DSB structures help determine the proteins that stably assemble (Reynolds et al. 2012).
None of the above observations reveal the extent to which c-NHEJ repair steps are executed in an ordered fashion. The iterative processing model emphasizes the importance of independent operations on the two strands (Ma et al. 2005). An implication is that many enzymes have access to the DSB, with repair proceeding as dictated by their simultaneous binding equilibria and kinetics. This idea, supported by joint patterns at uniform DSB substrates in vitro and in vivo (Wilson and Lieber 1999; Ma et al. 2004), recognizes that without an information template to guide repair, it may not be possible to order a c-NHEJ reaction meaningfully beyond simple reaction thermodynamics.
DISPOSITION OF DSBs BETWEEN c-NHEJ AND HR
c-NHEJ use relative to HR must be regulated, a relationship often seen as a competition. However, loss of most HR proteins does not increase c-NHEJ efficiency (Karathanasis and Wilson 2002). The exception is mutants that abolish the redundant mechanisms that resect the 5′-terminated strand to create a substrate for Rad51 binding in HR, which do increase c-NHEJ yield (Ira et al. 2004; Symington and Gautier 2011). Loss of Ku, in turn, allows more rapid resection, whereas overexpression delays the onset of resection (Clerici et al. 2008). These observations emphasize that c-NHEJ and HR are sequential reactions, with c-NHEJ becoming impossible once DSB resection begins and HR being necessary only if c-NHEJ fails (Fig. 5). Disposition of DSBs between pathways is therefore determined by the rate of c-NHEJ relative to the onset of 5′ resection. This fact leads to the concept that c-NHEJ and HR cooperate rather than compete to drive the greatest likelihood of successful repair.
HR works best in late S and G2, when the sister chromatid is available for repair and c-NHEJ-incompatible one-ended DSBs arise at collapsed replication forks. Consistently, 5′ resection is under cell cycle control, with increased cyclin-dependent kinase (CDK) activity in S phase leading to potentiation of resection (Fig. 5) by mechanisms that include phosphorylation of proresection proteins such as CtIP (Ira et al. 2004; Aylon and Kupiec 2005; Huertas and Jackson 2009). Importantly, c-NHEJ is still possible in S/G2 and remains a predominant repair pathway for DSBs in mammalian G2 (Beucher et al. 2009; Karanam et al. 2012). Regulation simply dictates the window of opportunity afforded to c-NHEJ, effectively placing it on a cell cycle-dependent timer.
Notably, normal Ku levels inhibit resection in G1, when CDK activity is low, but not S/G2 (Clerici et al. 2008). This may reveal in part a less potent binding of Ku in S/G2 (Zhang et al. 2009). Ku disappears from unrepaired DSBs in a manner that depends on MRN (Wu et al. 2008). This disappearance may result from an unknown active Ku removal mechanism that is necessary for resection to begin (Zhang et al. 2009; Langerak et al. 2011; Shao et al. 2012) but could also be a simple consequence of MRN-dependent resection, where the dynamic on–off rate of Ku might be sufficient to allow the entry of activated resection enzymes in S/G2 (Fig. 5) (Mari et al. 2006).
The nature of the DSB itself is the major determinant of whether c-NHEJ will be completed before the onset of resection. At many DSBs, c-NHEJ is considerably faster than HR, with the pathways finishing in 30 min and >7 h in mammalian cells, respectively (Mao et al. 2008). However, complex DSB structures delay c-NHEJ, as revealed by the stratification of IR-induced DSBs into fast and slow repair phases (Shibata et al. 2011; Reynolds et al. 2012). The dependence of the slow NHEJ phase on Artemis shows the importance of limited end processing to complete repair in G1, whereas in S/G2, HR can take over at breaks that are too slow to repair by NHEJ (Woodbine et al. 2011). Notably, MRN is required for the slow NHEJ phase but dispensable for fast repair, again showing its more restricted NHEJ role in mammalian cells (Riballo et al. 2009). Heterochromatic location is another important factor that delays c-NHEJ and promotes HR (Goodarzi et al. 2008).
In vertebrates, DNA-PKcs helps to regulate DSB repair pathway choice (Cui et al. 2005). The DNA-PK assembly can block further processing of DSB ends, but activation of the kinase leads to conformational changes and remodeling of DNA-PKcs through autophosphorylation that can facilitate its disassembly and regulate the transition to subsequent c-NHEJ steps (Dobbs et al. 2010; Hammel et al. 2010b; Morris et al. 2011). The situation is more complicated, however, because DNA-PKcs enzymatic activity is required for it to inhibit HR, whereas specific DNA-PKcs autophosphorylation sites, in fact, impede NHEJ and promote HR (Neal and Meek 2011).
A final axis that controls NHEJ/HR balance is an antagonistic binding of 53BP1 and BRCA1 proteins to DSB regions. 53BP1 enrichment in IR-induced foci is most prominent in G0/G1, where it inhibits HR by blocking resection by an unclear mechanism, where BRCA1 binding in S/G2 tends to exclude 53BP1 and promote resection and HR (Bunting et al. 2010; Chapman et al. 2012).
ALTERNATIVE NHEJ AND PATHWAYS TO MUTATION
Consequences of c-NHEJ Dysfunction
As a conserved DNA repair pathway dependent on many dedicated proteins, it is not surprising that c-NHEJ deficiency is strongly deleterious. Impairment of c-NHEJ proteins confers profound cellular sensitivity to IR and other DSB-inducing agents. Mouse models and genetic diseases, including DNA-PKcs, XLF, and Artemis mutations in humans, further show that c-NHEJ dysfunction leads to immunodeficiency and B- and T-cell loss due to failed V(D)J recombination as well as varying degrees of dysmorphology, growth retardation, and developmental delay that correlate with the degree of c-NHEJ dysfunction (Frank et al. 2000; Moshous et al. 2001; Buck et al. 2006; Li et al. 2008a; Kerzendorfer and O’Driscoll 2009; van der Burg et al. 2009). One notable case is the ligase IV syndrome caused by hypomorphic mutations in Lig4 (Chistiakov 2010), which, like mouse models (Frank et al. 2000; Rucci et al. 2010), suggests that complete loss of this obligatory and specific c-NHEJ factor is incompatible with mammalian life. Apoptosis of developing neurons and progressive loss of hematopoietic stem cells are two specific features of hypomorphic mouse Lig4 mutation that reveal its importance to viability of different cell systems (Nijnik et al. 2007; Gatz et al. 2011).
A distinct consequence of c-NHEJ deficiency is an increase in DSB-associated chromosomal mutagenesis. Concomitant with inefficient V(D)J recombination is a predisposition to B- and T-cell lymphomas arising from persistent DSBs, in addition to medulloblastoma formation (Ferguson and Alt 2001; Li et al. 2008a; Jacobs et al. 2011). Interestingly, hypomorphic mutations in the Artemis carboxyl terminus promote aberrant V(D)J recombination, chromosomal translocation, and thymic lymphoma in a manner that is distinct from complete Artemis loss (Huang et al. 2009; Jacobs et al. 2011). It has also been observed experimentally that loss of c-NHEJ leads to an increase in chromosomal translocations between two independent DSBs outside of V(D)J recombination (Weinstock et al. 2007; Simsek and Jasin 2010). Thus, c-NHEJ is clearly a protector of genome integrity. Although c-NHEJ might create some local mutations through imprecise joining, this appears preferable to the persistence of lethal and dangerous DSB intermediates.
Alt-NHEJ
In every system studied, there is a residual amount of NHEJ observed when c-NHEJ is disabled, referred to as alt-NHEJ. Of interest is the increased propensity of alt-NHEJ to create mutations, because joints often harbor local deletions with relatively long stretches of microhomology, itself often called MMEJ. However, it is more meaningful to characterize NHEJ pathways by enzymology than by joint structure because the DNA outcomes of different pathways can be the same (Fig. 5). Limited precise religation of overhangs is observed even in the absence of Lig4, which is influenced by features such as overhang length in a manner that suggests an equilibrium between DSB and ligatable SSB states (Daley and Wilson 2005). Similarly, MMEJ events can arise by both c-NHEJ and alt-NHEJ, albeit to a greater extent with alt-NHEJ.
