Abstract
The packaging of mitochondrial DNA (mtDNA) into DNA-protein assemblies called nucleoids provides an efficient segregating unit of mtDNA, coordinating mtDNA’s involvement in cellular metabolism. From the early discovery of mtDNA as “extranuclear” genetic material, its organization into nucleoids and integration into both the mitochondrial organellar network and the cell at large via a variety of signal transduction pathways, mtDNA is a crucial component of the cell’s homeostatic network. The mitochondrial nucleoid is composed of a set of DNA-binding core proteins involved in mtDNA maintenance and transcription, and a range of peripheral factors, which are components of signaling pathways controlling mitochondrial biogenesis, metabolism, apoptosis, and retrograde mitochondria-to-nucleus signaling. The molecular interactions of nucleoid components with the organellar network and cellular signaling pathways provide exciting clues to the dynamic integration of mtDNA into cellular metabolic homeostasis.
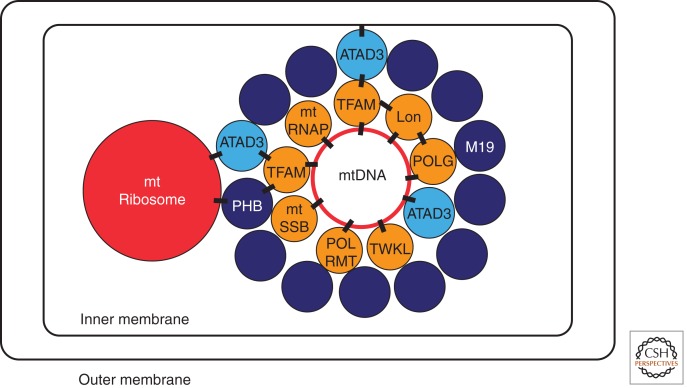
The mitochondrial nucleoid appears to consist of two regions: a core domain (with transcription and replication factors) and a peripheral zone (with proteins that signal to the rest of the cell).
ORIGINS: “EXTRANUCLEAR DNA” AND BIOENERGETICS
Historically, it has been clear that mitochondria play by their own rules. Beginning with the observations that mitochondria independently synthesize protein (McLean et al. 1958) and RNA (Wintersberger 1964), followed by the discovery of DNA within the organelle (Nass et al. 1965; Nass 1969), mitochondria have emerged as a highly distinct organellar system for providing ATP to the cell. The then-controversial endosymbiont theory (Sagan 1967), proposing that mitochondria are descended from proteobacteria engulfed by eukaryotic cells, was later confirmed by the similarity of mtDNA to bacterial, rather than human nuclear, DNA. This “extranuclear” DNA is in fact essential for mitochondrial ATP production (DiMauro and Schon 2003).
The 16,569 bp circular human mtDNA encodes two tRNAs, 22 rRNAs, and 13 polypeptides necessary for the proper assembly and function of the mitochondrial complexes of oxidative phosphorylation (OXPHOS) (Anderson et al. 1981), which are located in the mitochondrial inner membrane. Complexes I, II, III, and IV transfer electrons, ultimately donating them to molecular oxygen, to create the chemiosmotic proton-motive gradient (Δψm) which is used by complex V, the F1F0 ATP synthase, to synthesize ATP from ADP and Pi. These complexes are large, multisubunit macromolecular assemblies, in which all but a small handful of polypeptides are encoded by nuclear genes. The assembly of these complexes requires the coordination of both nuclear and mitochondrial transcription and translation, followed by complex assembly in the mitochondria and insertion into the inner membrane. The necessity of producing and assembling complexes of polypeptides derived from both chromosomal and organellar genomes means that mitochondria are susceptible to mutations in either.
Defects of both nuclear DNA and mtDNA have been shown to have devastating consequences for bioenergetics in human health. Mutations in nuclear-encoded factors such as mitochondrial DNA polymerase γ (POLG) and thymidine kinase 2 (TK2) cause depletion of mtDNA and increased mtDNA mutation, frequently manifesting as neuromuscular disease (Oskoui et al. 2006; Akman et al. 2008). Mutations or depletion of mtDNA cause a range of neuromuscular disorders, such as the classical mitochondrial diseases Kearns-Sayre syndrome (KSS), which is caused by large-scale deletions of mtDNA (Δ-mtDNAs), and myoclonus epilepsy with lactic acidosis and stroke-like episodes (MELAS), which is caused by point mutations in mtDNA-encoded tRNAs (reviewed in DiMauro and Schon 2003; Greaves et al. 2012). Although each of the mitochondrial neuromuscular disorders is individually rare, pathogenic mtDNA mutations as a whole occur at a high frequency of 1:200 individuals (Elliott et al. 2008; Greaves et al. 2012). The study of mitochondrial neuromuscular disorders has uncovered fundamental concepts of genotype-phenotype relationships for mtDNA mutations and mitochondrial function. Typically, wild-type and mutant mtDNAs exist side by side within the same cell and tissue, a situation called heteroplasmy. The relative proportions of the two mtDNA variants is a crucial determinant of mitochondrial function in cells carrying mutant mtDNAs: Cells can typically withstand a very high overall mtDNA mutation load (often ∼80%–90%, depending on the nature of the mutation), but when the mutation load exceeds the threshold of function, the cell can no longer maintain mitochondrial function (DiMauro and Schon 2003).
