Abstract
During bacterial DNA replication, DnaG primase and the χ subunit of DNA polymerase III compete for binding to single-stranded DNA-binding protein (SSB), thus facilitating the switch between priming and elongation. SSB proteins play an essential role in DNA metabolism by protecting single-stranded DNA and by mediating several important protein–protein interactions. Although an interaction of SSB with primase has been previously reported, it was unclear which domains of the two proteins are involved. This study identifies the C-terminal helicase-binding domain of DnaG primase (DnaG-C) and the highly conserved C-terminal region of SSB as interaction sites. By ConSurf analysis, it can be shown that an array of conserved amino acids on DnaG-C forms a hydrophobic pocket surrounded by basic residues, reminiscent of known SSB-binding sites on other proteins. Using protein–protein cross-linking, site-directed mutagenesis, analytical ultracentrifugation and nuclear magnetic resonance spectroscopy, we demonstrate that these conserved amino acid residues are involved in the interaction with SSB. Even though the C-terminal domain of DnaG primase also participates in the interaction with DnaB helicase, the respective binding sites on the surface of DnaG-C do not overlap, as SSB binds to the N-terminal subdomain, whereas DnaB interacts with the ultimate C-terminus.
INTRODUCTION
DNA replication is a fundamental process of life. In Eubacteria, DnaB helicase (1) is loaded onto the template DNA (2,3) where it recruits DnaG primase. Both proteins interact and form part of the primosome (4,5), a protein complex in which DnaB unwinds the DNA double-strand in an adenosine triphosphate (ATP)-dependent fashion (1) and primase generates short oligoribonucleotides (RNA primers) (6,7) as starting points of replication. After finishing primer synthesis, DnaG primase detaches from the helicase and stays bound to the primed template (8,9). Owing to the antiparallel nature of double-stranded DNA and the fact that DNA polymerases synthesize DNA exclusively in the 5′–3′ direction, only the leading strand can be synthesized continuously, whereas the lagging strand has to be synthesized in Okazaki fragments of ∼1000 nucleotides in length (10).
The main bacterial replication enzyme, DNA polymerase III holoenzyme (11), can be subdivided into three distinct substructures (12). The core contains the polymerase and proofreading activities (13), the homodimeric β sliding clamp ties the polymerase to its substrate and ensures processive DNA synthesis (14), and the [(γ/τ)3δδ′χΨ] clamp loader complex opens the ring-shaped β clamp and loads it onto the DNA using ATP hydrolysis (15). Whereas the accessory subunits χ and Ψ are not necessary for clamp loading itself (16), the χ protein provides the only direct link between DNA polymerase III and SSB, the single-stranded DNA-binding protein (17,18). SSB protects single-stranded DNA (ssDNA) at the lagging strand from nucleolytic attack and configures it for efficient replication by DNA polymerase III.
The primary structure of Escherichia coli SSB (EcoSSB) consists of three different regions. The N-terminal part of the protein comprises the DNA-binding domain (OB-fold) (19–21), which is followed by a largely unstructured region, rich in glycine and proline residues. The protein sequence is terminated by a highly conserved C-terminal region harbouring the amphipathic consensus sequence DDDIPF. Apart from binding to the χ subunit of DNA polymerase III, EcoSSB has been shown to interact with more than a dozen other proteins, including exonuclease I, RecQ and DnaG primase (17,22–25). In all cases investigated so far, protein–protein interaction requires the conserved C-terminal part of EcoSSB (26), which is essential for cellular viability, as SSB truncation variants lacking C-terminal amino acids cannot complement for wild-type protein (27). An exchange of the penultimate amino acid of EcoSSB from proline to serine produces the ssb-113 strain, which shows temperature-sensitive conditional lethality (28), and extension of EcoSSB by a C-terminal glycine residue (SSB+Gly) results in slower cell growth, indicating impaired protein function in vivo (29).