The enzymatic mechanisms of alt-NHEJ are less well defined than for c-NHEJ. It is not even clear how many distinct mechanisms alt-NHEJ encompasses. Like c-NHEJ, alt-NHEJ demands DSB synapsis. PARP-1 has been implicated in this function in alt-NHEJ, reminiscent of the role of DNA-PKcs in c-NHEJ (Audebert et al. 2008; Robert et al. 2009; Mansour et al. 2010). Not only is alt-NHEJ independent of DNA-PK, but also it is suppressed by Ku, implying a competition of factors for DSB ends that is typically won by c-NHEJ (Audebert et al. 2004; Wang et al. 2006). In addition to its possible synapsis function, PARP-1 may also serve as a platform for directly or indirectly recruiting alt-NHEJ repair factors (Table 1) (Audebert et al. 2004, 2006; Sallmyr et al. 2008; Della-Maria et al. 2011). PARP-1 action is pleiotropic, however, and has sometimes been seen to support NHEJ, including that mammalian SIRT6 can stimulate PARP-1, resulting in promotion of both NHEJ and HR (Mao et al. 2011).
As an obligatorily Lig4-independent pathway, other enzyme(s) must catalyze strand ligation in alt-NHEJ. Studies in mammalian cells have implicated DNA ligase III (Lig3) as the major alt-NHEJ ligase (Audebert et al. 2004; Wang et al. 2005; Sallmyr et al. 2008; Simsek et al. 2011; Chiruvella et al. 2012). As a cofactor of Lig3, XRCC1 is also suggested to be involved (Audebert et al. 2004; Saribasak et al. 2011), although more recent studies suggest that XRCC1 may be dispensable (Boboila et al. 2012; Han et al. 2012). A possible contribution to such discrepancies is that Lig1 (the only other mammalian ligase) has also been shown to support some level of alt-NHEJ (Liang et al. 2008; Simsek et al. 2011). It is unknown whether Lig3 and Lig1 are simply redundant or if their usage is somehow regulated.
Microhomology is an important feature of many alt-NHEJ joints that enhances the stability of PARP-1-mediated DNA synapsis (Audebert et al. 2008). One mechanism creates this base-pairing potential by locally templated extensions of the 3′ DSB strand (Yu and McVey 2010; Simsek et al. 2011), but more generally, internal microhomologies are exposed by resection of DSB ends, evident as deletions in final joints. Alt-NHEJ likely uses limited resection based on the size of these deletions, consistent with two-step models of DSB resection that transition from an initial local resection to a faster and more processive extended resection to support fully efficient HR (Fig. 5) (Symington and Gautier 2011; Grabarz et al. 2012). Thus, alt-NHEJ and HR appear to share a common initial resection mechanism promoted by the Mre11 nuclease and CtIP (Dinkelmann et al. 2009; Rass et al. 2009; Xie et al. 2009; Lee-Theilen et al. 2011; Zhang and Jasin 2011).
If alt-NHEJ and HR do rely on a common resection mechanism, alt-NHEJ will be subjected to the same regulatory influences as HR. However, limited resection in G1 can be seen as a potentially valuable action to support alt-NHEJ, or c-NHEJ-dependent MMEJ, independently of HR (Yun and Hiom 2009). ATM activity is likely important for alt-NHEJ regulation, as seen in studies of class switch recombination in mammalian cells (Bothmer et al. 2010) and genome rearrangement in budding yeast (Smith et al. 2005; Lee et al. 2008), where ATM influenced the Mre11 nuclease to modulate alt-NHEJ. Finally, a recently proposed end protection mediated by 53BP1 and H2AX that suppresses alt-NHEJ emphasizes the still incompletely understood nature of the c-NHEJ to alt-NHEJ transition (Bothmer et al. 2010, 2011).
c-NHEJ-Independent Genome Rearrangement
Unlike c-NHEJ, it is uncertain to what extent alt-NHEJ is an evolutionarily conserved mode of genome protection. Alt-NHEJ is described as a backup pathway when c-NHEJ is deficient (Wang et al. 2005), but this is only meaningful if some DSBs are processed by alt-NHEJ when c-NHEJ is intact. We can infer from c-NHEJ and HR mutants that alt-NHEJ is a low-frequency event. However, it might be disproportionately important because of its mutagenic nature. Experimental models suggest that chromosomal translocations might indeed be catalyzed by alt-NHEJ even when c-NHEJ is functional (Simsek and Jasin 2010; Simsek et al. 2011; Zhang and Jasin 2011). Moreover, c-NHEJ loss did not reduce the frequency of microhomology-mediated replication stress-induced intrachromosomal deletions and duplications (Arlt et al. 2012). These types of findings raise the interesting possibility that many, and perhaps most, homology-independent chromosomal alterations are mediated by mechanisms other than c-NHEJ in normal cells.
CONCLUDING REMARKS
Recent years have seen tremendous progress in the identification and description c-NHEJ proteins, providing us with “snapshots” of most (Figs. 1–4). Murkier at present is how the action of multiple proteins is coordinated in space and time to allow the joining of inherently bimolecular DSB substrates. We await the “movie version” of this interesting story. Even more work is required to articulate the mechanisms of alt-NHEJ and how significant a contributor it is to ongoing repair and mutagenesis. These tasks are important because different answers have very different implications for the etiology of human chromosome alterations and the potential consequences of therapeutic inhibition of c-NHEJ proteins such as Lig4.
Footnotes
Editors: Errol C. Friedberg, Stephen J. Elledge, Alan R. Lehmann, Tomas Lindahl, and Marco Muzi-Falconi
Additional Perspectives on DNA Repair, Mutagensis, and Other Responses to DNA Damage available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Ahnesorg P, Smith P, Jackson SP 2006. XLF interacts with the XRCC4-DNA ligase IV complex to promote DNA nonhomologous end-joining. Cell 124: 301–313 [DOI] [PubMed] [Google Scholar]
- Andres SN, Junop MS 2011. Crystallization and preliminary X-ray diffraction analysis of the human XRCC4–XLF complex. Acta Crystallogr Sect F Struct Biol Cryst Commun 67: 1399–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andres SN, Modesti M, Tsai CJ, Chu G, Junop MS 2007. Crystal structure of human XLF: A twist in nonhomologous DNA end-joining. Mol Cell 28: 1093–1101 [DOI] [PubMed] [Google Scholar]
- Andres SN, Vergnes A, Ristic D, Wyman C, Modesti M, Junop M 2012. A human XRCC4–XLF complex bridges DNA. Nucleic Acids Res 40: 1868–1878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlt MF, Rajendran S, Birkeland SR, Wilson TE, Glover TW 2012. De novo CNV formation in mouse embryonic stem cells occurs in the absence of Xrcc4-dependent nonhomologous end joining. PLoS Genet 8: e1002981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audebert M, Salles B, Calsou P 2004. Involvement of poly(ADP-ribose) polymerase-1 and XRCC1/DNA ligase III in an alternative route for DNA double-strand breaks rejoining. J Biol Chem 279: 55117–55126 [DOI] [PubMed] [Google Scholar]
- Audebert M, Salles B, Weinfeld M, Calsou P 2006. Involvement of polynucleotide kinase in a poly(ADP-ribose) polymerase-1-dependent DNA double-strand breaks rejoining pathway. J Mol Biol 356: 257–265 [DOI] [PubMed] [Google Scholar]
- Audebert M, Salles B, Calsou P 2008. Effect of double-strand break DNA sequence on the PARP-1 NHEJ pathway. Biochem Biophys Res Commun 369: 982–988 [DOI] [PubMed] [Google Scholar]
- Aylon Y, Kupiec M 2005. Cell cycle-dependent regulation of double-strand break repair: A role for the CDK. Cell Cycle 4: 259–261 [PubMed] [Google Scholar]
