Abstract
Bacteriophage T5 has a 120 kb double-stranded linear DNA genome encoding most of the genes required for its own replication. This lytic bacteriophage has a burst size of ∼500 new phage particles per infected cell, demonstrating that it is able to turn each infected bacterium into a highly efficient DNA manufacturing machine. To begin to understand DNA replication in this prodigious bacteriophage, we have characterized a putative helicase encoded by gene D2. We show that bacteriophage T5 D2 protein is the first viral helicase to be described with bipolar DNA unwinding activities that require the same core catalytic residues for unwinding in either direction. However, unwinding of partially single- and double-stranded DNA test substrates in the 3′–5′ direction is more robust and can be distinguished from the 5′–3′ activity by a number of features including helicase complex stability, salt sensitivity and the length of single-stranded DNA overhang required for initiation of helicase action. The presence of D2 in an early gene cluster, the identification of a putative helix-turn-helix DNA-binding motif outside the helicase core and homology with known eukaryotic and prokaryotic replication initiators suggest an involvement for this unusual helicase in DNA replication initiation.
INTRODUCTION
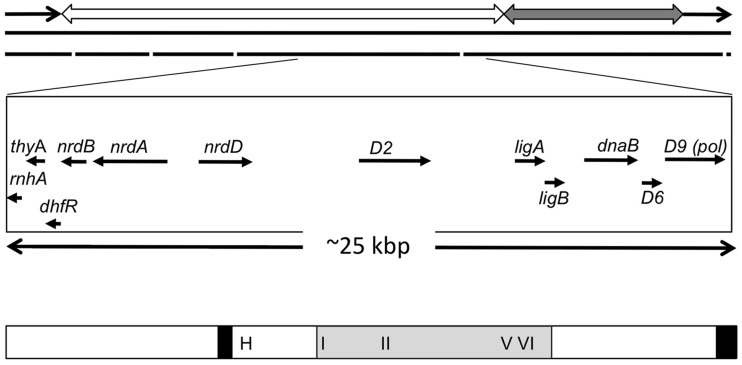
Bacteriophage T5 has an unusual and highly efficient replication cycle compared with the well-characterized T4 and T7 phages (1,2). T5 infection proceeds via a two-step DNA transfer mechanism (3) whereby the first 8% of the linear genome corresponding to a terminal repeat is initially injected into the host cell (Figure 1). Phage-encoded ‘pre-early’ genes are transcribed and translated leading to inactivation of host cell transcription and destruction of host-specific DNA. The remaining phage DNA is then injected into the cell allowing temporally distinct expression of early and late genes (2). T5 infection ultimately results in the production of up to 600 new infectious particles per bacterial cell (4). Despite this highly efficient process, few components of the bacteriophage T5 replication machinery have been characterized at the protein level. Analysis of the T5 genome indicates the presence of several ‘early’ genes likely to encode proteins involved in DNA replication and repair processes (Figure 1). The most extensively studied of these proteins is the product of the D15 gene. It is a 5′ nuclease sharing homology with the small fragment of Escherichia coli (E. coli) DNA polymerase I (PolI), an enzyme involved in processing Okazaki fragments (5–7). The nearby D9 gene encodes a DNA polymerase, equivalent to the Klenow domain of DNA polymerase I (8), as well as RNase H and DNA ligase homologues (9). The early region also contains the D2 gene encoding a putative helicase, a class of enzymes crucial for DNA replication.
Figure 1.
Schematic diagram showing bacteriophage T5 D2 gene in context of the viral genome. The upper panel shows the linear dsDNA T5 genome, which contains direct terminal repeats of ∼10 200 bp (black arrows) carrying the pre-early genes. The ‘early’ and ‘late’ genes are located on contiguous segments indicated by the open and grey double-headed arrows, respectively. The middle panel shows genes flanking D2 that are involved in DNA replication, with direction of transcription indicated by the arrows. These include genes encoding ribonuclease H (rnhA, AAU05228), thymidylate synthase (thyA, protein accession number AAU05229), ribonucleoside-diphosphate reductase alpha and beta chains (nrdA, AAU05233; nrdB, AAU05231), anaerobic ribonucleoside-triphosphate reductase (nrdD, AAU05236), D2 (AAU05245), NAD-dependent DNA ligase subunit A and B (ligA, AAU05253, ligB AAU05254), a second putative replicative DNA helicase (dnaB, AAU05256) and the D9 DNA polymerase (D9, AAU05259). The lower panel shows a schematic of the D2 protein. Two regions of the protein 270–289 and 907–928 predicted to have a >80% probability of forming coiled-coil regions are shown as black rectangles. The region sharing conservation with replication initiation proteins from bacteria and eukaryotic viruses is shaded in grey (residues 389–690), a Walker A motif (motif I, residues 396–406), Walker B motif (motif II, residues 475–489), motif V (residues 632–653) and arginine finger (motif VI, residues 668–682) are indicated. A potential HTH DNA-binding domain was identified (residues 308–329), indicated by H in the figure.
Helicases are nucleic acid translocases that use the energy of nucleotide hydrolysis to unwind duplex nucleic acid structures. As a result, they are players in almost all cellular processes involving double-stranded nucleic acids and are often involved in replication initiation (10–12). Replicative DNA helicases typically form stable hexameric rings on DNA and unwind long (kilobase) stretches of double-stranded DNA (dsDNA), while helicases involved in DNA repair tend to function in a low oligomeric state (monomer/dimer) and unwind short stretches of DNA. Helicases involved in DNA replication and repair usually recognize specific DNA structures, and this, rather than specific base sequence recognition, underlies substrate specificity (13–16). Many helicases are also modular and, as well as a catalytic helicase core, they contain accessory modules that contribute to functional diversity. For example, the human Werner syndrome helicase has nuclease and strand-annealing domains required for its genome stabilizing function (17), while the replicative DNA helicases of the small DNA tumour viruses such as SV40 and papillomavirus are fused to dsDNA-replication-origin–binding domains that allow them to function in replication initiation (18).
A classification system for helicases based largely on primary sequence analysis has been defined (10). Members of six superfamilies (SF1–6) are recognized according to sets of conserved motifs. For example, superfamily 1 (SF1), whose members include the bacterial helicases Rep, PcrA and UvrD, contains 11 distinct signature motifs. SF3, a small group of helicases from RNA and DNA viruses, contains 5 motifs. A more recent and in-depth analysis of the sequence and structure of the largest superfamilies, SF1 and SF2, identified discrete families within each division (19). However, regardless of the individual superfamily groupings, there are three core motifs common to all helicases: The Walker A (GxxxxGK[ST], x denotes any amino acid) and Walker B (hhhhDE, where h is a hydrophobic residue) boxes of the P-loop NTPase fold and an arginine finger domain with a conserved arginine residue. Walker A and B residues bind the phosphates (β and γ) of the bound nucleotide and the conjugated Mg2+ ion, respectively, while the arginine finger residue interacts with the γ-phosphate. Together, these coordinating residues stabilize the NTP:protein complex during hydrolysis, promote catalysis and communicate the resulting protein conformational changes that drive helicase action (10).
Helicases have also been subgrouped according to various structural and mechanistic differences such as those described above. One property of helicases that are also single-stranded DNA (ssDNA) translocases is that they generally move in either the 5′ to 3′ or 3′ to 5′ direction on the tracking strand (10). This operational definition arises from the ability of the helicase to bind to and initiate unwinding from partially single- and double-stranded test substrates with either 5′ (5′–3′ helicase) or 3′ (3′–5′ helicase) ssDNA overhangs. However, a small number of helicases have been shown to possess both 5′–3′ and 3′–5′ unwinding activities encoded within a single polypeptide, suggesting that they could be involved in several biochemical pathways [for example, (13)]. Here we have performed the first biochemical characterization of a helicase encoded by the D2 gene from bacteriophage T5 and show that it is one of this rare group of bipolar helicases. Our initial bioinformatics analysis identified the human Herpes Virus (HSV1) replication initiator protein UL9 and prokaryotic RepA replication initiation helicases as the closest characterized sequence homologues in the sequence databank. The D2 protein possesses consensus Walker A and B boxes and helicase motifs V and VI (arginine finger) that are most similar to the consensus sequences of SF2 helicases. There is also a putative helix-turn-helix (HTH) motif outside the helicase catalytic core. D2’s robust but atypical catalytic activities, combined with the absence of easily identifiable relatives among prokaryotic viruses outside the T5 genus, suggest that this protein may be evolutionarily streamlined to the highly efficient replication process of bacteriophage T5, in which each infected bacterial cell produces up to 50 times more DNA in the form of viral T5 genomes than was initially present in the cell on infection (4).
MATERIALS AND METHODS
Sequence analysis of bacteriophage T5 D2
The bacteriophage T5 D2 protein sequence (accession number AAU05245) was analysed using online bioinformatic software. The potential functional domains and conserved motifs of the D2 protein were predicted by InterPro (20), PROSCAN (21), the Coils server (22) and HTH motif prediction (23). Homologues of the D2 protein were identified using BLASTP (24), and additional helicase motifs in D2 were identified following a ClustalW alignment (25) with available data for sequence homologues in the database, human herpesvirus UL9 and RepA-family proteins from Halomonas phage and Pseudomonas putida.
Generation of expression constructs
The D2 open reading frame (ORF, GenBank accession number AY692264, bp 51489–54275) was amplified from bacteriophage T5 genomic DNA (isolated as described in (26) by primer extension with Pfu DNA polyermase (Promega) using the following primers: ATGCGGTCTCTTATGGTGTTTTCTATCCTCCAAGG (sense) and TCGCGGATCCTCATGCATCTTCATTAGTTG (antisense). Digestion with BsaI and BamHI (underlined) followed by incorporation of dCTP and dATP with Klenow created NdeI and BamHI cohesive ends for cloning into the vector pHisMAL, a modified pET vector carrying an N-terminal 6× histidine (His) tag in frame with the maltose-binding protein (MBP) ORF and a thrombin cleavage site (C-terminal to MBP). The K405E mutant was generated by overlapping primer extension using mutagenic primers (27). The sequence of all constructs was verified by BigDye Terminator v3.1 Cycle Sequencing Kit and ABI PRISM™ 3730 DNA Analyzer (Applied Biosystems) at the Genetics Core Facility, the University of Sheffield.