Proteolytic digestion of DnaG primase has shown that the protein is composed of three different domains (30). Whereas no 3D structure of full-length DnaG is available to date, the structures of all three domains have been solved separately, either from E. coli or Geobacillus stearothermophilus: the N-terminal zinc-binding domain, which is involved in template recognition (31), the central RNA polymerase domain, which is responsible for the actual primer synthesis (32), and the C-terminal helicase-binding domain (DnaG-C), which is essential for coupling primer synthesis to replication fork progression (33). It has been shown that the last eight amino acids of the C-terminal helix hairpin of DnaG-C are responsible for interaction with DnaB helicase (5,9,34). The domain of DnaG mediating the interaction with SSB has not been identified so far.
During lagging strand replication, the χ subunit of DNA polymerase III and DnaG primase compete for binding to EcoSSB to enable a primase-to-polymerase switch. Each time the synthesis of a new Okazaki fragment is started, the χ subunit detaches primase from the newly synthesized RNA primer, so that a new β sliding clamp can be placed onto the primed template (22). Whereas it is known that the SSB/χ interaction involves the C-terminal region of EcoSSB and their binding site has been characterized recently (29,35), not much is known about the interaction of EcoSSB and DnaG primase (26).
Using protein–protein cross-linking, site-directed mutagenesis, isothermal titration calorimetry (ITC), analytical ultracentrifugation and nuclear magnetic resonance (NMR) analysis, this study shows for the first time that the SSB/primase interaction is mediated by the highly conserved C-terminus of EcoSSB. Moreover, the SSB-binding site is mapped to the C-terminal helicase-binding domain of DnaG primase (8) and shares characteristic features with known SSB-binding sites of other proteins.
MATERIALS AND METHODS
Buffers and reagents
All materials were of the highest purity available and were obtained from Sigma, Pierce and J.T. Baker. The SSB-Carb peptide containing the last nine amino acids of EcoSSB and comprising the sequence WMDFDDDIPF was synthesized by Thermo Fisher, Germany. The N-terminal tryptophan residue was added to determine peptide concentration [extinction coefficient 5690 M−1 cm−1 at 280 nm (36)].
Protein concentrations were determined using the following extinction coefficients at 280 nm: 45 840 M−1 cm−1 for full-length DnaG, 33 350 M−1 cm−1 for DnaG-N (amino acid residues 1–433 of DnaG), 12 490 M−1 cm−1 for DnaG-C (amino acid residues 434–581 of DnaG) and all its mutants; all of these extinction coefficients were calculated from amino acid composition using the program Sednterp (36,37). An extinction coefficient at 280 nm of 113 000 M−1 cm−1 was used for EcoSSB wild-type and SSB+Gly (38). SSB concentrations are given in tetramers throughout the text.
Site-directed mutagenesis
Site-directed mutagenesis was performed with the QuikChange site-directed mutagenesis kit of Stratagene (LaJolla, USA) and vectors pET-15bDnaG (see below) or pKL1176 (kindly provided by Dr. N. E. Dixon, University of Wollongong) containing DnaG-C (39). The point mutations were introduced using the following oligonucleotides (MWG Biotech.), the antisense primers for each mutant have the reverse complementary sequence:
| DnaG-C K447A | 5′ | CCTGTTCCGCAGCTAGCACGTAC GACCATGC | 3′ |
| DnaG-C T450A | 5′ | CCGCAGCTAAAACGCACGGCCAT GCGTATACTTATAG | 3′ |
| DnaG-C R452A | 5′ | CGCACGACCATGGCCATACTTA TAGGG | 3′ |
| DnaG-C K478A | 5′ | GAATCTGGATGAAAATGCACTC CCGGGACTTGGC | 3′ |
| DnaG-C K518A | 5′ | GCTGCCACCCTTGAAGCCTTAAG TATGTGGGACGATATAGC | 3′ |
| DnaG-C K528A | 5′ | GGGACGATATAGCAGATGCAAA CATTGCTGAGCAAAC | 3′ |
| DnaG-N | 5′ | GAACGATTAATGCCAAAATAAC TCGAGAGCGGCGTTTCTCGC | 3′ |
For the DnaG-C mutants, the amino acid residue mentioned in the primer name was replaced by a different amino acid [e.g. in the case of DnaG-C K447A, the lysine residue at position 447 of full-length DnaG was substituted by an alanine. DnaG-C, however, starts at position 434 of DnaG (8)]. For the DnaG-N mutant, a stop codon was inserted at position 434 of DnaG in vector pET-15bDnaG. All mutated constructs were checked for errors by sequencing the complete gene (GATC Biotech. AG, Germany).