- Bahmed K, Nitiss KC, Nitiss JL 2010. Yeast Tdp1 regulates the fidelity of nonhomologous end joining. Proc Natl Acad Sci 107: 4057–4062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bebenek K, Garcia-Diaz M, Zhou RZ, Povirk LF, Kunkel TA 2010. Loop 1 modulates the fidelity of DNA polymerase λ. Nucleic Acids Res 38: 5419–5431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett SM, Woods DS, Pawelczak KS, Turchi JJ 2012. Multiple protein–protein interactions within the DNA–PK complex are mediated by the C-terminus of Ku 80. Int J Biochem Mol Biol 3: 36–45 [PMC free article] [PubMed] [Google Scholar]
- Beucher A, Birraux J, Tchouandong L, Barton O, Shibata A, Conrad S, Goodarzi AA, Krempler A, Jeggo PA, Lobrich M 2009. ATM and Artemis promote homologous recombination of radiation-induced DNA double-strand breaks in G2. EMBO J 28: 3413–3427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boboila C, Oksenych V, Gostissa M, Wang JH, Zha S, Zhang Y, Chai H, Lee CS, Jankovic M, Saez LM, et al. 2012. Robust chromosomal DNA repair via alternative end-joining in the absence of X-ray repair cross-complementing protein 1 (XRCC1). Proc Natl Acad Sci 109: 2473–2478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bothmer A, Robbiani DF, Feldhahn N, Gazumyan A, Nussenzweig A, Nussenzweig MC 2010. 53BP1 regulates DNA resection and the choice between classical and alternative end joining during class switch recombination. J Exp Med 207: 855–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bothmer A, Robbiani DF, Di Virgilio M, Bunting SF, Klein IA, Feldhahn N, Barlow J, Chen HT, Bosque D, Callen E, et al. 2011. Regulation of DNA end joining, resection, and immunoglobulin class switch recombination by 53BP1. Mol Cell 42: 319–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck D, Malivert L, de Chasseval R, Barraud A, Fondaneche MC, Sanal O, Plebani A, Stephan JL, Hufnagel M, le Deist F, et al. 2006. Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell 124: 287–299 [DOI] [PubMed] [Google Scholar]
- Bunting SF, Callen E, Wong N, Chen HT, Polato F, Gunn A, Bothmer A, Feldhahn N, Fernandez-Capetillo O, Cao L, et al. 2010. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell 141: 243–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capp JP, Boudsocq F, Bertrand P, Laroche-Clary A, Pourquier P, Lopez BS, Cazaux C, Hoffmann JS, Canitrot Y 2006. The DNA polymerase λ is required for the repair of non-compatible DNA double strand breaks by NHEJ in mammalian cells. Nucleic Acids Res 34: 2998–3007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capp JP, Boudsocq F, Besnard AG, Lopez BS, Cazaux C, Hoffmann JS, Canitrot Y 2007. Involvement of DNA polymerase μ in the repair of a specific subset of DNA double-strand breaks in mammalian cells. Nucleic Acids Res 35: 3551–3560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CY, Galli A, Schiestl RH 2008. Pol3 is involved in nonhomologous end-joining in Saccharomyces cerevisiae. DNA Repair (Amst) 7: 1531–1541 [DOI] [PubMed] [Google Scholar]
- Chapman JR, Sossick AJ, Boulton SJ, Jackson SP 2012. BRCA1-associated exclusion of 53BP1 from DNA damage sites underlies temporal control of DNA repair. J Cell Sci 125: 3529–3534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell C, Hanakahi LA, Karimi-Busheri F, Weinfeld M, West SC 2002. Involvement of human polynucleotide kinase in double-strand break repair by non-homologous end joining. EMBO J 21: 2827–2832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Tomkinson AE 2011. Yeast Nej1 is a key participant in the initial end binding and final ligation steps of nonhomologous end joining. J Biol Chem 286: 4931–4940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Trujillo KM, Van Komen S, Roh DH, Krejci L, Lewis LK, Resnick MA, Sung P, Tomkinson AE 2005. Effect of amino acid substitutions in the rad50 ATP binding domain on DNA double strand break repair in yeast. J Biol Chem 280: 2620–2627 [DOI] [PubMed] [Google Scholar]
- Chiruvella KK, Sebastian R, Sharma S, Karande AA, Choudhary B, Raghavan SC 2012. Time-dependent predominance of nonhomologous DNA end-joining pathways during embryonic development in mice. J Mol Biol 417: 197–211 [DOI] [PubMed] [Google Scholar]
- Chistiakov DA 2010. Ligase IV syndrome. Adv Exp Med Biol 685: 175–185 [DOI] [PubMed] [Google Scholar]
- Clements PM, Breslin C, Deeks ED, Byrd PJ, Ju L, Bieganowski P, Brenner C, Moreira MC, Taylor AM, Caldecott KW 2004. The ataxia-oculomotor apraxia 1 gene product has a role distinct from ATM and interacts with the DNA strand break repair proteins XRCC1 and XRCC4. DNA Repair (Amst) 3: 1493–1502 [DOI] [PubMed] [Google Scholar]
- Clerici M, Mantiero D, Guerini I, Lucchini G, Longhese MP 2008. The Yku70–Yku80 complex contributes to regulate double-strand break processing and checkpoint activation during the cell cycle. EMBO Rep 9: 810–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coquelle N, Havali-Shahriari Z, Bernstein N, Green R, Glover JN 2011. Structural basis for the phosphatase activity of polynucleotide kinase/phosphatase on single- and double-stranded DNA substrates. Proc Natl Acad Sci 108: 21022–21027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantini S, Woodbine L, Andreoli L, Jeggo PA, Vindigni A 2007. Interaction of the Ku heterodimer with the DNA ligase IV/Xrcc4 complex and its regulation by DNA-PK. DNA Repair (Amst) 6: 712–722 [DOI] [PubMed] [Google Scholar]
- Covo S, de Villartay JP, Jeggo PA, Livneh Z 2009. Translesion DNA synthesis-assisted non-homologous end-joining of complex double-strand breaks prevents loss of DNA sequences in mammalian cells. Nucleic Acids Res 37: 6737–6745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X, Yu Y, Gupta S, Cho YM, Lees-Miller SP, Meek K 2005. Autophosphorylation of DNA-dependent protein kinase regulates DNA end processing and may also alter double-strand break repair pathway choice. Mol Cell Biol 25: 10842–10852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley JM, Wilson TE 2005. Rejoining of DNA double-strand breaks as a function of overhang length. Mol Cell Biol 25: 896–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley JM, Wilson TE 2008. Evidence that base stacking potential in annealed 3′ overhangs determines polymerase utilization in yeast nonhomologous end joining. DNA Repair (Amst) 7: 67–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley JM, Laan RL, Suresh A, Wilson TE 2005. DNA joint dependence of Pol X family polymerase action in nonhomologous end joining. J Biol Chem 280: 29030–29037 [DOI] [PubMed] [Google Scholar]
- Daley JM, Wilson TE, Ramotar D 2010. Genetic interactions between HNT3/Aprataxin and RAD27/FEN1 suggest parallel pathways for 5′ end processing during base excision repair. DNA Repair (Amst) 9: 690–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFazio LG, Stansel RM, Griffith JD, Chu G 2002. Synapsis of DNA ends by DNA-dependent protein kinase. EMBO J 21: 3192–3200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delarue M, Boule JB, Lescar J, Expert-Bezancon N, Jourdan N, Sukumar N, Rougeon F, Papanicolaou C 2002. Crystal structures of a template-independent DNA polymerase: Murine terminal deoxynucleotidyltransferase. EMBO J 21: 427–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Della-Maria J, Zhou Y, Tsai MS, Kuhnlein J, Carney JP, Paull TT, Tomkinson AE 2011. Human Mre11/human Rad50/Nbs1 and DNA ligase IIIα/XRCC1 protein complexes act together in an alternative nonhomologous end joining pathway. J Biol Chem 286: 33845–33853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deriano L, Stracker TH, Baker A, Petrini JH, Roth DB 2009. Roles for NBS1 in alternative nonhomologous end-joining of V(D)J recombination intermediates. Mol Cell 34: 13–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRose EF, Clarkson