Protein expression and purification
E. coli BL21(DE3) was transformed with expression constructs and cultures grown in 2× YT medium supplemented with 100 μg/ml of ampicillin at 37°C. At an A600nm of 1.0, isopropyl-β-D-1-thiogalactopyranoside (IPTG) was added to a final concentration of 0.5 mM to induce His-MBP-tagged D2 [wild type (WT) or K405E] protein expression at 25°C for 8 h. Bacterial pellets were obtained by centrifugation at 10 000g for 20 min and stored at −80°C until use. Thawed pellets (∼60 g) were resuspended in 1.5 ml/g of lysis buffer + 0.1 M NaCl (25 mM Tris–HCl, pH 7.5, 1 mM ethylenediaminetetraacetic acid (EDTA), 1 mM phenylmethylsulphonyl fluoride (PMSF), 5 mM 1,4-dithio-DL-threitol (DTT), 10% v/v glycerol and treated with lysozyme (1 mg/ml, stirring at 4°C for 30 min), followed by the addition of a further 1.5 ml/g of lysis buffer + 1.9 M NaCl. Cells were sonicated to reduce viscosity and the extract cleared by centrifugation at 40 000g for 30 min. Polyethylenimine (5% w/v, pH 8.0, Sigma-Aldrich) was added to the supernatant to a final concentration of 0.5% w/v, and after incubation for 5 min, the mixture was centrifuged at 40 000g for 5 min. The supernatant was then incubated with amylose agarose beads (1 ml per 20 g of E. coli cells, New England BioLabs) for 12–16 h. The beads were washed in batch (50 bead volumes of lysis buffer), loaded into a column and washed with 50 bead volumes of lysis buffer with 0.5 M NaCl, no PMSF. The protein was eluted with elution buffer (25 mM Tris–HCl, pH 7.5, 0.5 M NaCl, 1 mM DTT, 20 mM maltose, 10% v/v glycerol). The protein extract was then diluted with 5 volumes of HisTrap buffer (25 mM Tris–HCl, pH 7.5, 0.5 M NaCl, 10% v/v glycerol) and applied to a 1-ml HisTrapTM HP affinity column (GE Healthcare). Protein was eluted with a linear gradient of 0.01–0.25 M imidazole in HisTrap buffer. D2 was further purified by anion exchange chromatography (Source Q, GE Healthcare, 25 mM Tris–HCl, pH 8.5, 1 mM EDTA, 5 mM DTT, 10% v/v glycerol, 0.1–0.4 M NaCl gradient). Peak fractions were pooled, concentrated, aliquoted and stored at −80°C. The protein concentration of His-MBP-tagged D2 WT and K405E was determined by Bradford assays (28), according to the manufacturers’ instructions (BioRad), using bovine serum albumin (BSA) as a standard.
Enzymatic assays
ATPase assays were performed in 20 mM 4-(2-hydroxyethyl)-l-piperazineethanesulfonic acid (HEPES)-NaOH, pH 7.5, 20 mM NaCl, 0.1% v/v NP40 alternative (Calbiochem), 0.1 mg/ml of BSA, 2 mM DTT, 0.0125 μM [γ-32P]ATP (6000 Ci/mmol), 5 mM MgCl2, 5 mM adenosine triphosphate (ATP) at 37°C for 15 min unless stated otherwise. The release of radioactive phosphate was determined using the charcoal-binding assay of Iggo and Lane (29). Helicase substrates were generated with one or more component oligonucleotides 5′-end labelled using T4 polynucleotide kinase (New England BioLabs) and [γ-32P]ATP (7.5 μM, 6000 Ci/mmol). To generate the test DNA substrate with short 20 bp dsDNA portions (the sequences of the oligonucleotides used are shown in Supplementary Figure S1), the labelled oligonucleotide and the appropriate complementary oligonucleotides (labelled or not) were annealed and resolved and purified from 8% (19:1) polyacrylamide gels [1× (90 mM Tris-borate (pH 8.3)-2.5 mM EDTA) TBE buffer]. The series of substrates with a 3′ T55 ssDNA tail and 25, 76 and 153 bp dsDNA portions were generated as described in (30). Briefly, the substrates with 76 and 153 bp of dsDNA were generated from polymerase chain reaction (PCR) products that incorporated a site for the nicking endonuclease Nt.BbvCI close to a DNA end, while the ds25 substrate was generated by annealing and ligating three oligonucleotides. PCR product were first nicked, heated to displace a short oligonucleotide and then ligated to an oligonucleotide with a complementary stretch of bases and 55 T residues. The matching series of substrates with a 5′ T55 tail used here were made in a similar way; Nb.BbvCI and the oligonucleotide 5′-(T)55CGCGC were used to generate the substrates with 5′ T55 tails and 76 or 153 bp of dsDNA, and the oligonucleotides 5′-(T)55CGCGC, 5′-[phos]TGAGGTGCGGTGTGAAATAC and 5′-GTATTTCACACCGCACCTCAGCGCG were annealed and ligated to generate the substrate with 25 bp of dsDNA. The concentration of the labelled substrates was determined on the basis of the specific activity measured for the labelled component oligonucleotides.
Helicase reactions (20 μl, 0.1 nM substrates) were performed in 20 mM HEPES-NaOH, pH 7.5, various NaCl concentrations as indicated, 0.1% v/v NP40 alternative, 0.1 mg/ml of BSA, 2 mM DTT, 5 mM MgCl2, 5 mM ATP at 37°C for 15 min and stopped by the addition of 6× termination buffer [120 mM EDTA, 0.6% w/v sodium dodecyl sulphate (SDS), 1% w/v bromophenol blue, 60% v/v glycerol, with 2 mg/ml of proteinase K]. Where indicated, ATP was replaced with either adenylyl-imidodiphosphate (AMP-PNP) or adenosine 5′diphosphate (ADP) (5 mM). Reaction products were resolved on 8% (19:1) polyacrylamide gels containing 0.05% w/v SDS (1× TBE/0.05% w/v SDS running buffer), visualized and quantified by phosphorimaging.
Gel-shift assays
The substrates for DNA-binding reactions were end labelled with [γ-32P]ATP and purified as described above. The reactions (20 μl, 0.05 nM substrates) were performed in 20 mM HEPES-NaOH, pH 7.5, 100 mM NaCl, 0.1% v/v NP40 alternative, 0.1 mg/ml of BSA, 2 mM DTT, 1 mM EDTA, 10% v/v glycerol at 22°C for 15 min. The binding reactions were separated on 5% (80:1) polyacrylamide gels in 0.25× TBE buffer, visualized and quantified as above.
RESULTS
Sequence analysis of bacteriophage T5 D2
The D2 amino acid sequence was analysed for known motifs using a number of web-based sequence tools. InterProScan identified one conserved domain (PF02399, Pfam database; amino acids 392–532), which is also found in herpesvirus origin-binding proteins and is probably involved in origin-dependent DNA replication. PROSCAN found a single classical ATP-binding signature, the Walker A motif (motif I) at amino acids 399–406 within this putative conserved domain. The sequence also contained two regions potentially able to form coiled coils. BLASTP analysis of the D2 sequence revealed homology with DNA replication origin-binding proteins and helicases from viruses including herpesvirus UL9 (AER38017), and replication proteins from Halomonas phage (YP_001686772) and P. putida (ADR59589, see Supplementary Figure S2). Moreover, Walker B (DEAD box), motif V and potential arginine-finger residues were identified by the ClustalW sequence alignments of D2 with UL9 (Supplementary Figure S2). Finally, HTH prediction software detected a HTH DNA-binding motif located at amino acids 308–329 (Figure 1).
Expression, purification and initial characterization of recombinant D2
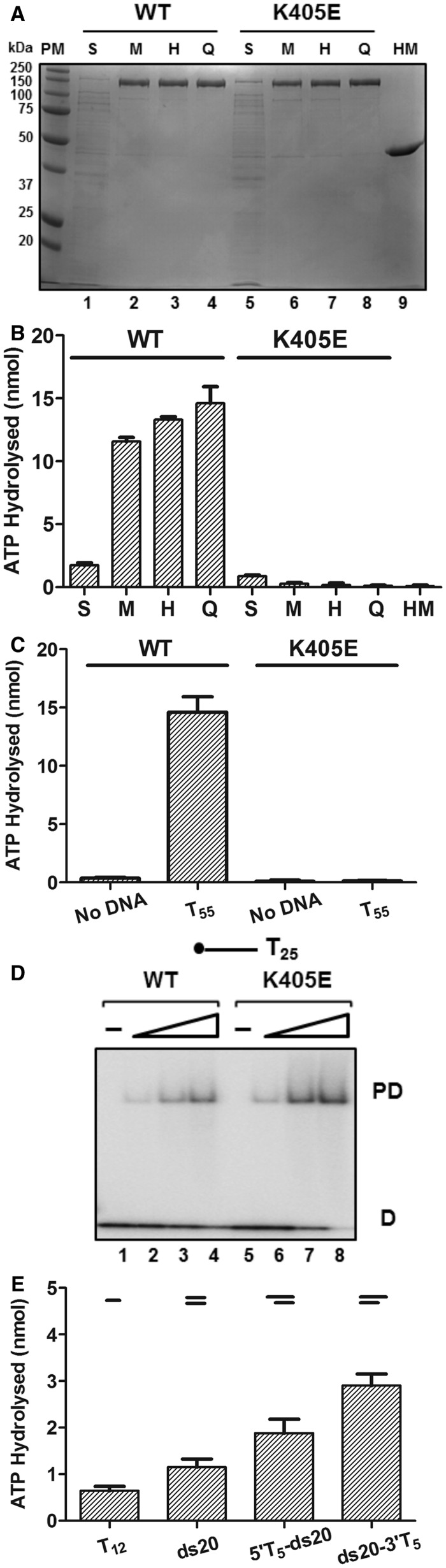
WT D2 and a Walker A K450E mutant were expressed as His-MBP-tagged fusion proteins in E. coli BL21(DE3) for subsequent purification. Figure 2A shows an SDS-polyacrylamide gel electrophoresis (PAGE) analysis of His-MBP-tagged D2 WT and K405E after each purification step. The molecular weight of the His-MBP-tagged D2 WT and K405E major expression products were ∼150 kDa, as predicted for the full-length recombinant ORFs (148 kDa). The purity of the recombinant expression product samples at each step was estimated by densitometric analysis of the SDS-PAGE gel and each was finally obtained to ∼98% purity (Supplementary Table S1 and Figure 2A). Attempts to cleave D2 from the His-MBP affinity tag using intervening sites for the site-specific proteases thrombin, enterokinase, Prescission Protease™ and TEV all resulted in low levels of specific cleavage and degradation of full-length D2 (not shown). For simplicity, the His-MBP tagged proteins used for analysis are referred to below as D2 WT and mutant.
