Abstract
In recent years, an increasing number of studies have shown that prokaryotes and eukaryotes are armed with sophisticated mechanisms to restart stalled or collapsed replication forks. Although these processes are better understood in bacteria, major breakthroughs have also been made to explain how fork restart mechanisms operate in eukaryotic cells. In particular, repriming on the leading strand and fork regression are now established as critical for the maintenance and recovery of stalled forks in both systems. Despite the lack of conservation between the factors involved, these mechanisms are strikingly similar in eukaryotes and prokaryotes. However, they differ in that fork restart occurs in the context of chromatin in eukaryotes and is controlled by multiple regulatory pathways.
The recovery of stalled replication forks involves fork regression and repriming on the leading strand. In eukaryotes, this occurs in the context of chromatin and is controlled by multiple regulatory pathways.
For DNA replication to be accurately completed, the replication fork must frequently overcome a multitude of structurally unrelated obstacles such as DNA lesions, transcribing RNA polymerases, and tightly bound protein–DNA complexes. As a consequence, numerous diverse mechanisms have evolved that either help minimize the frequency or impact of collisions or repair the damage that is left behind. This work will focus solely on the mechanisms that exist in prokaryotes and eukaryotes to facilitate replication on template DNA containing either leading- or lagging-strand polymerization-blocking lesions. Lesions of this type are generated frequently under normal growth conditions (Lindahl 1993), as well as being induced by exogenous genotoxic agents. While cells have mechanisms such as nucleotide excision repair (NER) and base excision repair that target and repair a vast array of DNA modifications (Freidberg 2005), it is inevitable that some damage will persist long enough to be encountered by the DNA replication machinery. To achieve the high fidelity required for genome duplication, the architecture and mechanism of replicative polymerases efficiently discriminate against the incorporation of mismatched bases (Kunkel 2004). As a consequence, even DNA lesions that do not significantly alter DNA structure often inhibit nascent chain elongation. Should the replisome encounter such damage, the template strand in which the damage is located impacts significantly on the mechanism by which it is overcome. It is generally accepted that lagging-strand template lesions present few obstacles to replication fork progression. The situation with leading-strand template damage is more complex, and as such, the events that occur following replisome collision remain the subject of considerable debate.
LAGGING-STRAND TEMPLATE LESIONS
Multiple studies, both in vitro and in vivo, have shown that bacterial replisomes efficiently bypass lagging-strand template damage provided that progression of the replicative helicase—which translocates on the lagging-strand template—is not inhibited. Rolling circle replication assays on templates containing site-specific lagging-strand abasic sites, using both the Escherichia coli (McInerney and O’Donnell 2004) and bacteriophage T4 replisomes (Nelson and Benkovic 2010), showed that leading-strand replication was not affected by the lesion’s presence. The ratio of leading- to lagging-strand replication products was also not altered significantly, indicating that coupled leading- and lagging-strand synthesis was maintained on the damage-containing templates. This is believed to be because the lagging strand is primed repeatedly for Okazaki fragment synthesis, providing an obvious mechanism by which lagging-strand reinitiation can occur. With the lagging-strand polymerase stalled at the site of damage, template unwinding and leading-strand synthesis continue. Lagging-strand synthesis is resumed downstream from the lesion once the stalled polymerase has dissociated and rebounded to a newly synthesized primer. Bypass of lesions in this manner leaves single-stranded (ss) DNA gaps in the lagging strand, which, using an oriC-based replication assay on a damage-containing template, have been estimated to be approximately 1–2 kb (Higuchi et al. 2003). Similarly, ssDNA gaps have been observed on the lagging strand in budding yeast and SV40 DNA after UV exposure (Mezzina et al. 1988; Lopes et al. 2006). Interestingly, these gaps are much smaller in eukaryotes (<400 nucleotides in budding yeast) than in E. coli, which may reflect differences in Okazaki fragment length. It is worth noting that unlike its bacterial counterpart, the eukaryotic replicative helicase complex translocates on the leading-strand template (Fu et al. 2011), which may facilitate the bypass of bulky DNA lesions on the lagging strand.
LEADING-STRAND TEMPLATE LESIONS
The debate surrounding leading-strand template lesions centers on whether the template damage presents an absolute block to replication that must be removed for replication to proceed or if it can be bypassed by reinitiating leading-strand synthesis downstream and then repaired postreplicatively. Following UV irradiation of NER-deficient (Rupp and Howard-Flanders 1968) or -proficient E. coli cells (Khidhir et al. 1985; Witkin et al. 1987; Courcelle et al. 2005; Belle et al. 2007; Rudolph et al. 2007), replication rates immediately postirradiation are reduced significantly (approximately 80%–90%) but do not appear to come to a complete halt. Replication then recovers in NER-proficient strains to the pre-UV rates over a period of time that correlates well with the time taken to remove the majority of pyrimidine dimers from the DNA (Courcelle et al. 1999; Rudolph et al. 2007). These data have been interpreted to mean that leading-strand lesions present a block to replication that must first be removed if replication is to continue. Consistent with these ideas, multiple accessory proteins that are involved in replisome remodeling and recombination are required for replication to recover following UV treatment (McGlynn and Lloyd 2002; Courcelle and Hanawalt 2003), some of which will be discussed later in this work.
Several of the above experiments were conducted using UV intensities sufficient to induce several hundred pyrimidine dimers per E. coli chromosome, equating to one dimer every 10–25 kbp (Rudolph et al. 2007). As the replisome travels at 500–1000 bp/sec (Chandler et al. 1975), and assuming that lagging-strand lesions are efficiently bypassed, replication forks would likely have encountered a leading-strand lesion within the first minute postirradiation, yet replication is seen to continue, albeit at a reduced rate, for considerably longer. Rudolph et al. argued that the majority of replication occurring after UV irradiation was in fact because of new rounds of replication initiation (Rudolph et al. 2007). While replication did take longer to recover in temperature-sensitive DnaA mutants unable to reinitiate replication at oriC, it did not stop completely following UV exposure. These observations therefore suggest that leading-strand template lesions may not form an absolute block to replication. Rupp and Howard-Flanders analyzed the DNA synthesized in E. coli immediately following UV irradiation. Their studies revealed that the newly synthesized DNA contained single-stranded gaps that they estimated to be 1–2 kb (Iyer and Rupp 1971) and that all the nascent chains were considerably shorter than those in untreated controls (Rupp and Howard-Flanders 1968). To explain these observations, they hypothesized that replication could be reinitiated downstream from lesions in both the leading- and lagging-strand templates (Rupp and Howard-Flanders 1968; Rupp 1996).
The leading-strand reinitiation model initially received little support, as the consensus view was that leading-strand priming was restricted to the origin of replication. Furthermore, two studies of the E. coli replication machinery (Higuchi et al. 2003; Pages and Fuchs 2003) concluded that replication becomes uncoupled following an encounter with a leading-strand template lesion, with template unwinding but only lagging-strand synthesis continuing beyond the damage. Similar results were also observed with the bacteriophage T4 replisome (Nelson and Benkovic 2010). However, the studies conducted in E. coli were not designed to visualize leading-strand replication products generated downstream from the damage, and as such, they may have failed to observe leading-strand reinitiation (Higuchi et al. 2003; Pages and Fuchs 2003).
