Abstract
Telomeres are the physical ends of eukaryotic chromosomes. They protect chromosome ends from DNA degradation, recombination, and DNA end fusions, and they are important for nuclear architecture. Telomeres provide a mechanism for their replication by semiconservative DNA replication and length maintenance by telomerase. Through telomerase repression and induced telomere shortening, telomeres provide the means to regulate cellular life span. In this review, we introduce the current knowledge on telomere composition and structure. We then discuss in depth the current understanding of how telomere components mediate their function during semiconservative DNA replication and how telomerase is regulated at the end of the chromosome. We focus our discussion on the telomeres from mammals and the yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe.
Most telomeric DNA is replicated by semiconservative DNA replication. The leading-strand telomere shortens (to generate the 3′ overhang required by telomerase); it is unclear whether the lagging-strand telomere shortens.
Herman Muller and Barbara McClintock were probably the first scientists to recognize that the ends of eukaryotic chromosomes have special properties and crucial functions (reviewed by Blackburn 2006). Herman Muller studied Drosophila chromosomes and found that X rays could induce chromosome rearrangements. However, he never observed chromosomes carrying terminal deletions (Muller 1938). These results led him to conclude that “the terminal gene must have a special function, that of sealing the end of the chromosome, so to speak,” and that “for some reason, a chromosome cannot persist indefinitely without having its ends thus sealed.” Muller coined the term “telomere” for this terminal gene (from the Greek words telos = end and meros = part). Barbara McClintock analyzed maize strains in which dicentric chromosomes were produced with high frequency. Dicentric chromosomes break when the two centromeres are pulled toward opposite poles of the mitotic spindle during anaphase of the cell cycle. The broken chromosomal ends were unstable and fused with other broken ends with which they came in contact. However, when dicentric chromosomes were present in embryonic cells, the broken ends were somehow “healed” (McClintock 1941).
The molecular sequence of telomeric DNA was first determined by Elizabeth Blackburn and Joseph Gall in the ciliated protozoan Tetrahymena thermophila (Blackburn and Gall 1978). These experiments revealed that the DNA ends in this organism consist of 5′-T2G4-3′/5′-C4A2-3′ telomeric repeats with the G-rich strand running toward the 3′ end of the chromosome. Cloning and sequencing of telomeres from other eukaryotes revealed that the repetitive nature of telomeric repeats and the presence of a G-rich strand are common features of nearly all eukaryotes. Furthermore, it has been established that the 3′-end-containing strand protrudes at both ends of the chromosome. However, notable exceptions do exist. The roundworm Caenorhabditis elegans contains both 3′ G-overhangs as well as 5′ C-overhangs (Raices et al. 2008). Several genera of insects and plants do contain long DNA repeats (Martinez et al. 2001). Drosophila contains, instead of short telomeric repeats, retrotransposons at chromosome ends that have overtaken telomere functions (Pardue and Debaryshe 2011).
The formulation of the DNA end-replication problem by Watson and Olovnikov inferred that specialized mechanisms must exist to maintain telomere length (Olovnikov 1971; Watson 1972). Greider and Blackburn discovered telomerase activity in extracts from Tetrahymena (Greider and Blackburn 1985). They also identified the telomerase RNA moiety in this organism, which contains a sequence complementary to Tetrahymena telomeric repeats (Greider and Blackburn 1989). Mutation of this sequence proved that the telomerase RNA moiety provides the template for DNA repeat synthesis and that the telomerase functions as reverse transcriptase (Yu et al. 1990). Around the same time, Lundblad and Szostak were the first to discover an essential telomere maintenance protein gene in Saccharomyces cerevisiae. Dysfunction of the identified gene gave rise to an ever shorter telomeres (est) phenotype culminating in cellular senescence (Lundblad and Szostak 1989). The discovery of EST1 was followed by the identification of EST2, EST3, and EST4 in the Lundblad laboratory (Lendvay et al. 1996). Est2 turned out to be orthologous with the p123 telomerase reverse transcriptase (Tert) protein subunit (Lingner et al. 1997) that was identified in the ciliate Euplotes aediculatus upon biochemical purification of the telomerase enzyme (Lingner and Cech 1996). The identification of the first Tert subunits opened the door to identify the corresponding genes in humans and other organisms, allowing the study of telomerase during development and in cancer. Significantly, it was shown that ectopic expression of human Tert in various primary human cells is sufficient to reconstitute telomerase, rendering them immortal (Bodnar et al. 1998).
Ciliated protozoa were also instrumental in purifying and cloning the first eukaryotic telomere-binding proteins (Gottschling and Zakian 1986; Hicke et al. 1990; Gray et al. 1991). However, orthologs of these proteins were at first not recognized in other eukaryotes, and independent approaches were undertaken to identify the first telomeric proteins in various eukaryotes including yeasts and humans. Only much later was it realized that the ciliate telomere-binding proteins contain counterparts in a broad range of eukaryotes (Baumann and Cech 2001).
In this review, we introduce known basic components of telomeres and review the mechanisms of semiconservative DNA replication of telomeric DNA and the regulation of telomerase. For the discussion of telomere protection and the three-dimensional structures of telomere components, the reader is referred to excellent recent reviews (de Lange 2009; Jain and Cooper 2010; Lewis and Wuttke 2012).
TELOMERE COMPONENTS
In the following section, the stable constituents of telomeres that are implicated in regulating telomere replication are discussed. Abundant telomere-binding proteins as well as telomerase subunits from S. cerevisiae, Schizosaccharomyces pombe, and Homo sapiens described in detail in the text are summarized in Table 1.
Table 1.
Abundant telomere-binding proteins and telomerase proteins in S. cerevisiae, S. pombe, and H. sapiens
| Organism | Subcomplex | Protein | Interaction partner | Ortholog | Function |
|---|---|---|---|---|---|
| S. cerevisiae | Rap1 | Sir3, Sir4, or Rif1, Rif2 | Rap1 (S. p., H. s.) | Essential; major double-stranded telomere-binding protein; transcriptional regulation of telomeres and protein-coding genes; protection from NHEJ; negative regulator of telomere length | |
| Sirtuins | Sir2 | Sir3, Sir4 | NAD+-dependent histone deacetylase; telomeric silencing | ||
| Sir3 | Sir2, Sir4, Rap1 | Telomeric silencing | |||
| Sir4 | Sir2, Sir3, Rap1 | Telomeric silencing | |||
| Rifs | Rif1 | Rif2, Rap1 | Negative regulator of telomere length | ||
| Rif2 | Rif1, Rap1 | Negative regulator of telomere length | |||
| CST | Cdc13 | Ten1, Stn1, Est1, Pol1 | Ctc1 (H. s.; limited sequence similarity) | Essential G-strand single-stranded telomere-binding protein; protects from C-strand loss; recruits telomerase through interaction with Est1; essential for telomerase activity in vivo but not in vitro; interacts with Pol1, the catalytic subunit of DNA polymerase α-primase, promoting fill-in synthesis of telomerase-elongated telomeres | |
| Stn1 | Ten1, Cdc13, Pol12 | Stn1 (S. p., H. s.) | Essential; protects from C-strand loss; negative regulator of telomere length; interacts with Pol12, the B subunit of DNA polymerase α-primase, promoting fill-in synthesis of telomerase-elongated telomeres | ||
| Ten1 | Stn1, Cdc13 | Ten1 (S. p., H. s.) | Essential; protects from C-strand loss; negative regulator of telomere length | ||
| Telomerase | Est1 | Tlc1, Est2, Cdc13 | Est1 (S. p., H. s. Est1A has limited sequence similarity) | est phenotype; binds Tlc1 and G-strand single-stranded telomeric DNA; recruits telomerase through interaction with Cdc13; essential for telomerase activity in vivo but not in vitro | |
| Est2 | Tlc1, Est1 | Tert (H. s.), Trt1 (S. p.) | est phenotype; telomerase reverse transcriptase; essential for telomerase activity in vivo and in vitro | ||
| Est3 | Est1, Est2 | est phenotype; essential for telomerase activity in vivo but not in vitro | |||
| Sm proteins | Smb1, Smd1, Smd2, Smd3, Sme1, Smx3, Smx2 | Form heteroheptameric Sm ring that binds and stabilizes Tlc1 | |||
| S. pombe | Taz1 | Rif1, Rap1 | Trf1, Trf2 (H. s., S. c. Tbf1 has limited sequence similarity) | Major double-stranded telomere-binding protein; protection from NHEJ; negative regulator of telomere length; promotes telomere replication; telomeric silencing and TERRA repression; meiosis | |
| Rif1 | Taz1 | Negative regulator of telomere length; recruited by Taz1 | |||
| Rap1 | Taz1, Poz1 | Rap1 (S. c., H. s.) | Negative regulator of telomere length; recruited by Taz1; telomeric silencing and TERRA repression | ||
| Poz1 | Rap1, Tpz1 | Pot1 (H. s.) | Negative regulator of telomere length | ||
| Tpz1 | Poz1, Pot1, Ccq1 | Tpp1 (H. s.) | Protects telomeric 3′ overhang; telomerase recruitment through interaction with Poz1 and Ccq1 | ||
| Pot1 | Tpz1 | Pot1 (H. s.) | Essential; single-stranded G-strand telomere-binding protein; protects telomeres from nucleolytic degradation | ||
| Ccq1 | Tpz1, Est1 | Telomerase recruitment through interaction with Est1; meiosis | |||
| Ten1 | Stn1 | Ten1 (S. c., H. s.) | Telomere-capping protein; protects from telomere loss | ||
| Stn1 | Ten1 | Stn1 (S. c., H. s.) | Telomere-capping protein; needed for telomere maintenance | ||
| Telomerase | Est1 | Ter1, Ccq1, Trt1 | Est1 (S. c.) | est phenotype; binds Ter1, which has an Sm protein-binding motif; recruits telomerase through interaction with Ccq1; essential for telomerase activity in vivo but not in vitro | |
| Trt1 | Ter1, Est1 | est phenotype; telomerase reverse transcriptase; essential for telomerase activity in vivo and in vitro | |||
| Sm proteins | SmB, SmD1, SmD2, SmD3, SmE, SmF, SmG | Form heteroheptameric Sm ring that binds Ter1 precursor and promotes spliceosomal cleavage; promotes hypermethylation of the Ter1 5′ cap by Tgs1 | |||
| Lsm proteins | Lsm2–8 | Bind mature Ter1 and promote association with Trt1; protect Ter1 from nucleolytic degradation | |||
| H. sapiens | Shelterin | Trf1 | Tin2 | Taz1 (S. p.) | Double-stranded homodimeric telomere-binding protein; negative regulator of telomere length; promotes telomere replication |
| Trf2 | Rap1, Tin2 | Trf1 (H. s.), Taz1 (S. p.) | Double-stranded homodimeric telomere-binding protein; protection from NHEJ; repression of Atm | ||
| Rap1 | Trf2 | Rap1 (S. c., S. p.) | Recruited by Trf2; repression of telomere recombination and homology-directed repair | ||
| Tin2 | Trf1, Trf2, Tpp1 | Stabilization of shelterin complex; recruits telomerase through interaction with Tpp1 | |||
| Tpp1 | Pot1, Tin2, Tert | Tpz1 (S. p.) | Increased affinity of Pot1 for single-stranded telomeric DNA; stimulation of telomerase processivity in conjunction with Pot1; recruits telomerase through interaction with Tin2 and telomerase | ||
| Pot1 | Tpp1 | Pot1 (S. p.) | Binds single-stranded G-strand telomeric DNA; inhibits telomerase in absence of Tpp1; Pot1–Tpp1 promotes telomerase processivity; repression of Atr | ||
| CST | Ctc1 | Stn1, Ten1 | Cdc13 (S. c.; limited sequence similarity) | When assembled as CST, it binds single-stranded DNA with preference for telomeric G-strand sequence; inhibits telomerase activity through direct interaction with Pot1/Tpp1 and primer sequestration; increased telomere binding upon telomerase-mediated telomere elongation; competes with Pot1/Tpp1 for binding of telomeric DNA; with Stn1, Ctc1 stimulates affinity of DNA polymerase α-primase for template DNA; semiconservative DNA replication of specialized DNA sequences | |
| Stn1 | Ten1, Ctc1 | Stn1 (S. p., S. c.) | See Ctc1 | ||
| Ten1 | Ctc1, Stn1 | Ten1 (S. p., S. c.) | See Ctc1 | ||
| Telomerase | Tert | Terc, Tpp1 | Telomerase reverse transcriptase; essential for telomerase activity in vivo and in vitro | ||
| Tcab1 | Telomerase localization to Cajal bodies and to telomeres in S phase; essential for telomerase activity in vivo but not in vitro | ||||
| Dyskerin | Nop10, Nhp2, Gar1 | Binds H/ACA RNA motif–containing RNAs including Terc; Terc stability | |||
| Nop10 | Dyskerin, Nhp2 | See Dyskerin | |||
| Nhp2 | Dyskerin, Nop10 | See Dyskerin | |||
| Gar1 | Dyskerin | See Dyskerin | |||
| Est1a | Terc, Tert | Est1 (S. p. and S. c. Est1 have limited sequence similarity; no evidence for conserved functional role in telomerase recruitment) | Negative regulator of TERRA at telomeres; depletion leads to stochastic telomere loss; overexpression leads to telomere uncapping and telomere fusions; involved in nonsense-mediated mRNA decay |
NHEJ, nonhomologous end joining; TERRA, telomeric repeat–containing RNA; CST, Cdc13–Stn1–Ten1.