MW, Gilmore SA, Galban CJ, Tripathy A, Havener JM, Mueller GA, Ramsden DA, London RE, Lee AL 2007. Solution structure of polymerase μ’s BRCT domain reveals an element essential for its role in nonhomologous end joining. Biochemistry 46: 12100–12110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande RA, Wilson TE 2007. Modes of interaction among yeast Nej1, Lif1 and Dnl4 proteins and comparison to human XLF, XRCC4 and Lig4. DNA Repair (Amst) 6: 1507–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinkelmann M, Spehalski E, Stoneham T, Buis J, Wu Y, Sekiguchi JM, Ferguson DO 2009. Multiple functions of MRN in end-joining pathways during isotype class switching. Nat Struct Mol Biol 16: 808–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Virgilio M, Gautier J 2005. Repair of double-strand breaks by nonhomologous end joining in the absence of Mre11. J Cell Biol 171: 765–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbs TA, Tainer JA, Lees-Miller SP 2010. A structural model for regulation of NHEJ by DNA-PKcs autophosphorylation. DNA Repair (Amst) 9: 1307–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dore AS, Furnham N, Davies OR, Sibanda BL, Chirgadze DY, Jackson SP, Pellegrini L, Blundell TL 2006. Structure of an Xrcc4–DNA ligase IV yeast ortholog complex reveals a novel BRCT interaction mode. DNA Repair (Amst) 5: 362–368 [DOI] [PubMed] [Google Scholar]
- Douglas P, Gupta S, Morrice N, Meek K, Lees-Miller SP 2005. DNA-PK-dependent phosphorylation of Ku70/80 is not required for non-homologous end joining. DNA Repair (Amst) 4: 1006–1018 [DOI] [PubMed] [Google Scholar]
- Ellenberger T, Tomkinson AE 2008. Eukaryotic DNA ligases: Structural and functional insights. Annu Rev Biochem 77: 313–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falck J, Coates J, Jackson SP 2005. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature 434: 605–611 [DOI] [PubMed] [Google Scholar]
- Feng L, Chen J 2012. The E3 ligase RNF8 regulates KU80 removal and NHEJ repair. Nat Struct Mol Biol 19: 201–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson DO, Alt FW 2001. DNA double strand break repair and chromosomal translocation: Lessons from animal models. Oncogene 20: 5572–5579 [DOI] [PubMed] [Google Scholar]
- Frank KM, Sharpless NE, Gao Y, Sekiguchi JM, Ferguson DO, Zhu C, Manis JP, Horner J, DePinho RA, Alt FW 2000. DNA ligase IV deficiency in mice leads to defective neurogenesis and embryonic lethality via the p53 pathway. Mol Cell 5: 993–1002 [DOI] [PubMed] [Google Scholar]
- Gapud EJ, Sleckman BP 2011. Unique and redundant functions of ATM and DNA-PKcs during V(D)J recombination. Cell Cycle 10: 1928–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garces F, Pearl LH, Oliver AW 2011. The structural basis for substrate recognition by mammalian polynucleotide kinase 3′ phosphatase. Mol Cell 44: 385–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Diaz M, Bebenek K, Krahn JM, Kunkel TA, Pedersen LC 2005. A closed conformation for the Pol λ catalytic cycle. Nat Struct Mol Biol 12: 97–98 [DOI] [PubMed] [Google Scholar]
- Gatz SA, Ju L, Gruber R, Hoffmann E, Carr AM, Wang ZQ, Liu C, Jeggo PA 2011. Requirement for DNA ligase IV during embryonic neuronal development. J Neurosci 31: 10088–10100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y, Zhu D, Ding J, Dou CN, Ren X, Gu L, Jiang T, Wang DC 2011. Crystal structures of aprataxin ortholog Hnt3 reveal the mechanism for reversal of 5′-adenylated DNA. Nat Struct Mol Biol 18: 1297–1299 [DOI] [PubMed] [Google Scholar]
- Goodarzi AA, Yu Y, Riballo E, Douglas P, Walker SA, Ye R, Harer C, Marchetti C, Morrice N, Jeggo PA, et al. 2006. DNA-PK autophosphorylation facilitates Artemis endonuclease activity. EMBO J 25: 3880–3889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodarzi AA, Noon AT, Deckbar D, Ziv Y, Shiloh Y, Lobrich M, Jeggo PA 2008. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol Cell 31: 167–177 [DOI] [PubMed] [Google Scholar]
- Grabarz A, Barascu A, Guirouilh-Barbat J, Lopez BS 2012. Initiation of DNA double strand break repair: Signaling and single-stranded resection dictate the choice between homologous recombination, non-homologous end-joining and alternative end-joining. Am J Cancer Res 2: 249–268 [PMC free article] [PubMed] [Google Scholar]
- Grawunder U, Wilm M, Wu X, Kulesza P, Wilson TE, Mann M, Lieber MR 1997. Activity of DNA ligase IV stimulated by complex formation with XRCC4 protein in mammalian cells. Nature 388: 492–495 [DOI] [PubMed] [Google Scholar]
- Gu Y, Seidl KJ, Rathbun GA, Zhu C, Manis JP, van der Stoep N, Davidson L, Cheng HL, Sekiguchi JM, Frank K, et al. 1997. Growth retardation and leaky SCID phenotype of Ku70-deficient mice. Immunity 7: 653–665 [DOI] [PubMed] [Google Scholar]
- Gu Y, Sekiguchi J, Gao Y, Dikkes P, Frank K, Ferguson D, Hasty P, Chun J, Alt FW 2000. Defective embryonic neurogenesis in Ku-deficient but not DNA-dependent protein kinase catalytic subunit-deficient mice. Proc Natl Acad Sci 97: 2668–2673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J, Lu H, Tippin B, Shimazaki N, Goodman MF, Lieber MR 2007a. XRCC4:DNA ligase IV can ligate incompatible DNA ends and can ligate across gaps. EMBO J 26: 1010–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J, Lu H, Tsai AG, Schwarz K, Lieber MR 2007b. Single-stranded DNA ligation and XLF-stimulated incompatible DNA end ligation by the XRCC4–DNA ligase IV complex: Influence of terminal DNA sequence. Nucleic Acids Res 35: 5755–5762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammarsten O, DeFazio LG, Chu G 2000. Activation of DNA-dependent protein kinase by single-stranded DNA ends. J Biol Chem 275: 1541–1550 [DOI] [PubMed] [Google Scholar]
- Hammel M, Yu Y, Fang S, Lees-Miller SP, Tainer JA 2010a. XLF regulates filament architecture of the XRCC4·ligase IV complex. Structure 18: 1431–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammel M, Yu Y, Mahaney BL, Cai B, Ye R, Phipps BM, Rambo RP, Hura GL, Pelikan M, So S, et al. 2010b. Ku and DNA-dependent protein kinase dynamic conformations and assembly regulate DNA binding and the initial non-homologous end joining complex. J Biol Chem 285: 1414–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammel M, Rey M, Yu Y, Mani RS, Classen S, Liu M, Pique ME, Fang S, Mahaney BL, Weinfeld M, et al. 2011. XRCC4 protein interactions with XRCC4-like factor (XLF) create an extended grooved scaffold for DNA ligation and double strand break repair. J Biol Chem 286: 32638–32650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L, Mao W, Yu K 2012. X-ray repair cross-complementing protein 1 (XRCC1) deficiency enhances class switch recombination and is permissive for alternative end joining. Proc Natl Acad Sci 109: 4604–4608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Shi LZ, Truong LN, Lu CS, Razavian N, Li Y, Negrete A, Shiloach J, Berns MW, Wu X 2012. Rad50 zinc hook is important for the Mre11 complex to bind chromosomal DNA double-stranded breaks and initiate various DNA damage responses. J Biol Chem 287: 31747–31756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmink BA, Bredemeyer AL, Lee BS, Huang CY, Sharma GG, Walker LM, Bednarski JJ, Lee WL, Pandita TK, Bassing CH, et al. 2009. MRN complex function in the repair of chromosomal Rag-mediated DNA double-strand breaks. J Exp Med 206: 669–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohl M, Kwon Y, Galvan SM, Xue X, Tous C, Aguilera A, Sung P, Petrini JH 2011. The Rad50 coiled-coil domain is indispensable for Mre11 complex functions. Nat Struct Mol Biol 18: 1124–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopfner KP, Karcher A, Shin DS, Craig L, Arthur LM, Carney JP, Tainer JA 2000. Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell 101: 789–800 [DOI] [PubMed] [Google Scholar]