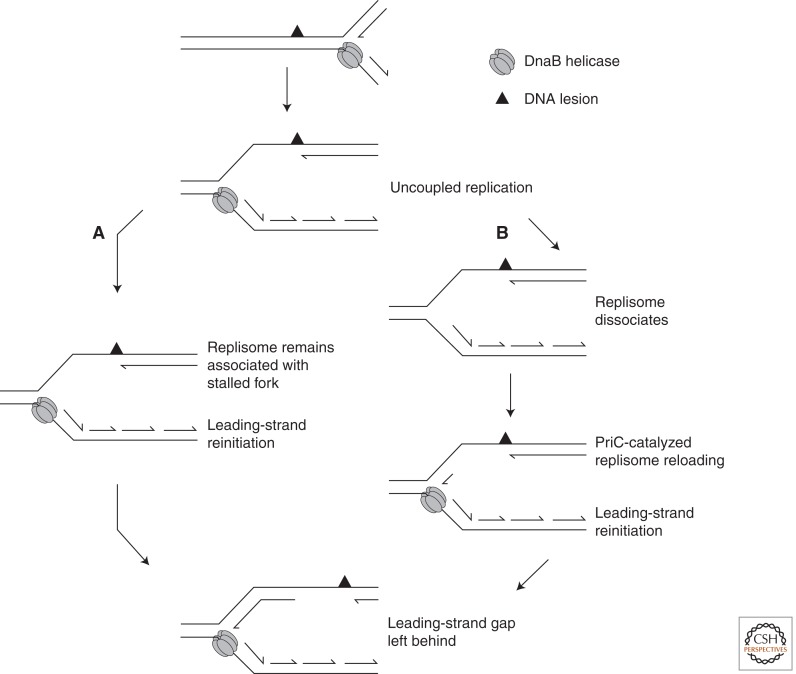
The discovery that primase could in fact prime the leading-strand template on model fork structures provided the first mechanistic evidence that leading-strand synthesis could be reinitiated outside of the origin of replication (Heller and Marians 2006a). This idea was developed in a recent study. Using an oriC-based replication system, the outcomes of collisions between the E. coli replisome and either a single, site-specific pyrimidine dimer or abasic site were investigated (Yeeles and Marians 2011). The data revealed that the replisome transiently stalls upon collision with the lesion but does not dissociate, remaining stably associated with the DNA template (Fig. 1A). Following a short lag, leading- and lagging-strand synthesis were reinitiated downstream from the damage via a de novo, DnaG-dependent priming event on the leading-strand template. The reaction proceeded independently of the replication restart proteins, demonstrating that the replisome has the inherent capacity to replicate beyond leading-strand template lesions by synthesizing the leading strand discontinuously. The precise details of this reaction are yet to be fully elucidated. It is presumed that template unwinding continues for some distance beyond the damage to expose a region of ssDNA on the leading-strand template where primer synthesis occurs. This region is estimated to be anywhere between tens of bases and several hundred, based on the distribution of leading-strand restart products that were observed (Yeeles and Marians 2011). Leading-strand synthesis would resume once the stalled polymerase had dissociated from the stall site and rebounded to the new leading-strand primer. Alternatively, as it has recently been shown that the E. coli replisome can contain three polymerases (McInerney et al. 2007; Reyes-Lamothe et al. 2010; Lia et al. 2012), the possibility exists that a third polymerase that is not bound to a template strand may bind to the new leading-strand primer to reinitiate leading-strand synthesis. The sequences with which these events take place and the rate-limiting step in the reaction are an interesting subject for future investigation.
Figure 1.
Bypass of leading-strand template damage by leading-strand repriming in E. coli. Following a collision with a leading-strand lesion, template unwinding and lagging-strand synthesis are believed to continue beyond the site of damage. (A) Repriming of the leading strand can occur downstream from the damage, which enables replication to continue without the replisome dissociating from the DNA (Yeeles and Marians 2011). (B) Should the replisome dissociate following the collision, the replication restart protein PriC can reload DnaB, which enables replication to be reinitiated by priming the leading strand downstream from the damage (Heller and Marians 2006b).
Leading-strand reinitiation without replisome dissociation provides a potential explanation for the continued replication that is observed immediately post-UV irradiation in vivo. However, the observed leading-strand reinitiation was not 100% efficient, and in some instances template unwinding and only lagging-strand synthesis continued to the end of the template (4 kb). Thus it seems that although the replisome is able to bypass leading-strand template lesions, such lesions will also lead to the breakdown of the replication fork. Under conditions of replication stress—for example, following UV irradiation—multiple lesions in the chromosome will increase the chance of replisome dissociation, eventually leading to a replication arrest. In the absence of exogenous DNA-damaging agents, when template lesions are infrequently encountered, the ability to reinitiate replication downstream from lesions may be considered a general housekeeping function that prevents significant delays to replication fork movement. This will be particularly advantageous during periods of rapid bacterial growth when multiple replication forks are traversing the chromosome, while at the same time chromosome decatenation and cell division are also occurring. Furthermore, induction of the SOS response should be avoided when low levels of DNA damage are present, which in turn will prevent a cell division arrest and the induction of mutagenic translesion DNA polymerases (Goodman and Woodgate 2013).
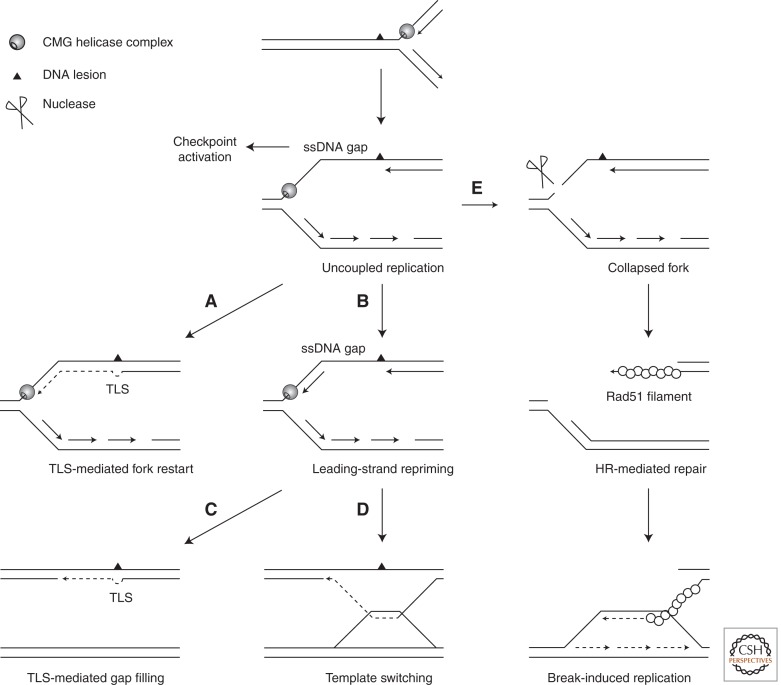
In eukaryotes, it was long believed that leading-strand synthesis is continuous and is exclusively initiated at replication origins. Continuous synthesis through DNA lesions is ensured by specialized DNA polymerases called lesion-bypass DNA polymerases (Goodman and Woodgate 2013) through a process called translesion DNA synthesis (TLS). Recruitment of TLS polymerases to stalled forks depends on a conserved mechanism involving the Rad6- and Rad18-dependent ubiquitination of proliferating cell nuclear antigen (PCNA) (Fig. 2A). TLS polymerases are replaced with replicative polymerases once the damage is passed (Kannouche et al. 2004; Moldovan et al. 2007). Interestingly, discontinuities on both strands have been reported in budding yeast after UV exposure (Lopes et al. 2006), suggesting that repriming also occurs on the leading strand in eukaryotes, as may be the case in bacteria (Fig. 2B). These gaps are repaired postreplicatively by translesion synthesis (Fig. 2C) or by recombination-dependent template switch mechanisms (Fig. 2D) (Lehmann and Fuchs 2006; Branzei and Foiani 2010). Repriming and postreplicative gap filling is supported by recent studies showing that the Saccharomyces cerevisiae Rad6 pathway can be uncoupled from bulk DNA synthesis and can act in G2/M to bypass UV lesions (Daigaku et al. 2010; Karras and Jentsch 2010; Ulrich 2011). In vertebrates, it is generally believed that replicative and postreplicative translesion syntheses coexist. Studies in chicken DT40 cells have shown that postreplicative TLS depends on PCNA ubiquitination, whereas TLS at stalled forks is independent of PCNA modifications but is promoted by a noncatalytic function of the Rev1 polymerase (Edmunds et al. 2008).
Figure 2.