Protein preparation
Expression and purification of EcoSSB and EcoSSB+Gly were performed as described previously (29).
Vector pET-15bDnaG carries the E. coli primase gene under control of the T7 promoter. It was constructed by amplifying the respective genomic region of E. coli strain LK111λ (40) with the following oligonucleotides: 5′-AGGACCATGGCTGGACGAATCCCACGCG-3′ and 5′-AGGACTCGAGTCACTTTTTCGCCAGCTCCTGG-3′. The PCR product was cut by NcoI and XhoI and ligated into pET-15b treated with the same enzymes. pET-15bDnaG was used to transform E. coli strain Rosetta (DE3) pLysS (Novagen). Protein expression was induced with 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) and cells were harvested after 4.5 h at 37°C. The cell pellet was washed in 50 mM tris(hydroxymethyl)aminomethane (Tris), pH 8.3, 200 mM NaCl, 1 mM ethylenediaminetetraacetic acid (EDTA), 10% (w/v) sucrose and then resuspended in the same buffer containing 15 mM spermidine and flash-frozen in N2 (liq). For purification of DnaG, the cell suspension was thawed and ‘Complete EDTA free Protease Inhibitor Tablets’ (Roche) were added according to volume as well as 0.1 mM phenylmethanesulfonyl fluoride (PMSF), 1 mM dithiothreitol (DTT) and 0.04% sodium desoxycholate. After cell lysis by sonication, the sample was centrifuged for 45 min at 120 000g and the protein was precipitated from the supernatant with 194 g/l (NH4)2SO4. The pellet was resuspended in 20 mM Tris, pH 7.2, 50 mM NaCl, 10% (v/v) glycerol, 1 mM DTT, 1 mM EDTA and dialysed extensively against the same buffer. The sample was applied to a Q-Sepharose FF (GE Healthcare) column equilibrated with dialysis buffer and a linear gradient up to 400 mM NaCl was used to elute the protein. Fractions containing DnaG were pooled and the protein was precipitated by adding 250 g/l (NH4)2SO4. The pellet was dissolved in 20 mM potassium phosphate, pH 7.4, 0.3 M NaCl, 10% (v/v) glycerol, 1 mM DTT (buffer A) and applied to a HiLoad-26/60-Superdex-75-prepgrade column (GE Healthcare) equilibrated with the same buffer. Fractions of pure DnaG were pooled and precipitated as before. The pellet was dissolved in buffer A containing 50 mM NaCl and 1 mM EDTA, dialysed against the same buffer, flash-frozen in N2 (liq) and stored at −80°C.
DnaG-N expression was induced by adding 1 mM IPTG to a Rosetta (DE3) pLysS culture transformed with pET-15bDnaG-N. Cells were harvested after 5 h at 37°C. The cell pellet was washed and resuspended in 50 mM Tris, pH 7.5, 10% (w/v) sucrose and flash-frozen in N2 (liq). For purification, an equal volume of 20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), pH 8.0, 150 mM NaCl, 10% (v/v) glycerol, 1 mM DTT and 1 mM EDTA (buffer B) was added to the cell suspension together with protease inhibitors as described above. After cell lysis, the sample was centrifuged for 45 min at 120 000g and the protein was precipitated from the supernatant with 300 g/l (NH4)2SO4. The pellet was resuspended in and dialysed extensively against buffer B. The sample was applied to a Blue-Sepharose (41) column equilibrated with buffer B, and a linear gradient up to 3 M NaCl was used to elute the protein. Fractions containing DnaG-N were pooled, and the protein was precipitated by adding 400 g/l (NH4)2SO4. The pellet was dissolved in 20 mM HEPES, pH 8.0, 20 mM NaCl, 10% (v/v) glycerol, 1 mM DTT and 0.5 mM EDTA (buffer C) and dialysed extensively against the same buffer. The sample was applied to a Q-Sepharose FF column equilibrated with dialysis buffer and a linear gradient up to 500 mM NaCl was used. Fractions containing DnaG-N were pooled, and the protein was precipitated as before. The pellet was dissolved in buffer C containing 200 mM NaCl and applied to a HiLoad-26/60-Superdex-75-prepgrade column equilibrated with the same buffer. Fractions containing pure DnaG-N were pooled and precipitated with 500 g/l (NH4)2SO4. The pellet was dissolved in and dialysed against buffer C containing 200 mM NaCl, flash-frozen in N2 (liq) and stored at −80°C.