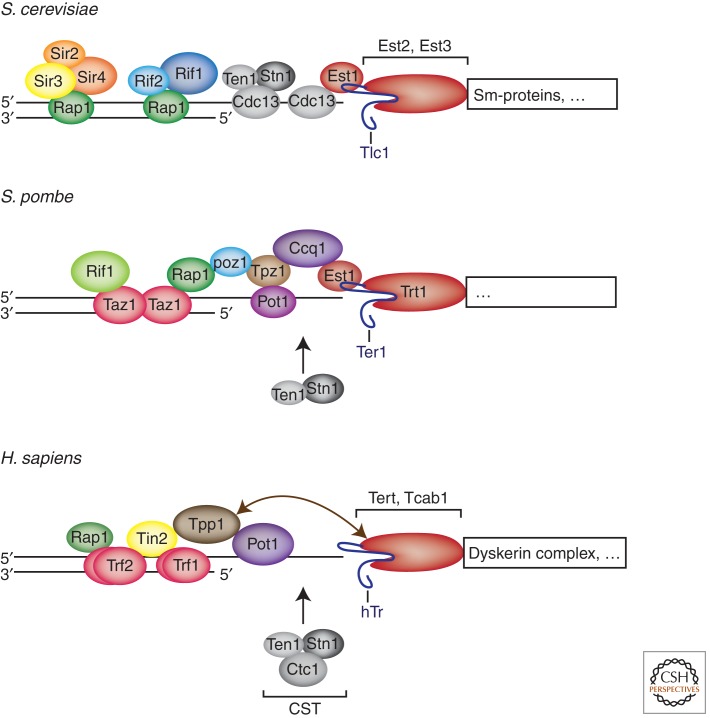
S. cerevisiae
The telomeric DNA repeat consensus sequence for the 3′-end-containing strand from S. cerevisiae is 5′-(TG)0–6TGGGTGTG(G)-3′ (Forstemann and Lingner 2001). The telomere length is around 300 bp. Native chromosome ends in S. cerevisiae have a number of subtelomeric repeat elements termed X and Y′, adjacent to the telomeric repeats, which vary between ends and strains (Louis and Borts 1995). The 473-bp core X is found at all chromosome ends. It contains an ACS (ARS consensus sequence, the binding site for the yeast origin recognition complex) and an Abf1p-binding site at 31 out of 32 ends. There are also binding sites for the telomere-binding protein Tbf1 (Bilaud et al. 1996). So-called Y′ elements with a length of 6.7 kb or 5.2 kb are present at approximately half of S. cerevesiae telomeres.
The telomeric repeat sequence is bound by the double-stranded DNA-binding protein Rap1 (repressor activator protein 1) (Fig. 1) (Lustig et al. 1990). Rap1 also binds chromosome internal sites acting as positive transcriptional regulator of genes involved in growth control. Rap1 has two central Myb domains responsible for DNA binding and a Rap1 carboxy-terminal domain (RCT). RCT recruits the Rif proteins, which repress telomerase (see below), and it recruits the Sir3 and Sir4 proteins, which in turn interact with the Sir2 NAD+-dependent histone deacetylase involved in transcriptional repression. Rap1 is also important for protection from end fusions (Marcand et al. 2008).
Figure 1.
Telomeric proteins and telomerases at yeast and human telomeres. Only abundant telomere-binding proteins and telomerase components are indicated. Trf1 and Trf2 bind as homodimers to the double-stranded telomeric DNA repeats. Homologous proteins are shown in the same color.
The telomeric 3′ overhang is bound by a trimeric complex consisting of Cdc13, Stn1, and Ten1 (CST) (Fig. 1) (Gao et al. 2007). Despite binding to the G-strand, the CST complex protects the C-strand from 5′-3′ resection. In addition, CST regulates telomere length through telomerase (see below). CST has been proposed to resemble in structure the replication protein A (RPA) complex. CST contains as RPA multiple oligosaccharide/oligonucleotide binding (OB)-folds. In particular, Stn1 and Ten1 proteins contain structural similarities with Rpa2 and Rpa3 (Gao et al. 2007).
Telomeres in S. cerevisiae are transcribed by RNA polymerase II into the long noncoding telomeric repeat-containing RNA (TERRA) (Luke et al. 2008). TERRA transcription starts in the subtelomeric DNA and proceeds approximately 100 nucleotides into the telomeric tract. S. cerevisiae TERRA carries a cap structure at the 5′ end and is polyadenylated. TERRA is degraded by the 5′-3′ exonuclease Rat1. TERRA transcription is negatively regulated by the Sir proteins at telomeres that contain only the X subtelomeric element at their chromosome ends. At Y′ elements containing telomeres, TERRA transcription is negatively regulated by the Rif proteins (Iglesias et al. 2011).
Telomeres in S. cerevisiae have been inferred to form fold-back or looped structures because Rap1 is associated with subtelomeric chromatin as well as with telomeric DNA (reviewed by Ottaviani et al. 2008). This fold-back structure has been proposed to be critical for the so-called telomere position effect, which refers to the repression of reporter genes inserted in the subtelomeric region (Gottschling et al. 1990). Silencing at telomeres is mediated by the silent information regulators Sir3, Sir4, and Sir2.
S. pombe
The telomeric DNA sequence from S. pombe is unusually heterogeneous with the consensus 5′-C1–8G0–1T0–2GTA1–3-3′ (Sugawara 1989). Telomere length, as in S. cerevisiae, is ∼300 bp. The subtelomeric DNA in S. pombe contains specialized repeats (dg and dh), which are also found at other heterochromatic loci contributing to formation of heterochromatin by the RNA interference (RNAi)-induced transcriptional silencing machinery (Kanoh et al. 2005). The first identified telomere-binding protein, Taz1, was identified in a one-hybrid screen using the double-stranded telomeric DNA as bait (Cooper et al. 1997). Taz1 is orthologous with the mammalian telomere-binding proteins Trf1 and Trf2. Taz1 recruits Rap1 to telomeres (Fig. 1). Rap1 binds Poz1, Poz1 binds Tpz1, and Tpz1 interacts with Ccq1 and Pot1 (Miyoshi et al. 2008). Pot1 also binds the telomeric single-stranded 3′ overhang (Baumann and Cech 2001). Thus, multiple protein interactions bridge the double-stranded part of the telomeric DNA to the single-stranded 3′ overhang, which is reminiscent of the situation in mammals. S. pombe Pot1 also plays crucial roles in protecting chromosome ends from degradation, Ccq1 interacts with the telomerase subunit Est1, recruiting telomerase to telomeres (see below). In addition, Stn1 and Ten1 orthologs but not Cdc13 orthologs have been identified in fission yeast (Martin et al. 2007). They protect from rapid loss of telomeric DNA. TERRA and other telomeric transcripts have been recently characterized in fission yeast (Bah et al. 2012; Greenwood and Cooper 2012). Their expression is negatively regulated by Taz1 and Rap1.
Mammals
Vertebrate telomeric DNA consists of 5′-TTAGGG-3′ repeats. The telomere length in humans is considerably shorter (5–15 kb) than in Mus musculus (>40 kb), and the human subtelomeric sequences are not well annotated. They contain repetitive sequence variants of the 5′-TTAGGG-3′ repeat sequence and a patchwork of segmentally duplicated DNA tracts that are shared in some instances between different telomeres and are highly polymorphic (Riethman 2008). Biochemical purification identified the first human telomere-binding protein, Trf1 (Chong et al. 1995). Dimeric Trf1 binds to double-stranded telomeric DNA (Fig. 1). A second homologous protein termed Trf2 also binds directly to the double-stranded telomeric DNA as a dimer (Bilaud et al. 1997; Broccoli et al. 1997). Trf1 and Trf2 interact with Tin2, which in turn binds Tpp1. Tpp1 also binds the single telomere-binding protein Pot1 (de Lange 2005). Notably, whereas in humans there is only one Pot1 protein, the mouse contains two Pot1-like proteins termed Pot1a and Pot1b (Hockemeyer et al. 2006; Wu et al. 2006). Trf2 also recruits Rap1 to telomeres. Thus, as in fission yeast, Rap1 is tethered to telomeres via protein interaction, whereas in budding yeast it directly binds the double-stranded telomeric DNA. No mammalian protein that is orthologous with S. pombe Ccq1 has yet been identified. However, a human CST complex was identified that is present at telomeres and at other regions in the genome (Miyake et al. 2009; Surovtseva et al. 2009).