Bypass of leading-strand template damage in eukaryotes. As in bacteria, template unwinding and lagging-strand synthesis are believed to continue beyond the site of damage, resulting in the formation of an ssDNA gap. (A) Fork-associated lesion bypass allows the restart of leading-strand synthesis upon PCNA modification and the transient recruitment of a mutagenic TLS polymerase. (B) Repriming of the leading strand downstream from the lesion leaves an ssDNA gap. (C) This gap can be repaired postreplicatively by TLS polymerases. (D) Error-free lesion bypass can also be performed through a recombination-mediated mechanism called a template switch, which uses the newly synthesized sister chromatid as a template for primer elongation. Note that template switching can also occur after fork regression, as illustrated in Figure 4B. (E) Incomplete nucleotide excision repair of the DNA lesion or cleavage of the fork by endonucleases may also lead to the formation of a one-ended DSB, which can be repaired by an HR-related process called break-induced replication.
REPLICATION FORK BREAKDOWN
Failure of the bacterial replication machinery to reinitiate replication downstream from a leading-strand lesion results in arrest of replication fork progression and will likely lead to replisome dissociation, although it is worth noting that the fates of the original replisome components during such an event have yet to be described. It is also currently unclear how frequently this process occurs in the absence of exogenous DNA-damaging agents or precisely what structure the arrested replication fork adopts. Currently the consensus view is that lagging-strand replication continues beyond the damage for up to several thousand base pairs, generating an extensive region of ssDNA on the leading-strand template (Fig. 1B) (Higuchi et al. 2003; Pages and Fuchs 2003). However, as it has been recently shown that the replisome transiently stalls in response to leading-strand template damage (Yeeles and Marians 2011), an alternative possibility is that the replisome may dissociate close to the site of damage, before extensive template unwinding and uncoupled replication. In this scenario, little ssDNA would be generated on the leading-strand template, and the 3′ end of the blocked nascent leading strand would be situated in close proximity to the fork junction.
In eukaryotic cells, replication fork breakdown represents a major source of genomic instability and has been directly implicated in cancer development (Aguilera and Gomez-Gonzalez 2008; Hastings et al. 2009). To restrain this instability, eukaryotes have evolved complex mechanisms to maintain and signal stalled replisomes, which are absent in bacteria. The Mec1/ATR pathway plays a central role in this process and acts in various ways to protect stalled forks (Lopes et al. 2001; Tercero and Diffley 2001; see Makarova and Koonin 2013 for details). Although this crucial function has been extensively studied over the past decade, the underlying mechanisms remain poorly understood. Based on chromatin immunoprecipitation studies in budding yeast, it has been proposed that the replisome disassembles when checkpoint mutants of the Mec1 pathway are exposed to hydroxyurea (Katou et al. 2003; Cobb et al. 2005; Branzei and Foiani 2010). However, this view has recently been challenged by a biochemical study showing that the replisome remains associated with DNA in hydroxyurea-treated checkpoint mutants (De Piccoli et al. 2012). One way to reconcile these data is to assume that under these conditions, the replisome does not disassemble but moves away from sites of DNA synthesis. This would expose newly synthesized DNA to exonucleases (Cotta-Ramusino et al. 2005; Segurado and Diffley 2008) or to structure-specific endonucleases (Kai et al. 2005; Froget et al. 2008), leading to irreversible fork collapse. Further work is needed to clarify this important issue.
Besides checkpoint factors, homologous recombination (HR) proteins are also implicated in the protection of arrested forks, in a recombination-independent manner (Costanzo 2011). Studies in vertebrates have shown that the Rad51 recombinase is recruited to stalled forks through a mechanism that depends on Mre11 and Brca2 (Hashimoto et al. 2010; Schlacher et al. 2011; Sirbu et al. 2011). In current models the exonucleolytic activity of Mre11 enlarges ssDNA gaps left behind the replisome to facilitate postreplicative repair, while Brca2 loads Rad51 to limit the extension of ssDNA gaps and protect arrested forks (Costanzo 2011).
Multiple pathways for replication fork reactivation have been described. In bacteria the key step, common to all models, is the origin-independent assembly of the replisome, catalyzed by either the PriA- or PriC-dependent replisome loading systems that are defined by the different DNA structures that they recognize (Heller and Marians 2005). The reactivation pathways differ, however, when describing the sequence of events that occurs before replisome loading. Whereas some models envisage minimal fork processing, others posit extensive remodeling to enable the original lesion to be either repaired or bypassed before the resumption of replication. Recent evidence indicates that similar mechanisms exist in eukaryotes, despite the absence of PriA and PriC orthologs.
DIRECT REPLICATION RESTART
The first biochemical evidence for direct replication restart in bacteria was provided when primase was shown to catalyze the de novo synthesis of a primer on the leading-strand template (Fig. 1B) (Heller and Marians 2006a). Using a model forked substrate lacking nascent leading and lagging strands, and therefore mimicking a stalled fork generated by uncoupled replication downstream from a lesion, it was shown that PriC could direct the assembly of a replisome, which in turn led to the initiation of coupled replication via leading-strand priming. PriC facilitates the loading of DnaB to single-stranded DNA binding protein coated ssDNA on the lagging-strand template, with approximately 20 bp of ssDNA required for efficient loading. However, such a region of ssDNA may not always be available if initiation of the last Okazaki fragment occurs close to the fork junction. In this scenario the 3′ → 5′ helicases Rep or PriA function to unwind the nascent lagging stand, exposing the region of ssDNA required for PriC-directed DnaB loading (Heller and Marians 2005a).
If the blocked nascent leading strand is located in close proximity to the fork junction, replication could be reinitiated via the PriA-dependent replication restart pathway (recently reviewed in Gabbai and Marians 2010), as PriA preferentially targets forked structures where the nascent leading strand is located at or close to the fork junction (Heller and Marians 2005b). Following PriA-directed replisome assembly, a leading-strand priming event would enable replication to proceed downstream from the original blockage in an analogous manner to the PriC-dependent pathway. A similar reaction has been fully reconstituted in vitro, in which replication was reinitiated downstream from a leading strand blocked with a chain-terminating dideoxynucleotide (Heller and Marians 2006a). Direct replication restart generates ssDNA gaps in the leading-strand template, in much the same way as those generated when the leading strand is reinitiated without replisome dissociation. To date these are the only models that are able to account for the discontinuous replication that has been observed following UV irradiation in vivo (Rupp and Howard-Flanders 1968).
Unlike bacterial genomes, eukaryotic chromosomes contain a large excess of licensed replication origins that can be used as backup origins to rescue terminally arrested forks (Ge et al. 2007; Ibarra et al. 2008). Consequently, the importance of replication restart pathways has long been disregarded in eukaryotes. With the emergence of DNA combing and other related DNA fiber assays (Tourrière et al. 2005; Petermann and Helleday 2010), it has become clearer that fork restart mechanisms operate in eukaryotic cells and are important for viability under replication stress conditions. For instance, it has been recently reported that human cells briefly exposed to hydroxyurea are able to restart paused forks through a Rad51-dependent mechanism that is distinct from classical double-strand break (DSB) repair (Petermann et al. 2010). Similarly, fork restart by recombination-dependent pathways has been reported in fission yeast, independently of DSB formation (Mizuno et al. 2009; Lambert et al. 2010). In budding yeast hydroxyurea induces a 10- to 20-fold reduction of fork rate (Sogo et al. 2002; Poli et al. 2012) and increases the amount of ssDNA gaps at paused forks by ∼100 nucleotides (Sogo et al. 2002), which could be sufficient to allow repriming on the leading strand (Lambert et al. 2007). In higher eukaryotes much larger ssDNA gaps are generated as a consequence of extensive uncoupling between helicase and polymerase activities (Byun et al. 2005). In Xenopus egg extracts Pol α-primase is recruited to these ssDNA gaps and undergoes continued primer synthesis (Van et al. 2010). This recruitment depends on TopBP1, a replication initiation factor also involved in the activation of the ATR/Mec1 pathway (Yan and Michael 2009), indicating that the events signaling and reactivating arrested forks are functionally linked. Upon initiation, Pol α-primase interacts with other replisome components such as Mcm10 and Ctf4/And-1 (Ricke and Bielinsky 2004; Zhu et al. 2007; Gambus et al. 2009). These interactions are conserved from yeast to human and are important for both initiation and elongation (Tanaka et al. 2009; Kanke et al. 2012; van Deursen et al. 2012; Watase et al. 2012). Whether Pol α-primase binding to Ctf4 and Mcm10 is also required for repriming at stalled forks is currently unknown. However, recent evidence indicates that S. cerevisiae mutants that are unable to tether Pol α-primase to the replisome depend on the Mec1/ATR pathway for viability (Kilkenny et al. 2012).