Vector pKL1176 containing DnaG-C or mutants thereof under control of the λpL and λpR promoters (39) was used to transform Rosetta (DE3) pLysS. Protein expression was induced by a temperature shift from 30°C to 42°C, and cells were harvested 4 h after induction. DnaG-C protein and its mutants were purified essentially as in (39). However, the supernatant of the cell lysate was precipitated with 270 g/l (NH4)2SO4, and anion exchange chromatography was performed on a Q-Sepharose FF column. For size exclusion chromatography, a HiLoad-26/60-Superdex-75-prepgrade column was used. After purification, the protein was precipitated with 500 g/l (NH4)2SO4, dissolved in and dialysed against buffer A. The protein was flash-frozen in N2 (liq) and stored at −80°C.
Uniformly 15N and 13C,15N-labelled DnaG-C were obtained by growing Rosetta (DE3) pLysS harbouring pKL1176 in M9 minimal medium (42) containing 1 g/l 15NH4Cl (Sigma) and either 12C6-Glucose or 13C6-Glucose (Cambridge Isotope Laboratories) at 4 g/l, respectively, as sole nitrogen and carbon sources at 42°C. The expression and purification were performed in the same way as for unlabelled DnaG-C. The pure protein was dialysed extensively against 5 mM potassium phosphate, pH 7.4, 5 mM NaCl, 2 mM DTT, then flash-frozen in N2 (liq) and stored at −80°C.
After purification, the proteins were checked by analytical ultracentrifugation for homogeneity. All mutants showed essentially the same c(s) distributions as wild-type protein, no hint for changes in tertiary structure due to the introduced mutations could be found.
Cross-linking experiments
DnaG and SSB were used in cross-linking reactions with the zero-length cross-linker 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride (EDC, Pierce), which is able to covalently link carboxyl groups to closely neighbouring amino groups under the formation of an amide bond. The reactions were performed in a buffer containing 1 mM potassium phosphate, pH 7.4, 1 mM NaCl, 8% (w/v) glycerol and 1 mM DTT (extra low salt buffer) with 25 µM DnaG full-length, DnaG-C or DnaG-N, respectively, 5 µM SSB and 50 mM EDC. Samples and controls (lacking the respective interaction partner in absence or presence of EDC) were incubated for 2 h at room temperature. Then, the samples were diluted 1:5 in sodium dodecyl sulphate (SDS) loading buffer [50% (v/v) glycerol, 0.16 M Tris/HCl, pH 6.8, 5% (v/v) β-mercaptoethanol, 2% (w/v) SDS, 0.01% (w/v) bromophenol blue] and heated to 95°C for 10 min. Samples and controls were analysed on a 12 or 13.5% SDS-polyacrylamide gel (SDS-PAGE), respectively (43).
Isothermal titration calorimetry
ITC experiments were performed at 25°C using a Nano ITC microcalorimeter (TA instruments, USA) in a buffer containing 5 mM potassium phosphate, pH 7.4, and 8% (w/v) glycerol, supplemented with 5 mM or 150 mM NaCl, respectively. After protein dialysis, the SSB-Carb peptide was dissolved in the buffer used for dialysis. In a typical experiment, 1.3 ml of DnaG-C solution (∼50 µM) were filled into the sample cell and titrated with a solution of ∼1 mM SSB-Carb peptide. An initial injection of 2.1 µl was followed by 19 injections of 4.9 µl SSB-Carb solution. Data were collected for 500 s after each injection. After baseline subtraction, the area under each peak was determined to yield the integrated heat response per injection. Each data set was corrected for heats of dilution measured by a reference titration of SSB-Carb peptide into sample buffer. Binding stoichiometry n, association constant KA and binding enthalpy ΔH were obtained by fitting a model of n identical independent binding sites for the peptide on DnaG-C to the ITC titration curves using the instrumental software package NanoAnalyze.