Human TERRA is transcribed from several or possibly all chromosome ends (Azzalin et al. 2007; Schoeftner and Blasco 2008). Transcription starts in the subtelomeric region and proceeds up to ∼400 nucleotides into the pure TTAGGG tract (Porro et al. 2010). Poly(A) tails containing and lacking TERRA fractions can be distinguished. Approximately half of the poly(A)– TERRA fraction is chromatin-associated, colocalizing with telomeres. The less abundant poly(A)+ TERRA is all nucleoplasmic (Porro et al. 2010).
Mammalian telomeric DNA has been found to fold into so-called T-loop structures in which the 3′ overhang of telomeres invades through strand displacement the double-stranded part of telomeric repeats (Griffith et al. 1999). T-loops have been proposed to provide a mechanism for telomere capping, but this awaits experimental testing.
TELOMERASE
The catalytic core of telomerase consists of Tert and the telomerase RNA. The Tert subunit reverse transcribes the template sequence of telomerase RNA in an iterative fashion at the ends of chromosomes. When the 3′ end of the telomere is extended to the 5′ end of the RNA template, telomerase may reposition the substrate to the other side of the template and add the next telomere repeat, or it may dissociate from the substrate. The propensity of some telomerases to add several repeats without dissociation is referred to as repeat addition processivity. The Tert subunit is well conserved throughout evolution. It contains canonical reverse transcriptase motifs also found in retroelements (Lingner et al. 1997; Nakamura and Cech 1998). In addition, Tert contains a telomerase essential amino-terminal domain (TEN), a telomerase RNA-binding domain (TRBD), and a carboxy-terminal extension (reviewed by Lewis and Wuttke 2012). Other telomerase subunits are less well conserved. These subunits are not involved per se in catalysis, but they are important for telomerase assembly or the recruitment to telomeres. The S. cerevisiae Est1, which recruits telomerase to telomeres (discussed below), has a clear counterpart in S. pombe (Beernink et al. 2003; Webb and Zakian 2012), but the orthologous relationship to human Est1-like proteins (referred to as Est1A/Smg6, Est1B/Smg5, and Est1C/Smg7) is uncertain (Reichenbach et al. 2003; Snow et al. 2003). Est1A interacts as the budding and fission yeast Est1 proteins with telomerase, but a role in telomerase recruitment has not been shown (Redon et al. 2007). Est1A depletion has a strong effect on TERRA abundance at telomeres (Azzalin et al. 2007). Furthermore, Est1A to -C are also involved in the nonsense-mediated mRNA decay in the cytoplasm (Isken and Maquat 2008).
S. cerevisiae Est3 is essential for telomerase action at chromosome ends, but its contribution is not clear. Est3 contains an OB-fold, which is similar to an OB-fold present in the mammalian Tpp1 protein (Lee et al. 2008). Sm proteins associate with S. cerevisiae telomerase RNA, contributing to telomerase maturation and stability (Seto et al. 1999). S. pombe telomerase RNA (Ter1) also contains an Sm consensus site (Leonardi et al. 2008; Webb and Zakian 2008). The canonical Sm ring and the related Lsm2–8 complex sequentially associate with S. pombe Ter1 (Tang et al. 2012). The Sm ring binds the Ter1 precursor, stimulating its maturation by spliceosomal cleavage. Subsequently, the Lsm2–8 complex associates with Ter1, promoting the association with catalytic moiety Trt1 (Tang et al. 2012). In mammals, the dyskerin protein complex may fulfill similar functions in telomerase assembly (Mitchell et al. 1999; Pogacic et al. 2000). Another crucial component of human telomerase is Tcab1, which is discussed further below (Venteicher et al. 2009).
TELOMERE REPLICATION
Semiconservative Replication of Telomeric DNA
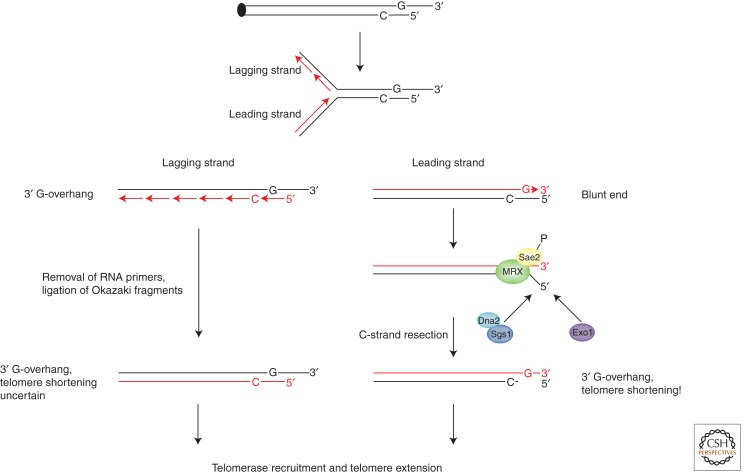
The largest part of the telomeric DNA is replicated by semiconservative DNA replication. The G-strand serves as a template for lagging-strand synthesis and the C-strand for leading-strand synthesis (Fig. 2). Only in recent years did it become apparent that even the semiconservative replication of telomeric DNA is difficult, requiring specialized helicases and telomere-binding proteins. In S. cerevisiae, the Rrm3 5′-3′ helicase prevents replication fork stalling at telomeres (Ivessa et al. 2002; Makovets et al. 2004). In fission yeast, it was noted that efficient replication of telomeric DNA requires the telomere-binding protein Taz1 (Miller et al. 2006). In its absence, replication forks frequently stall and collapse, leading to rapid loss of entire telomeric tracts. Taz1-deleted cells can only survive because telomerase very efficiently reextends the transiently truncated telomeric sequences. Evidence for truncated and by telomerase reextended telomeres was also obtained in wild-type budding yeast cells, supporting the notion that the semiconservative replication of telomeric DNA is error-prone (Chang et al. 2007).
Figure 2.
The end-replication problem and DNA end resection. The G-rich strand serves as a template for lagging-strand synthesis and the C-rich strand for leading-strand synthesis. The presumed blunt end intermediate at the leading-strand telomere is processed by 5′ end resection in order to recreate a 3′ overhang (right). Therefore, the leading-strand telomere shortens.
In mouse cells, it was shown that efficient replication requires the Taz1 ortholog Trf1 (Martinez et al. 2009; Sfeir et al. 2009). Upon Trf1 deletion in mouse cells, replication forks stall and telomeric DNA splits in multiple signals at metaphase chromosomes, indicative of telomeric DNA breakage or lack of condensation. The mechanism by which Trf1 promotes telomere replication involves recruitment of the Blm or Rtel1 helicases (Sfeir et al. 2009; Vannier et al. 2012). Both of these helicases belong to the highly conserved RecQ 3′-5′ helicase family, which use ATP hydrolysis to drive the unwinding of RNA and DNA. Another member of this family, the Werner helicase (Wrn), is required for efficient telomere lagging-strand synthesis (Crabbe et al. 2004; Arnoult et al. 2009). Interestingly, the stochastic telomere loss in Wrn-deficient cells and mice can be healed by the expression of telomerase (Chang et al. 2004; Crabbe et al. 2004). Indeed, in mice, the Wrn phenotype, which is characterized by multiple signs of premature aging, only manifests itself upon concomitant deletion of telomerase (Chang et al. 2004). S. cerevisiae Pif1 is another DNA helicase important for replication of G-rich sequences being enriched at telomeres and other G-rich sequences throughout the genome that may form G-quadruplex structures (Paeschke et al. 2011).
The Upf1 DNA- or RNA-dependent ATPase and 5′-to-3′ helicase is best known for its roles in cytoplasmic RNA quality control. However, human Upf1 binds to telomeres in vivo (Azzalin et al. 2007), interacting with the shelterin component Tpp1 (Chawla et al. 2011). Upf1 depletion leads to frequent loss of the telomeres replicated by leading-strand synthesis (Chawla et al. 2011). Upf1 depletion also leads to an increase of the TERRA signal at telomeres, suggesting a link between this RNA and telomere loss.
The crucial functions of helicases during replication of telomeric DNA appear to be multiple. First, helicases may unfold G-quadruplex DNA structures (Vannier et al. 2012) that may form during the lagging-strand replication of telomeric DNA, during which single-stranded G-rich DNA forms transiently. The structures involve guanine tetrads that associate through Hoogsteen hydrogen bonding to form a square planar structure, and several tetrads stack on top of each other to form a G-quadruplex (Parkinson et al. 2002). Second, in the T-loop structures, the 3′ overhang of telomeres invades through strand displacement the double-stranded part of telomeric repeats (Griffith et al. 1999). It is conceivable that helicases are necessary to unwind T-loops during semiconservative DNA replication (Vannier et al. 2012). Third, TERRA is associated with telomeres. If TERRA and telomeric DNA form R-loops, which are characterized by RNA–DNA hybrids and the displacement of single-stranded DNA, they may need to be resolved by helicases in order to avoid formation of double-stranded DNA breaks (Huertas and Aguilera 2003; Chawla et al. 2011; Wahba et al. 2011).
Telomere End Resection
Telomere leading-strand synthesis is predicted to yield a double-stranded blunt end intermediate (Fig. 2, right panel). This presumed intermediate is not detectable, as it will be quickly processed to regenerate a 3′ overhang. Thus, the 5′-end-containing parental C-rich strand is resected (Fig. 2, right panel). The generated G-rich 3′ overhang is important in order to generate a substrate for telomerase in addition to providing a platform for the binding of telomeric proteins and the ability to invade the double-stranded part of the telomere in the T-loop configuration. Thus, the telomere will shorten at the leading-strand end because the parental C-rich strand is resected during each round of replication (Lingner et al. 1995). During telomere lagging-strand synthesis, short RNA primers of 8–14 nucleotides are synthesized by primase and extended (Fig. 2, left panel). It is unknown where on the G-rich strand the most telomere-proximal primer is laid down. Because the parental G-rich strand forms a 3′ overhang that is longer than the RNA primer, the end-replication problem may not manifest itself at this end of the chromosome. However, telomerase binds leading- and lagging-strand telomeres (see below). Telomere lagging-strand replication also generates a 3′ overhang. Whether this involves simply RNA primer removal or in addition nucleolytic processing is currently unknown.