In addition to the classical mitochondrial diseases, mtDNA mutations and defects are emerging in a variety of prevalent human diseases in disparate tissues, such as Parkinson disease (Bender et al. 2006; Kraytsberg et al. 2006), multiple sclerosis (Campbell et al. 2012), heart failure (Karamanlidis et al. 2010), diabetes (Ritov et al. 2010), and aging (Bender et al. 2006). The crucial role of mtDNA in cellular metabolism and bioenergetics, as well as its profound importance to human health, motivates a better understanding of the elegant complexity of mtDNA’s macromolecular organization and integration into both the organellar network and cell at large. Despite the bioenergetic importance and medical relevance of mtDNA, a number of misconceptions regarding its organization have persisted. MtDNA has often been thought to exist in a “naked” or unprotected state. However, mtDNA is efficiently packaged into a macromolecular assembly that allows for dynamic genetic maintenance and integration into cellular signaling.
THE NUCLEOID: A MACROMOLECULAR ASSEMBLY FOR MITOCHONDRIAL GENETICS
Given their endosymbiotic bacterial origins, it is not surprising that the organization of mtDNA is similar to that of bacterial DNA. The bacterial genome undergoes a 104-fold compaction of its volume to form the bacterial nucleoid (de Vries 2010). Similarly, the mitochondrial nucleoid typically has a diameter of ∼100 nm (Kukat et al. 2011), whereas a completely relaxed 16.6 kb circular DNA would have a length on the order of ∼2 µm within the mitochondrial matrix. Beginning with the original fluorescence micrographs of human mtDNA within the cell (Satoh and Kuroiwa 1991), nucleoids have been clearly evident by microscopy as discrete punctae distributed throughout the mitochondrial network (Alam et al. 2003; Legros et al. 2004). This punctate appearance reveals the unique organization of mtDNA within the organellar network. The mitochondrial nucleoid is composed of a range of proteins with a diverse array of functions, from DNA packaging and transcription to factors with broader signaling functions, facilitating mtDNA’s integration into crucial cell-wide metabolic and proliferative signaling networks. Nucleoid packaging provides an efficient means of ensuring that mitochondrial genetic material is distributed throughout the cell’s mitochondria and is responsive to cellular metabolic needs.
The packaging of mtDNA into nucleoids is mediated by the nucleoid’s protein components. As the different individual protein factors of the nucleoid have been cataloged, the macromolecular structure of the nucleoid has been suggested to consist of a core domain comprised of key mtDNA-associated proteins, and a peripheral zone, consisting of factors which are less constitutively bound, many of which have important signaling roles with the rest of the cell (Fig. 1). The core nucleoid components center around transcription and replication factors such as transcription factor A, mitochondrial (TFAM), mitochondrial single-strand binding protein (mtSSB), POLG, and mtRNA polymerase (POLRMT), which were found in both cross-linked and native nucleoid fractions of HeLa cells (Bogenhagen et al. 2008). TFAM is a 24-kDa protein with two high-mobility group (HMG)-box domains. HMG-family proteins are able to bend, wrap, and unwind DNA. Although it was early identified as a transcription factor for mtDNA (Fisher and Clayton 1985), it has become clear that TFAM plays an equally important role as a packaging protein for mtDNA (Alam et al. 2003). TFAM packages DNA in vitro (Kaufman et al. 2007) and colocalizes with mtDNA in nucleoids by immunomicroscopy (Iborra et al. 2004; Legros et al. 2004). Crystallographic studies have shown that TFAM induces U-turn bends in DNA (Ngo et al. 2011; Rubio-Cosials et al. 2011). Nucleoids tightly package mtDNA, to the extent that freely diffusible matrix protein is excluded from the nucleoid (Brown et al. 2011). Collectively, these studies show that TFAM is a crucial packaging factor for mtDNA, in addition to its role as a key transcription factor for mtDNA. Although TFAM is crucial to mtDNA maintenance, overexpression of TFAM can actually result in excessive binding of mtDNA, effectively choking off transcription of mtDNA (Maniura-Weber et al. 2004). Thus, although TFAM appears to be the major mtDNA packaging protein, the other core constituents of the nucleoid appear to function primarily in replication, transcription, and translation. POLG, POLG2, mtSSB, and the DNA helicase Twinkle (all nucleoid core factors), are the minimal factors necessary for mitochondrial replication (reviewed in Pohjoismaki and Goffart 2011), whereas POLRMT and mtTFB are the minimal factors necessary for mitochondrial transcription. Intriguingly, Shadel and coworkers have found that TFAM is in fact dispensable for transcription in vitro (Shutt et al. 2011), further indicating that TFAM’s primary importance is in mtDNA packaging. The Lon protease has been found to be an integral nucleoid core factor (Lu et al. 2007; Bogenhagen et al. 2008). Similarly, the POLGb subunit has been shown to play a role in the organization of the nucleoid (Di Re et al. 2009), as has the newly discovered CSB factor (associated with Cockayne syndrome) (Berquist et al. 2012). The nucleoid’s core factors thus both package mtDNA to bring together the factors necessary for replication and transcription.