Analytical ultracentrifugation
Sedimentation velocity experiments were performed in a ProteomeLab™ XL-I analytical ultracentrifuge (Beckman Coulter, USA), using an An50Ti rotor. During the experiment, the absorption of the sample at 280 nm was recorded as a function of radial position. Programming of the centrifuge and data recording was done using the computer software ProteomeLab™ XL-I GUI v6.0 (Beckman Coulter, USA). Samples with a volume of 400 µl were centrifuged in 12 mm standard double sector cells at 20°C and 50 000 rpm. All experiments were carried out in a buffer containing 5 mM potassium phosphate, pH 7.4, 5 mM NaCl, 8% (w/v) glycerol and 0.5 mM DTT (low salt buffer).
To check whether the binding of DnaG is mediated by the C-terminus of SSB, 2 µM SSB or SSB+Gly were titrated with increasing concentrations of full-length DnaG (4 µM to 14 µM). In the case of DnaG-N, 2.5 µM of SSB and 5 to 25 µM of DnaG-N were used. For DnaG-C, 2.5 µM SSB were titrated with increasing concentrations of DnaG-C wild-type or mutant, ranging from 5 to 42.5 µM, representing stoichiometries of 1:2 to 1:17. The measured data were evaluated with the program package SEDFIT (44). This program provides a model for diffusion-deconvoluted differential sedimentation coefficient distributions [c(s) distributions]. As the areas under the separate peaks in c(s) distributions are a measure of the absorbance of the species represented by the peaks (45), this information can be used to determine binding isotherms (29,46,47).
NMR experiments
Samples for NMR-spectroscopy (600 µl) were in a buffer containing 5 mM potassium phosphate, pH 7.4, 5 mM NaCl, 2 mM DTT, supplemented with 10% (v/v) D2O. For the titration experiment, 15N-labelled DnaG-C was used; assignment of the protein backbone was obtained with 13C,15N-labelled protein. The peptide was added from a stock solution of 30 mM SSB-Carb in dimethyl sulfoxide (DMSO) to a sample of 300 µM 15N-labelled protein to yield final concentrations of 0, 0.1, 0.3, 0.6 and 1 mM of peptide. For the assignment of backbone resonances, samples with or without 1 mM SSB-Carb were measured.
NMR spectra were recorded at 600 MHz (1H frequency) on a Bruker AV-III spectrometer (Bruker Biospin, Germany) using a cryogenically cooled 5-mm TXI-triple resonance probe equipped with a one-axis self-shielded gradient. All spectra were recorded at 295 K. The titration was performed recording SOFAST-HMQC (48) spectra, each spectrum was recorded using 16 scans and 512* × 256* data points (where n* refers to the number of complex points). For the assignment of the protein without peptide, a full set of six triple-resonance experiments (49) was performed: HNCA, HN(CO)CA, HNCO, HN(CA)CO, HNCACB, HN(CO)CACB; all were run as BEST (50) experiments with 32 scans and 512* × 48* × 64* data points, only the HNCO was run with 16 scans. The HNCA and HN(CO)CA experiments were repeated using identical parameters after addition of 1 mM peptide.
Data were processed using tospin 3.1 (Bruker Biospin, Germany) and subsequently transferred to CCPN (51) for analysis. After the assignment had been obtained, the shift caused by SSB-Carb was calculated from the spectra without peptide and the one with a peptide concentration of 1 mM using the formula shift =sqrt {[Δδ(1H)]2 + [Δδ(15N)/10]2} (where Δδ is the difference in chemical shift in the respective dimensions).
RESULTS AND DISCUSSION
Complex formation between DnaG primase and SSB is important for the primase-to-polymerase switch at the lagging strand of the DNA replication fork, where a competition of the χ subunit of DNA polymerase III with DnaG primase for binding to SSB ensures primer hand-off to the polymerase (22). Whereas it is known that the SSB/χ interaction involves the C-terminal region of EcoSSB and their binding site was characterized recently (29,35), no details are known about the interaction of SSB and DnaG. To identify the interacting domains of SSB and primase and to characterize their binding site, different mutants of DnaG and EcoSSB were analysed for their interaction.