- Hopfner KP, Craig L, Moncalian G, Zinkel RA, Usui T, Owen BA, Karcher A, Henderson B, Bodmer JL, McMurray CT, et al. 2002. The Rad50 zinc-hook is a structure joining Mre11 complexes in DNA recombination and repair. Nature 418: 562–566 [DOI] [PubMed] [Google Scholar]
- Huang J, Dynan WS 2002. Reconstitution of the mammalian DNA double-strand break end-joining reaction reveals a requirement for an Mre11/Rad50/NBS1-containing fraction. Nucleic Acids Res 30: 667–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Giblin W, Kubec M, Westfield G, St Charles J, Chadde L, Kraftson S, Sekiguchi J 2009. Impact of a hypomorphic Artemis disease allele on lymphocyte development, DNA end processing, and genome stability. J Exp Med 206: 893–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas P, Jackson SP 2009. Human CtIP mediates cell cycle control of DNA end resection and double strand break repair. J Biol Chem 284: 9558–9565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iles N, Rulten S, El-Khamisy SF, Caldecott KW 2007. APLF (C2orf13) is a novel human protein involved in the cellular response to chromosomal DNA strand breaks. Mol Cell Biol 27: 3793–3803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ira G, Pellicioli A, Balijja A, Wang X, Fiorani S, Carotenuto W, Liberi G, Bressan D, Wan L, Hollingsworth NM, et al. 2004. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature 431: 1011–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs C, Huang Y, Masud T, Lu W, Westfield G, Giblin W, Sekiguchi JM 2011. A hypomorphic Artemis human disease allele causes aberrant chromosomal rearrangements and tumorigenesis. Hum Mol Genet 20: 806–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juarez R, Ruiz JF, Nick McElhinny SA, Ramsden D, Blanco L 2006. A specific loop in human DNA polymerase μ allows switching between creative and DNA-instructed synthesis. Nucleic Acids Res 34: 4572–4582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karanam K, Kafri R, Loewer A, Lahav G 2012. Quantitative live cell imaging reveals a gradual shift between DNA repair mechanisms and a maximal use of HR in mid S phase. Mol Cell 47: 320–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karathanasis E, Wilson TE 2002. Enhancement of Saccharomyces cerevisiae end-joining efficiency by cell growth stage but not by impairment of recombination. Genetics 161: 1015–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi-Busheri F, Rasouli-Nia A, Allalunis-Turner J, Weinfeld M 2007. Human polynucleotide kinase participates in repair of DNA double-strand breaks by nonhomologous end joining but not homologous recombination. Cancer Res 67: 6619–6625 [DOI] [PubMed] [Google Scholar]
- Kerzendorfer C, O’Driscoll M 2009. Human DNA damage response and repair deficiency syndromes: Linking genomic instability and cell cycle checkpoint proficiency. DNA Repair (Amst) 8: 1139–1152 [DOI] [PubMed] [Google Scholar]
- Koch CA, Agyei R, Galicia S, Metalnikov P, O’Donnell P, Starostine A, Weinfeld M, Durocher D 2004. Xrcc4 physically links DNA end processing by polynucleotide kinase to DNA ligation by DNA ligase IV. EMBO J 23: 3874–3885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosawa A, Adachi N 2010. Functions and regulation of Artemis: A goddess in the maintenance of genome integrity. J Radiat Res 51: 503–509 [DOI] [PubMed] [Google Scholar]
- Kysela B, Chovanec M, Jeggo PA 2005. Phosphorylation of linker histones by DNA-dependent protein kinase is required for DNA ligase IV-dependent ligation in the presence of histone H1. Proc Natl Acad Sci 102: 1877–1882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammens K, Bemeleit DJ, Mockel C, Clausing E, Schele A, Hartung S, Schiller CB, Lucas M, Angermuller C, Soding J, et al. 2011. The Mre11:Rad50 structure shows an ATP-dependent molecular clamp in DNA double-strand break repair. Cell 145: 54–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langerak P, Mejia-Ramirez E, Limbo O, Russell P 2011. Release of Ku and MRN from DNA ends by Mre11 nuclease activity and Ctp1 is required for homologous recombination repair of double-strand breaks. PLoS Genet 7: e1002271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Zhang Y, Lee SE 2008. Saccharomyces cerevisiae ATM orthologue suppresses break-induced chromosome translocations. Nature 454: 543–546 [DOI] [PubMed] [Google Scholar]
- Lee-Theilen M, Matthews AJ, Kelly D, Zheng S, Chaudhuri J 2011. CtIP promotes microhomology-mediated alternative end joining during class-switch recombination. Nat Struct Mol Biol 18: 75–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman JA, Hoelz DJ, Turchi JJ 2008. DNA-dependent conformational changes in the Ku heterodimer. Biochemistry 47: 4359–4368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lempiainen H, Halazonetis TD 2009. Emerging common themes in regulation of PIKKs and PI3Ks. EMBO J 28: 3067–3073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Alt FW, Cheng HL, Brush JW, Goff PH, Murphy MM, Franco S, Zhang Y, Zha S 2008a. Lymphocyte-specific compensation for XLF/cernunnos end-joining functions in V(D)J recombination. Mol Cell 31: 631–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Chirgadze DY, Bolanos-Garcia VM, Sibanda BL, Davies OR, Ahnesorg P, Jackson SP, Blundell TL 2008b. Crystal structure of human XLF/Cernunnos reveals unexpected differences from XRCC4 with implications for NHEJ. EMBO J 27: 290–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Kanno S, Watanabe R, Ogiwara H, Kohno T, Watanabe G, Yasui A, Lieber MR 2011. Polynucleotide kinase and aprataxin-like Forkhead-associated protein (PALF) acts as both a single-stranded DNA endonuclease and a single-stranded DNA 3′ exonuclease and can participate in DNA end joining in a biochemical system. J Biol Chem 286: 36368–36377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang L, Deng L, Nguyen SC, Zhao X, Maulion CD, Shao C, Tischfield JA 2008. Human DNA ligases I and III, but not ligase IV, are required for microhomology-mediated end joining of DNA double-strand breaks. Nucleic Acids Res 36: 3297–3310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Pannicke U, Schwarz K, Lieber MR 2007. Length-dependent binding of human XLF to DNA and stimulation of XRCC4.DNA ligase IV activity. J Biol Chem 282: 11155–11162 [DOI] [PubMed] [Google Scholar]
- Ma Y, Pannicke U, Schwarz K, Lieber MR 2002. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell 108: 781–794 [DOI] [PubMed] [Google Scholar]
- Ma Y, Lu H, Tippin B, Goodman MF, Shimazaki N, Koiwai O, Hsieh CL, Schwarz K, Lieber MR 2004. A biochemically defined system for mammalian nonhomologous DNA end joining. Mol Cell 16: 701–713 [DOI] [PubMed] [Google Scholar]
- Ma Y, Lu H, Schwarz K, Lieber MR 2005. Repair of double-strand DNA breaks by the human nonhomologous DNA end joining pathway: The iterative processing model. Cell Cycle 4: 1193–1200 [DOI] [PubMed] [Google Scholar]
- Macrae CJ, McCulloch RD, Ylanko J, Durocher D, Koch CA 2008. APLF (C2orf13) facilitates nonhomologous end-joining and undergoes ATM-dependent hyperphosphorylation following ionizing radiation. DNA Repair (Amst) 7: 292–302 [DOI] [PubMed] [Google Scholar]