Figure 1.
Schematic of interactions at the mitochondrial nucleoid. Core nucleoid factors (gold) have known mtDNA-binding activity. Peripheral factors (blue) do not directly interact with mtDNA, but associate with the nucleoid via protein–protein interactions. ATAD3 (light blue) associates with mtDNA, the mitochondrial ribosome, and the mitochondrial inner membrane. Shown protein–protein and DNA–protein interactions are designated by black rectangles.
Although a great deal of research has explored the protein composition of the nucleoid, its DNA content has been a crucial question of nucleoid biology. The number of mtDNAs contained within a single nucleoid is a fundamental question of this genetic unit. The studies surrounding this issue have recently come full circle, suggesting that nucleoids carry either one or two mtDNA molecules in a single nucleoid. The early studies of Satoh and Kuroiwa (1991) quantitated the fluorescence of individual nucleoids by ethidium bromide and calculating the quantity of mtDNA contained within it, calibrated against the fluorescence of known quantities of phage DNA. Using this direct, elegant method, they found that a single mitochondrion, on average, carried 4.6 mtDNAs. Further, each mitochondrion carried an average of 3.2 nucleoids, with most mitochondria carrying a single nucleoid. Thus, an individual nucleoid contains 1.4 mtDNAs per nucleoid, suggesting that nucleoids contain either one or two mtDNAs. Subsesquent studies have taken approaches in which the number of nucleoid foci within a cell was counted, followed by quantitation of mtDNA copy number per cell, then obtaining an average value of mtDNAs per nucleoid. Using these estimates, a number of researchers found that rapidly dividing cells, such as HeLa and osteosarcoma cell lines, maintain 2–10 mtDNAs per nucleoid (Iborra et al. 2004; Legros et al. 2004; Gilkerson et al. 2008). More recently, however, investigators have used next-generation imaging methods (Kukat et al. 2011), allowing higher-resolution imaging of the mitochondrial nucleoid in situ. Strikingly, these studies reveal that nucleoids are smaller than previously thought (and often aggregate). These small nucleoids contain an average of 1.4 mtDNAs per nucleoid (Kukat et al. 2011). Intriguingly, this value is identical to that first found by Satoh and Kuroiwa (above), suggesting that this issue has come “full circle” and that a single nucleoid likely consists of only one or two mtDNA molecules.
Integration and Dynamics within the Organellar Network
The highly dynamic, responsive nature of mitochondria as an organellar network has been a revolutionary new frontier in mitochondrial biology. The dynamics of the mitochondrial network involve a complex set of interactions that modulate mitochondrial morphology and shape the organellar network (Nunnari and Suomalainen 2012). Although mitochondria have traditionally been regarded as a population of individual football-shaped organelles, it is clear that mitochondria actually balance between fragmented and networked states of organization in response to both fusion and fission events. Mitochondrial fusion is mediated by mitofusins 1 and 2 (MFN1 and MFN2) and OPA1, whereas fission events are mediated by FIS1, DRP1, and MFF (Twig et al. 2008; Otera et al. 2010). These two pathways allow for modulation of mitochondrial organization in response to a variety of cellular cues. Mitochondrial morphology is linked to function, as a loss of mitochondrial bioenergetic capacity results in an inability to maintain the fused, networked mitochondrial organization (Santra et al. 2004); this loss of interconnectivity is likely owing to the collapse of the mitochondrial transmembrane potential across the inner membrane (Legros et al. 2004). Thus, mitochondrial morphology can provide a visual indication of the relative “health” of a cell’s mitochondria, as cells with genetic or pharmacologic mitochondrial dysfunction show an inability to maintain a united mitochondrial network (Legros et al. 2004; Gilkerson et al. 2008). Defects in mitochondrial organization and trafficking are appearing as a common manifestation of mitochondrial dysfunction in diseases such as Parkinson’s disease and Huntington’s disease (Schon and Przedborski 2011).