The 5′ end resection of leading-strand telomeres involves redundant activities consisting of nucleases, DNA helicases, and associated proteins that also resect 5′ ends from DNA double-strand breaks, preparing them for homologous recombination (Mimitou and Symington 2009). The Mre11–Rad50–Xrs2 (MRX) complex plays a role in end processing, as in its absence the G-overhangs at S. cerevisiae telomeres that have a length of 12–14 bases are shorter (Larrivee et al. 2004). Furthermore, the longer G-overhangs that accumulate in yeast in late S phase are also diminished. However, Mre11 has 3′-5′ exonuclease activity as well as endonuclease nicking activity but not the 5′-3′ exonuclease activity that should be required to generate a telomeric 3′ overhang. Furthermore, although MRX is important for wild-type telomere length maintenance, this function may at least partially relate to its activity as a Tel1 recruiter (see below). Indeed, a nuclease-deficient Mre11 mutant was proficient in telomere maintenance (Tsukamoto et al. 2001). This is in contrast to results obtained with an endonuclease-induced cut at telomere repeats at which resection and telomerase-mediated healing depends on MRX (Diede and Gottschling 2001; Bonetti et al. 2009). Sae2 and Sgs1 define parallel pathways required for processing of telomere ends (Bonetti et al. 2009). The Sae2 nuclease interacts with MRX and may start telomere end resection (Fig. 2). The Sgs1 helicase functions in conjunction with the Dna2 Flap endonuclease/helicase. Concomitant absence of Sae2 and Sgs1 triggered shorter telomeres but not cellular senescence, indicating that telomerase is still able to act (Bonetti et al. 2009). Indeed, exonuclease 1 (Exo1) can also contribute to telomere end resection, and telomere shortening in sae2Δ/sgs1Δ cells is partially suppressed by Exo1 overexpression (Bonetti et al. 2009). Cdk1 activity is required to promote 3′ overhang formation at S. cevevisiae telomeres (Frank et al. 2006; Vodenicharov and Wellinger 2006). This involves phosphorylation of Sae2 and possibly additional substrates. How phosphorylation activates Sae2 is not known.
In normal human fibroblasts, 3′ overhang length at leading-stand telomeres is around 60 nucleotides, versus 105 nucleotides at lagging-strand telomeres (Chai et al. 2006). Eighty percent of the C-rich strands end with the sequence 3′-CCAATC-5′, whereas the sequence at the 3′ terminus is less precise (Sfeir et al. 2005). The enzymatic activities that are involved in telomere and double-strand break end resection in S. cerevisiae are conserved throughout evolution. The MRN complex (Mre11–Rad50–Nbs1) interacts with Trf2 and binds to telomeres (Zhu et al. 2000). The role of MRN and the other processing activities discussed above in the processing of mammalian telomeres is not established. However, the Apollo 5′-3′ exonuclease is a member of the Snm1/Pso2 family of nucleases, binding to telomeres through its interaction with Trf2 (Lenain et al. 2006; van Overbeek and de Lange 2006). Apollo–/– cells have reduced 3′ overhangs at telomeres and they lose telomere protection at the leading-strand telomeres (Lam et al. 2010; Wu et al. 2010). Whether other nucleases in addition to Apollo contribute to telomere end processing at mammalian telomeres, as in yeast, is unclear. Notably also, Apollo has been proposed to facilitate semiconservative replication of telomeric DNA, relieving topological stress in conjunction with topoisomerase 2α (Ye et al. 2010).
TELOMERASE RECRUITMENT, TELOMERE EXTENSION, AND LENGTH HOMEOSTASIS
S. cerevisiae
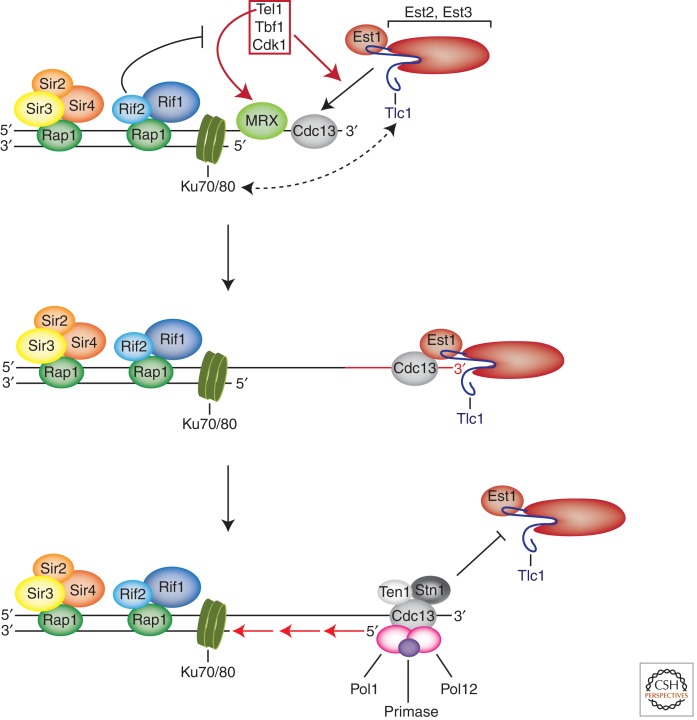
S. cerevisiae telomerase extends telomeres in late S phase, presumably after semiconservative DNA replication and telomere end processing. Single-telomere extension analysis (STEX) revealed that telomerase extends only a small fraction (∼7%) of telomeres in a given cell cycle and that extension occurs most frequently at the shortest telomeres (Teixeira et al. 2004). Recognition of the shortest telomeres is mediated on one side by Tel1 (ortholog of mammalian Atm), which is recruited specifically to the shortest telomeres in an MRX complex-dependent manner (Fig. 3) (Bianchi and Shore 2007b; Hector et al. 2007; Sabourin et al. 2007). Tel1 function can be partially compensated for by Mec1 (ortholog of mammalian Atr) as tel1Δ/mec1Δ double-mutant cells but not the single mutants senesce (Ritchie et al. 1999). A second pathway that mediates recognition of the shortest telomeres requires Tbf1, a protein that carries sequence similarity with fission yeast Taz1 and mammalian Trf proteins (Arneric and Lingner 2007). Tbf1 binds to 5′-TTAGGG-3′ repeats that are present in the subtelomeric region of yeast telomeres (Koering et al. 2000). Third, short telomeres replicate earlier in S phase. This therefore might provide more time for telomerase to mediate extension of the shortest telomeres (Bianchi and Shore 2007a).
Figure 3.
Telomerase recruitment and telomere extension in S. cerevisiae. MRX recruits Tel1 to the shortest telomeres, which promotes telomerase recruitment through the interaction of Cdc13 with Est1. Tbf1 can also promote preferential extension of the shortest telomeres in tel1Δ cells. Cdk1 contributes to the cell-cycle-dependent recruitment of telomerase through phosphorylation of Cdc13. Telomerase extends telomeres in a nonprocessive manner in S. cerevisiae. Human telomerase, on the other hand, adds ∼60 nucleotides per telomere in a single binding and extension event (not shown; see text). Telomerase extension is terminated upon formation of the CST complex, which promotes recruitment of DNA Pol α-primase for fill-in synthesis.
The recruitment to telomeres in late S phase and telomere extension depend on a physical interaction between Cdc13 that binds to the telomeric 3′ overhang and the telomerase subunit Est1 (Fig. 3). The cdc13-2 allele gives an est phenotype (Evans and Lundblad 1999). Cdc13-2 carries a single point mutation (E252L). Intriguingly, this mutation can be suppressed by a single point mutation in Est1 (L444E) (Pennock et al. 2001). Because the suppression involves the restoration of opposite charges in Cdc13 and Est1, this provides excellent genetic evidence that Cdc13 and Est1 interact directly and that the interaction involves these two amino acids. A second telomerase recruitment pathway in S. cerevisiae is mediated by the Ku70/80 heterodimer, which binds telomeres and other DNA ends via a preformed channel (Fig. 3). In addition, Ku80 binds the telomerase RNA subunit Tlc1 (Peterson et al. 2001; Fisher et al. 2004). Ku enriches telomerase near telomeres throughout the cell cycle as assessed by chromatin immunoprecipitation and positively contributes to telomere length maintenance. Disruption of the Tlc1–Ku80 interaction reduces telomere length while not eliciting an est1 phenotype. When assessing telomere association of telomerase by live-cell imaging, it seemed that the Tlc1–Ku80 interaction might only promote a transient interaction with telomeres, whereas the stable interaction in late S phase is mediated by Cdc13–Est1 (Gallardo et al. 2011). The Ku-mediated stimulation of telomerase may also involve its role in nuclear accumulation of Tlc1, which is reduced in Ku-deficient cells (Gallardo et al. 2008; Pfingsten et al. 2012). How Tel1 and Tbf1 promote the interaction of Cdc13 and Est1 and the preferential extension of the shortest telomeres is unclear. Cdc13 contains several consensus S/TQ sites for Tel1, but mutation of all these sites did not substantially impact on telomere length when the cdc13 mutants were tested in their chromosomal context (Gao et al. 2010). However, opposite results were reported in a previous study by another group, in which the mutant cdc13 alleles were expressed from a plasmid (Tseng et al. 2006). However, it seems that crucial Tel1 targets remain to be discovered. Cdk1 contributes to the cell-cycle-dependent recruitment of telomerase. The phosphorylation of Cdc13 at T308 by Cdk1 in late S phase and G2 favors the interaction of Cdc13 with Est1 rather than the competing interaction of Cdc13 with Stn1–Ten1 (Li et al. 2009). Another modification, Cdc13 SUMOylation, peaks in early to mid S phase (Hang et al. 2011). Mutation of the SUMO site reduced the interaction with Stn1, promoting telomere elongation (Hang et al. 2011). Ku70/80 and Sir4 are other telomeric proteins modified by Siz2 E3 ligase-dependent SUMOylation, promoting perinuclear position of budding yeast telomeres (Ferreira et al. 2011). Because telomeres shift away from the nuclear envelope when elongating, this modification possibly links nuclear position with telomerase regulation.
STEX analysis of yeast cells that expressed two RNA subunits that differed in the telomeric sequence revealed that S. cerevisiae telomerase acts mostly in a nonprocessive manner in terms of telomere repeat addition (Chang et al. 2007). However, when very short telomeres, <125 nucleotides, are elongated, yeast telomerase gains in repeat addition processivity. The increased processivity requires Tel1. The telomere-shortening rate in the absence of telomerase is length-independent (Marcand et al. 1999). The observed increased frequency of telomere elongation by telomerase with telomere shortening is sufficient to explain telomere length homeostasis. The steady-state telomere length corresponds to the point at which the product of frequency of elongation and average extension length equals the shortening rate. The frequency of elongation is regulated by the number of Rap1/Rif1/Rif2 proteins that are bound to telomeres in dependence of their length (Marcand et al. 1997). Mechanistically, this may involve the competition of Rif1/2 with Tel1 for binding of the carboxyl terminus of Xrs2 (Hirano et al. 2009). Thus, at short telomeres that bind less Rap1/Rif/Rif2, Tel1 recruitment to the MRX complex may be favored, leading to preferential recruitment and activation of telomerase at short telomeres (Fig. 3).
Telomerase-mediated telomere extension must be followed by fill-in synthesis of the C-strand, presumably by classical replication enzymes involved in lagging-strand synthesis. Importantly, the CST complex may play also an important role here, as physical interactions have been detected between Cdc13 and Pol1, the catalytic subunit of DNA Pol α-primase (Qi and Zakian 2000); and between Stn1 and Pol12, the B subunit of DNA Pol α-primase (Fig. 3) (Grossi et al. 2004). Whether CST substitutes the related RPA stimulating the fill-in reaction is unknown.