REPLICATION FORK REMODELING
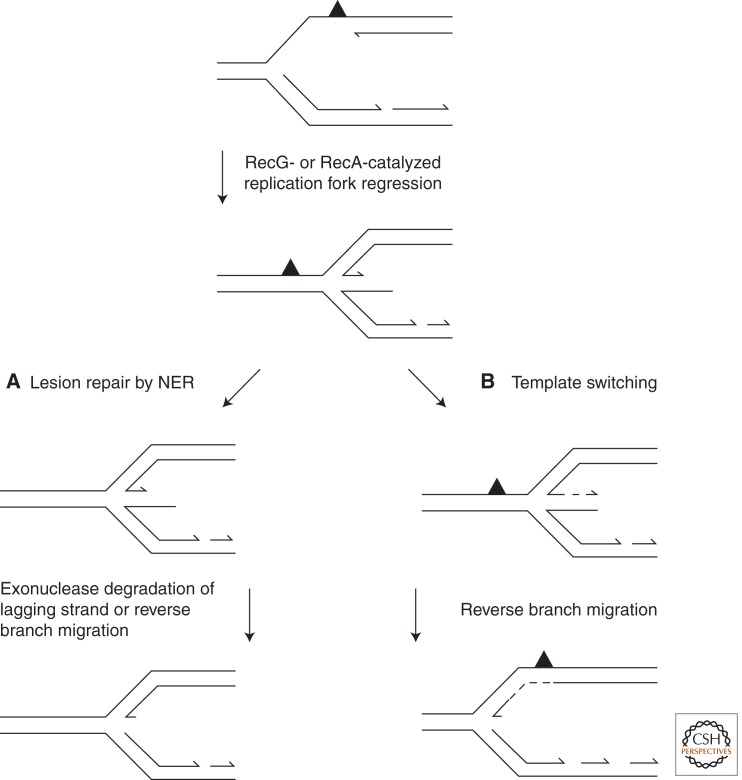
In contrast to models of direct replication restart, additional pathways for fork reactivation in E. coli have been proposed that require extensive remodeling to enable the original lesion to be either removed or bypassed before PriA-dependent replisome reloading (McGlynn and Lloyd 2002; Courcelle and Hanawalt 2003). Two of the major pathways describe the regression of the replication fork, either catalyzed by the helicase RecG (McGlynn and Lloyd 2000; McGlynn et al. 2001) or promoted by the strand exchange protein RecA (Fig. 3) (Robu et al. 2001; Lusetti and Cox 2002; Courcelle et al. 2003). Regression of a replication fork results in rewinding of the parental DNA and displacement of the nascent leading and lagging strands, which can subsequently base pair to form a four-way Holliday junction. Consequently, the original replication-blocking lesion is returned to a region of duplex DNA, enabling it to be removed by NER. Reverse branch migration, catalyzed by RecG (McGlynn and Lloyd 2000), or processing of the extruded nascent strands (Courcelle et al. 2003) enables a replication fork structure to be regenerated (Fig. 3A). Alternatively, the pairing of the extruded nascent leading and lagging strands can provide a template for extension of the leading strand across the site of damage in a reaction termed template switching (Fig. 3B) (Higgins et al. 1976). Resetting of the replication fork places the 3′ OH of the nascent leading strand beyond the site of damage so that replication can be resumed without a need for leading-strand repriming.
Figure 3.
Models for replication fork regression at E. coli replication forks stalled by leading-strand template damage. The helicase RecG binds with high affinity to forks containing a leading-strand gap and unwinds the structure to generate a Holliday junction. A second pathway of fork regression involves RecA binding to the single-stranded region of the fork to drive fork regression and potentially form a Holliday junction. (A) Regression of the fork places the original lesion in a region of double-stranded DNA, enabling it to be repaired by NER. Exonucleolytic degradation of the lagging-strand extension resets the fork to enable replisome loading. (B) The nascent leading strand that is displaced by the regression reaction can be extended using the lagging strand as template. Rewinding of the Holliday junction, possibly by RecG, will place the nascent 3′ end of the leading strand beyond the site of damage to enable replication restart to occur without a need to reprime the leading strand.
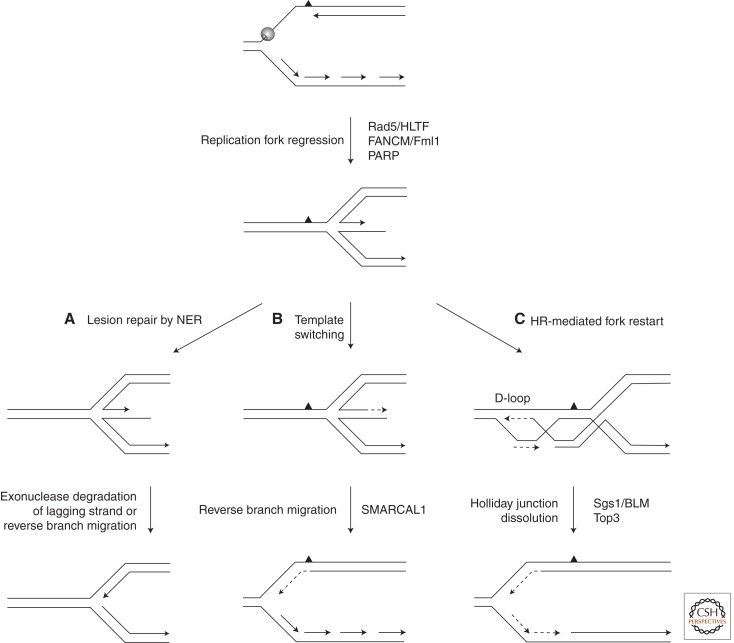
Whether fork regression also occurs in eukaryotic cells is a highly debated issue (Atkinson and McGlynn 2009). Because the unwinding of replication intermediates to form a four-way junction implies a disengagement of the replisome, it was initially believed that fork reversal is a pathological situation leading to irreversible fork breakdown, as observed in yeast checkpoint mutants exposed to hydroxyurea (Sogo et al. 2002; Bermejo et al. 2011). However, a recent study indicates that fork reversal is a physiological process that occurs at 15%–40% of the forks in different organisms on exposure to sublethal doses of Top1 inhibitors, such as camptothecin and topotecan. Interestingly, this process is mediated by poly(ADP-ribose) polymerase (PARP). PARP inactivation with chemical inhibitors or PARP1 genetic ablation impairs fork reversal and leads rather to the formation of DSBs, indicating that fork reversal prevents fork collapse at camptothecin-induced lesions (Ray Chaudhuri et al. 2012). An increasing number of eukaryotic factors able to drive fork reversal in vitro have been identified. These include the yeast Rad5 helicase (Blastyak et al. 2007; Minca and Kowalski 2010) and its human ortholog HLTF (Achar et al. 2011), as well as the Fanconi anemia helicase FANCM (Gari et al. 2008) and its fission yeast ortholog Fml1 (Sun et al. 2008). As illustrated in Figure 4, reversed forks can be restarted by exonucleolytic degradation, reverse branch migration catalyzed by the annealing helicase SMARCAL1 (Driscoll and Cimprich 2009; Betous et al. 2012), or HR-mediated mechanisms. How these mechanisms operate in vivo remains largely unexplored.
Figure 4.