The molecular interactions that integrate the nucleoid into this dynamic organellar network are beginning to emerge. Within the organelle, the nucleoid is located within the protein-dense matrix compartment. The factors associated with the nucleoid’s peripheral zone, such as the ATPase AAA domain-containing protein 3 (ATAD3) and prohibitins 1 and 2 (PHB1 and PHB2), do not appear to directly bind mtDNA, but provide molecular scaffolding, help facilitate mitochondrial translation, and interact with important cell-wide signaling pathways. The mitochondrial M19 protein (mitochondrial nucleoid factor 1 [MNF1]) has been identified as a nucleoid-associated protein, and appears to reside in the peripheral zone of the nucleoid (Sumitani et al. 2009). M19 appears to play a role in modulating mitochondrial ATP production, providing a crucial link between mtDNA and mitochondrial bioenergetic function (Cambier et al. 2012). Prohibitin appears to play an important role in stabilizing TFAM at the mitochondrial nucleoid (Kasashima et al. 2008), and is strongly implicated in mitochondrial protein synthesis (He et al. 2012). Prohibitin forms oligomeric rings in the inner mitochondrial membrane that are necessary for the maintenance of mitochondrial morphology (Merkwirth and Langer 2009). Researchers have long postulated that mitochondrial DNA may be “tethered” to the inner membrane (Albring et al. 1977; Van Tuyle and McPherson 1979); however, the actual tethering interactions have remained elusive. More recently, the ATAD3 protein has been shown to associate with both the mitochondrial nucleoid and the inner membrane ( He et al. 2007; Bogenhagen et al. 2008). He et al. (2012) propose that ATAD3 may actually provide a scaffold to which mitochondrial nucleoids and ribosomes may bind to within the matrix (He et al. 2012), tethering both within close proximity to each other at the inner membrane. ATAD3 may play a unique central role in nucleoid organization, as it associates with both the inner membrane and the mitochondrial ribosome, and also binds to D-loop sequences of mtDNA (He et al. 2007, 2012; Bogenhagen et al. 2008). ATAD3 and prohibitin are both required for mitochondrial protein synthesis, and appear to bring the mitochondrial nucleoid and translational machinery into close physical proximity (He et al. 2012), allowing for an efficient coordination of mtDNA-encoded mitochondrial protein synthesis and physical attachment to the mitochondrial inner membrane (Fig. 1). Accordingly, nucleoids do not appear to freely diffuse within the matrix compartment of the mitochondrion. Although it has been shown that fusions of wild-type cells with cells lacking mtDNA (ρ0 cells) result in repopulation of the mtDNA-depleted cells, nucleoids were found to migrate more slowly than GFP targeted to the mitochondrial outer membrane (Legros et al. 2004). This finding indicates that nucleoid movement is limited by their attachment to the mitochondrial inner membrane, rather than freely diffusing through the matrix compartment. Iborra et al. (2004) found that nucleoids move in a “constrained walk,” consistent with more recent findings via superresolution fluorescence microscopy (Brown et al. 2011).
An immediately striking feature of nucleoid organization in fluorescence microscopy is the regularity at which nucleoids appear along mitochondrial filaments, in which mtDNA is distributed evenly throughout the cell by appearing at discrete intervals throughout mitochondria (Iborra et al. 2004; Legros et al. 2004). More striking yet was the observation that, when in a fragmented morphology, all mitochondria within a cell will contain at least one nucleoid (Margineantu et al. 2002)! These observations strongly suggest that mitochondrial nucleoid maintenance is a regulated process, coordinated with the mitochondrial fission and fusion machinery to ensure the efficient distribution of mtDNA throughout the cell, while simultaneously protecting the nucleoid from fission events at the nucleoid itself. Genetic inhibition of mitochondrial fission, via RNAi of DRP1, results in loss of nucleoid content and mtDNA copy number (Parone et al. 2008), whereas disruption of mitochondrial fusion through knockout of MFN1 and MFN2 similarly results in loss of mtDNA copy number (Chen et al. 2011). Live cell imaging of nucleoids revealed that mitochondrial division typically occurs just to the side of nucleoids (Iborra et al. 2004). Thus, it appears very likely that nucleoid factors are intricately connected to the fission and fusion machinery to ensure nucleoid propagation.
Conversations beyond the Organelle: MtDNA and Cellular Signaling
As the organizing unit of mtDNA, the nucleoid is likely to play a crucial role in coordinating organellar response to cellular signaling cues. As mitochondria are increasingly found to participate in crucial cellular signaling networks, a number of nucleoid-associated factors are found to have signaling functions that may determine mitochondrial response to nuclear-encoded signaling factors, particularly those involving metabolism and apoptosis. In addition, there is growing evidence that retrograde mitochondria-to-nucleus cross talk may be mediated by mtDNA and nucleoid-associated factors. Collectively, the nucleoid integrates mtDNA into cell-wide signaling networks, playing a direct role in mitochondrial participation in cellular signaling.
The regulation of mitochondrial content by metabolic signaling pathways has revealed a vital mechanism of nuclear-mitochondrial cross talk. Transcription of the nuclear-encoded mitochondrial OXPHOS complex subunits is controlled by the nuclear respiratory factors 1 and 2 (NRF-1 [Evans and Scarpulla 1990] and NRF-2 [Virbasius et al. 1993]). These NRFs are modulated by the peroxisome proliferation γ-related coactivator (PGC)-1α master regulator (Puigserver et al. 1998) and its related factor, PRC (Scarpulla 2008). Increased PGC-1α signaling leads to elevated levels of nuclear-encoded, mitochondrially localized proteins and thus increased mitochondrial content within the cell. Interestingly, a number of nucleoid-associated factors play direct roles in the modulation of mtDNA copy number, providing direct biogenesis signaling from within the nucleoid. TFAM plays a key role in modulating mtDNA copy number, and has in fact been shown to accurately reflect the overall level of mtDNA within the cell (Ekstrand et al. 2004; Kanki et al. 2004). TFAM, Lon, mtSSB, and NRF-1 and -2 have all been found to be coordinately up-regulated in response to myogenic differentiation (Wagatsuma et al. 2011). Importantly, however, it must be noted that this is a delicate coordination, as overexpression of TFAM can actually inhibit transcription of mtDNA (Matsushima et al. 2010a) and cause mitochondrial dysfunction (Ylikallio et al. 2010), though overexpression of TFAM has been shown to ameliorate mitochondrial dysfunction in neuronal and cardiac settings (Ikeuchi et al. 2005; Hokari et al. 2010). It was shown previously that cells will accurately regulate mtDNA content on the basis of mass of mtDNA, rather than number of copies per se (Tang et al. 2000); this seems consistent with a role for TFAM as the principal means by which the cell regulates mtDNA content. Interestingly, Lon-mediated proteolytic degradation of TFAM is emerging as a major mechanism of mtDNA copy control, in which TFAM levels are regulated by the Lon protease to modulate the overall copy number of mtDNA within the cell (Matsushima et al. 2010b; Ambro et al. 2012; Matsushima and Kaguni 2012). The Lon protease has been identified as a core nucleoid component (Bogenhagen et al. 2008) with the ability to bind mtDNA (Liu et al. 2004). The dynamics of TFAM’s association with mtDNA are still being elucidated, but it is clear that TFAM plays a very elegant role in maintaining mtDNA in nucleoids. Thus, the presence of known metabolic signaling molecules at the nucleoid allows for the efficient, precise modulation of mtDNA content, transcription, translation (via the nearby mitochondrial ribosomes), and thus mitochondrial function, in response to cellular signaling cues.