S. pombe
Telomerase recruitment to telomeres in S. pombe occurs through the interaction of the telomerase subunit Est1 with Ccq1, which is tethered to telomeres through the interaction with Tpz1, which in turn interacts with Pot1 and Poz1 (Miyoshi et al. 2008; Tomita and Cooper 2008; Webb and Zakian 2012). Importantly, S. pombe Tel1 (ortholog of mammalian Atm) and Rad3 (ortholog of mammalian Atr) promote the direct interaction between Ccq1 and the Est1 subunit of telomerase (Moser et al. 2011; Yamazaki et al 2012). Ccq1 is phosphorylated on T93 by Tel1 and Rad3, and mutation of this site abolishes telomerase recruitment. A Cdc13 homolog has not been identified in S. pombe, and there is no known Ccq1 counterpart outside of fission yeast. Thus, telomerase recruitment seems to depend on different factors in fission and budding yeast. Telomere length regulation in S. pombe involves Taz1, Rap1, and Poz1, whose deletions lead to very substantial telomere elongation.
Mammals
Telomerase in humans is enriched in so-called Cajal bodies (CBs), dynamic subnuclear structures that are characterized by the coilin protein and that contain a variety of proteins and RNAs involved in the assembly and modification of small nuclear and nucleolar ribonucleoproteins (snRNPs and snoRNPs). CBs also harbor a class of CB-specific RNAs (scaRNAs) that are involved in methylation and pseudouridylation of small nuclear RNAs. In S phase of the cell cycle, a subset of CBs together with telomerase RNA is seen in association with telomeres in human cells (Jady et al. 2006; Tomlinson et al. 2006). As with other scaRNAs, human telomerase RNA contains a CAB-box motif that mediates association with CBs (Jady et al. 2004). Mutation of this motif does not prevent association of Tert with telomerase RNA (hTR), as catalytic telomerase activity is not impaired (Cristofari et al. 2007). However, CAB-box mutant telomerase does not efficiently associate with telomeres, and telomere extension is reduced (Cristofari et al. 2007). Tcab1 (Venteicher et al. 2009), also referred to as Wdr79 (Tycowski et al. 2009), was identified as a CAB-box binding protein and telomerase subunit. Tcab1 depletion from human cells leads to continuous telomere shortening (Venteicher et al. 2009). These results underline the importance of the CAB box and its interaction with Tcab1 for telomerase recruitment to telomeres. The CB association of telomerase has, in contrast to human cells, not been seen in mouse cells (Tomlinson et al. 2010). Thus, it will be interesting to determine if Tcab1 is also important for telomerase recruitment to telomeres in mice.
At the telomere, Tpp1 and Tin2 are important for telomerase recruitment or its retention, as depletion of either of these factors reduces (but does not abolish) telomerase association with telomeres (Abreu et al. 2010). Depletion and rescue studies further identified the amino-terminal OB-fold of Tpp1 as important for telomerase recruitment (Abreu et al. 2010), a domain that was found to also interact with telomerase in extracts (Xin et al. 2007). Whether telomerase recruitment is regulated in mammals by Atm or Atr, as in fission and budding yeast, is unknown, nor is it clear if in addition to Tpp1 other factors contribute to recruitment. The single-stranded telomere-binding protein Pot1 is an inhibitor of telomerase per se when binding to the 3′ end of the substrate (Kelleher et al. 2005; Lei et al. 2005). When associated with Tpp1, however, Pot1–Tpp1 decreases the dissociation of primer from telomerase and increases the efficiency of translocation (Wang et al. 2007; Latrick and Cech 2010). Therefore, the Pot1–Tpp1 complex increases repeat addition processivity of human telomerase severalfold. Thus, with the assistance of Pot1–Tpp1, telomerase can synthesize in vitro roughly with one or two binding and extension events the ∼60 nucleotides that are typically added to a human telomere by telomerase in vivo (see below).
Using sophisticated molecular biological methods, the frequency of telomerase-mediated telomere elongation was measured to be 70%–100% at lagging-strand telomeres in HeLa and H1299 lung adenocarcinoma cells, and similar conclusions were drawn for the leading-strand telomeres (Zhao et al. 2009). This result was unexpected, as at lagging-strand telomeres the G-rich strand is parental and thus this strand is not predicted to shorten from incomplete end replication (Fig. 2, left panel). However, because this end is extended by telomerase, this finding implies that the parental G-rich strand must be shortened again subsequent to its elongation by telomerase, if not in every cell cycle at least in every couple of cell cycles. If not, the parental G-strand should suffer from continuous telomere elongation. The telomerase-mediated extension length was determined to be ∼60 nucleotides, and experimental inhibition or depletion of telomerase reduced the frequency of extension events but not extension length (Zhao et al. 2011). This supports the notion that one single telomerase molecule acts at every telomere in every cell cycle.
The restriction of human telomerase to a single telomere binding and extension event may involve the human CST complex, which was identified recently (Miyake et al. 2009; Surovtseva et al. 2009). Human Stn1 and Ten1 carry clear sequence similarity with their budding yeast counterparts. Ctc1 (conserved telomere maintenance components 1), also referred to as AAF-132 (α-accessory factor) (Casteel et al. 2009), which assembles with Stn1 and Ten1 in the trimeric CST complex, bears little sequence similarity with S. cerevisiae Cdc13. Like budding yeast CST components, those of human CST contain putative OB-fold domains involved in binding single-stranded DNA and promoting protein interactions. Human CST inhibits telomerase activity through primer sequestration and physical interaction with the Pot1–Tpp1 telomerase processivity factor (Chen et al. 2012). CST binding at telomeres increases during late S/G2 phase only upon telomerase action, coinciding with telomerase shutoff. CST depletion unleashes excessive telomerase activity, promoting telomere elongation. Increased binding of CST upon telomerase-mediated telomere elongation coincides with a switch from Pot1–Tpp1 binding with concomitant telomerase stimulation to CST binding and concomitant telomerase repression. Through binding of the telomerase-extended telomere, CST may limit telomerase action at individual telomeres to approximately one binding and extension event per cell cycle (Chen et al. 2012).
C-strand fill-in synthesis in human cells follows telomerase extension at the end of S phase (Zhao et al. 2009). Whether Pot1–Tpp1 or RPA stimulates this process is unclear. Possibly more relevant for this is the CST complex, which binds to the telomerase-extended telomeric 3′ end (Chen et al. 2012). Indeed, human Ctc1 and Stn1 stimulate DNA polymerase α-primase, increasing its affinity for template DNA (Casteel et al. 2009), and CST has been implicated in assisting lagging-strand synthesis at telomeres and other special DNA sequences throughout the genome (Miyake et al. 2009; Price et al. 2010; Gu et al. 2012). How the binding of single-stranded telomeric DNA by telomere-binding proteins, which include Pot1–Tpp1, CST, RPA, and possibly hnRNPA1, is regulated is unclear, nor is it known how T-loop formation may be influenced by these factors, possibly in a cell-cycle-dependent manner. It has been proposed that a switch from RPA binding in S phase to Pot1–Tpp1 after S phase is triggered by hnRNPA1 and TERRA (Flynn et al. 2011). In vitro, hnRNPA1 can compete with RPA for the binding of the single-stranded G-strand (Flynn et al. 2011). An increase of TERRA levels after S phase may sequester hnRNPA1 from telomeric DNA, as it has higher binding affinity for the 5′-UUAGGG-3′ repeats in TERRA than for the same 5′-TTAGGG-3′ telomeric DNA sequence. Binding of hnRNPA1 to TERRA may in turn allow binding of the telomeric 3′ overhang by Pot1–Tpp1. On the other hand, it has been argued that the higher local concentration of Pot1–Tpp1 near telomeres is sufficient to trigger Pot1–Tpp1 binding once semiconservative telomere replication is completed (Takai et al. 2011).
The above results on telomere extension in vivo cannot on their own explain telomere length homeostasis. Telomere length homeostasis requires that the frequency of telomere elongation, the extension length, or the shortening of telomeres occurs in a length-dependent manner. Telomerase preferentially elongated critically short telomeres in the offspring of mice to which telomerase was reintroduced after their ancestors grew in the absence of the telomerase RNA gene for several generations (Hemann et al. 2001). Thus, under nonhomeostatic conditions, telomerase is regulated in a telomere length-dependent manner, at least in mice. Telomere length-dependent regulation of telomerase in mammals is also supported by studies on the shelterin components, whose abundance at telomeres is length-dependent and whose experimental manipulation leads to telomere length changes (Smogorzewska and de Lange 2004; Hug and Lingner 2006). Generally, it has been observed that depletion of shelterin components by RNAi promotes telomerase-mediated telomere elongation, whereas their overexpression reduces telomere length. This may be at odds when considering the above-discussed positive roles of at least Tpp1 for telomerase recruitment and stimulation of processivity. However, it is important to consider that many telomere-binding proteins function like Swiss army knives. They may have multiple tasks in several processes at the chromosome end, such as telomere replication, regulation of telomerase, protection from nucleases, recombination and repair, and suppression of checkpoint signaling. Thus, depletion of a factor may highlight one function but may not allow detection of the multiple tools it may be equipped with to contribute to the correct functioning of chromosome ends.
CONCLUDING REMARKS
Elucidation of the complexity of telomere replication and the regulation of telomerase is a fascinating basic problem of biological research. It is clear that a better understanding of telomere replication and the regulation of telomerase in the future will come from studies in biochemistry, structural biology, genetics, and cell biology. Single-celled eukaryotes as well as animal model systems will continue to be of important value. For human telomere replication, the studies in induced pluripotent stem cells may become particularly revealing, as this system may allow learning about telomerase regulation and telomere replication in stem cells and during cellular differentiation. It is hoped that a much more thorough understanding of telomere maintenance may identify new targets that may become useful to inhibit telomerase action in cancer. After all these years (Kim et al. 1994), it has still not been possible to test if telomerase inhibition in human cancer is a valuable strategy to inhibit tumor growth. In addition, an increasing number of diseases have been linked to short telomeres (reviewed by Lansdorp 2009). Short-telomere diseases include dyskeratosis congenita (DC), idiopathic pulmonary fibrosis, the Werner syndrome, and the ICF (immunodeficiency, centromeric instability, facial anomalies) syndrome. DC patients die prematurely of bone marrow failure. ICF syndrome patients suffer from reduced immunoglobulin levels. Werner patients age prematurely and are predisposed to cancer. Although in some cases defective telomerase could be linked to short-telomere diseases (particularly in DC), in other cases the molecular mechanisms that underlie abnormal telomere shortening remain to be defined. In conclusion, it seems safe to assume that telomeres will not cease to unveil many hidden treasures.