The interaction of SSB and primase is mediated by the C-terminus of SSB
For EcoSSB, at least 14 different proteins involved in DNA replication, repair and recombination have been identified as interaction partners. In all cases investigated so far, protein–protein interaction requires the conserved C-terminal region of SSB (26). To find out whether the SSB/primase interaction also relies on this region, the binding of DnaG primase to EcoSSB wild-type and a C-terminal extension mutant of EcoSSB, SSB+Gly, was examined by analytical ultracentrifugation. SSB+Gly carries an additional glycine residue at the ultimate C-terminus and is severely impaired in binding to the χ subunit of DNA polymerase III, as the final phenylalanine residue of SSB can no longer fit into the well-defined binding pocket on χ (29).
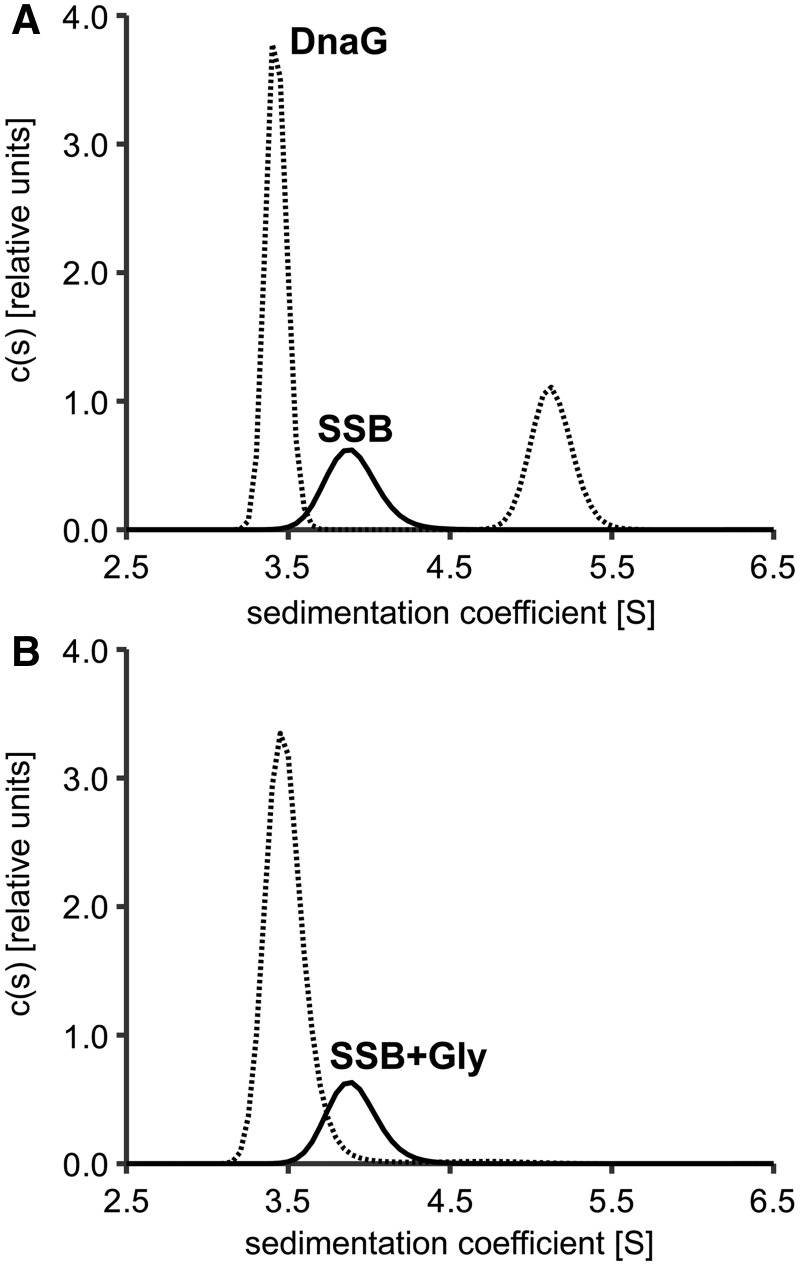
When two proteins interact, they form a complex of a larger mass, which usually sediments faster than both proteins alone. Figure 1 shows diffusion-deconvoluted sedimentation coefficient distributions [c(s) distributions] obtained from analytical ultracentrifugation experiments for the interaction of DnaG primase with wild-type EcoSSB or the SSB+Gly mutant under low salt conditions. Whereas free DnaG showed a sedimentation coefficient of 3.4 S under these conditions, free EcoSSB and SSB+Gly sedimented with an s-value of 3.9 S (Figure 1). In the case of EcoSSB wild-type, this value increased to 5.1 S on addition of a 7-fold molar excess of DnaG, indicating complex formation (Figure 1A). However, when SSB+Gly was used in the experiment, no protein complex could be detected (Figure 1B). Therefore, also for the interaction of SSB and primase, the C-terminus of SSB is essential.