Figure 2.
Biochemical analysis of D2. (A) A 12% SDS-PAGE analysis of His-MBP-D2 WT (lanes 1–4) and K405E mutant (lanes 5–8) proteins at each purification step: supernatant from bacterial lysate (S), eluate from amylose column (M), eluate from HisTrapTM column (H), eluate from Source Q column (Q); His-MBP tag protein (HM) control; 1 μg total protein. (B) The ATPase activity of His-MBP-D2 WT and K405E protein samples from each purification step and His-MBP tag (74.5 ng total protein per assay) was determined in the presence of oligonucleotide T55 (25 nM). The experiment was repeated three times [n = 3, mean and standard deviation (SD)]. (C) ATPase activities of D2 WT and K405E (25 nM) were determined in the absence or presence of T55 (25 nM). The value for ATP hydrolysed at 15 min is given. Further detailed characterization of the ATPase activity of purified D2 indicated that the rate of ATP hydrolysis under the standard assay conditions is outside the linear range after 20 min (Supplementary Figure S3). (D) Binding of D2 WT and K405E (Lanes 2–4 and 6–8, 0.01, 0.05 and 0.1 nM protein) to 5′ 32P-end-labelled T25 ssDNA (0.05 nM substrate) determined by EMSA and visualized by phosphorimaging (PD, protein–DNA complex; D, free DNA). A small increase in ssDNA-binding activity was observed repeatedly with D2 K405E. (E) ATPase activity of D2 WT (25 nM) measured in the presence of various DNA substrates (25 nM): ssDNA, T12; blunt-ended dsDNA, ds20; partial duplexes with a 20 bp and a 5′ or 3′ 5-base oligo-dT overhang, 5′T5-ds20 and ds20-3′T5.
The ATPase activity of D2 WT and K405E protein samples from each purification step were determined in the presence of T55 ssDNA and for D2 WT protein samples a robust ATPase activity was obtained that increased in specific activity through each purification step (Figure 2B; see also Supplementary Table S1 detailing the purity and specific ATPase activity at each step). For D2 K405E protein samples, the ATPase activity decreased after each purification step (Figure 2B), and the final purified D2 K405E protein sample lacked any significant ATPase activity, as did a control His-MBP protein (HM, Figure 2A, lane 9) after only the first two affinity steps (Supplementary Table S1). Figure 2C demonstrates that the ATPase activity of WT D2 after the final purification step was largely dependent on the addition of exogenous T55 ssDNA, while the K405A mutant lacked activity, with or without T55.
D2-ssDNA binding (without ATP/Mg2+) was also tested directly using a 32P-end-labelled T25 substrate and an electrophoretic gel mobility shift assay (EMSA). The K405E mutation did not reduce D2 ssDNA-binding activity (Figure 2D, lanes 5–8 compared with 1–4), indicating that this mutation does not have a wider disruptive effect on protein function. No DNA-binding activity was detected for the control protein His-MBP (data not shown). A comparison of the ability of a series of short ds-, ss- and partially single- and double-stranded DNAs to stimulate the ATPase activity is also shown in Figure 2E (see also below). The data demonstrate that a 20 bp duplex with 3′ T5 tail (ds20-3′T5) stimulates the ATPase activity of D2 better than the same duplex sequence with a 5′ T5 tail (5′T5-ds20), and significantly better than the blunt- ended 20 bp duplex sequence or T12 ssDNA.
D2 is a bipolar DNA helicase
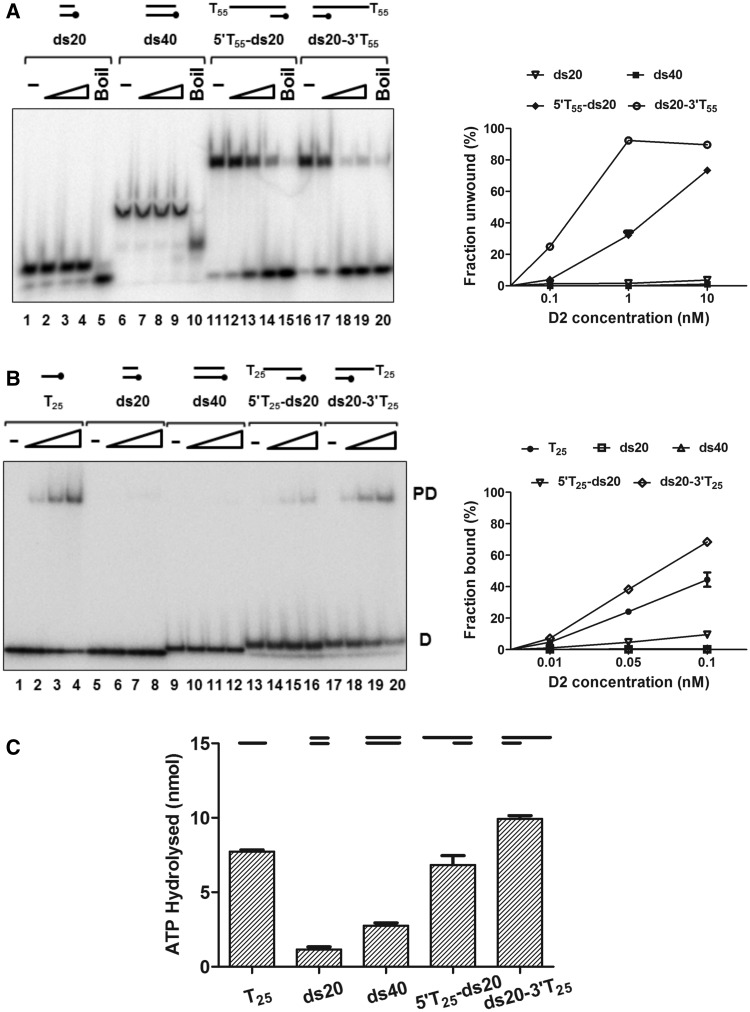
Given that D2 bound ssDNA and that stimulation of ATPase activity preferred partially single- and double-stranded substrates, we next asked if D2 had helicase activity (Figure 3). D2 did not unwind blunt-ended dsDNA substrates (ds20 and ds40, Figure 3A, lanes 1–10), but it could unwind the 20 bp dsDNA substrates with a 5′ or 3′ oligo-dT55 ssDNA overhang (substrates 5′T55-ds20 and ds20-3′T55, lanes 11–20). However, the extent of unwinding of test substrates in the 3′–5′ direction (substrate ds20-3′T55) was greater than that in the 5′–3′ direction (substrate 5′T55-ds20). As shown in the graph on the right of Figure 3A, the ratio of the extent of unwinding 3′–5′ /5′–3′ was ∼3-fold at the intermediate concentration of 1 nM protein and ∼6-fold at the lower protein concentration (0.1 nM). Furthermore, time course experiments show that unwound products accumulate at a faster rate for unwinding in the 3′–5′ compared with 5′–3′ direction (Supplementary Figure S4).
Figure 3.
D2 is a bipolar DNA helicase. (A) Helicase reactions (0.1 nM substrate) were incubated at 37°C for 15 min in reaction buffer with 20 mM NaCl. The reaction products were resolved on 8% (19:1) polyacrylamide gels, visualized and quantified by phosphorimaging. The quantitative data (n = 3, mean and SD) for the extents of unwinding are shown in the graph on the right. Lanes 1, 6, 11 and 16, no protein (-); lanes 5, 10, 15 and 20, heat-denatured substrate (Boil); lanes 2–4, lanes 7–9, lanes 12–14 and lanes 17–19, D2 protein titrations (0.1, 1, 10 nM) and indicated substrates. (B) Protein-DNA–binding reactions (0.05 nM substrate) were assessed by EMSA in the absence of ATP/Mg2+. Lanes 1, 5, 9, 13 and 17, no protein control (-); lanes 2–4, lanes 6–8, lanes 10–12, lanes 14–16 and lanes 18–20, with D2 protein (0.01, 0.05 and 0.1 nM) and the substrates indicated. Quantitative data (fraction bound, n = 3, mean and SD) are shown in the graph on the right. (C) Stimulation of D2 (25 nM) ATPase activity by the same substrates (25 nM) used for the EMSA assays.
To complement the helicase assays, we also performed DNA-binding EMSA in the absence of ATP/Mg2+ with D2 and the 32P-end-labelled helicase/ATPase substrates (Figure 3B) and ATPase assays (Figure 3C). As above, a single discrete complex for D2 binding to T25 ssDNA was observed in EMSA (Figure 3B, lanes 1–4), but stable binding to the blunt-ended duplex substrates ds20 and ds40 was not observed (lanes 5–12), even though they can stimulate ATPase activity (Figure 3C). D2 could also bind to 5′T25-ds20 and ds20-3′T25 (substrates with a 20 bp duplex and T25 tails) to form a discrete protein–DNA complex (Figure 3B, lanes 13–20). As with the preferential unwinding of the substrate with a 3′T overhang, D2 showed a higher binding affinity for ds20-3′T25 compared with 5′T25-ds20 (>8-fold, at 0.05 nM D2, lane 19 compared with lane 15 and graphed data to the right in Figure 3B) and a higher stimulation of ATPase activity (Figure 3C). Furthermore, mutant D2 K405E was inactive as either a helicase or ATPase, while retaining full DNA-binding activity (Supplementary Figure S5), indicating that the catalytic residues (Walker A) for ATP hydrolysis are required for unwinding DNA in either direction.