Nucleoid “Give-and-Take” in Cellular Signaling
Although it is clear that nuclear signaling networks, particularly those involving mitochondrial biogenesis via PGC-1α and related factors, can impact mitochondrial content function, there is also an accumulation of evidence suggesting that mitochondria can communicate with the rest of the cell via retrograde signaling to the nucleus. Experiments in Drosophila melanogaster have shown that genetically induced ablation of mitochondrial genes leads to cell-cycle arrest via either reactive oxygen species (ROS)-induced JNK signaling, or activation of AMP kinase via low ATP levels (Owusu-Ansah et al. 2008). Studies in mammalian systems indicate that either genetically or pharmacologically induced mitochondrial dysfunction results in altered expression of nuclear-encoded signaling factors (Gilkerson et al. 2012). Accordingly, although the loss of mitochondrial function in mtDNA-depleted cells is certainly detrimental to cellular homeostasis, the resultant down-regulation of nuclear transcription of prosurvival signaling molecules, even those not associated with mitochondrial bioenergetics per se, is potentially an even more devastating mechanism of cellular dysfunction. This suggests that nucleoid-associated factors may alter their localization to participate in crucial metabolic signaling networks, consistent with findings from yeast, in which the mitochondrial nucleoid undergoes remodeling in response to metabolic stress (Kucej et al. 2008). Thus, there are likely a range of nucleoid factors that undergo dynamic changes in localization based on cellular signaling cues. Taken as a whole, these interactions reveal the dynamic interplay and integral involvement of the nucleoid in metabolic and apoptotic signaling. Although a range of important signaling functions have been assigned to nucleoid-associated factors, the dynamics of the interactions mediating the nucleoid’s role in crucial signaling networks is only beginning to be understood. mtDNA and the nucleoid may thus be at the front line of environmental interactions.
ACKNOWLEDGMENTS
We thank Dr. Eric A. Schon for comments on the manuscript. This work is supported by a University of Texas-Pan American Faculty Research Council Award (R.G.) and the National Science Foundation Undergraduate Research Mentoring program DBI 0934013 (N.G. and B.Q.).
Footnotes
Editors: Douglas C. Wallace and Richard J. Youle
Additional Perspectives on Mitochondria available at www.cshperspectives.org
REFERENCES
- Akman HO, Dorado B, Lopez LC, Garcia-Cazorla A, Vila MR, Tanabe LM, Dauer WT, Bonilla E, Tanji K, Hirano M 2008. Thymidine kinase 2 (H126N) knockin mice show the essential role of balanced deoxynucleotide pools for mitochondrial DNA maintenance. Hum Mol Genet 17: 2433–2440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam TI, Kanki T, Muta T, Ukaji K, Abe Y, Nakayama H, Takio K, Hamasaki N, Kang D 2003. Human mitochondrial DNA is packaged with TFAM. Nucleic Acids Res 31: 1640–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albring M, Griffith J, Attardi G 1977. Association of a protein structure of probable membrane derivation with HeLa cell mitochondrial DNA near its origin of replication. Proc Natl Acad Sci 74: 1348–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambro L, Pevala V, Bauer J, Kutejova E 2012. The influence of ATP-dependent proteases on a variety of nucleoid-associated processes. J Struct Biol 179: 181–192 [DOI] [PubMed] [Google Scholar]
- Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, et al. 1981. Sequence and organization of the human mitochondrial genome. Nature 290: 457–465 [DOI] [PubMed] [Google Scholar]
- Bender A, Krishnan KJ, Morris CM, Taylor GA, Reeve AK, Perry RH, Jaros E, Hersheson JS, Betts J, Klopstock T, et al. 2006. High levels of mitochondrial DNA deletions in substantia nigra neurons in aging and Parkinson disease. Nat Genet 38: 515–517 [DOI] [PubMed] [Google Scholar]
- Berquist BR, Canugovi C, Sykora P, Wilson DM III, Bohr VA 2012. Human Cockayne syndrome B protein reciprocally communicates with mitochondrial proteins and promotes transcriptional elongation. Nucl Acids Res 40: 8392–8405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogenhagen DF, Rousseau D, Burke S 2008. The layered structure of human mitochondrial DNA nucleoids. J Biol Chem 283: 3665–3675 [DOI] [PubMed] [Google Scholar]