Figure 1.
Addition of a C-terminal glycine residue to EcoSSB abolishes the interaction with DnaG primase. (A) In the absence of DnaG (solid line), EcoSSB (2 µM) sediments with an s-value of 3.9 S. When a 7-fold molar excess of DnaG primase is applied (dashed line), the sedimentation coefficient of EcoSSB increases to 5.1 S, indicating complex formation. (B) In contrast, when the same excess of DnaG is added to 2 µM of the C-terminal extension mutant SSB+Gly (dashed line), s-values higher than that of DnaG and EcoSSB cannot be observed.
DnaG-C contains the SSB-binding site
The results from analytical ultracentrifugation showed that the C-terminus of SSB mediates the SSB/primase interaction. However, amino acid residues of the primase involved in complex formation remained to be identified. As the C-terminal region of SSB contains several aspartate residues that may interact with basic residues of the primase, chemical cross-linking reactions with EDC were performed. This zero-length cross-linker forms amide bonds between closely adjacent carboxyl and amino groups (see ‘Material and Methods’ section). Incubation of DnaG with SSB and EDC resulted in the appearance of additional protein bands at higher apparent molecular weights in SDS-PAGE (Supplementary Figure S1). Therefore, a surface lysine of DnaG is most probably involved in the cross-link and reacts with one of the aspartates in the C-terminal region of SSB or its C-terminal carboxyl group, respectively.
To find out which residues had reacted with EDC, cross-linked protein samples were analysed by mass spectrometry, but, unfortunately, the experiments failed to give clear results. Because DnaG is relatively large and contains many lysine residues, it became necessary to divide the full-length protein into two parts for further analysis: on the one hand, the N-terminal two-thirds of the sequence (DnaG-N, amino acid residues 1–433 of DnaG), containing the zinc-binding and the RNA polymerase domain and on the other hand the C-terminal helicase-binding domain (DnaG-C, amino acid residues 434–581 of DnaG). Both constructs were originally derived from partial proteolysis of full-length DnaG and have been thoroughly analysed biochemically and structurally (9,33,34,39).
When DnaG-N and DnaG-C were used in cross-linking experiments, additional protein bands of higher apparent molecular weights only appeared upon incubation of DnaG-C with EcoSSB and EDC (Supplementary Figure S2). This result indicates that it is the C-terminal helicase-binding domain that is involved in the interaction of SSB and primase. However, a participation of DnaG-N could not be excluded, as EDC cross-linking relies on closely neighbouring residues, one of which has to be a lysine.