ACKNOWLEDGMENTS
V.P. is supported by a long-term European Molecular Biology Organization postdoctoral fellowship. J.L.’s lab is supported by the Swiss National Science Foundation, a European Research Council advanced investigator grant (grant agreement number 232812), the Swiss Cancer League, and EPFL. We apologize to the researchers whose work could not be discussed and cited here because of tight space restrictions.
Footnotes
Editors: Stephen D. Bell, Marcel Méchali, and Melvin L. DePamphilis
Additional Perspectives on DNA Replication available at www.cshperspectives.org
REFERENCES
- Abreu E, Aritonovska E, Reichenbach P, Cristofari G, Culp B, Terns RM, Lingner J, Terns MP 2010. TIN2-tethered TPP1 recruits human telomerase to telomeres in vivo. Mol Cell Biol 30: 2971–2982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arneric M, Lingner J 2007. Tel1 kinase and subtelomere-bound Tbf1 mediate preferential elongation of short telomeres by telomerase in yeast. EMBO Rep 8: 1080–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnoult N, Saintome C, Ourliac-Garnier I, Riou JF, Londono-Vallejo A 2009. Human POT1 is required for efficient telomere C-rich strand replication in the absence of WRN. Genes Dev 23: 2915–2924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J 2007. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science 318: 798–801 [DOI] [PubMed] [Google Scholar]
- Bah A, Wischnewski H, Shchepachev V, Azzalin CM 2012. The telomeric transcriptome of Schizosaccharomyces pombe. Nucleic Acids Res 40: 2995–3005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann P, Cech T 2001. Pot1, the putative telomere end-binding protein in fission yeast and humans. Science 292: 1171–1175 [DOI] [PubMed] [Google Scholar]
- Beernink HT, Miller K, Deshpande A, Bucher P, Cooper JP 2003. Telomere maintenance in fission yeast requires an Est1 ortholog. Curr Biol 13: 575–580 [DOI] [PubMed] [Google Scholar]
- Bianchi A, Shore D 2007a. Early replication of short telomeres in budding yeast. Cell 128: 1051–1062 [DOI] [PubMed] [Google Scholar]
- Bianchi A, Shore D 2007b. Increased association of telomerase with short telomeres in yeast. Genes Dev 21: 1726–1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilaud T, Koering CE, Binet-Brasselet E, Ancelin K, Pollice A, Gasser SM, Gilson E 1996. The telobox, a Myb-related telomeric DNA binding motif found in proteins from yeast, plants and human. Nucleic Acids Res 24: 1294–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilaud T, Brun C, Ancelin K, Koering CE, Laroche T, Gilson E 1997. Telomeric localization of TRF2, a novel human telobox protein. Nat Genet 17: 236–239 [DOI] [PubMed] [Google Scholar]
- Blackburn EH 2006. A history of telomere biology. In Telomeres, 2nd ed. (ed. de Lange T, Lundblad V, Blackburn EH), pp. 1–19 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Blackburn E, Gall J 1978. A tandemly repeated sequence at the termini of the extrachromosomal ribosomal RNA genes in Tetrahymena. J Mol Biol 120: 33–53 [DOI] [PubMed] [Google Scholar]
- Bodnar AG, Ouellette M, Frolkis M, Holt SE, Chiu C-P, Morin GB, Harley CB, Shay JW, Lichtsteinter S, Wright WE 1998. Extension of life-span by introduction of telomerase into normal human cells. Science 279: 349–352 [DOI] [PubMed] [Google Scholar]
- Bonetti D, Martina M, Clerici M, Lucchini G, Longhese MP 2009. Multiple pathways regulate 3′ overhang generation at S. cerevisiae telomeres. Mol Cell 35: 70–81 [DOI] [PubMed] [Google Scholar]
- Broccoli D, Smogorzewska A, Chong L, de Lange T 1997. Human telomeres contain two distinct Myb-related proteins, TRF1 and TRF2. Nat Genet 17: 231–235 [DOI] [PubMed] [Google Scholar]
- Casteel DE, Zhuang S, Zeng Y, Perrino FW, Boss GR, Goulian M, Pilz RB 2009. A DNA polymerase-α · primase cofactor with homology to replication protein A-32 regulates DNA replication in mammalian cells. J Biol Chem 284: 5807–5818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai W, Sfeir AJ, Hoshiyama H, Shay JW, Wright WE 2006. The involvement of the Mre11/Rad50/Nbs1 complex in the generation of G-overhangs at human telomeres. EMBO Rep 7: 225–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S, Multani AS, Cabrera NG, Naylor ML, Laud P, Lombard D, Pathak S, Guarente L, DePinho RA 2004. Essential role of limiting telomeres in the pathogenesis of Werner syndrome. Nat Genet 36: 877–882 [DOI] [PubMed] [Google Scholar]
- Chang M, Arneric M, Lingner J 2007. Telomerase repeat addition processivity is increased at critically short telomeres in a Tel1-dependent manner in Saccharomyces cerevisiae. Genes Dev 21: 2485–2494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawla R, Redon S, Raftopoulou C, Wischnewski H, Gagos S, Azzalin CM 2011. Human UPF1 interacts with TPP1 and telomerase and sustains telomere leading-strand replication. EMBO J 30: 4047–4058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LY, Redon S, Lingner J 2012. The human CST complex is a terminator of telomerase activity. Nature 488: 540–544 [DOI] [PubMed] [Google Scholar]
- Chong L, van Steensel B, Broccoli D, Erdjument-Bromage H, Hanish J, Tempst P, de Lange T 1995. A human telomeric protein. Science 270: 1663–1667 [DOI] [PubMed] [Google Scholar]
- Cooper JP, Nimmo ER, Allshire RC, Cech TR 1997. Regulation of telomere length and function by a Myb-domain protein in fission yeast. Nature 385: 744–747 [DOI] [PubMed] [Google Scholar]
- Crabbe L, Verdun RE, Haggblom CI, Karlseder J 2004. Defective telomere lagging strand synthesis in cells lacking WRN helicase activity. Science 306: 1951–1953 [DOI] [PubMed] [Google Scholar]
- Cristofari G, Adolf E, Reichenbach P, Sikora K, Terns RM, Terns MP, Lingner J 2007. Human telomerase RNA accumulation in Cajal bodies facilitates telomerase recruitment to telomeres and telomere elongation. Mol Cell 27: 882–889 [DOI] [PubMed] [Google Scholar]
- de Lange T 2005. Shelterin: The protein complex that shapes and safeguards human telomeres. Genes Dev 19: 2100–2110 [DOI] [PubMed] [Google Scholar]
- de Lange T 2009. How telomeres solve the end-protection problem. Science 326: 948–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diede SJ, Gottschling DE 2001. Exonuclease activity is required for sequence addition and Cdc13p loading at a de novo telomere. Curr Biol 11: 1336–1340 [DOI] [PubMed] [Google Scholar]
- Evans SK, Lundblad V 1999. Est1 and Cdc13 as comediators of telomerase access. Science 286: 117–120 [DOI] [PubMed] [Google Scholar]
- Ferreira HC, Luke B, Schober H, Kalck V, Lingner J, Gasser SM 2011. The PIAS homologue Siz2 regulates perinuclear telomere position and telomerase activity in budding yeast. Nat Cell Biol 13: 867–874 [DOI] [PubMed] [Google Scholar]
- Fisher TS, Taggart AK, Zakian VA 2004. Cell cycle-dependent regulation of yeast telomerase by Ku. Nat Struct Mol Biol 11: 1198–1205 [DOI] [PubMed] [Google Scholar]
- Flynn RL, Centore RC, O’Sullivan RJ, Rai R, Tse A, Songyang Z, Chang S, Karlseder J, Zou L 2011. TERRA and hnRNPA1 orchestrate an RPA-to-POT1 switch on telomeric single-stranded DNA. Nature 471: 532–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forstemann K, Lingner J 2001. Molecular basis for telomere repeat divergence in budding yeast. Mol Cell Biol 21: 7277–7286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank CJ, Hyde M, Greider CW 2006. Regulation of telomere elongation by the cyclin-dependent kinase CDK1. Mol Cell 24: 423–432 [DOI] [PubMed] [Google Scholar]
- Gallardo F, Olivier C, Dandjinou AT, Wellinger RJ, Chartrand P 2008. TLC1 RNA nucleo-cytoplasmic trafficking links telomerase biogenesis to its recruitment to telomeres. EMBO J 27: 748–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo F, Laterreur N, Cusanelli E, Ouenzar F, Querido E, Wellinger RJ, Chartrand P 2011. Live cell imaging of telomerase RNA dynamics reveals cell cycle-dependent clustering of telomerase at elongating telomeres. Mol Cell 44: 819–827 [DOI] [PubMed] [Google Scholar]
- Gao H, Cervantes RB, Mandell EK, Otero JH, Lundblad V 2007. RPA-like proteins mediate yeast telomere function. Nat Struct Mol Biol 14: 208–214 [DOI] [PubMed] [Google Scholar]
- Gao H, Toro TB, Paschini M, Braunstein-Ballew B, Cervantes RB, Lundblad V 2010. Telomerase recruitment in Saccharomyces cerevisiae is not dependent on Tel1-mediated phosphorylation of Cdc13. Genetics 186: 1147–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschling D, Zakian V 1986. Telomere proteins: Specific recognition and protection of the natural termini of Oxytricha macronuclear DNA. Cell 47: 195–205 [DOI] [PubMed] [Google Scholar]
- Gottschling DE, Aparicio OM, Billington BL, Zakian VA 1990. Position effect at S. cerevisiae telomeres: Reversible repression of Pol II transcription. Cell 63: 751–762 [DOI] [PubMed] [Google Scholar]
- Gray JT, Celander DW, Price CM, Cech TR 1991. Cloning and expression of genes for the Oxytricha telomere-binding protein: Specific subunit interactions in the telomeric complex. Cell 67: 807–814 [DOI] [PubMed] [Google Scholar]
- Greenwood J, Cooper JP 2012. Non-coding telomeric and subtelomeric transcripts are differentially regulated by telomeric and heterochromatin assembly factors in fission yeast. Nucleic Acids Res 40: 2956–2963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greider CW, Blackburn EH 1985. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell 43: 405–413 [DOI] [PubMed] [Google Scholar]
- Greider CW, Blackburn EH 1989. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature 337: 331–337 [DOI] [PubMed] [Google Scholar]
- Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, de Lange T 1999. Mammalian telomeres end in a large duplex loop. Cell 97: 503–514 [DOI] [PubMed] [Google Scholar]
- Grossi S, Puglisi A, Dmitriev PV, Lopes M, Shore D 2004. Pol12, the B subunit of DNA polymerase α, functions in both telomere capping and length regulation. Genes Dev 18: 992–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu P, Min JN, Wang Y, Huang C, Peng T, Chai W, Chang S 2012. CTC1 deletion results in defective telomere replication, leading to catastrophic telomere loss and stem cell exhaustion. EMBO J 31: 2309–2321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hang LE, Liu X, Cheung I, Yang Y, Zhao X 2011. SUMOylation regulates telomere length homeostasis by targeting Cdc13. Nat Struct Mol Biol 18: 920–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hector RE, Shtofman RL, Ray A, Chen BR, Nyun T, Berkner KL, Runge KW 2007. Tel1p preferentially associates with short telomeres to stimulate their elongation. Mol Cell 27: 851–858 [DOI] [PubMed] [Google Scholar]