Models for replication fork regression at eukaryotic forks stalled by leading-strand template damage. Several DNA helicases have been shown to promote fork regression in vitro in yeast and vertebrates, including Rad5/HLTF and FANCM/Fml1. In vertebrates, fork regression in vivo also depends on poly(ADP-ribose) polymerase (PARP) (Ray Chaudhuri et al. 2012). Resumption of DNA replication after repair of the lesion (A) or template switching (B) is mediated by nucleolytic degradation of branched structures or reverse branch migration, as described for bacteria. (C) Fork restart can also occur after invasion of the duplex ahead of the lesion by the 3′ overhang of the forked structure. The resulting Holliday junctions are resolved by resolvases or dissolved by the combined action of the RecQ helicases BLM/Sgs1 and the type I topoisomerase Top3, whose function is conserved from yeasts to human.
RECOMBINATION-MEDIATED FORK RESTART
When fork restart mechanisms fail or when the replisome encounters ssDNA nicks or gaps, replication forks can be converted into one-ended DSBs that are subsequently repaired by homology-mediated recombination mechanisms related to break-induced replication (Fig. 2, pathway E; see de la Paz Sanchez et al. 2012 for details). In eukaryotes, this process depends on all the factors involved in processive DNA replication, with the exception of components of the prereplication complex such as ORC and Cdc6 (Lydeard et al. 2010). Evidence from Xenopus egg extracts indicates that when forks encounter ssDNA lesions, Pol ε and the helicase component GINS detach from the replisome and are reloaded in an Mre11- and Rad51-dependent manner. As for break-induced replication, fork restart involves PCNA modifications allowing Pol η–dependent strand extension (Hashimoto et al. 2012). In yeasts, fork restart depends on Rad52 and replication factor C, but not on factors involved in nonhomologous end joining. Interestingly, fork restart can also occur in the absence of Rad51, through a mechanism that depends on the Pol δ subunit Pol32 (Moriel-Carretero and Aguilera 2010). Importantly, recombination-mediated fork restart also occurs in the absence of DSBs when the replisome encounters a replication fork barrier (Lambert et al. 2010).
Besides recombination factors, a wide variety of proteins have been implicated in the recombinational repair of stalled or broken replication forks. These include Slx4, a conserved scaffold protein that interacts with Mus81 and other structure-specific endonucleases (Flott et al. 2007; Andersen et al. 2009; Fekairi et al. 2009; Muñoz et al. 2009; Svendsen et al. 2009). The mechanisms by which Slx4 and Mus81 promote fork recovery are currently unclear. An attractive possibility could be that Mus81 cleaves terminally arrested forks to convert them into structures that are more amenable to recombinational repair (Hanada et al. 2007). However, recent evidence indicates that terminally arrested forks are not efficiently restarted by Mus81 and are rescued primarily by the activation of dormant replication origins (Petermann et al. 2010).
In budding yeast, fork restart depends on Rtt107 (Roberts et al. 2008), a scaffold protein with BRCT domains that interacts with fork repair complexes such as Smc5/6 and Rtt101–Mms1–Mms22 (Luke et al. 2006; Ohouo et al. 2010), which are potentially conserved from yeasts to humans (De Piccoli et al. 2009; Piwko et al. 2011). In budding yeast, the Rtt101 complex promotes the recovery of MMS-arrested forks by sister-chromatid recombination (Duro et al. 2008). Interestingly, Rtt101 function also depends on the acetylation of histone H3 on lysine 56, which is found on newly incorporated histones and may help cells discriminate between parental and newly replicated DNA (Han et al. 2007; Wurtele et al. 2011). Deacetylation of H3K56 is prevented by the Mec1 kinase in response to replication stress in order to facilitate fork recovery (Masumoto et al. 2005). Remarkably, Mec1 also promotes the interaction between Dpb11/TopBP1 and the scaffold proteins Rtt107 and Slx4, which may target these factors to stalled forks (Ohouo et al. 2010). Finally, recent evidence indicates that fork restart also depends on cohesin (Tittel-Elmer et al. 2009) and on the chromatin remodeling complex INO80 (Papamichos-Chronakis and Peterson 2008; Shimada et al. 2008; Falbo et al. 2009). How these multiple factors cooperate to promote fork restart is currently unclear.
As is the case in eukaryotes, the bacterial homologous recombination machinery plays a critical role in salvaging broken replication forks—generated following collisions between the replisome and template nicks or via the cleavage of regressed forks (McGlynn and Lloyd 2002)—in a process termed recombination-dependent replication (Asai et al. 1994). The first step of the pathway involves the resection of the double-stranded DNA end by either an AddAB- or RecBCD-type helicase–nuclease to generate a 3′ ssDNA tail onto which the strand exchange protein RecA is loaded in a sequence-regulated manner (reviewed in Yeeles and Dillingham 2010). Subsequent strand invasion and homologous pairing with the intact chromosome arm generates a D-loop structure, which is then targeted by PriA for origin-independent replisome loading, leading to the establishment of a fully functional replisome (Xu and Marians 2003).
CONCLUDING REMARKS
Studies from many laboratories over the past 20 years have shown convincingly that stalling of replication fork progression can be a dangerous genome-destabilizing event, leading to unscheduled, toxic recombination; degradation of the nascent DNA; and the loss, rearrangement, and alteration of genetic information. The importance of stabilizing and restarting stalled replication forks is underscored by the many proteins involved in these processes that, when mutated, lead to DNA damage syndromes and cancer predisposition. Our understanding of these processes has improved considerably, but there is still much to do, particularly in modeling these events in vitro with the eukaryotic replisome.
ACKNOWLEDGMENTS
We thank Angelos Constantinou, Sarah Lambert, Massimo Lopes, and Benjamin Pardo for discussions and critical comments on the manuscript. Studies from K.J.M.’s laboratory were supported by National Institutes of Health grant GM34557. Studies from P.P.’s laboratory were supported by ANR and Fondation pour la Recherche Médicale (Equipe FRM).