- Majka J, Alford B, Ausio J, Finn RM, McMurray CT 2012. ATP hydrolysis by RAD50 protein switches MRE11 enzyme from endonuclease to exonuclease. J Biol Chem 287: 2328–2341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malu S, De Ioannes P, Kozlov M, Greene M, Francis D, Hanna M, Pena J, Escalante CR, Kurosawa A, Erdjument-Bromage H, et al. 2012. Artemis C-terminal region facilitates V(D)J recombination through its interactions with DNA Ligase IV and DNA-PKcs. J Exp Med 209: 955–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani RS, Yu Y, Fang S, Lu M, Fanta M, Zolner AE, Tahbaz N, Ramsden DA, Litchfield DW, Lees-Miller SP, et al. 2010. Dual modes of interaction between XRCC4 and polynucleotide kinase/phosphatase: Implications for nonhomologous end joining. J Biol Chem 285: 37619–37629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolis KG, Nimmo ER, Hartsuiker E, Carr AM, Jeggo PA, Allshire RC 2001. Novel functional requirements for non-homologous DNA end joining in Schizosaccharomyces pombe. EMBO J 20: 210–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour WY, Rhein T, Dahm-Daphi J 2010. The alternative end-joining pathway for repair of DNA double-strand breaks requires PARP1 but is not dependent upon microhomologies. Nucleic Acids Res 38: 6065–6077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Z, Bozzella M, Seluanov A, Gorbunova V 2008. Comparison of nonhomologous end joining and homologous recombination in human cells. DNA Repair (Amst) 7: 1765–1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Z, Hine C, Tian X, Van Meter M, Au M, Vaidya A, Seluanov A, Gorbunova V 2011. SIRT6 promotes DNA repair under stress by activating PARP1. Science 332: 1443–1446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mari PO, Florea BI, Persengiev SP, Verkaik NS, Bruggenwirth HT, Modesti M, Giglia-Mari G, Bezstarosti K, Demmers JA, Luider TM, et al. 2006. Dynamic assembly of end-joining complexes requires interaction between Ku70/80 and XRCC4. Proc Natl Acad Sci 103: 18597–18602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin MJ, Juarez R, Blanco L 2012. DNA-binding determinants promoting NHEJ by human Polμ. Nucleic Acids Res 10.1093/nar/gks896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki K, Shinohara A, Shinohara M 2008. Forkhead-associated domain of yeast Xrs2, a homolog of human Nbs1, promotes nonhomologous end joining through interaction with a ligase IV partner protein, Lif1. Genetics 179: 213–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne GT, Jin S, Shannon KB, Weaver DT 1996. Mutations in two Ku homologs define a DNA end-joining repair pathway in Saccharomyces cerevisiae. Mol Cell Biol 16: 4189–4198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockel C, Lammens K, Schele A, Hopfner KP 2012. ATP driven structural changes of the bacterial Mre11:Rad50 catalytic head complex. Nucleic Acids Res 40: 914–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon AF, Garcia-Diaz M, Bebenek K, Davis BJ, Zhong X, Ramsden DA, Kunkel TA, Pedersen LC 2007. Structural insight into the substrate specificity of DNA Polymerase mu. Nat Struct Mol Biol 14: 45–53 [DOI] [PubMed] [Google Scholar]
- Moreno-Herrero F, de Jager M, Dekker NH, Kanaar R, Wyman C, Dekker C 2005. Mesoscale conformational changes in the DNA-repair complex Rad50/Mre11/Nbs1 upon binding DNA. Nature 437: 440–443 [DOI] [PubMed] [Google Scholar]
- Morris EP, Rivera-Calzada A, da Fonseca PC, Llorca O, Pearl LH, Spagnolo L 2011. Evidence for a remodelling of DNA-PK upon autophosphorylation from electron microscopy studies. Nucleic Acids Res 39: 5757–5767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moshous D, Callebaut I, de Chasseval R, Corneo B, Cavazzana-Calvo M, Le Deist F, Tezcan I, Sanal O, Bertrand Y, Philippe N, et al. 2001. Artemis, a novel DNA double-strand break repair/V(D)J recombination protein, is mutated in human severe combined immune deficiency. Cell 105: 177–186 [DOI] [PubMed] [Google Scholar]
- Mueller GA, Moon AF, Derose EF, Havener JM, Ramsden DA, Pedersen LC, London RE 2008. A comparison of BRCT domains involved in nonhomologous end-joining: Introducing the solution structure of the BRCT domain of polymerase λ. DNA Repair (Amst) 7: 1340–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neal JA, Meek K 2011. Choosing the right path: Does DNA-PK help make the decision? Mutat Res 711: 73–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nick McElhinny SA, Havener JM, Garcia-Diaz M, Juarez R, Bebenek K, Kee BL, Blanco L, Kunkel TA, Ramsden DA 2005. A gradient of template dependence defines distinct biological roles for family X polymerases in nonhomologous end joining. Mol Cell 19: 357–366 [DOI] [PubMed] [Google Scholar]
- Nijnik A, Woodbine L, Marchetti C, Dawson S, Lambe T, Liu C, Rodrigues NP, Crockford TL, Cabuy E, Vindigni A, et al. 2007. DNA repair is limiting for haematopoietic stem cells during ageing. Nature 447: 686–690 [DOI] [PubMed] [Google Scholar]
- Ochi T, Wu Q, Chirgadze DY, Grossmann JG, Bolanos-Garcia VM, Blundell TL 2012. Structural insights into the role of domain flexibility in human DNA ligase IV. Structure 20: 1212–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksenych V, Alt FW, Kumar V, Schwer B, Wesemann DR, Hansen E, Patel H, Su A, Guo C 2012. Functional redundancy between repair factor XLF and damage response mediator 53BP1 in V(D)J recombination and DNA repair. Proc Natl Acad Sci 109: 2455–2460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmbos PL, Daley JM, Wilson TE 2005. Mutations of the Yku80 C terminus and Xrs2 FHA domain specifically block yeast nonhomologous end joining. Mol Cell Biol 25: 10782–10790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmbos PL, Wu D, Daley JM, Wilson TE 2008. Recruitment of Saccharomyces cerevisiae Dnl4–Lif1 complex to a double-strand break requires interactions with Yku80 and the Xrs2 FHA domain. Genetics 180: 1809–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascal JM, O’Brien PJ, Tomkinson AE, Ellenberger T 2004. Human DNA ligase I completely encircles and partially unwinds nicked DNA. Nature 432: 473–478 [DOI] [PubMed] [Google Scholar]
- Pawelczak KS, Turchi JJ 2008. A mechanism for DNA-PK activation requiring unique contributions from each strand of a DNA terminus and implications for microhomology-mediated nonhomologous DNA end joining. Nucleic Acids Res 36: 4022–4031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawelczak KS, Turchi JJ 2010. Purification and characterization of exonuclease-free Artemis: Implications for DNA-PK-dependent processing of DNA termini in NHEJ-catalyzed DSB repair. DNA Repair (Amst) 9: 670–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawelczak KS, Andrews BJ, Turchi JJ 2005. Differential activation of DNA-PK based on DNA strand orientation and sequence bias. Nucleic Acids Res 33: 152–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry JJ, Cotner-Gohara E, Ellenberger T, Tainer JA 2010. Structural dynamics in DNA damage signaling and repair. Curr Opin Struct Biol 20: 283–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postow L, Ghenoiu C, Woo EM, Krutchinsky AN, Chait BT, Funabiki H 2008. Ku80 removal from DNA through double strand break-induced ubiquitylation. J Cell Biol 182: 467–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Povirk LF, Zhou T, Zhou R, Cowan MJ, Yannone SM 2007. Processing of 3′-phosphoglycolate-terminated DNA double strand breaks by Artemis nuclease. J Biol Chem 282: 3547–3558 [DOI] [PubMed] [Google Scholar]
- Ramsden DA, Asagoshi K 2012. DNA polymerases in nonhomologous end joining: Are there any benefits to standing out from the crowd? Environ Mol Mutagen 53: 741–751 [DOI] [PubMed] [Google Scholar]
- Rass E, Grabarz A, Plo I, Gautier J, Bertrand P, Lopez BS 2009. Role of Mre11 in chromosomal nonhomologous end joining in mammalian cells. Nat Struct Mol Biol 16: 819–824 [DOI] [PubMed] [Google Scholar]