- Hemann MT, Strong MA, Hao LY, Greider CW 2001. The shortest telomere, not average telomere length, is critical for cell viability and chromosome stability. Cell 107: 67–77 [DOI] [PubMed] [Google Scholar]
- Hicke BJ, Celander DW, MacDonald GH, Price CM, Cech TR 1990. Two versions of the gene encoding the 41-kilodalton subunit of the telomere binding protein of Oxytricha nova. Proc Natl Acad Sci 87: 1481–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano Y, Fukunaga K, Sugimoto K 2009. Rif1 and Rif2 inhibit localization of Tel1 to DNA ends. Mol Cell 33: 312–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D, Daniels JP, Takai H, de Lange T 2006. Recent expansion of the telomeric complex in rodents: Two distinct POT1 proteins protect mouse telomeres. Cell 126: 63–77 [DOI] [PubMed] [Google Scholar]
- Huertas P, Aguilera A 2003. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol Cell 12: 711–721 [DOI] [PubMed] [Google Scholar]
- Hug N, Lingner J 2006. Telomere length homeostasis. Chromosoma 115: 413–425 [DOI] [PubMed] [Google Scholar]
- Iglesias N, Redon S, Pfeiffer V, Dees M, Lingner J, Luke B 2011. Subtelomeric repetitive elements determine TERRA regulation by Rap1/Rif and Rap1/Sir complexes in yeast. EMBO Rep 12: 587–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isken O, Maquat LE 2008. The multiple lives of NMD factors: Balancing roles in gene and genome regulation. Nat Rev Genet 9: 699–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivessa AS, Zhou JQ, Schulz VP, Monson EK, Zakian VA 2002. Saccharomyces Rrm3p, a 5′ to 3′ DNA helicase that promotes replication fork progression through telomeric and subtelomeric DNA. Genes Dev 16: 1383–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jady BE, Bertrand E, Kiss T 2004. Human telomerase RNA and box H/ACA scaRNAs share a common Cajal body–specific localization signal. J Cell Biol 164: 647–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jady BE, Richard P, Bertrand E, Kiss T 2006. Cell cycle-dependent recruitment of telomerase RNA and Cajal bodies to human telomeres. Mol Biol Cell 17: 944–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain D, Cooper JP 2010. Telomeric strategies: Means to an end. Annu Rev Genet 44: 243–269 [DOI] [PubMed] [Google Scholar]
- Kanoh J, Sadaie M, Urano T, Ishikawa F 2005. Telomere binding protein Taz1 establishes Swi6 heterochromatin independently of RNAi at telomeres. Curr Biol 15: 1808–1819 [DOI] [PubMed] [Google Scholar]
- Kelleher C, Kurth I, Lingner J 2005. Human protection of telomeres 1 (POT1) is a negative regulator of telomerase activity in vitro. Mol Cell Biol 25: 808–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PL, Coviello GM, Wright WE, Weinrich SL, Shay JW 1994. Specific association of human telomerase activity with immortal cells and cancer. Science 266: 2011–2015 [DOI] [PubMed] [Google Scholar]
- Koering CE, Fourel G, Binet-Brasselet E, Laroche T, Klein F, Gilson E 2000. Identification of high affinity Tbf1p-binding sites within the budding yeast genome. Nucleic Acids Res 28: 2519–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam YC, Akhter S, Gu P, Ye J, Poulet A, Giraud-Panis MJ, Bailey SM, Gilson E, Legerski RJ, Chang S 2010. SNMIB/Apollo protects leading-strand telomeres against NHEJ-mediated repair. EMBO J 29: 2230–2241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lansdorp PM 2009. Telomeres and disease. EMBO J 28: 2532–2540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larrivee M, LeBel C, Wellinger RJ 2004. The generation of proper constitutive G-tails on yeast telomeres is dependent on the MRX complex. Genes Dev 18: 1391–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latrick CM, Cech TR 2010. POT1–TPP1 enhances telomerase processivity by slowing primer dissociation and aiding translocation. EMBO J 29: 924–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Mandell EK, Tucey TM, Morris DK, Lundblad V 2008. The Est3 protein associates with yeast telomerase through an OB-fold domain. Nat Struct Mol Biol 15: 990–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei M, Zaug AJ, Podell ER, Cech TR 2005. Switching human telomerase on and off with hPOT1 protein in vitro. J Biol Chem 280: 20449–20456 [DOI] [PubMed] [Google Scholar]
- Lenain C, Bauwens S, Amiard S, Brunori M, Giraud-Panis MJ, Gilson E 2006. The Apollo 5′ exonuclease functions together with TRF2 to protect telomeres from DNA repair. Curr Biol 16: 1303–1310 [DOI] [PubMed] [Google Scholar]
- Lendvay TS, Morris DK, Sah J, Balasubramanian B, Lundblad V 1996. Senescence mutants of Saccharomyces cerevisiae with a defect in telomere replication identify three additional EST genes. Genetics 144: 1399–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonardi J, Box JA, Bunch JT, Baumann P 2008. TER1, the RNA subunit of fission yeast telomerase. Nat Struct Mol Biol 15: 26–33 [DOI] [PubMed] [Google Scholar]
- Lewis KA, Wuttke DS 2012. Telomerase and telomere-associated proteins: Structural insights into mechanism and evolution. Structure 20: 28–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Makovets S, Matsuguchi T, Blethrow JD, Shokat KM, Blackburn EH 2009. Cdk1-dependent phosphorylation of Cdc13 coordinates telomere elongation during cell-cycle progression. Cell 136: 50–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner J, Cech TR 1996. Purification of telomerase from Euplotes aediculatus: Requirement of a primer 3′ overhang. Proc Natl Acad Sci 93: 10712–10717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner J, Cooper JP, Cech TR 1995. Telomerase and DNA end replication: No longer a lagging strand problem? Science 269: 1533–1534 [DOI] [PubMed] [Google Scholar]
- Lingner J, Hughes TR, Shevchenko A, Mann M, Lundblad V, Cech TR 1997. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science 276: 561–567 [DOI] [PubMed] [Google Scholar]
- Louis EJ, Borts RH 1995. A complete set of marked telomeres in Saccharomyces cerevisiae for physical mapping and cloning. Genetics 139: 125–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke B, Panza A, Redon S, Iglesias N, Li Z, Lingner J 2008. The Rat1p 5′ to 3′ exonuclease degrades telomeric repeat-containing RNA and promotes telomere elongation in Saccharomyces cerevisiae. Mol Cell 32: 465–477 [DOI] [PubMed] [Google Scholar]
- Lundblad V, Szostak JW 1989. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell 57: 633–643 [DOI] [PubMed] [Google Scholar]
- Lustig AJ, Kurtz S, Shore D 1990. Involvement of the silencer and UAS binding protein RAP1 in regulation of telomere length. Science 250: 549–553 [DOI] [PubMed] [Google Scholar]
- Makovets S, Herskowitz I, Blackburn EH 2004. Anatomy and dynamics of DNA replication fork movement in yeast telomeric regions. Mol Cell Biol 24: 4019–4031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcand S, Gilson E, Shore D 1997. A protein-counting mechanism for telomere length regulation in yeast. Science 275: 986–990 [DOI] [PubMed] [Google Scholar]
- Marcand S, Brevet V, Gilson E 1999. Progressive cis-inhibition of telomerase upon telomere elongation. EMBO J 18: 3509–3519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcand S, Pardo B, Gratias A, Cahun S, Callebaut I 2008. Multiple pathways inhibit NHEJ at telomeres. Genes Dev 22: 1153–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin V, Du LL, Rozenzhak S, Russell P 2007. Protection of telomeres by a conserved Stn1–Ten1 complex. Proc Natl Acad Sci 104: 14038–14043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez JL, Edstrom JE, Morcillo G, Diez JL 2001. Telomeres in Chironomus thummi are characterized by different subfamilies of complex DNA repeats. Chromosoma 110: 221–227 [DOI] [PubMed] [Google Scholar]
- Martinez P, Thanasoula M, Munoz P, Liao C, Tejera A, McNees C, Flores JM, Fernandez-Capetillo O, Tarsounas M, Blasco MA 2009. Increased telomere fragility and fusions resulting from TRF1 deficiency lead to degenerative pathologies and increased cancer in mice. Genes Dev 23: 2060–2075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B 1941. The stability of broken ends of chromosomes in Zea mays. Genetics 41: 234–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller KM, Rog O, Cooper JP 2006. Semi-conservative DNA replication through telomeres requires Taz1. Nature 440: 824–828 [DOI] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS 2009. DNA end resection: Many nucleases make light work. DNA Repair (Amst) 8: 983–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell JR, Cheng J, Collins K 1999. A box H/ACA small nucleolar RNA-like domain at the human telomerase RNA 3′ end. Mol Cell Biol 19: 567–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake Y, Nakamura M, Nabetani A, Shimamura S, Tamura M, Yonehara S, Saito M, Ishikawa F 2009. RPA-like mammalian Ctc1-Stn1-Ten1 complex binds to single-stranded DNA and protects telomeres independently of the Pot1 pathway. Mol Cell 36: 193–206 [DOI] [PubMed] [Google Scholar]
- Miyoshi T, Kanoh J, Saito M, Ishikawa F 2008. Fission yeast Pot1-Tpp1 protects telomeres and regulates telomere length. Science 320: 1341–1344 [DOI] [PubMed] [Google Scholar]
- Moser BA, Chang YT, Kosti J, Nakamura TM 2011. Tel1ATM and Rad3ATR kinases promote Ccq1-Est1 interaction to maintain telomeres in fission yeast. Nat Struct Mol Biol 18: 1408–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller HJ 1938. The remaking of chromosomes. Collecting Net (Woods Hole) 13: 181–198 [Google Scholar]
- Nakamura TM, Cech TR 1998. Reversing time—origin of telomerase. Cell 92: 587–590 [DOI] [PubMed] [Google Scholar]
- Olovnikov AM 1971. Principle of marginotomy in template synthesis of polynucleotides [in Russian]. Dokl Akad Nauk SSSR 201: 1496–1499 [PubMed] [Google Scholar]
- Ottaviani A, Gilson E, Magdinier F 2008. Telomeric position effect: From the yeast paradigm to human pathologies? Biochimie 90: 93–107 [DOI] [PubMed] [Google Scholar]