Initiation of 5′–3′ and 3′–5′ helicase activity as a function of substrate ssDNA tail length
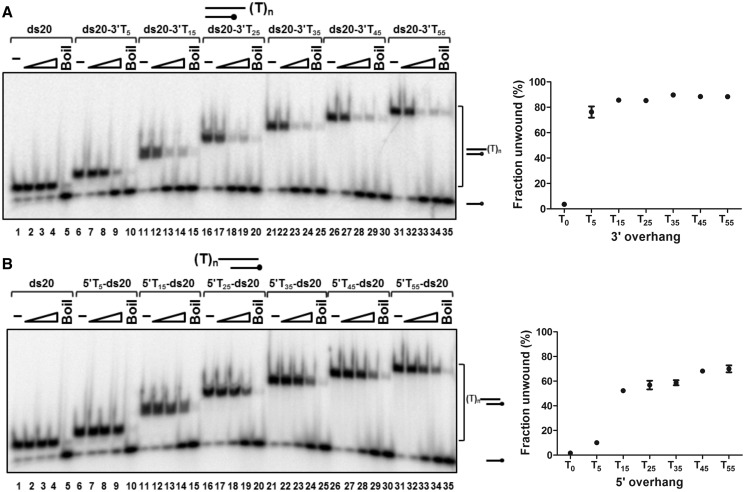
As described above (Figure 3A), D2 only unwound duplex DNA (20 bp) with a T55 5′ or 3′ ssDNA overhang (tail). The effect of the length of the 5′ or 3′ single-stranded tail on helicase action was investigated using substrates with a 20-bp dsDNA component and oligo dT tails from 0 to 55 bases (Figure 4). Again, blunt-ended dsDNA (ds20, without a tail) was not unwound (Figure 4A and B, lanes 1–5). The results for substrates with a 3′ tail (Figure 4A, lanes 6–35 and graphed data on the right) demonstrate that D2 requires only a 5-base 3′ tail for initiation of unwinding. Approximately 76% of the substrate ds20-3′T5 was unwound (10 nM D2), and when the 3′ tail length was extended from 5 to 35 bases (10-base increments), the extent of unwinding activity increased only slightly, reaching a plateau of ∼90% substrate unwound (10 nM D2).
Figure 4.
Unwinding of partially single- and double-stranded substrates with 3′ (A) or 5′ (B) overhangs by D2. Substrates were 20 bp with oligo-dT ssDNA tail from 0 to 55 bases. Helicase reactions (0.1 nM substrate; 20 mM NaCl) were incubated at 37°C for 15 min and products were resolved on 8% (19:1) polyacrylamide gels, visualized and quantified by phosphorimaging. Lanes 1, 6, 11, 16, 21, 26 and 31, no protein (-); lanes 5, 10, 15, 20, 25, 30 and 35, heat-denatured substrate (Boil); lanes 2–4, lanes 7–9, lanes 12–14, lanes 17–19, lanes 22–24, lanes 27–29 and lanes 32–34, D2 protein titrations (0.1, 1, 10 nM) and the indicated substrates. The graphs show the statistical data for the extents of unwinding for 10 nM D2, three repeats, mean and SD.
For substrates with 5′ oligo-T tails (Figure 4B), the substrate with a 5-base 5′ tail, (5′T5-ds20) was a poorer substrate for D2, with ∼10% unwinding at 10 nM D2 compared with 76% for ds20-3′T5 as described above. The proportion of substrate unwound by D2 increased sharply as the 5′ tail length was extended from 5 to 15 bases (∼50% of 5′T15-ds20 unwound) and rose slightly thereafter when the 5′ tail length was increased to 35 bases (∼60% unwound with 10 nM D2). A further increase in the efficiency of initiation of unwinding was observed when the 5′ tail length was increased to 45 bases (∼70% of 5′T45-ds20 unwound), but no further increase in the extent of unwinding was observed when the 5′ tail length was extended to 55 bases. In summary, compared with the 5′ tail substrates, substrates with shorter 3′ tails were better substrates for the initiation of D2 unwinding.
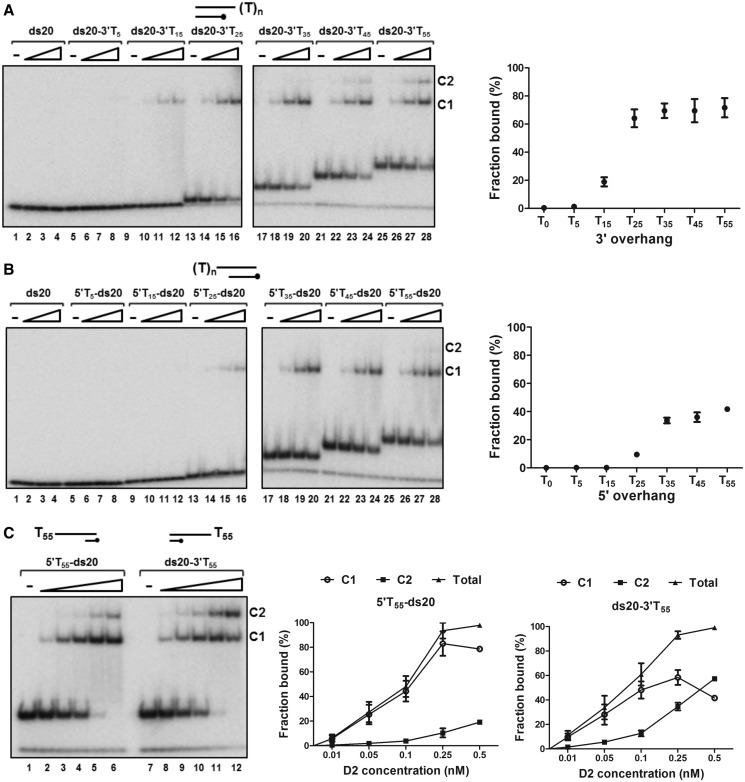
D2 binding to partially single- and double-stranded helicase substrates
We also analysed DNA binding to the same 5′ and 3′ ssDNA tailed duplex helicase substrates by EMSA, for binding reactions without ATP/Mg2+ (Figure 5). For the 3′-tailed substrates, a stable D2–DNA complex was not observed under these conditions for substrates with a 3′ tail of 5 bases or less (ds20 and ds20-3′T5, Figure 5A, lanes 1–8 and the graph to the right). The extent of binding increased sharply as the 3′ tail was extended from 15 to 25 bases (from ∼20% of ds20-3′T15 bound to ∼65% of ds20-3′T25 bound, 0.1 nM D2) and only one predominant protein–DNA complex (C1) was observed (lanes 9–16). A further increase in the tail length (35 to 55 residues) resulted in the appearance of a second complex (C2) first with substrate ds20-3′T35. In contrast, with the 5′ tail substrates (Figure 5B), there was no detectable D2 binding to substrates with a 5′ tail of ≤15 bases (lanes 1–12 and the graphed data). Although less than that observed with the 3′-overhang, significant quantities of product were observed with substrates possessing 5′-overhangs of 25 nucleotides (5′T25-ds20). When the 5′ tail length was increased from 25 to 55 bases (5′T25 to 5′T55-ds20), D2 was bound with gradually increasing affinity (from ∼10% of 5′T25-ds20 bound to ∼42% of 5′T55-ds20 bound, 0.1 nM D2) and there was one main complex observed (C1). A second complex (C2) was only detected with substrate 5′T55-ds20 at the higher D2 concentrations (lanes 27 and 28).
Figure 5.
D2 binding to partially single- and double-stranded DNA substrates. Reactions (0.05 nM DNA substrate, 0.01, 0.05 and 0.1 nM D2) were assembled and analysed by EMSA (n = 3, mean and SD). (A) Binding to 20 bp DNA substrates with 3′ T tails of increasing length. Complex formation was first observed with substrate ds20-3′T15 and increased significantly in extent when the tail length was increased. The increase in 3′ tail length from 35 to 55 residues promoted the formation of a second complex, C2. Quantitation of the binding data is shown for 0.1 nM D2 (total fraction of substrate bound, n = 3). (B) Binding to substrates with 5′ T tails. (C) Complex formation with D2 and a 20 bp DNA substrate with either a 5′ or 3′ 55-bp poly T overhang. D2 concentrations were increased up to 0.5 nM (0.01, 0.05, 0.1, 0.25 and 0.5 nM). The graphed data show total binding and the individual C1 and C2 components.
The smallest ssDNA tail length where we observed C2 formation on the helicase substrates was 35 nucleotides. C2 is also first detectable with ssDNA poly T substrates of 35 nucleotides (Supplementary Figure S6). When we quantified accurately, in an extended protein titration series, the extent of complex formation on the helicase substrates with 55 base 5′ and 3′ extensions (Figure 5C), the fraction of DNA bound was similar at each protein concentration. However, for ds20-3′T55, as the protein concentration increased, the ratio of C1:C2 decreased progressively, while for 5′T55-ds20, complex C1 remained the dominant species that formed, only until the higher protein concentrations were reached. Therefore, C2 forms more efficiently on the substrates with a 3′ tail, suggesting that C2 is derived from C1 and is influenced by the polarity of DNA binding.