Footnotes
Editors: Stephen D. Bell, Marcel Méchali, and Melvin L. DePamphilis
Additional Perspectives on DNA Replication available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Achar YJ, Balogh D, Haracska L 2011. Coordinated protein and DNA remodeling by human HLTF on stalled replication fork. Proc Natl Acad Sci 108: 14073–14078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilera A, Gomez-Gonzalez B 2008. Genome instability: A mechanistic view of its causes and consequences. Nat Rev Genet 9: 204–217 [DOI] [PubMed] [Google Scholar]
- Andersen SL, Bergstralh DT, Kohl KP, LaRocque JR, Moore CB, Sekelsky J 2009. Drosophila MUS312 and the vertebrate ortholog BTBD12 interact with DNA structure-specific endonucleases in DNA repair and recombination. Mol Cell 35: 128–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai T, Bates DB, Kogoma T 1994. DNA replication triggered by double-stranded breaks in E. coli: Dependence on homologous recombination functions. Cell 78: 1051–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson J, McGlynn P 2009. Replication fork reversal and the maintenance of genome stability. Nucleic Acids Res 37: 3475–3492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belle JJ, Casey A, Courcelle CT, Courcelle J 2007. Inactivation of the DnaB helicase leads to the collapse and degradation of the replication fork: A comparison to UV-induced arrest. J Bacteriol 189: 5452–5462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermejo R, Capra T, Jossen R, Colosio A, Frattini C, Carotenuto W, Cocito A, Doksani Y, Klein H, Gomez-Gonzalez B, et al. 2011. The replication checkpoint protects fork stability by releasing transcribed genes from nuclear pores. Cell 146: 233–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betous R, Mason AC, Rambo RP, Bansbach CE, Badu-Nkansah A, Sirbu BM, Eichman BF, Cortez D 2012. SMARCAL1 catalyzes fork regression and Holliday junction migration to maintain genome stability during DNA replication. Genes Dev 26: 151–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blastyak A, Pinter L, Unk I, Prakash L, Prakash S, Haracska L 2007. Yeast Rad5 protein required for postreplication repair has a DNA helicase activity specific for replication fork regression. Mol Cell 28: 167–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branzei D, Foiani M 2010. Maintaining genome stability at the replication fork. Nat Rev Mol Cell Biol 11: 208–219 [DOI] [PubMed] [Google Scholar]
- Byun TS, Pacek M, Yee M-C, Walter JC, Cimprich KA 2005. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev 19: 1040–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler M, Bird RE, Caro L 1975. The replication time of the Escherichia coli K12 chromosome as a function of cell doubling time. J Mol Biol 94: 127–132 [DOI] [PubMed] [Google Scholar]
- Cobb JA, Schleker T, Rojas V, Bjergbaek L, Tercero JA, Gasser SM 2005. Replisome instability, fork collapse, and gross chromosomal rearrangements arise synergistically from Mec1 kinase and RecQ helicase mutations. Genes Dev 19: 3055–3069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo V 2011. Brca2, Rad51 and Mre11: Performing balancing acts on replication forks. DNA Repair (Amst) 10: 1060–1065 [DOI] [PubMed] [Google Scholar]
- Cotta-Ramusino C, Fachinetti D, Lucca C, Doksani Y, Lopes M, Sogo J, Foiani M 2005. Exo1 processes stalled replication forks and counteracts fork reversal in checkpoint-defective cells. Mol Cell 17: 153–159 [DOI] [PubMed] [Google Scholar]
- Courcelle J, Hanawalt PC 2003. RecA-dependent recovery of arrested DNA replication forks. Annu Rev Genet 37: 611–646 [DOI] [PubMed] [Google Scholar]
- Courcelle J, Crowley DJ, Hanawalt PC 1999. Recovery of DNA replication in UV-irradiated Escherichia coli requires both excision repair and RecF protein function. J Bacteriol 181: 916–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courcelle J, Donaldson JR, Chow KH, Courcelle CT 2003. DNA damage-induced replication fork regression and processing in Escherichia coli. Science 299: 1064–1067 [DOI] [PubMed] [Google Scholar]
- Courcelle CT, Belle JJ, Courcelle J 2005. Nucleotide excision repair or polymerase V-mediated lesion bypass can act to restore UV-arrested replication forks in Escherichia coli. J Bacteriol 187: 6953–6961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigaku Y, Davies AA, Ulrich HD 2010. Ubiquitin-dependent DNA damage bypass is separable from genome replication. Nature 465: 951–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.de la Paz Sanchez M, Costas C, Sequeira-Mendes J, Gutierrez C 2012. Regulating DNA replication in plants. Cold Spring Harb Perspect Biol 4: a010140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Piccoli G, Torres-Rosell J, Aragon L 2009. The unnamed complex: What do we know about Smc5-Smc6? Chromosome Res 17: 251–263 [DOI] [PubMed] [Google Scholar]
- De Piccoli G, Katou Y, Itoh T, Nakato R, Shirahige K, Labib K 2012. Replisome stability at defective DNA replication forks is independent of S phase checkpoint kinases. Mol Cell 45: 696–704 [DOI] [PubMed] [Google Scholar]
- Driscoll R, Cimprich KA 2009. HARPing on about the DNA damage response during replication. Genes Dev 23: 2359–2365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duro E, Vaisica JA, Brown GW, Rouse J 2008. Budding yeast Mms22 and Mms1 regulate homologous recombination induced by replisome blockage. DNA Repair (Amst) 7: 811–818 [DOI] [PubMed] [Google Scholar]
- Edmunds CE, Simpson LJ, Sale JE 2008. PCNA ubiquitination and REV1 define temporally distinct mechanisms for controlling translesion synthesis in the avian cell line DT40. Mol Cell 30: 519–529 [DOI] [PubMed] [Google Scholar]
- Falbo KB, Alabert C, Katou Y, Wu S, Han J, Wehr T, Xiao J, He X, Zhang Z, Shi Y, et al. 2009. Involvement of a chromatin remodeling complex in damage tolerance during DNA replication. Nat Struct Mol Biol 16: 1167–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fekairi S, Scaglione S, Chahwan C, Taylor ER, Tissier A, Coulon S, Dong M-Q, Ruse C, Yates Iii JR, Russell P, et al. 2009. Human SLX4 is a Holliday junction resolvase subunit that binds multiple DNA repair/recombination endonucleases. Cell 138: 78–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flott S, Alabert C, Toh GW, Toth R, Sugawara N, Campbell DG, Haber JE, Pasero P, Rouse J 2007. Phosphorylation of Slx4 by Mec1 and Tel1 regulates the single-strand annealing mode of DNA repair in budding yeast. Mol Cell Biol 27: 6433–6445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freidberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T 2005. DNA repair and mutagenesis, 2nd ed. ASM Press, Washington [Google Scholar]
- Froget B, Blaisonneau J, Lambert S, Baldacci G 2008. Cleavage of stalled forks by fission yeast Mus81/Eme1 in absence of DNA replication checkpoint. Mol Biol Cell 19: 445–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu YV, Yardimci H, Long DT, Ho TV, Guainazzi A, Bermudez VP, Hurwitz J, van Oijen A, Scharer OD, Walter JC 2011. Selective bypass of a lagging strand roadblock by the eukaryotic replicative DNA helicase. Cell 146: 931–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabbai CB, Marians KJ 2010. Recruitment to stalled replication forks of the PriA DNA helicase and replisome-loading activities is essential for survival. DNA Repair (Amst) 9: 202–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambus A, van Deursen F, Polychronopoulos D, Foltman M, Jones RC, Edmondson RD, Calzada A, Labib K 2009. A key role for Ctf4 in coupling the MCM2-7 helicase to DNA polymerase α within the eukaryotic replisome. EMBO J 28: 2992–3004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gari K, Decaillet C, Stasiak AZ, Stasiak A, Constantinou A 2008. The Fanconi anemia protein FANCM can promote branch migration of Holliday junctions and replication forks. Mol Cell 29: 141–148 [DOI] [PubMed] [Google Scholar]
- Ge XQ, Jackson DA, Blow JJ 2007. Dormant origins licensed by excess Mcm2–7 are required for human cells to survive replicative stress. Genes Dev 21: 3331–3341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Goodman MF, Woodgate R 2013. Translesion DNA polymerases. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a010363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Zhou H, Horazdovsky B, Zhang K, Xu RM, Zhang Z 2007. Rtt109 acetylates histone H3 lysine 56 and functions in DNA replication. Science 315: 653–655 [DOI] [PubMed] [Google Scholar]