- Paeschke K, Capra JA, Zakian VA 2011. DNA replication through G-quadruplex motifs is promoted by the Saccharomyces cerevisiae Pif1 DNA helicase. Cell 145: 678–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardue ML, Debaryshe PG 2011. Retrotransposons that maintain chromosome ends. Proc Natl Acad Sci 108: 20317–20324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson GN, Lee MP, Neidle S 2002. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature 417: 876–880 [DOI] [PubMed] [Google Scholar]
- Pennock E, Buckley K, Lundblad V 2001. Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell 104: 387–396 [DOI] [PubMed] [Google Scholar]
- Peterson S, Stellwagen A, Diede S, Singer M, Haimberger Z, Johnson C, Tzoneva M, Gottschling D 2001. The function of a stem-loop in telomerase RNA is linked to the DNA repair protein Ku. Nat Genet 27: 64–67 [DOI] [PubMed] [Google Scholar]
- Pfingsten JS, Goodrich KJ, Taabazuing C, Ouenzar F, Chartrand P, Cech TR 2012. Mutually exclusive binding of telomerase RNA and DNA by Ku alters telomerase recruitment model. Cell 148: 922–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogacic V, Dragon F, Filipowicz W 2000. Human H/ACA small nucleolar RNPs and telomerase share evolutionarily conserved proteins NHP2 and NOP10. Mol Cell Biol 20: 9028–9040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porro A, Feuerhahn S, Reichenbach P, Lingner J 2010. Molecular dissection of TERRA biogenesis unveils the presence of distinct and multiple regulatory pathways. Mol Cell Biol 30: 4808–4817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price CM, Boltz KA, Chaiken MF, Stewart JA, Beilstein MA, Shippen DE 2010. Evolution of CST function in telomere maintenance. Cell Cycle 9: 3157–3165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi H, Zakian VA 2000. The Saccharomyces telomere-binding protein Cdc13p interacts with both the catalytic subunit of DNA polymerase α and the telomerase-associated Est1 protein. Genes Dev 14: 1777–1788 [PMC free article] [PubMed] [Google Scholar]
- Raices M, Verdun RE, Compton SA, Haggblom CI, Griffith JD, Dillin A, Karlseder J 2008. C. elegans telomeres contain G-strand and C-strand overhangs that are bound by distinct proteins. Cell 132: 745–757 [DOI] [PubMed] [Google Scholar]
- Redon S, Reichenbach P, Lingner J 2007. Protein RNA and protein protein interactions mediate association of human EST1A/SMG6 with telomerase. Nucleic Acids Res 35: 7011–7022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichenbach P, Hoss M, Azzalin CM, Nabholz M, Bucher P, Lingner J 2003. A human homolog of yeast Est1 associates with telomerase and uncaps chromosome ends when overexpressed. Curr Biol 13: 568–574 [DOI] [PubMed] [Google Scholar]
- Riethman H 2008. Human telomere structure and biology. Annu Rev Genomics Hum Genet 9: 1–19 [DOI] [PubMed] [Google Scholar]
- Ritchie KB, Mallory JC, Petes TD 1999. Interactions of TLC1 (which encodes the RNA subunit of telomerase), TEL1, and MEC1 in regulating telomere length in the yeast Saccharomyces cerevisiae. Mol Cell Biol 19: 6065–6075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabourin M, Tuzon CT, Zakian VA 2007. Telomerase and Tel1p preferentially associate with short telomeres in S. cerevisiae. Mol Cell 27: 550–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeftner S, Blasco MA 2008. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat Cell Biol 10: 228–236 [DOI] [PubMed] [Google Scholar]
- Seto AG, Zaug AJ, Sobel SG, Wolin SL, Cech TR 1999. Saccharomyces cerevisiae telomerase is an Sm small nuclear ribonucleoprotein particle. Nature 401: 177–180 [DOI] [PubMed] [Google Scholar]
- Sfeir AJ, Chai W, Shay JW, Wright WE 2005. Telomere-end processing the terminal nucleotides of human chromosomes. Mol Cell 18: 131–138 [DOI] [PubMed] [Google Scholar]
- Sfeir A, Kosiyatrakul ST, Hockemeyer D, MacRae SL, Karlseder J, Schildkraut CL, de Lange T 2009. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell 138: 90–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smogorzewska A, de Lange T 2004. Regulation of telomerase by telomeric proteins. Annu Rev Biochem 73: 177–208 [DOI] [PubMed] [Google Scholar]
- Snow BE, Erdmann N, Cruickshank J, Goldman H, Gill RM, Robinson MO, Harrington L 2003. Functional conservation of the telomerase protein Est1p in humans. Curr Biol 13: 698–704 [DOI] [PubMed] [Google Scholar]
- Sugawara N 1989. DNA sequences at the telomeres of the fission yeast S. pombe. PhD thesis, Harvard University, Cambridge, MA [Google Scholar]
- Surovtseva YV, Churikov D, Boltz KA, Song X, Lamb JC, Warrington R, Leehy K, Heacock M, Price CM, Shippen DE 2009. Conserved telomere maintenance component 1 interacts with STN1 and maintains chromosome ends in higher eukaryotes. Mol Cell 36: 207–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai KK, Kibe T, Donigian JR, Frescas D, de Lange T 2011. Telomere protection by TPP1/POT1 requires tethering to TIN2. Mol Cell 44: 647–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Kannan R, Blanchette M, Baumann P 2012. Telomerase RNA biogenesis involves sequential binding by Sm and Lsm complexes. Nature 484: 260–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira MT, Arneric M, Sperisen P, Lingner J 2004. Telomere length homeostasis is achieved via a switch between telomerase-extendible and -nonextendible states. Cell 117: 323–335 [DOI] [PubMed] [Google Scholar]
- Tomita K, Cooper JP 2008. Fission yeast Ccq1 is telomerase recruiter and local checkpoint controller. Genes Dev 22: 3461–3474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson RL, Ziegler TD, Supakorndej T, Terns RM, Terns MP 2006. Cell cycle-regulated trafficking of human telomerase to telomeres. Mol Biol Cell 17: 955–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson RL, Li J, Culp BR, Terns RM, Terns MP 2010. A Cajal body-independent pathway for telomerase trafficking in mice. Exp Cell Res 316: 2797–2809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng SF, Lin JJ, Teng SC 2006. The telomerase-recruitment domain of the telomere binding protein Cdc13 is regulated by Mec1p/Tel1p-dependent phosphorylation. Nucleic Acids Res 34: 6327–6336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto Y, Taggart AK, Zakian VA 2001. The role of the Mre11-Rad50-Xrs2 complex in telomerase-mediated lengthening of Saccharomyces cerevisiae telomeres. Curr Biol 11: 1328–1335 [DOI] [PubMed] [Google Scholar]
- Tycowski KT, Shu MD, Kukoyi A, Steitz JA 2009. A conserved WD40 protein binds the Cajal body localization signal of scaRNP particles. Mol Cell 34: 47–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Overbeek M, de Lange T 2006. Apollo, an Artemis-related nuclease, interacts with TRF2 and protects human telomeres in S phase. Curr Biol 16: 1295–1302 [DOI] [PubMed] [Google Scholar]
- Vannier JB, Pavicic-Kaltenbrunner V, Petalcorin MI, Ding H, Boulton SJ 2012. RTEL1 dismantles T loops and counteracts telomeric G4-DNA to maintain telomere integrity. Cell 149: 795–806 [DOI] [PubMed] [Google Scholar]
- Venteicher AS, Abreu EB, Meng Z, McCann KE, Terns RM, Veenstra TD, Terns MP, Artandi SE 2009. A human telomerase holoenzyme protein required for Cajal body localization and telomere synthesis. Science 323: 644–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vodenicharov MD, Wellinger RJ 2006. DNA degradation at unprotected telomeres in yeast is regulated by the CDK1 (Cdc28/Clb) cell-cycle kinase. Mol Cell 24: 127–137 [DOI] [PubMed] [Google Scholar]
- Wahba L, Amon JD, Koshland D, Vuica-Ross M 2011. RNase H and multiple RNA biogenesis factors cooperate to prevent RNA:DNA hybrids from generating genome instability. Mol Cell 44: 978–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Podell ER, Zaug AJ, Yang Y, Baciu P, Cech TR, Lei M 2007. The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature 445: 506–510 [DOI] [PubMed] [Google Scholar]
- Watson J 1972. Origin of concatemeric T7 DNA. Nat New Biol 239: 197–201 [DOI] [PubMed] [Google Scholar]
- Webb CJ, Zakian VA 2008. Identification and characterization of the Schizosaccharomyces pombe TER1 telomerase RNA. Nat Struct Mol Biol 15: 34–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb CJ, Zakian VA 2012. Schizosaccharomyces pombe Ccq1 and TER1 bind the 14-3-3-like domain of Est1, which promotes and stabilizes telomerase-telomere association. Genes Dev 26: 82–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Multani AS, He H, Cosme-Blanco W, Deng Y, Deng JM, Bachilo O, Pathak S, Tahara H, Bailey SM, et al. 2006. Pot1 deficiency initiates DNA damage checkpoint activation and aberrant homologous recombination at telomeres. Cell 126: 49–62 [DOI] [PubMed] [Google Scholar]
- Wu P, van Overbeek M, Rooney S, de Lange T 2010. Apollo contributes to G overhang maintenance and protects leading-end telomeres. Mol Cell 39: 606–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin H, Liu D, Wan M, Safari A, Kim H, Sun W, O’Connor MS, Songyang Z 2007. TPP1 is a homologue of ciliate TEBP-β and interacts with POT1 to recruit telomerase. Nature 445: 559–562 [DOI] [PubMed] [Google Scholar]
- Yamazaki H, Tarumoto Y, Ishikawa F 2012. Tel1(ATM) and Rad3(ATR) phosphorylate the telomere protein Ccq1 to recruit telomerase and elongate telomeres in fission yeast. Genes Dev 26: 241–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Lenain C, Bauwens S, Rizzo A, Saint-Leger A, Poulet A, Benarroch D, Magdinier F, Morere J, Amiard S, et al. 2010. TRF2 and Apollo cooperate with topoisomerase 2α to protect human telomeres from replicative damage. Cell 142: 230–242 [DOI] [PubMed] [Google Scholar]
- Yu GL, Bradley JD, Attardi LD, Blackburn EH 1990. In vivo alteration of telomere sequences and senescence caused by mutated Tetrahymena telomerase RNAs. Nature 344: 126–132 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Sfeir AJ, Zou Y, Buseman CM, Chow TT, Shay JW, Wright WE 2009. Telomere extension occurs at most chromosome ends and is uncoupled from fill-in in human cancer cells. Cell 138: 463–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Abreu E, Kim J, Stadler G, Eskiocak U, Terns MP, Terns RM, Shay JW, Wright WE 2011. Processive and distributive extension of human telomeres by telomerase under homeostatic and nonequilibrium conditions. Mol Cell 42: 297–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu XD, Kuster B, Mann M, Petrini JH, Lange T 2000. Cell-cycle-regulated association of RAD50/MRE11/NBS1 with TRF2 and human telomeres. Nat Genet 25: 347–352 [DOI] [PubMed] [Google Scholar]