Duplex DNA unwinding in the presence of ADP and AMP-PNP
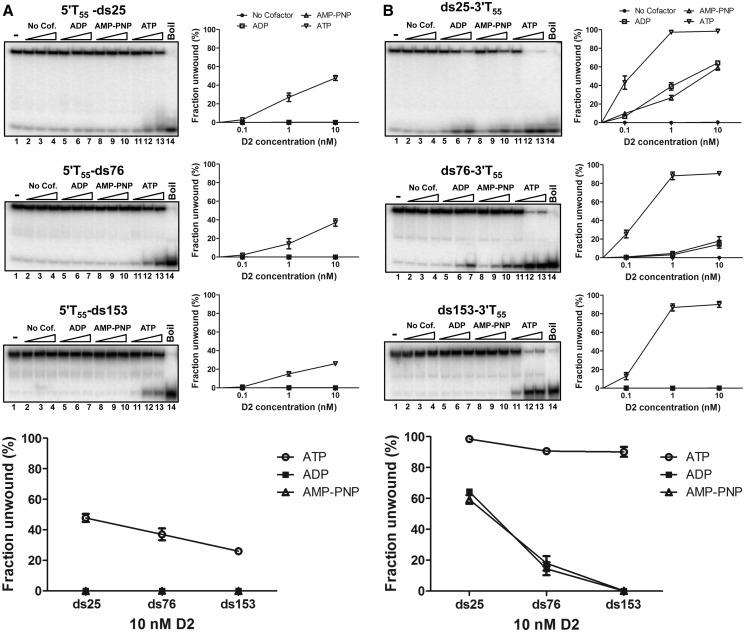
DNA unwinding by D2 in the absence of nucleotide cofactors and with ADP and the non-hydrolysable ATP analogue AMP-PNP present was tested using a series of substrates with either 3′ or 5′ T55 overhangs and dsDNA components of 25, 76 or 153 bp (Figure 6). In the presence of ATP (lanes 11–13), substrates with the 5′ T55 tail were unwound with progressively decreasing efficiency as the length of the dsDNA component increased (Figure 6A, panels top to bottom). Approximately 50% of the substrate 5′T55-ds25 was unwound, and this decreased to ∼25% for substrate 5′T55-ds153 (at 10 nM D2, Figure 6A). Without nucleotide cofactor (lanes 2–4) or with ADP (lanes 5–7) or AMP-PNP (lanes 8–10), no unwinding of the 5′T-tailed substrates was observed. In contrast, in the presence of ATP, all the 3′ T55-tailed substrates were unwound efficiently with little decrease as the dsDNA length was increased, (Figure 6B and integral graph). However, both ADP and AMP-PNP were able to support limited unwinding of substrate ds25-3′T55 and ds76-3′T55, but no unwinding was observed in the absence of cofactor.
Figure 6.
Unwinding of extended duplex DNA up to 153 bp, with and without nucleotide cofactors. Helicase assays were performed as described (see ‘Materials and Methods’ section) (0.1 nM DNA substrate, 0.1, 1 and 10 nM D2, 5 mM nucleotide cofactor, as indicated) but with substrates containing 25, 76 and 153 bp of DNA and 3′ or 5′ T55 extensions. All reactions contained 5 mM MgCl2. As indicated, ATP (lanes 11–13) was replaced with AMP-PNP (lanes 8–10), ADP (lanes 5–7) or nucleotide co-factor was omitted (lanes 2–4). (A) Unwinding of substrates with a 5′ T55 tail. (B) Unwinding of substrates with a 3′ T55 tail.
Stable D2 unwinding complexes assemble on substrates with 3′ but not 5′ ssDNA extensions
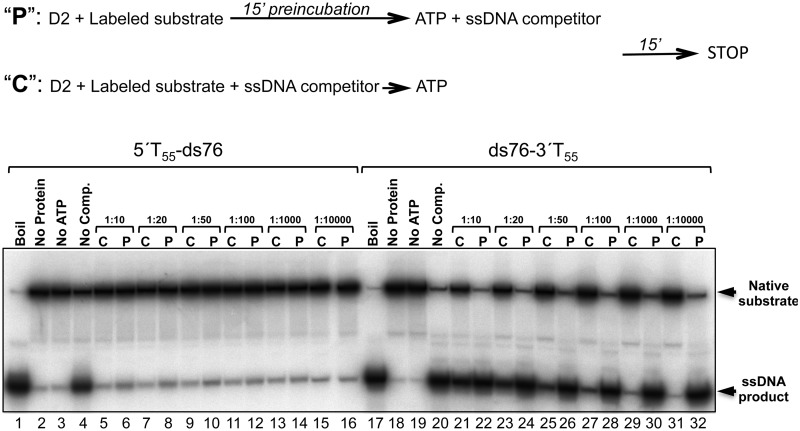
We next tested whether stable D2-DNA unwinding complexes could assemble on substrates with either 5′ or 3′ T55 ssDNA extensions. Substrates with either 20 or 76 bp dsDNA gave identical results, and data for 5′T55-ds76 and ds76-3′T55 are shown in Figure 7. D2 was pre-incubated with 32P-radiolabelled substrates in the absence of ATP (15 min) before the addition of excess unlabelled T55 competitor ssDNA and the ATP required to initiate helicase action. In parallel control reactions D2, labelled substrate and competitor DNA were premixed before the addition of ATP. For substrate, 5′T55-ds76 (lanes 1–16), ∼34% of the substrate was unwound under the standard assay procedure (lane 4). In control reactions (indicated ‘C’), all concentrations of competitor DNA, from 10- to 10 000-fold excess over D2 protein, abolished unwinding. When D2 was pre-incubated with labelled substrate before the addition of ATP and competitor ssDNA (indicated ‘P’), only low levels of substrate unwinding were observed, (up to ∼1.3%; for example, lane 10 compared with 9). In contrast, for substrate ds76-3′T55 (lanes 17–32), competitor DNA concentrations exceeding a 1000-fold excess over D2 protein were required to completely abolish unwinding in control reactions (compare lane 20 with lanes 29 and 31). However, at these same concentrations of competitor ssDNA (≥1000-fold excess), when substrate and D2 were pre-incubated before the addition of ATP and competitor, the extents of unwinding observed were similar to the control reaction with no competitor DNA (compare lane 20 with lanes 30 and 32). These data indicate that stable helicase complexes assemble on the substrates with 3′ but not 5′ ssDNA extensions and that the processivity of 3′–5′ helicase action exceeds that in the 5′–3′ direction.
Figure 7.
Stable unwinding complexes assemble on substrates with 3′ but not 5′ extension. Unwinding reactions (10 nM D2) were assembled as described (see ‘Materials and Methods’ section) except the substrate concentration used was 1 nM and the order of addition of ATP and excess T55 ssDNA competitor was varied, as depicted above. Lanes 1–16, substrate 5′T55-ds76; lanes 17–32, substrate ds76-3′T55. Lanes 1–4 and 17–20, control reactions. Lanes 5–16 and 21–32, reactions pre-incubated with (‘C’) or without (‘P’) competitor, before the addition of ATP. D2-DNA unwinding complexes pre-formed on substrate ds76-3′T55 but not 5′T55-ds76 were resistant to challenge with excess competitor DNA (fold molar excess over D2 protein indicated).
Salt sensitivity of D2 DNA-stimulated ATPase and 5′ and 3′ helicase activity
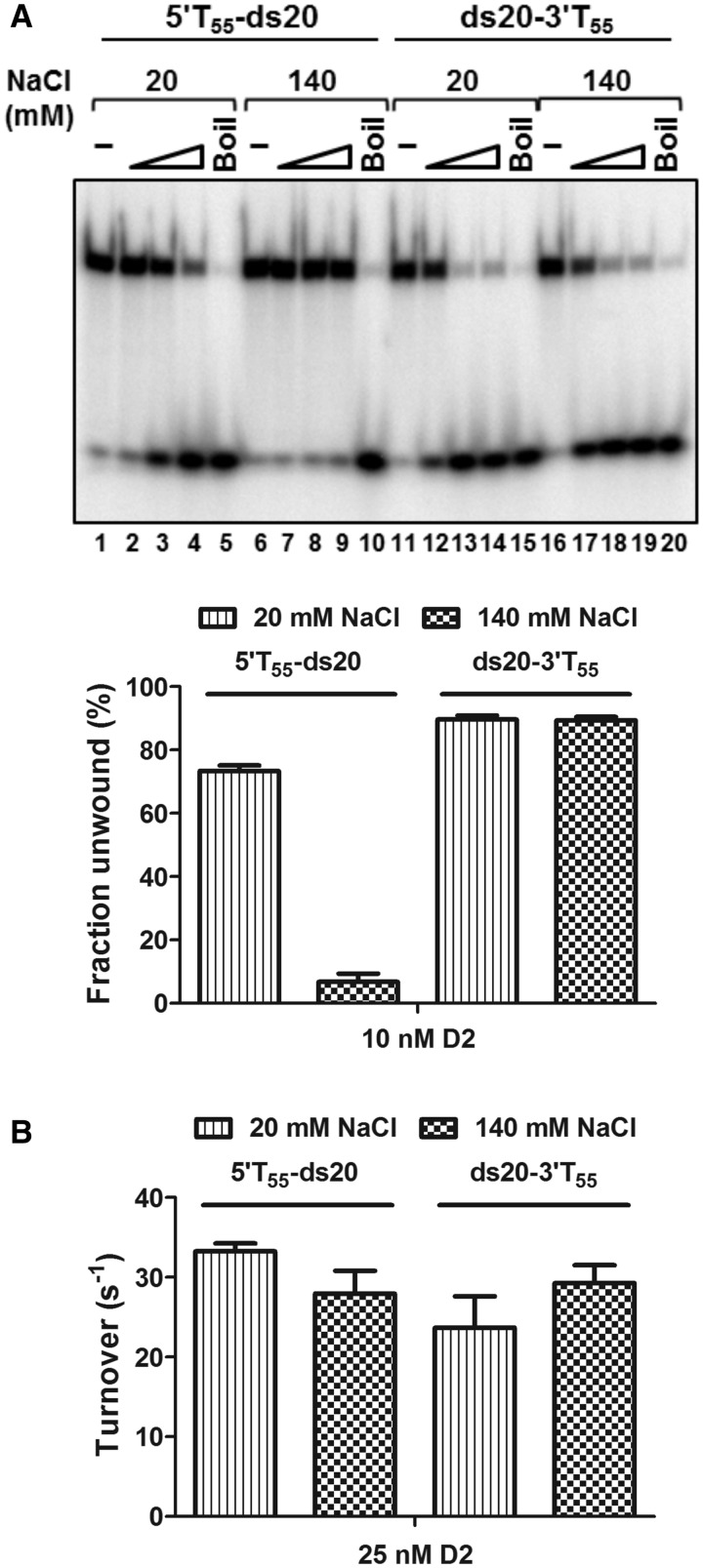
To further investigate the DNA helicase substrate specificity of D2, we determined how NaCl concentration influenced unwinding activity. Using the dsDNA substrates with 55 base 5′ or 3′ T overhangs (5′T55-ds20 and ds20-3′T55), the salt concentration in helicase reaction was varied from 20 to 140 mM in 20 mM increments (Supplementary Figure S7). As summarized in Figure 8A, the 3′–5′ unwinding activity was not affected by changes in NaCl concentration over the entire range tested (lanes 11–20). In contrast, however, the 5′–3′ unwinding activity decreased progressively as the salt concentration was increased, to ∼10-fold inhibition at 140 mM NaCl (Figure 8A, lanes 1–10, and Supplementary Figure S7).