- Brown TA, Tkachuk AN, Shtengel G, Kopek BG, Bogenhagen DF, Hess HF, Clayton DA 2011. Superresolution fluorescence imaging of mitochondrial nucleoids reveals their spatial range, limits, and membrane interaction. Mol Cell Biol 31: 4994–5010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambier L, Rassam P, Chabi B, Mezghenna K, Gross R, Eveno E, Auffray C, Wrutniak-Cabello C, Lajoix AD, Pomies P 2012. M19 modulates skeletal muscle differentiation and insulin secretion in pancreatic β-cells through modulation of respiratory chain activity. PLoS ONE 7: e31815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell GR, Kraytsberg Y, Krishnan KJ, Ohno N, Ziabreva I, Reeve A, Trapp BD, Newcombe J, Reynolds R, Lassmann H, et al. 2012. Clonally expanded mitochondrial DNA deletions within the choroid plexus in multiple sclerosis. Acta Neuropath 124: 209–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Liu Y, Dorn GW II 2011. Mitochondrial fusion is essential for organelle function and cardiac homeostasis. Circ Res 109: 1327–1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries R 2010. DNA condensation in bacteria: Interplay between macromolecular crowding and nucleoid proteins. Biochimie 92: 1715–1721 [DOI] [PubMed] [Google Scholar]
- DiMauro S, Schon EA 2003. Mitochondrial respiratory-chain diseases. N Engl J Med 348: 2656–2668 [DOI] [PubMed] [Google Scholar]
- Di Re M, Sembongi H, He J, Reyes A, Yasukawa T, Martinsson P, Bailey LJ, Goffart S, Boyd-Kirkup JD, Wong TS, et al. 2009. The accessory subunit of mitochondrial DNA polymerase γ determines the DNA content of mitochondrial nucleoids in human cultured cells. Nucl Acids Res 37: 5701–5713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekstrand MI, Falkenberg M, Rantanen A, Park CB, Gaspari M, Hultenby K, Rustin P, Gustafsson CM, Larsson NG 2004. Mitochondrial transcription factor A regulates mtDNA copy number in mammals. Hum Mol Gen 13: 935–944 [DOI] [PubMed] [Google Scholar]
- Elliott HR, Samuels DC, Eden JA, Relton CL, Chinnery PF 2008. Pathogenic mitochondrial DNA mutations are common in the general population. Am J Hum Genet 83: 254–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans MJ, Scarpulla RC 1990. NRF-1: A trans-activator of nuclear-encoded respiratory genes in animal cells. Genes Dev 4: 1023–1034 [DOI] [PubMed] [Google Scholar]
- Fisher RP, Clayton DA 1985. A transcription factor required for promoter recognition by human mitochondrial RNA polymerase. Accurate initiation at the heavy- and light-strand promoters dissected and reconstituted in vitro. J Biol Chem 260: 11330–11338 [PubMed] [Google Scholar]
- Gilkerson RW, Schon EA, Hernandez E, Davidson MM 2008. Mitochondrial nucleoids maintain genetic autonomy but allow for functional complementation. J Cell Biol 181: 1117–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilkerson RW, De Vries RL, Lebot P, Wikstrom JD, Torgyekes E, Shirihai OS, Przedborski S, Schon EA 2012. Mitochondrial autophagy in cells with mtDNA mutations results from synergistic loss of transmembrane potential and mTORC1 inhibition. Hum Mol Genet 21: 978–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaves LC, Reeve AK, Taylor RW, Turnbull DM 2012. Mitochondrial DNA and disease. J Pathol 226: 274–286 [DOI] [PubMed] [Google Scholar]
- He J, Mao CC, Reyes A, Sembongi H, Di Re M, Granycome C, Clippingdale AB, Fearnley IM, Harbour M, Robinson AJ, et al. 2007. The AAA+ protein ATAD3 has displacement loop binding properties and is involved in mitochondrial nucleoid organization. J Cell Biol 176: 141–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Cooper HM, Reyes A, Di Re M, Sembongi H, Litwin TR, Gao J, Neuman KC, Fearnley IM, Spinazzola A, et al. 2012. Mitochondrial nucleoid interacting proteins support mitochondrial protein synthesis. Nucl Acids Res 40: 6109–6121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hokari M, Kuroda S, Kinugawa S, Ide T, Tsutsui H, Iwasaki Y 2010. Overexpression of mitochondrial transcription factor A (TFAM) ameliorates delayed neuronal death due to transient forebrain ischemia in mice. Neuropathol 30: 401–407 [DOI] [PubMed] [Google Scholar]
- Iborra FJ, Kimura H, Cook PR 2004. The functional organization of mitochondrial genomes in human cells. BMC Biol 2: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeuchi M, Matsusaka H, Kang D, Matsushima S, Ide T, Kubota T, Fujiwara T, Hamasaki N, Takeshita A, Sunagawa K, et al. 2005. Overexpression of mitochondrial transcription factor A ameliorates mitochondrial deficiencies and cardiac failure after myocardial infarction. Circulation 112: 683–690 [DOI] [PubMed] [Google Scholar]