- Reynolds P, Anderson JA, Harper JV, Hill MA, Botchway SW, Parker AW, O’Neill P 2012. The dynamics of Ku70/80 and DNA-PKcs at DSBs induced by ionizing radiation is dependent on the complexity of damage. Nucleic Acids Res 10.1093/nar/gks879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riballo E, Woodbine L, Stiff T, Walker SA, Goodarzi AA, Jeggo PA 2009. XLF-Cernunnos promotes DNA ligase IV–XRCC4 re-adenylation following ligation. Nucleic Acids Res 37: 482–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert I, Dantzer F, Reina-San-Martin B 2009. Parp1 facilitates alternative NHEJ, whereas Parp2 suppresses IgH/c-myc translocations during immunoglobulin class switch recombination. J Exp Med 206: 1047–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts SA, Ramsden DA 2007. Loading of the nonhomologous end joining factor, Ku, on protein-occluded DNA ends. J Biol Chem 282: 10605–10613 [DOI] [PubMed] [Google Scholar]
- Roberts SA, Strande N, Burkhalter MD, Strom C, Havener JM, Hasty P, Ramsden DA 2010. Ku is a 5′-dRP/AP lyase that excises nucleotide damage near broken ends. Nature 464: 1214–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ropars V, Drevet P, Legrand P, Baconnais S, Amram J, Faure G, Marquez JA, Pietrement O, Guerois R, Callebaut I, et al. 2011. Structural characterization of filaments formed by human Xrcc4-Cernunnos/XLF complex involved in nonhomologous DNA end-joining. Proc Natl Acad Sci 108: 12663–12668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosidi B, Wang M, Wu W, Sharma A, Wang H, Iliakis G 2008. Histone H1 functions as a stimulatory factor in backup pathways of NHEJ. Nucleic Acids Res 36: 1610–1623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Rothstein R 2013. Repair of strand breaks by homologous recombination. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a012740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rucci F, Notarangelo LD, Fazeli A, Patrizi L, Hickernell T, Paganini T, Coakley KM, Detre C, Keszei M, Walter JE, et al. 2010. Homozygous DNA ligase IV R278H mutation in mice leads to leaky SCID and represents a model for human LIG4 syndrome. Proc Natl Acad Sci 107: 3024–3029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rulten SL, Cortes-Ledesma F, Guo L, Iles NJ, Caldecott KW 2008. APLF (C2orf13) is a novel component of poly(ADP-ribose) signaling in mammalian cells. Mol Cell Biol 28: 4620–4628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rulten SL, Fisher AE, Robert I, Zuma MC, Rouleau M, Ju L, Poirier G, Reina-San-Martin B, Caldecott KW 2011. PARP-3 and APLF function together to accelerate nonhomologous end-joining. Mol Cell 41: 33–45 [DOI] [PubMed] [Google Scholar]
- Sallmyr A, Tomkinson AE, Rassool FV 2008. Up-regulation of WRN and DNA ligase IIIα in chronic myeloid leukemia: Consequences for the repair of DNA double-strand breaks. Blood 112: 1413–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saribasak H, Maul RW, Cao Z, McClure RL, Yang W, McNeill DR, Wilson DM 3rd, Gearhart PJ 2011. XRCC1 suppresses somatic hypermutation and promotes alternative nonhomologous end joining in Igh genes. J Exp Med 208: 2209–2216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiller CB, Lammens K, Guerini I, Coordes B, Feldmann H, Schlauderer F, Mockel C, Schele A, Strasser K, Jackson SP, et al. 2012. Structure of Mre11–Nbs1 complex yields insights into ataxia-telangiectasia-like disease mutations and DNA damage signaling. Nat Struct Mol Biol 19: 693–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Z, Davis AJ, Fattah KR, So S, Sun J, Lee KJ, Harrison L, Yang J, Chen DJ 2012. Persistently bound Ku at DNA ends attenuates DNA end resection and homologous recombination. DNA Repair (Amst) 11: 310–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata A, Conrad S, Birraux J, Geuting V, Barton O, Ismail A, Kakarougkas A, Meek K, Taucher-Scholz G, Lobrich M, et al. 2011. Factors determining DNA double-strand break repair pathway choice in G2 phase. EMBO J 30: 1079–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim EY, Ma JL, Oum JH, Yanez Y, Lee SE 2005. The yeast chromatin remodeler RSC complex facilitates end joining repair of DNA double-strand breaks. Mol Cell Biol 25: 3934–3944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim EY, Hong SJ, Oum JH, Yanez Y, Zhang Y, Lee SE 2007. RSC mobilizes nucleosomes to improve accessibility of repair machinery to the damaged chromatin. Mol Cell Biol 27: 1602–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibanda BL, Chirgadze DY, Blundell TL 2010. Crystal structure of DNA-PKcs reveals a large open-ring cradle comprised of HEAT repeats. Nature 463: 118–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simsek D, Jasin M 2010. Alternative end-joining is suppressed by the canonical NHEJ component Xrcc4–ligase IV during chromosomal translocation formation. Nat Struct Mol Biol 17: 410–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simsek D, Brunet E, Wong SY, Katyal S, Gao Y, McKinnon PJ, Lou J, Zhang L, Li J, Rebar EJ, et al. 2011. DNA ligase III promotes alternative nonhomologous end-joining during chromosomal translocation formation. PLoS Genet 7: e1002080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Sirbu BM, Cortez D 2013. DNA damage response: Three levels of DNA repair regulation. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a012724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S, Gupta A, Kolodner RD, Myung K 2005. Suppression of gross chromosomal rearrangements by the multiple functions of the Mre11–Rad50–Xrs2 complex in Saccharomyces cerevisiae. DNA Repair (Amst) 4: 606–617 [DOI] [PubMed] [Google Scholar]
- Strande N, Roberts SA, Oh S, Hendrickson EA, Ramsden DA 2012. Specificity of the dRP/AP lyase of Ku promotes nonhomologous end joining (NHEJ) fidelity at damaged ends. J Biol Chem 287: 13686–13693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington LS, Gautier J 2011. Double-strand break end resection and repair pathway choice. Annu Rev Genet 45: 247–271 [DOI] [PubMed] [Google Scholar]
- Takahashi T, Tada M, Igarashi S, Koyama A, Date H, Yokoseki A, Shiga A, Yoshida Y, Tsuji S, Nishizawa M, et al. 2007. Aprataxin, causative gene product for EAOH/AOA1, repairs DNA single-strand breaks with damaged 3′-phosphate and 3′-phosphoglycolate ends. Nucleic Acids Res 35: 3797–3809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujillo KM, Yuan SS, Lee EY, Sung P 1998. Nuclease activities in a complex of human recombination and DNA repair factors Rad50, Mre11, and p95. J Biol Chem 273: 21447–21450 [DOI] [PubMed] [Google Scholar]
- Tsai CJ, Kim SA, Chu G 2007. Cernunnos/XLF promotes the ligation of mismatched and noncohesive DNA ends. Proc Natl Acad Sci 104: 7851–7856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng SF, Gabriel A, Teng SC 2008. Proofreading activity of DNA polymerase Pol2 mediates 3′-end processing during nonhomologous end joining in yeast. PLoS Genet 4: e1000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukuda T, Fleming AB, Nickoloff JA, Osley MA 2005. Chromatin remodelling at a DNA double-strand break site in Saccharomyces cerevisiae. Nature 438: 379–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumbale P, Appel CD, Kraehenbuehl R, Robertson PD, Williams JS, Krahn J, Ahel I, Williams RS 2011. Structure of an aprataxin–DNA complex with insights into AOA1 neurodegenerative disease. Nat Struct Mol Biol 18: 1189–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uematsu N, Weterings E, Yano K, Morotomi-Yano K, Jakob B, Taucher-Scholz G, Mari PO, van Gent DC, Chen BP, Chen DJ 2007. Autophosphorylation of DNA-PKCS regulates its dynamics at DNA double-strand breaks. J Cell Biol 177: 219–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Burg M, van Dongen JJ, van Gent DC 2009. DNA-PKcs deficiency in human: Long predicted, finally found. Curr Opin Allergy Clin Immunol 9: 503–509 [DOI] [PubMed] [Google Scholar]