Figure 8.
Effect of NaCl concentration on the enzymatic activities of D2. (A) The unwinding of 5′T55-ds20 and ds20-3′T55 at 20 and 140 mM NaCl (n = 3, mean and SD). At 20 mM NaCl, D2 showed bipolar unwinding activity although with preferential unwinding of the substrate with a 3′ overhang (lanes 1–5 and lanes 11–15). At 140 mM NaCl, the 5′–3′ unwinding activity was almost completely inhibited (lanes 6–10), but not the 3′–5′ unwinding activity (lanes 16–20). The graphed data are for the highest concentration of D2 tested, 10 nM. (B) ATPase activity stimulated by 5′T55-ds20 and ds20-3′T55 (25 nM D2 and DNA, n = 3, mean and SD). Turnover numbers were calculated from the 5-min time point by dividing the amount of product formed by the molar quantity of enzyme and time in seconds: 5′T55-ds20, 33.3 ± 1 s −1 and 28 ± 2.9 s−1; ds20-3′T55, 23.7 ± 3.9 s−1 and 29.3 ± 2.2 s−1 at 20 and 140 mM NaCl, respectively.
When the ATPase activity stimulated by the helicase substrates with either 3′ or 5′ T55 tails was compared at 20 or 140 mM NaCl, it was observed that at 20 mM NaCl, ATPase activity increased in the presence of 5′T55-ds20 compared with ds20-3′T55, although it is the poorer helicase substrate (Figure 8B). This is in contrast to helicase substrates with short 5′ oligo T extensions (e.g. 5 base, Figure 2, and 25 bases, Figure 3) that trigger lower levels of ATPase activity compared with the substrates with similar 3′ overhangs. However, at 140 mM NaCl, the ATPase activity of D2 was lower in the presence of both helicase substrates, but the difference between them was small (29.3 s−1 for substrate ds20-3′T55 and 28.0 s−1 for 5′T55-ds20).
DISCUSSION
The D2 ORF resides in the ‘early’ gene region of the bacteriophage T5 genome and is flanked by genes encoding a number of replication proteins, including DNA polymerase, RNaseH and two putative helicases, D6 and D10. The T5 D2 amino acid sequence was predicted to contain a HTH DNA-binding motif, potential coiled-coil- forming helices and four principal conserved helicase motifs (Figure 1). Because the sequence of motifs V and VI belong to SF2, D2 is most likely an SF2 helicase. Homology searches using the D2 protein sequence identified significant similarity with several replication-origin DNA-binding helicases. The best characterized is the herpesvirus UL9 protein, also an SF2 helicase-family member, which binds specifically to the viral origin of replication (31). Recent evidence suggests that UL9 exists as reciprocal dimers, i.e. the N terminus of one molecule interacts with the C terminus of the second (32). UL9 helicase also interacts with two other virally encoded proteins, ICP8 and UL42, enhancing its ability to unwind DNA (33). While the well-characterized RepA of E. coli plasmid RSF1010 protein is representative of a group of hexameric SF4 helicases involved in the replication of IncQ plasmids (34,35), the larger Halomonas phage RepA-like protein, also identified in the sequence homology search, is a SF2-like helicase and a sequence-specific DNA-binding protein probably involved in replication of the phage’s linear genome (36). These findings suggested that D2 may be an initiator and helicase protein with a role in T5 replication.
Our attempts to express the native soluble D2 protein were unsuccessful (data not shown); however, we were able to produce biologically active His-MBP-D2 fusion proteins that were analysed in detail. Initial biochemical assay of purified D2 demonstrated a DNA-dependent ATPase activity stimulated by a variety of linear single– and partially double–stranded DNA substrates, consistent with the prediction that D2 is a helicase. Furthermore, the kinetics of these activities (∼23–33 s−1 depending on the DNA cofactor, Figure 8) is comparable with the ATPase activities reported for other bacteriophage helicases, such as T7 gp4 dTTPase, 23.6 ± 4.1 s−1 (37), and T4 UvsW-ATPase, 50.1 ± 3.14 s−1 (38).
In general, DNA helicases show specific directionality of unwinding, defined as 5′–3′ or 3′–5′ depending on whether the test substrate unwound possesses a 5′ or 3′ ssDNA overhang. An analysis of the ability of D2 to unwind dsDNA substrates revealed a rare bipolar helicase activity, since D2 could unwind dsDNA substrates with either a 5′ or 3′ ssDNA tail. A single amino acid substitution (K405E) in the Walker A motif abolished D2 ATPase and bipolar helicase activities (but not DNA-binding activity), suggesting both 5′–3′ and 3′–5′ motor units of D2 share the same catalytic core. However, the degree of each directional unwinding by D2 is not equivalent: 3′–5′ unwinding is superior in extent compared with 5′–3′ unwinding activity, and the polarity can be distinguished by a number of other criteria. First, unwinding in the 3′–5′ direction is initiated on test substrates with short (5 base) ssDNA tails, compared with a requirement for longer tails (>45 bases) to achieve similar extents of unwinding at higher protein concentrations in the 5′–3′ direction. This is also reflected in D2 protein-DNA–binding affinity. Second, stable unwinding complexes form on substrates with 3′ ssDNA tails but not 5′ tails of equivalent length, manifesting as enhanced apparent 3′–5′ processivity, even without hydrolysable ATP. Third, the rates of accumulation of product are greater for the 3′–5′ unwinding activity, and finally, the polar unwinding activities show a marked difference in salt sensitivity. While the 3′–5′ unwinding activity is insensitive to salt concentration over the 20–140 mM range tested, the 5′–3′ activity decreases ∼10-fold. However, the ATPase stimulated by helicase substrates with 3′ or 5′ T55 extensions was relatively insensitive to changes in salt concentration. This could indicate that a catalytically productive interaction is made by D2 with T55-tailed substrates of either polarity, but with substrate 5′T55-ds20, the chemical energy of ATP hydrolysis is not channelled effectively to the mechanical events of unwinding. These data may indicate that the relative extent of the polar unwinding activities could be regulated in a physiological context.
Helicases can be classified simplistically based on whether they are hexameric rings in the active form or not (10). SF2 (and SF1) helicases are considered to be non-ring form helicases that are usually functional in a low oligomeric form (39–41). In some cases, SF1 and SF2 helicases function as monomers but multiple subunits can assemble and track together on DNA to increase the overall unwinding efficiency [such as T4 Dda (42,43), HCV NS3 (44) and Bacillus stearothermophilus PcrA (45)]. Rate- zonal ultracentrifugation experiments (glycerol gradient sedimentation) show D2 binding to ssDNA (T25) most likely as a monomer, with or without nucleotide cofactors present (Supplementary Figure S8). Because substrates with 20-bp dsDNA and 5′ extension as short as five nucleotides are effectively unwound by D2, D2 probably functions in a low oligomeric form as expected for SF2 helicases. As a complement to helicase assays, we performed DNA-binding EMSA in the absence of ATP/Mg2+, with D2 and the 32P-end-labelled helicase/ATPase substrates. An initial complex C1 that formed at low protein concentrations and a second complex C2 whose formation was ssDNA-length and protein-concentration dependent were observed. The observed binding and stoichiometries are also consistent with D2 binding in C1 as a low oligomeric form, less than hexameric. When binding to partially single- and double-stranded DNA helicase substrates, the 3′ ssDNA extensions and the initial formation of C1 promotes formation of C2 (Figure 5). Again, as unwinding is initiated efficiently on test substrates (20 bp dsDNA) with short 3′ overhangs (5–15 T bases) and the ssDNA length for formation of C2 is >35 bases, it is unlikely that formation of the higher order species C2 is obligatory for unwinding. Furthermore, C2 formation does not appear to be truly co-operative, as the results do not indicate that the D2-dimerization constant exceeds the DNA binding constant; C1 is always in excess over C2 until protein is saturating. Nonetheless, the observation that the polarity of the ssDNA overhang (3′) influences the efficiency of C2 formation could indicate that orientation-specific dimers form, as noted above for HSV UL9 (32). This could be a mechanism to increase 3′–5′ helicase activity in vivo.