- Kanki T, Ohgaki K, Gaspari M, Gustafsson CM, Fukuoh A, Sasaki N, Hamasaki N, Kang D 2004. Architectural role of mitochondrial transcription factor A in maintenance of human mitochondrial DNA. Mol Cell Biol 24: 9823–9834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karamanlidis G, Nascimben L, Couper GS, Shekar PS, del Monte F, Tian R 2010. Defective DNA replication impairs mitochondrial biogenesis in human failing hearts. Circ Res 106: 1541–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasashima K, Sumitani M, Satoh M, Endo H 2008. Human prohibitin 1 maintains the organization and stability of the mitochondrial nucleoids. Exp Cell Res 314: 988–996 [DOI] [PubMed] [Google Scholar]
- Kaufman BA, Durisic N, Mativetsky JM, Costantino S, Hancock MA, Grutter P, Shoubridge EA 2007. The mitochondrial transcription factor TFAM coordinates the assembly of multiple DNA molecules into nucleoid-like structures. Mol Biol Cell 18: 3225–3236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraytsberg Y, Kudryavtseva E, McKee AC, Geula C, Kowall NW, Khrapko K 2006. Mitochondrial DNA deletions are abundant and cause functional impairment in aged human substantia nigra neurons. Nat Genet 38: 518–520 [DOI] [PubMed] [Google Scholar]
- Kucej M, Kucejova B, Subramanian R, Chen XJ, Butow RA 2008. Mitochondrial nucleoids undergo remodeling in response to metabolic cues. J Cell Sci 121: 1861–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukat C, Wurm CA, Spahr H, Falkenberg M, Larsson NG, Jakobs S 2011. Super-resolution microscopy reveals that mammalian mitochondrial nucleoids have a uniform size and frequently contain a single copy of mtDNA. Proc Natl Acad Sci 108: 13534–13539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legros F, Malka F, Frachon P, Lombes A, Rojo M 2004. Organization and dynamics of human mitochondrial DNA. J Cell Sci 117: 2653–2662 [DOI] [PubMed] [Google Scholar]
- Liu T, Lu B, Lee I, Ondrovicova G, Kutejova E, Suzuki CK 2004. DNA and RNA binding by the mitochondrial lon protease is regulated by nucleotide and protein substrate. J Biol Chem 279: 13902–13910 [DOI] [PubMed] [Google Scholar]
- Lu B, Yadav S, Shah PG, Liu T, Tian B, Pukszta S, Villaluna N, Kutejova E, Newlon CS, Santos JH, et al. 2007. Roles for the human ATP-dependent Lon protease in mitochondrial DNA maintenance. J Biol Chem 282: 17363–17374 [DOI] [PubMed] [Google Scholar]
- Maniura-Weber K, Goffart S, Garstka HL, Montoya J, Wiesner RJ 2004. Transient overexpression of mitochondrial transcription factor A (TFAM) is sufficient to stimulate mitochondrial DNA transcription, but not sufficient to increase mtDNA copy number in cultured cells. Nucl Acids Res 32: 6015–6027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margineantu DH, Gregory Cox W, Sundell L, Sherwood SW, Beechem JM, Capaldi RA 2002. Cell cycle dependent morphology changes and associated mitochondrial DNA redistribution in mitochondria of human cell lines. Mitochondrion 1: 425–435 [DOI] [PubMed] [Google Scholar]
- Matsushima Y, Kaguni LS 2012. Matrix proteases in mitochondrial DNA function. Biochim Biophys Acta 1819: 1080–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushima Y, Goto Y, Kaguni LS 2010a. Mitochondrial Lon protease regulates mitochondrial DNA copy number and transcription by selective degradation of mitochondrial transcription factor A (TFAM). Proc Natl Acad Sci 107: 18410–18415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushima Y, Goto Y, Kaguni LS 2010b. Mitochondrial Lon protease regulates mitochondrial DNA copy number and transcription by selective degradation of mitochondrial transcription factor A (TFAM). Proc Natl Acad Sci 107: 18410–18415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean JR, Cohn GL, Brandt IK, Simpson MV 1958. Incorporation of labeled amino acids into the protein of muscle and liver mitochondria. J Biol Chem 233: 657–663 [PubMed] [Google Scholar]
- Merkwirth C, Langer T 2009. Prohibitin function within mitochondria: Essential roles for cell proliferation and cristae morphogenesis. Biochim Biophys Acta 1793: 27–32 [DOI] [PubMed] [Google Scholar]
- Nass S 1969. The significance of the structural and functional similarities of bacteria and mitochondria. Int Rev Cytol 25: 55–129 [DOI] [PubMed] [Google Scholar]
- Nass S, Nass MM, Hennix U 1965. Deoxyribonucleic acid in isolated rat-liver mitochondria. Biochim Biophys Acta 95: 426–435 [DOI] [PubMed] [Google Scholar]
- Ngo HB, Kaiser JT, Chan DC 2011. The mitochondrial transcription and packaging factor Tfam imposes a U-turn on mitochondrial DNA. Nat Struct Mol Biol 18: 1290–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunnari J, Suomalainen A 2012. Mitochondria: In sickness and in health. Cell 148: 1145–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oskoui M, Davidzon G, Pascual J, Erazo R, Gurgel-Giannetti J, Krishna S, Bonilla E, De Vivo DC, Shanske S, DiMauro S 2006. Clinical spectrum of mitochondrial DNA depletion due to mutations in the thymidine kinase 2 gene. Archiv Neurol 63: 1122–1126 [DOI] [PubMed] [Google Scholar]