- Walker JR, Corpina RA, Goldberg J 2001. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature 412: 607–614 [DOI] [PubMed] [Google Scholar]
- Wang YG, Nnakwe C, Lane WS, Modesti M, Frank KM 2004. Phosphorylation and regulation of DNA ligase IV stability by DNA-dependent protein kinase. J Biol Chem 279: 37282–37290 [DOI] [PubMed] [Google Scholar]
- Wang H, Rosidi B, Perrault R, Wang M, Zhang L, Windhofer F, Iliakis G 2005. DNA ligase III as a candidate component of backup pathways of nonhomologous end joining. Cancer Res 65: 4020–4030 [DOI] [PubMed] [Google Scholar]
- Wang M, Wu W, Wu W, Rosidi B, Zhang L, Wang H, Iliakis G 2006. PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucleic Acids Res 34: 6170–6182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinfeld M, Mani RS, Abdou I, Aceytuno RD, Glover JN 2011. Tidying up loose ends: The role of polynucleotide kinase/phosphatase in DNA strand break repair. Trends Biochem Sci 36: 262–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstock DM, Brunet E, Jasin M 2007. Formation of NHEJ-derived reciprocal chromosomal translocations does not require Ku70. Nat Cell Biol 9: 978–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RS, Moncalian G, Williams JS, Yamada Y, Limbo O, Shin DS, Groocock LM, Cahill D, Hitomi C, Guenther G, et al. 2008. Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair. Cell 135: 97–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RS, Dodson GE, Limbo O, Yamada Y, Williams JS, Guenther G, Classen S, Glover JN, Iwasaki H, Russell P, et al. 2009. Nbs1 flexibly tethers Ctp1 and Mre11–Rad50 to coordinate DNA double-strand break processing and repair. Cell 139: 87–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson TE, Lieber MR 1999. Efficient processing of DNA ends during yeast nonhomologous end joining. Evidence for a DNA polymerase β (Pol4)-dependent pathway. J Biol Chem 274: 23599–23609 [DOI] [PubMed] [Google Scholar]
- Woodbine L, Brunton H, Goodarzi AA, Shibata A, Jeggo PA 2011. Endogenously induced DNA double strand breaks arise in heterochromatic DNA regions and require ataxia telangiectasia mutated and Artemis for their repair. Nucleic Acids Res 39: 6986–6997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Topper LM, Wilson TE 2008. Recruitment and dissociation of nonhomologous end joining proteins at a DNA double-strand break in Saccharomyces cerevisiae. Genetics 178: 1237–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu PY, Frit P, Meesala S, Dauvillier S, Modesti M, Andres SN, Huang Y, Sekiguchi J, Calsou P, Salles B, et al. 2009. Structural and functional interaction between the human DNA repair proteins DNA ligase IV and XRCC4. Mol Cell Biol 29: 3163–3172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Ochi T, Matak-Vinkovic D, Robinson CV, Chirgadze DY, Blundell TL 2011. Non-homologous end-joining partners in a helical dance: Structural studies of XLF–XRCC4 interactions. Biochem Soc Trans 39: 1387–1392 [DOI] [PubMed] [Google Scholar]
- Xie A, Kwok A, Scully R 2009. Role of mammalian Mre11 in classical and alternative nonhomologous end joining. Nat Struct Mol Biol 16: 814–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Iwai Y, Sonoda E, Sasaki MS, Morrison C, Haraguchi T, Hiraoka Y, Yamashita YM, Yagi T, Takata M, Price C, et al. 1999. Mre11 is essential for the maintenance of chromosomal DNA in vertebrate cells. EMBO J 18: 6619–6629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y, Akhter S, Zhang X, Legerski R 2010. The multifunctional SNM1 gene family: Not just nucleases. Future Oncol 6: 1015–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaneva M, Kowalewski T, Lieber MR 1997. Interaction of DNA-dependent protein kinase with DNA and with Ku: Biochemical and atomic-force microscopy studies. EMBO J 16: 5098–5112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yannone SM, Khan IS, Zhou RZ, Zhou T, Valerie K, Povirk LF 2008. Coordinate 5′ and 3′ endonucleolytic trimming of terminally blocked blunt DNA double-strand break ends by Artemis nuclease and DNA-dependent protein kinase. Nucleic Acids Res 36: 3354–3365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano K, Chen DJ 2008. Live cell imaging of XLF and XRCC4 reveals a novel view of protein assembly in the non-homologous end-joining pathway. Cell Cycle 7: 1321–1325 [DOI] [PubMed] [Google Scholar]
- Yano K, Morotomi-Yano K, Wang SY, Uematsu N, Lee KJ, Asaithamby A, Weterings E, Chen DJ 2008. Ku recruits XLF to DNA double-strand breaks. EMBO Rep 9: 91–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano K, Morotomi-Yano K, Lee KJ, Chen DJ 2011. Functional significance of the interaction with Ku in DNA double-strand break recognition of XLF. FEBS Lett 585: 841–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu AM, McVey M 2010. Synthesis-dependent microhomology-mediated end joining accounts for multiple types of repair junctions. Nucleic Acids Res 38: 5706–5717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Mahaney BL, Yano K, Ye R, Fang S, Douglas P, Chen DJ, Lees-Miller SP 2008. DNA-PK and ATM phosphorylation sites in XLF/Cernunnos are not required for repair of DNA double strand breaks. DNA Repair (Amst) 7: 1680–1692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun MH, Hiom K 2009. CtIP-BRCA1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle. Nature 459: 460–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zha S, Guo C, Boboila C, Oksenych V, Cheng HL, Zhang Y, Wesemann DR, Yuen G, Patel H, Goff PH, et al. 2011. ATM damage response and XLF repair factor are functionally redundant in joining DNA breaks. Nature 469: 250–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Jasin M 2011. An essential role for CtIP in chromosomal translocation formation through an alternative end-joining pathway. Nat Struct Mol Biol 18: 80–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Paull TT 2005. The Mre11/Rad50/Xrs2 complex and non-homologous end-joining of incompatible ends in S. cerevisiae. DNA Repair (Amst) 4: 1281–1294 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Zhu L, Lin D, Chen F, Chen DJ, Chen Y 2001. The three-dimensional structure of the C-terminal DNA-binding domain of human Ku70. J Biol Chem 276: 38231–38236 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Hu W, Cano L, Lee TD, Chen DJ, Chen Y 2004. Solution structure of the C-terminal domain of Ku80 suggests important sites for protein–protein interactions. Structure 12: 495–502 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Hefferin ML, Chen L, Shim EY, Tseng HM, Kwon Y, Sung P, Lee SE, Tomkinson AE 2007. Role of Dnl4–Lif1 in nonhomologous end-joining repair complex assembly and suppression of homologous recombination. Nat Struct Mol Biol 14: 639–646 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Shim EY, Davis M, Lee SE 2009. Regulation of repair choice: Cdk1 suppresses recruitment of end joining factors at DNA breaks. DNA Repair (Amst) 8: 1235–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Akopiants K, Mohapatra S, Lin PS, Valerie K, Ramsden DA, Lees-Miller SP, Povirk LF 2009. Tyrosyl-DNA phosphodiesterase and the repair of 3′-phosphoglycolate-terminated DNA double-strand breaks. DNA Repair (Amst) 8: 901–911 [DOI] [PMC free article] [PubMed] [Google Scholar]