- Hanada K, Budzowska M, Davies SL, van Drunen E, Onizawa H, Beverloo HB, Maas A, Essers J, Hickson ID, Kanaar R 2007. The structure-specific endonuclease Mus81 contributes to replication restart by generating double-strand DNA breaks. Nat Struct Mol Biol 14: 1096–1104 [DOI] [PubMed] [Google Scholar]
- Hashimoto Y, Chaudhuri AR, Lopes M, Costanzo V 2010. Rad51 protects nascent DNA from Mre11-dependent degradation and promotes continuous DNA synthesis. Nat Struct Mol Biol 17: 1305–1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto Y, Puddu F, Costanzo V 2012. RAD51- and MRE11-dependent reassembly of uncoupled CMG helicase complex at collapsed replication forks. Nat Struct Mol Biol 19: 17–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings PJ, Ira G, Lupski JR 2009. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet 5: e1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller RC, Marians KJ 2005a. Unwinding of the nascent lagging strand by Rep and PriA enables the direct restart of stalled replication forks. J Biol Chem 280: 34143–34151 [DOI] [PubMed] [Google Scholar]
- Heller RC, Marians KJ 2005b. The disposition of nascent strands at stalled replication forks dictates the pathway of replisome loading during restart. Mol Cell 17: 733–743 [DOI] [PubMed] [Google Scholar]
- Heller RC, Marians KJ 2006a. Replication fork reactivation downstream of a blocked nascent leading strand. Nature 439: 557–562 [DOI] [PubMed] [Google Scholar]
- Heller RC, Marians KJ 2006b. Replisome assembly and the direct restart of stalled replication forks. Nat Rev Mol Cell Biol 7: 932–943 [DOI] [PubMed] [Google Scholar]
- Higgins NP, Kato K, Strauss B 1976. A model for replication repair in mammalian cells. J Mol Biol 101: 417–425 [DOI] [PubMed] [Google Scholar]
- Higuchi K, Katayama T, Iwai S, Hidaka M, Horiuchi T, Maki H 2003. Fate of DNA replication fork encountering a single DNA lesion during oriC plasmid DNA replication in vitro. Genes Cells 8: 437–449 [DOI] [PubMed] [Google Scholar]
- Ibarra A, Schwob E, Mendez J 2008. Excess MCM proteins protect human cells from replicative stress by licensing backup origins of replication. Proc Natl Acad Sci 105: 8956–8961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer VN, Rupp WD 1971. Usefulness of benzoylated naphthoylated DEAE-cellulose to distinguish and fractionate double-stranded DNA bearing different extents of single-stranded regions. Biochem Biophys Acta 228: 117–126 [DOI] [PubMed] [Google Scholar]
- Kai M, Boddy MN, Russell P, Wang TS 2005. Replication checkpoint kinase Cds1 regulates Mus81 to preserve genome integrity during replication stress. Genes Dev 19: 919–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanke M, Kodama Y, Takahashi TS, Nakagawa T, Masukata H 2012. Mcm10 plays an essential role in origin DNA unwinding after loading of the CMG components. EMBO J 31: 2182–2194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannouche PL, Wing J, Lehmann AR 2004. Interaction of human DNA polymerase η with monoubiquitinated PCNA: A possible mechanism for the polymerase switch in response to DNA damage. Mol Cell 14: 491–500 [DOI] [PubMed] [Google Scholar]
- Karras GI, Jentsch S 2010. The RAD6 DNA damage tolerance pathway operates uncoupled from the replication fork and is functional beyond S phase. Cell 141: 255–267 [DOI] [PubMed] [Google Scholar]
- Katou Y, Kanoh Y, Bando M, Noguchi H, Tanaka H, Ashikari T, Sugimoto K, Shirahige K 2003. S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature 424: 1078–1083 [DOI] [PubMed] [Google Scholar]
- Khidhir MA, Casaregola S, Holland IB 1985. Mechanism of transient inhibition of DNA synthesis in ultraviolet-irradiated E. coli: Inhibition is independent of recA whilst recovery requires RecA protein itself and an additional, inducible SOS function. Mol Gen Genet 199: 133–140 [DOI] [PubMed] [Google Scholar]
- Kilkenny ML, De Piccoli G, Perera RL, Labib K, Pellegrini L 2012. A conserved motif in the C-terminal tail of DNA polymerase α tethers primase to the eukaryotic replisome. J Biol Chem 287: 23740–23747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel TA 2004. DNA replication fidelity. J Biol Chem 279: 16895–16898 [DOI] [PubMed] [Google Scholar]
- Lambert S, Froget B, Carr AM 2007. Arrested replication fork processing: Interplay between checkpoints and recombination. DNA Repair (Amst) 6: 1042–1061 [DOI] [PubMed] [Google Scholar]
- Lambert S, Mizuno K, Blaisonneau J, Martineau S, Chanet R, Freon K, Murray JM, Carr AM, Baldacci G 2010. Homologous recombination restarts blocked replication forks at the expense of genome rearrangements by template exchange. Mol Cell 39: 346–359 [DOI] [PubMed] [Google Scholar]
- Lehmann AR, Fuchs RP 2006. Gaps and forks in DNA replication: Rediscovering old models. DNA Repair (Amst) 5: 1495–1498 [DOI] [PubMed] [Google Scholar]
- Lia G, Michel B, Allemand JF 2012. Polymerase exchange during Okazaki fragment synthesis observed in living cells. Science 335: 328–331 [DOI] [PubMed] [Google Scholar]
- Lindahl T 1993. Instability and decay of the primary structure of DNA. Nature 362: 709–715 [DOI] [PubMed] [Google Scholar]
- Lopes M, Foiani M, Sogo JM 2006. Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol Cell 21: 15–27 [DOI] [PubMed] [Google Scholar]
- Luke B, Versini G, Jaquenoud M, Zaidi IW, Kurz T, Pintard L, Pasero P, Peter M 2006. The cullin Rtt101p promotes replication fork progression through damaged DNA and natural pause sites. Curr Biol 16: 786–792 [DOI] [PubMed] [Google Scholar]
- Lusetti SL, Cox MM 2002. The bacterial RecA protein and the recombinational DNA repair of stalled replication forks. Annu Rev Biochem 71: 71–100 [DOI] [PubMed] [Google Scholar]
- Lydeard JR, Lipkin-Moore Z, Sheu Y-J, Stillman B, Burgers PM, Haber JE 2010. Break-induced replication requires all essential DNA replication factors except those specific for pre-RC assembly. Genes Dev 24: 1133–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Makarova KS, Koonin EV 2013. Archaeology of eukaryotic DNA replication. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a012963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masumoto H, Hawke D, Kobayashi R, Verreault A 2005. A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature 436: 294–298 [DOI] [PubMed] [Google Scholar]
- McGlynn P, Lloyd RG 2000. Modulation of RNA polymerase by (p)ppGpp reveals a RecG-dependent mechanism for replication fork progression. Cell 101: 35–45 [DOI] [PubMed] [Google Scholar]
- McGlynn P, Lloyd RG 2002. Recombinational repair and restart of damaged replication forks. Nat Rev Mol Cell Biol 3: 859–870 [DOI] [PubMed] [Google Scholar]
- McGlynn P, Lloyd RG, Marians KJ 2001. Formation of Holliday junctions by regression of nascent DNA in intermediates containing stalled replication forks: RecG stimulates regression even when the DNA is negatively supercoiled. Proc Natl Acad Sci 98: 8235–8240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInerney P, O’Donnell M 2004. Functional uncoupling of twin polymerases: Mechanism of polymerase dissociation from a lagging-strand block. J Biol Chem 279: 21543–21551 [DOI] [PubMed] [Google Scholar]
- McInerney P, Johnson A, Katz F, O’Donnell M 2007. Characterization of a triple DNA polymerase replisome. Mol Cell 27: 527–538 [DOI] [PubMed] [Google Scholar]
- Mezzina M, Menck CF, Courtin P, Sarasin A 1988. Replication of simian virus 40 DNA after UV irradiation: Evidence of growing fork blockage and single-stranded gaps in daughter strands. J Virol 62: 4249–4258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minca EC, Kowalski D 2010. Multiple Rad5 activities mediate sister chromatid recombination to bypass DNA damage at stalled replication forks. Mol Cell 38: 649–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno K, Lambert S, Baldacci G, Murray JM, Carr AM 2009. Nearby inverted repeats fuse to generate acentric and dicentric palindromic chromosomes by a replication template exchange mechanism. Genes Dev 23: 2876–2886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldovan G-L, Pfander B, Jentsch S 2007. PCNA, the maestro of the replication fork. Cell 129: 665–679 [DOI] [PubMed] [Google Scholar]