- Otera H, Wang C, Cleland MM, Setoguchi K, Yokota S, Youle RJ, Mihara K 2010. Mff is an essential factor for mitochondrial recruitment of Drp1 during mitochondrial fission in mammalian cells. J Cell Biol 191: 1141–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owusu-Ansah E, Yavari A, Mandal S, Banerjee U 2008. Distinct mitochondrial retrograde signals control the G1-S cell cycle checkpoint. Nat Genet 40: 356–361 [DOI] [PubMed] [Google Scholar]
- Parone PA, Da Cruz S, Tondera D, Mattenberger Y, James DI, Maechler P, Barja F, Martinou JC 2008. Preventing mitochondrial fission impairs mitochondrial function and leads to loss of mitochondrial DNA. PLoS ONE 3: e3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohjoismaki JL, Goffart S 2011. Of circles, forks and humanity: Topological organisation and replication of mammalian mitochondrial DNA. Bioessays 33: 290–299 [DOI] [PubMed] [Google Scholar]
- Puigserver P, Wu Z, Park CW, Graves R, Wright M, Spiegelman BM 1998. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell 92: 829–839 [DOI] [PubMed] [Google Scholar]
- Ritov VB, Menshikova EV, Azuma K, Wood R, Toledo FG, Goodpaster BH, Ruderman NB, Kelley DE 2010. Deficiency of electron transport chain in human skeletal muscle mitochondria in type 2 diabetes mellitus and obesity. Amer J Physiol 298: E49–E58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio-Cosials A, Sidow JF, Jimenez-Menendez N, Fernandez-Millan P, Montoya J, Jacobs HT, Coll M, Bernado P, Sola M 2011. Human mitochondrial transcription factor A induces a U-turn structure in the light strand promoter. Nat Struct Mol Biol 18: 1281–1289 [DOI] [PubMed] [Google Scholar]
- Sagan L 1967. On the origin of mitosing cells. J Theor Biol 14: 255–274 [DOI] [PubMed] [Google Scholar]
- Santra S, Gilkerson RW, Davidson M, Schon EA 2004. Ketogenic treatment reduces deleted mitochondrial DNAs in cultured human cells. Ann Neurol 56: 662–669 [DOI] [PubMed] [Google Scholar]
- Satoh M, Kuroiwa T 1991. Organization of multiple nucleoids and DNA molecules in mitochondria of a human cell. Exp Cell Res 196: 137–140 [DOI] [PubMed] [Google Scholar]
- Scarpulla RC 2008. Nuclear control of respiratory chain expression by nuclear respiratory factors and PGC-1-related coactivator. Ann NY Acad Sci 1147: 321–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schon EA, Przedborski S 2011. Mitochondria: The next (neurode)generation. Neuron 70: 1033–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shutt TE, Bestwick M, Shadel GS 2011. The core human mitochondrial transcription initiation complex: It only takes two to tango. Transcription 2: 55–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumitani M, Kasashima K, Ohta E, Kang D, Endo H 2009. Association of a novel mitochondrial protein M19 with mitochondrial nucleoids. J Biochem 146: 725–732 [DOI] [PubMed] [Google Scholar]
- Tang Y, Schon EA, Wilichowski E, Vazquez-Memije ME, Davidson E, King MP 2000. Rearrangements of human mitochondrial DNA (mtDNA): New insights into the regulation of mtDNA copy number and gene expression. Mol Biol Cell 11: 1471–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twig G, Elorza A, Molina AJ, Mohamed H, Wikstrom JD, Walzer G, Stiles L, Haigh SE, Katz S, Las G, et al. 2008. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J 27: 433–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Tuyle GC, McPherson ML 1979. A compact form of rat liver mitochondrial DNA stabilized by bound proteins. J Biol Chem 254: 6044–6053 [PubMed] [Google Scholar]
- Virbasius JV, Virbasius CA, Scarpulla RC 1993. Identity of GABP with NRF-2, a multisubunit activator of cytochrome oxidase expression, reveals a cellular role for an ETS domain activator of viral promoters. Genes Dev 7: 380–392 [DOI] [PubMed] [Google Scholar]
- Wagatsuma A, Kotake N, Yamada S 2011. Muscle regeneration occurs to coincide with mitochondrial biogenesis. Mol Cell Biochem 349: 139–147 [DOI] [PubMed] [Google Scholar]
- Wintersberger E 1964. [DNA-dependent RNA synthesis in rat liver mitochondria]. Hoppe-Seyler’s Zeit Physiol Chemie 336: 285–288 [DOI] [PubMed] [Google Scholar]
- Ylikallio E, Tyynismaa H, Tsutsui H, Ide T, Suomalainen A 2010. High mitochondrial DNA copy number has detrimental effects in mice. Hum Mol Genet 19: 2695–2705 [DOI] [PubMed] [Google Scholar]