Helicases normally demonstrate unidirectional unwinding activity, and bipolar helicases are rare. A literature search reveals that only 10 bipolar helicases have been discovered in all organisms to date, including E. coli RecBCD (46,47), Staphylococcus aureus and Bacillus anthracis PcrA (13,48), Sulfolobus acidocaldarius HerA (49), Plasmodium falciparum PfDH45 and PfDH60 (50,51), pea PDH47 (52), rabbit eIF4A (53), human p68 (54) and hChlR1 (55). Hence, to our knowledge, D2 is the first identified bipolar helicase encoded by a virus. As discussed above, D2 displayed a biased unwinding polarity preference (3′–5′ unwinding > 5′–3′ unwinding activity). Different levels of unwinding activity in either the 5′ or 3′ direction are common for most of the bipolar helicases characterized to date (13,46,48,51,52,55). Therefore, although it is conceivable that the presence of the MBP fusion tag influences D2 unwinding to generate the biased polarity, we consider this unlikely. In general, a major barrier to understanding the exact role of this feature of the bipolar helicases is that there is little information available on their biological functions. Furthermore, for D2, the identified homologues (UL9 and the RepA-like proteins of a Halomonas phage and P. putida) provide few additional clues as to the significance of the bipolar activity, as UL9 is unidirectional (3′–5′) and the RepA proteins have not been characterized biochemically. However, enticing speculation arises from the observation that non-hydrolysable adenine nucleotides support limited 3′–5′ helicase action. By definition, helicases couple protein conformational changes induced by ATP binding and hydrolysis to nucleic acid translocation and unwinding, although non-enzymatic strand displacement by some ssDNA-binding proteins such as human replication protein A (RPA) (56) can occur through stable ssDNA nucleoprotein filament formation. ATP-independent unwinding, albeit slow or limited in efficiency and extent, has been reported for only a small minority of characterized helicases [for example, the E. coli DEAD box helicase DpbA (57)]. More relevant though, HSV UL9 helicase, the protein with the most sequence similarity to D2 that we identified, can unwind the dsDNA HSV origin of replication (ori) in the absence of nucleotides. Although this UL9 initiator activity is dependent on interaction with the HSV single-stranded binding protein ICP8 (58), the D2 nucleotide dependent but ATP hydrolysis and accessory protein independent unwinding activity could, nonetheless, suggest a role for D2 in T5 replication initiation.
Recently, a model to explain how the polarity preference for unwinding relates to the function of the E. coli RecBCD bipolar helicase has been proposed. The recombinational repair enzyme RecBCD possesses two independent helicase subunits with opposite polarity (RecB: 3′–5′ helicase, RecD: 5′–3′ helicase) that translocate along both ssDNA strands of duplex DNA (59,60). The bidirectional unwinding biased towards one direction results in the two helicase subunits moving on opposite strands at different speeds during unwinding. The faster motor is a leading motor acting as a genuine helicase, while the slower motor is a trailing motor acting as an ssDNA translocase. Two motors with different speeds were reported to adjust the overall speed of RecBCD. Before Chi (χ, crossover hotspot instigator) sequence recognition, RecD is the faster motor and an ssDNA loop forms ahead of the slower RecB motor. When Chi is recognized and tightly bound by RecC, RecB becomes the leading motor and the speed of RecD is reduced to be below that of RecB. Consequently, the existing ssDNA loop shortens and another one forms behind RecB, where the RecA protein is loaded at a rate matched to the slow growth of the new ssDNA loop. The ssDNA strand is then prepared for recombination and repair (60–62).
The biological role of D2 helicase bipolarity is difficult to discern, as bacteriophage T5 replication is poorly understood at present. The genomic location of D2, on a segment of the DNA that is transcribed after second-step transfer, offers few clues beyond hinting at a possible role in replication. Nearby genes include RNaseH (AAU05228), an anaerobic ribonucleotide reductase (AAU05236), thymidylate synthase (AAU05229), two subunits of ribonucleoside-diphosphate reductase (AAU05233 and AAU05231), dihydrofolate reductase (AAU05230) and NAD-dependent DNA ligase subunits (AAU05253 and AAU05254) and DNA polymerase (AAU05259) lie within 12 kb of the D2 locus. Also, a previous study demonstrated that amber mutants in D2 showed no net DNA replication at 80 min post infection (63). As discussed above, as D2 shares some sequence similarity with DNA replication initiator proteins/helicases, the genomic location and observed biochemical activities are all consistent with the notion that D2 encodes a replication initiation protein that unwinds DNA at the replication fork. Furthermore, two D2 helicase motor units of opposite polarity and efficiency may lead to the formation of sterically separated ssDNA loops, as described in the RecBCD model. T5 DNA replicons consisting of multiple internal origins of bidirectional synthesis resembling theta and/or sigma structures have also been observed (64). Thus, two helicase motors with different speeds may act as a molecular ‘gearbox’ in DNA replication. The switch of the acting leading motor between the two helicases may adjust the overall speed of the replication machinery to coordinate continuous leading-strand and discontinuous lagging-strand DNA synthesis at the replication fork, preventing leading-strand synthesis from outpacing lagging-strand synthesis. Alternatively, D2 could function in distinct T5 replication pathways that require either unwinding polarity.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1, Supplementary Figures 1–8 and Supplementary Reference [65].
FUNDING
Sheffield University Doctoral Studentship (to I.N.W). Funding for open access charge: The University of Sheffield.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Weigel C, Seitz H. Bacteriophage replication modules. FEMS Microbiol. Rev. 2006;30:321–381. doi: 10.1111/j.1574-6976.2006.00015.x. [DOI] [PubMed] [Google Scholar]
- 2.Sayers JR. Bacteriophage T5. In: Calender R, editor. The Bacteriophages. 2nd edn. Oxford, UK: Oxford University Press; 2006. pp. 268–276. [Google Scholar]
- 3.Lanni YT. Invasion by bacteriophage T5. II. Dissociation of calcium-independent and calcium-dependent processes. Virology. 1960;10:514–529. doi: 10.1016/0042-6822(60)90133-1. [DOI] [PubMed] [Google Scholar]
- 4.Constantinou A, Voelkel-Meiman K, Sternglanz R, McCorquodale MM, McCorquodale DJ. Involvement of host DNA gyrase in growth of bacteriophage T5. J. Virol. 1986;57:875–882. doi: 10.1128/jvi.57.3.875-882.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tomlinson CG, Syson K, Sengerová B, Atack JM, Sayers JR, Swanson L, Tainer JA, Williams NH, Grasby JA. Neutralizing mutations of carboxylates that bind metal 2 in T5 flap endonuclease result in an enzyme that still requires two metal ions. J. Biol. Chem. 2011;286:30878–30887. doi: 10.1074/jbc.M111.230391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Allen LM, Hodskinson MR, Sayers JR. Active site substitutions delineate distinct classes of eubacterial flap endonuclease. Biochem. J. 2009;418:285–292. doi: 10.1042/BJ20081637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feng M, Patel P, Dervan JJ, Ceska T, Suck D, Haq I, Sayers JR. Roles of divalent metal ions in flap endonuclease-substrate interactions. Nat. Struct. Mol. Biol. 2004;11:450–456. doi: 10.1038/nsmb754. [DOI] [PubMed] [Google Scholar]
- 8.Andraos N, Tabor S, Richardson CC. The highly processive DNA polymerase of bacteriophage T5. Role of the unique N and C termini. J. Biol. Chem. 2004;279:50609–50618. doi: 10.1074/jbc.M408428200. [DOI] [PubMed] [Google Scholar]
- 9.Effantin G, Boulanger P, Neumann E, Letellier L, Conway JF. Bacteriophage T5 structure reveals similarities with HK97 and T4 suggesting evolutionary relationships. J. Mol. Biol. 2006;361:993–1002. doi: 10.1016/j.jmb.2006.06.081. [DOI] [PubMed] [Google Scholar]
- 10.Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- 11.Lohman TM, Tomko EJ, Wu CG. Non-hexameric DNA helicases and translocases: mechanisms and regulation. Nat. Rev. Mol. Cell Biol. 2008;9:391–401. doi: 10.1038/nrm2394. [DOI] [PubMed] [Google Scholar]
- 12.Patel SS, Picha KM. Structure and function of hexameric helicases. Annu. Rev. Biochem. 2000;69:651–697. doi: 10.1146/annurev.biochem.69.1.651. [DOI] [PubMed] [Google Scholar]
- 13.Anand SP, Khan SA. Structure-specific DNA binding and bipolar helicase activities of PcrA. Nucleic Acids Res. 2004;32:3190–3197. doi: 10.1093/nar/gkh641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brosh RM, Jr, Majumdar A, Desai S, Hickson ID, Bohr VA, Seidman MM. Unwinding of a DNA triple helix by the Werner and Bloom syndrome helicases. J. Biol. Chem. 2001;276:3024–3030. doi: 10.1074/jbc.M006784200. [DOI] [PubMed] [Google Scholar]
- 15.George T, Wen Q, Griffiths R, Ganesh A, Meuth M, Sanders CM. Human Pif1 helicase unwinds synthetic DNA structures resembling stalled DNA replication forks. Nucleic Acids Res. 2009;37:6491–6502. doi: 10.1093/nar/gkp671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Whitby MC, Lloyd RG. Targeting holliday junctions by the RecG branch migration protein of Escherichia coli. J. Biol. Chem. 1998;273:19729–19739. doi: 10.1074/jbc.273.31.19729. [DOI] [PubMed] [Google Scholar]
- 17.Bernstein KA, Gangloff S, Rothstein R. The RecQ DNA helicases in DNA repair. Annu. Rev. Genet. 2010;44:393–417. doi: 10.1146/annurev-genet-102209-163602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Simmons D. Initiation of DNA replication from closed circular DNA, fundamental aspects of DNA replication. In: Jelena Kušić-Tišma., editor. Fundamental aspects of DNA replication. Rijeka, Croatia: InTech; 2011. [Google Scholar]