- Moriel-Carretero M, Aguilera A 2010. A postincision-deficient TFIIH causes replication fork breakage and uncovers alternative Rad51- or Pol32-mediated restart mechanisms. Mol Cell 37: 690–701 [DOI] [PubMed] [Google Scholar]
- Muñoz IM, Hain K, Déclais A-C, Gardiner M, Toh GW, Sanchez-Pulido L, Heuckmann JM, Toth R, Macartney T, Eppink B, et al. 2009. Coordination of structure-specific nucleases by human SLX4/BTBD12 is required for DNA repair. Mol Cell 35: 116–127 [DOI] [PubMed] [Google Scholar]
- Nelson SW, Benkovic SJ 2010. Response of the bacteriophage T4 replisome to noncoding lesions and regression of a stalled replication fork. J Mol Biol 401: 743–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohouo PY, Bastos de Oliveira FM, Almeida BS, Smolka MB 2010. DNA damage signaling recruits the Rtt107-Slx4 scaffolds via Dpb11 to mediate replication stress response. Mol Cell 39: 300–306 [DOI] [PubMed] [Google Scholar]
- Pages V, Fuchs RP 2003. Uncoupling of leading- and lagging-strand DNA replication during lesion bypass in vivo. Science 300: 1300–1303 [DOI] [PubMed] [Google Scholar]
- Papamichos-Chronakis M, Peterson CL 2008. The Ino80 chromatin-remodeling enzyme regulates replisome function and stability. Nat Struct Mol Biol 15: 338–345 [DOI] [PubMed] [Google Scholar]
- Petermann E, Helleday T 2010. Pathways of mammalian replication fork restart. Nat Rev Mol Cell Biol 11: 683–687 [DOI] [PubMed] [Google Scholar]
- Petermann E, Orta ML, Issaeva N, Schultz N, Helleday T 2010. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol Cell 37: 492–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piwko W, Buser R, Peter M 2011. Rescuing stalled replication forks: MMS22L-TONSL, a novel complex for DNA replication fork repair in human cells. Cell Cycle 10: 1703–1705 [DOI] [PubMed] [Google Scholar]
- Poli J, Tsaponina O, Crabbe L, Keszthelyi A, Pantesco V, Chabes A, Lengronne A, Pasero P 2012. dNTP pools determine fork progression and origin usage under replication stress. EMBO J 31: 883–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray Chaudhuri A, Hashimoto Y, Herrador R, Neelsen KJ, Fachinetti D, Bermejo R, Cocito A, Costanzo V, Lopes M 2012. Topoisomerase I poisoning results in PARP-mediated replication fork reversal. Nat Struct Mol Biol 19: 417–423 [DOI] [PubMed] [Google Scholar]
- Reyes-Lamothe R, Sherratt DJ, Leake MC 2010. Stoichiometry and architecture of active DNA replication machinery in Escherichia coli. Science 328: 498–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricke RM, Bielinsky AK 2004. Mcm10 regulates the stability and chromatin association of DNA polymerase-α. Mol Cell 16: 173–185 [DOI] [PubMed] [Google Scholar]
- Roberts TM, Zaidi IW, Vaisica JA, Peter M, Brown GW 2008. Regulation of Rtt107 recruitment to stalled DNA replication forks by the cullin Rtt101 and the Rtt109 acetyltransferase. Mol Biol Cell 19: 171–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robu ME, Inman RB, Cox MM 2001. RecA protein promotes the regression of stalled replication forks in vitro. Proc Natl Acad Sci 98: 8211–8218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph CJ, Upton AL, Lloyd RG 2007. Replication fork stalling and cell cycle arrest in UV-irradiated Escherichia coli. Genes Dev 21: 668–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupp WD 1996. DNA repair mechanisms. In Escherichia coli and Salmonella typhimurium: Cellular and molecular biology (ed. Neidhardt FC), pp. 2277–2294 American Society for Microbiology, Washington, DC [Google Scholar]
- Rupp WD, Howard-Flanders P 1968. Discontinuities in the DNA synthesized in an excision-defective strain of Escherichia coli following ultraviolet irradiation. J Mol Biol 31: 291–304 [DOI] [PubMed] [Google Scholar]
- Schlacher K, Christ N, Siaud N, Egashira A, Wu H, Jasin M 2011. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell 145: 529–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segurado M, Diffley JFX 2008. Separate roles for the DNA damage checkpoint protein kinases in stabilizing DNA replication forks. Genes Dev 22: 1816–1827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada K, Oma Y, Schleker T, Kugou K, Ohta K, Harata M, Gasser SM 2008. Ino80 chromatin remodeling complex promotes recovery of stalled replication forks. Curr Biol 18: 566–575 [DOI] [PubMed] [Google Scholar]
- Sirbu BM, Couch FB, Feigerle JT, Bhaskara S, Hiebert SW, Cortez D 2011. Analysis of protein dynamics at active, stalled, and collapsed replication forks. Genes Dev 25: 1320–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogo JM, Lopes M, Foiani M 2002. Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science 297: 599–602 [DOI] [PubMed] [Google Scholar]
- Sun W, Nandi S, Osman F, Ahn JS, Jakovleska J, Lorenz A, Whitby MC 2008. The FANCM ortholog Fml1 promotes recombination at stalled replication forks and limits crossing over during DNA double-strand break repair. Mol Cell 32: 118–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svendsen JM, Smogorzewska A, Sowa ME, O’Connell BC, Gygi SP, Elledge SJ, Harper JW 2009. Mammalian BTBD12/SLX4 assembles a Holliday junction resolvase and is required for DNA repair. Cell 138: 63–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H, Katou Y, Yagura M, Saitoh K, Itoh T, Araki H, Bando M, Shirahige K 2009. Ctf4 coordinates the progression of helicase and DNA polymerase α. Genes Cells 14: 807–820 [DOI] [PubMed] [Google Scholar]
- Tittel-Elmer M, Alabert C, Pasero P, Cobb JA 2009. The MRX complex stabilizes the replisome independently of the S phase checkpoint during replication stress. EMBO J 28: 1142–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourrière H, Versini G, Cordón-Preciado V, Alabert C, Pasero P 2005. Mrc1 and Tof1 promote replication fork progression and recovery independently of Rad53. Mol Cell 19: 699–706 [DOI] [PubMed] [Google Scholar]
- Ulrich HD 2011. Timing and spacing of ubiquitin-dependent DNA damage bypass. FEBS Lett 585: 2861–2867 [DOI] [PubMed] [Google Scholar]
- Van C, Yan S, Michael WM, Waga S, Cimprich KA 2010. Continued primer synthesis at stalled replication forks contributes to checkpoint activation. J Cell Biol 189: 233–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Deursen F, Sengupta S, De Piccoli G, Sanchez-Diaz A, Labib K 2012. Mcm10 associates with the loaded DNA helicase at replication origins and defines a novel step in its activation. EMBO J 31: 2195–2206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watase G, Takisawa H, Kanemaki MT 2012. Mcm10 plays a role in functioning of the eukaryotic replicative DNA helicase, Cdc45-Mcm-GINS. Curr Biol 22: 343–349 [DOI] [PubMed] [Google Scholar]
- Witkin EM, Roegner-Maniscalco V, Sweasy JB, McCall JO 1987. Recovery from ultraviolet light-induced inhibition of DNA synthesis requires umuDC gene products in recA718 mutant strains but not in recA+ strains of Escherichia coli. Proc Natl Acad Sci 84: 6805–6809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtele H, Bacal J, Schalck Kaiser G, St-Hilaire E, Lee EH, Dorn JF, Maddox P, Lisby M, Pasero P, Verreault A 2011. Histone H3 lysine 56 acetylation and the response to DNA replication fork damage. Mol Cell Biol 32: 154–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Marians KJ 2003. PriA mediates DNA replication pathway choice at recombination intermediates. Mol Cell 11: 817–826 [DOI] [PubMed] [Google Scholar]
- Yan S, Michael WM 2009. TopBP1 and DNA polymerase-α directly recruit the 9-1-1 complex to stalled DNA replication forks. J Cell Biol 184: 793–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeeles JT, Dillingham MS 2010. The processing of double-stranded DNA breaks for recombinational repair by helicase–nuclease complexes. DNA Repair (Amst) 9: 276–285 [DOI] [PubMed] [Google Scholar]
- Yeeles JT, Marians KJ 2011. The Escherichia coli replisome is inherently DNA damage tolerant. Science 334: 235–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Ukomadu C, Jha S, Senga T, Dhar SK, Wohlschlegel JA, Nutt LK, Kornbluth S, Dutta A 2007. Mcm10 and And-1/CTF4 recruit DNA polymerase α to chromatin for initiation of DNA replication. Genes Dev 21: 2288–2299 [DOI] [PMC free article] [PubMed] [Google Scholar]