- 19.Fairman-Williams ME, Guenther UP, Jankowsky E. SF1 and SF2 helicases: family matters. Curr. Opin. Struct. Biol. 2010;20:313–324. doi: 10.1016/j.sbi.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goujon M, McWilliam H, Li W, Valentin F, Squizzato S, Paern J, Lopez R. A new bioinformatics analysis tools framework at EMBL–EBI. Nucleic Acids Res. 2010;38:W695–W699. doi: 10.1093/nar/gkq313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bairoch A, Bucher P, Hofmann K. The PROSITE database, its status in 1997. Nucleic Acids Res. 1997;25:217–221. doi: 10.1093/nar/25.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 23.Narasimhan G, Bu C, Gao Y, Wang X, Xu N, Mathee K. Mining protein sequences for motifs. J. Comput. Biol. 2002;9:707–720. doi: 10.1089/106652702761034145. [DOI] [PubMed] [Google Scholar]
- 24.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sayers JR, Eckstein F. Properties of overexpressed phage T5 D15 exonuclease. Similarities with Escherichia coli DNA polymerase I 5′-3′ exonuclease. J. Biol. Chem. 1990;265:18311–18317. [PubMed] [Google Scholar]
- 27.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 28.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 29.Iggo RD, Lane DP. Nuclear protein p68 is an RNA-dependent ATPase. EMBO J. 1989;8:1827–1831. doi: 10.1002/j.1460-2075.1989.tb03577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Whelan F, Stead JA, Shkumatov AV, Svergun DI, Sanders CM, Antson AA. A flexible brace maintains the assembly of a hexameric replicative helicase during DNA unwinding. Nucleic Acids Res. 2012;40:2271–2278. doi: 10.1093/nar/gkr906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arbuckle MI, Stow ND. A mutational analysis of the DNA-binding domain of the herpes simplex virus type 1 UL9 protein. J. Gen. Virol. 1993;74:1349–1355. doi: 10.1099/0022-1317-74-7-1349. [DOI] [PubMed] [Google Scholar]
- 32.Chattopadhyay S, Weller SK. Direct interaction between the N- and C-terminal portions of the herpes simplex virus type 1 origin binding protein UL9 implies the formation of a head-to-tail dimer. J. Virol. 2007;81:13659–13667. doi: 10.1128/JVI.01204-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trego KS, Parris DS. Functional interaction between the herpes simplex virus type 1 polymerase processivity factor and origin-binding proteins: enhancement of UL9 helicase activity. J Virol. 2003;77:12646–12659. doi: 10.1128/JVI.77.23.12646-12659.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rawlings DE, Tietze E. Comparative biology of IncQ and IncQ-like plasmids. Microbiol. Mol. Biol. Rev. 2001;65:481–496. doi: 10.1128/MMBR.65.4.481-496.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ziegelin G, Niedenzu T, Lurz R, Saenger W, Lanka E. Hexameric RSF1010 helicase RepA: the structural and functional importance of single amino acid residues. Nucleic Acids Res. 2003;31:5917–5929. doi: 10.1093/nar/gkg790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mobberley JM, Authement RN, Segall AM, Paul JH. The temperate marine phage PhiHAP-1 of Halomonas aquamarina possesses a linear plasmid-like prophage genome. J. Virol. 2008;82:6618–6630. doi: 10.1128/JVI.00140-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jeong YJ, Kim DE, Patel SS. Kinetic pathway of dTTP hydrolysis by hexameric T7 helicase-primase in the absence of DNA. J. Biol. Chem. 2002;277:43778–43784. doi: 10.1074/jbc.M208634200. [DOI] [PubMed] [Google Scholar]
- 38.Nelson SW, Benkovic SJ. The T4 phage UvsW protein contains both DNA unwinding and strand annealing activities. J. Biol. Chem. 2007;282:407–416. doi: 10.1074/jbc.M608153200. [DOI] [PubMed] [Google Scholar]
- 39.McGlynn P, Lloyd RG. Rescue of stalled replication forks by RecG: simultaneous translocation on the leading and lagging strand templates supports an active DNA unwinding model of fork reversal and Holliday junction formation. Proc. Natl. Acad. Sci. USA. 2001;98:8227–8234. doi: 10.1073/pnas.111008698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nelson SW, Perumal SK, Benkovic SJ. Processive and unidirectional translocation of monomeric UvsW helicase on single-stranded DNA. Biochemistry. 2009;48:1036–1046. doi: 10.1021/bi801792q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gyimesi M, Sarlos K, Kovacs M. Processive translocation mechanism of the human Bloom's syndrome helicase along single-stranded DNA. Nucleic Acids Res. 2010;38:4404–4414. doi: 10.1093/nar/gkq145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Byrd AK, Raney KD. Protein displacement by an assembly of helicase molecules aligned along single-stranded DNA. Nat. Struct. Mol. Biol. 2004;11:531–538. doi: 10.1038/nsmb774. [DOI] [PubMed] [Google Scholar]
- 43.Byrd AK, Raney KD. Increasing the length of the single-stranded overhang enhances unwinding of duplex DNA by bacteriophage T4 Dda helicase. Biochemistry. 2005;44:12990–12997. doi: 10.1021/bi050703z. [DOI] [PubMed] [Google Scholar]
- 44.Tackett AJ, Chen Y, Cameron CE, Raney KD. Multiple full-length NS3 molecules are required for optimal unwinding of oligonucleotide DNA in vitro. J. Biol. Chem. 2005;280:10797–10806. doi: 10.1074/jbc.M407971200. [DOI] [PubMed] [Google Scholar]
- 45.Dillingham MS, Wigley DB, Webb MR. Demonstration of unidirectional single-stranded DNA translocation by PcrA helicase: measurement of step size and translocation speed. Biochemistry. 2000;39:205–212. doi: 10.1021/bi992105o. [DOI] [PubMed] [Google Scholar]
- 46.Dillingham MS, Spies M, Kowalczykowski SC. RecBCD enzyme is a bipolar DNA helicase. Nature. 2003;423:893–897. doi: 10.1038/nature01673. [DOI] [PubMed] [Google Scholar]
- 47.Taylor AF, Smith GR. RecBCD enzyme is a DNA helicase with fast and slow motors of opposite polarity. Nature. 2003;423:889–893. doi: 10.1038/nature01674. [DOI] [PubMed] [Google Scholar]
- 48.Naqvi A, Tinsley E, Khan SA. Purification and characterization of the PcrA helicase of Bacillus anthracis. J. Bacteriol. 2003;185:6633–6639. doi: 10.1128/JB.185.22.6633-6639.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Constantinesco F, Forterre P, Koonin EV, Aravind L, Elie C. A bipolar DNA helicase gene, herA, clusters with rad50, mre11 and nurA genes in thermophilic archaea. Nucleic Acids Res. 2004;32:1439–1447. doi: 10.1093/nar/gkh283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pradhan A, Tuteja R. Plasmodium falciparum DNA helicase 60. dsRNA- and antibody-mediated inhibition of malaria parasite growth and downregulation of its enzyme activities by DNA-interacting compounds. FEBS J. 2006;273:3545–3556. doi: 10.1111/j.1742-4658.2006.05362.x. [DOI] [PubMed] [Google Scholar]
- 51.Pradhan A, Tuteja R. Bipolar, dual Plasmodium falciparum helicase 45 expressed in the intraerythrocytic developmental cycle is required for parasite growth. J. Mol. Biol. 2007;373:268–281. doi: 10.1016/j.jmb.2007.07.056. [DOI] [PubMed] [Google Scholar]
- 52.Vashisht AA, Pradhan A, Tuteja R, Tuteja N. Cold- and salinity stress-induced bipolar pea DNA helicase 47 is involved in protein synthesis and stimulated by phosphorylation with protein kinase C. Plant J. 2005;44:76–87. doi: 10.1111/j.1365-313X.2005.02511.x. [DOI] [PubMed] [Google Scholar]
- 53.Rozen F, Edery I, Meerovitch K, Dever TE, Merrick WC, Sonenberg N. Bidirectional RNA helicase activity of eucaryotic translation initiation factors 4A and 4F. Mol. Cell. Biol. 1990;10:1134–1144. doi: 10.1128/mcb.10.3.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang Y, Liu ZR. The ATPase, RNA unwinding, and RNA binding activities of recombinant p68 RNA helicase. J. Biol. Chem. 2002;277:12810–12815. doi: 10.1074/jbc.M200182200. [DOI] [PubMed] [Google Scholar]
- 55.Hirota Y, Lahti JM. Characterization of the enzymatic activity of hChlR1, a novel human DNA helicase. Nucleic Acids Res. 2000;28:917–924. doi: 10.1093/nar/28.4.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Treuner K, Ramsperger U, Knippers R. Replication protein A induces the unwinding of long double-stranded DNA regions. J. Mol. Biol. 1996;259:104–112. doi: 10.1006/jmbi.1996.0305. [DOI] [PubMed] [Google Scholar]
- 57.Boddeker N, Stade K, Franceschi F. Characterisation of DpbA, and Escherichia coli DEAD box protein with ATP independent RNA unwinding activity. Nucleic Acids Res. 1997;25:537–544. doi: 10.1093/nar/25.3.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.He X, Lehman IR. An initial ATP-independent step in the unwinding of a herpes simplex virus I origin of replication by a complex of the viral origin-binding protein and single-strand DNA-binding protein. Proc. Natl. Acad. Sci. USA. 2001;98:3024–3028. doi: 10.1073/pnas.061028298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dillingham MS, Webb MR, Kowalczykowski SC. Bipolar DNA translocation contributes to highly processive DNA unwinding by RecBCD enzyme. J. Biol. Chem. 2005;280:37069–37077. doi: 10.1074/jbc.M505520200. [DOI] [PubMed] [Google Scholar]
- 60.Dillingham MS, Kowalczykowski SC. RecBCD enzyme and the repair of double-stranded DNA breaks. Microbiol. Mol. Biol. Rev. 2008;72:642–671. doi: 10.1128/MMBR.00020-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Amundsen SK, Smith GR. Chi hotspot activity in Escherichia coli without RecBCD exonuclease activity: implications for the mechanism of recombination. Genetics. 2007;175:41–54. doi: 10.1534/genetics.106.065524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Spies M, Amitani I, Baskin RJ, Kowalczykowski SC. RecBCD enzyme switches lead motor subunits in response to χ recognition. Cell. 2007;131:694–705. doi: 10.1016/j.cell.2007.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hendrickson HE, McCorquodale DJ. Genetic and physiological studies of bacteriophage T5. 3. Patterns of deoxyribonucleic acid synthesis induced by mutants of T5 and the identification of genes influencing the appearance of phage-induced dihydrofolate reductase and deoxyribonuclease. J. Virol. 1972;9:981–989. doi: 10.1128/jvi.9.6.981-989.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bourguignon GJ, Sweeney TK, Delius H. Multiple origins and circular structures in replicating T5 bacteriophage DNA. J. Virol. 1976;18:245–259. doi: 10.1128/jvi.18.1.245-259.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li M, Desiderio S. Physical characterisation of DNA binding proteins in crude preparations. In: Harris ELV, Angal S, editors. Transcription Factors: A Practical Approach. Oxford, UK: IRL Press at Oxford University Press; 1993. pp. 185–196. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.