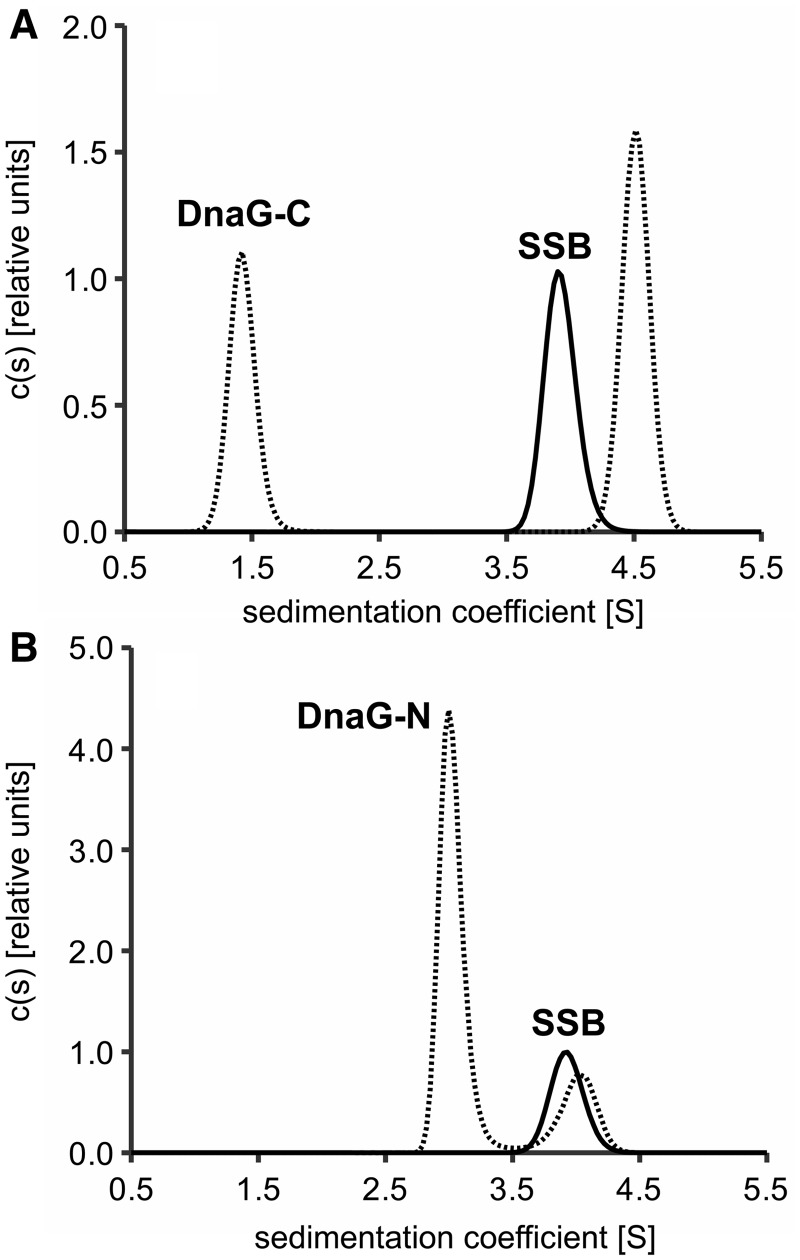
Therefore, both parts of DnaG were checked for their binding to EcoSSB by analytical ultracentrifugation. Figure 2 shows c(s) distributions obtained from sedimentation velocity experiments under low salt conditions. Whereas free DnaG-C and free DnaG-N sedimented with an s-value of 1.4 and 2.9 S, respectively (Figure 2), free EcoSSB showed a sedimentation coefficient of 3.9 S under these conditions. Upon addition of a 10-fold molar excess of DnaG-C, this value rose to 4.5 S, indicating complex formation (Figure 2A). Although DnaG-N has approximately three times the molecular weight of DnaG-C, addition of the same excess of DnaG-N to EcoSSB did not increase the sedimentation coefficient of SSB significantly (Figure 2B). Moreover, addition of DnaG-C resulted in a clear increase in the peak area of EcoSSB (reaction boundary, see below) in the c(s) distribution (Figure 2A). This is caused by an increase in absorbance due to DnaG-C co-sedimenting in complexes with SSB, an effect which is not observed in the case of DnaG-N. Thus, in the concentration range used, an interaction of DnaG-N with EcoSSB can be excluded and it is the C-terminal helicase-binding domain of primase that harbours the SSB-binding site.
Figure 2.
The C-terminal helicase-binding domain of primase contains the SSB-interaction site. EcoSSB (2.5 µM) was sedimented in the absence and presence of a 10-fold molar excess of (A) DnaG-C or (B) DnaG-N. Whereas the sedimentation coefficient of SSB increases from 3.9 S in the free state (A, solid line) to about 4.5 S upon addition of DnaG-C (A, dashed line), the s-value of SSB does not change significantly when DnaG-N is present (B).
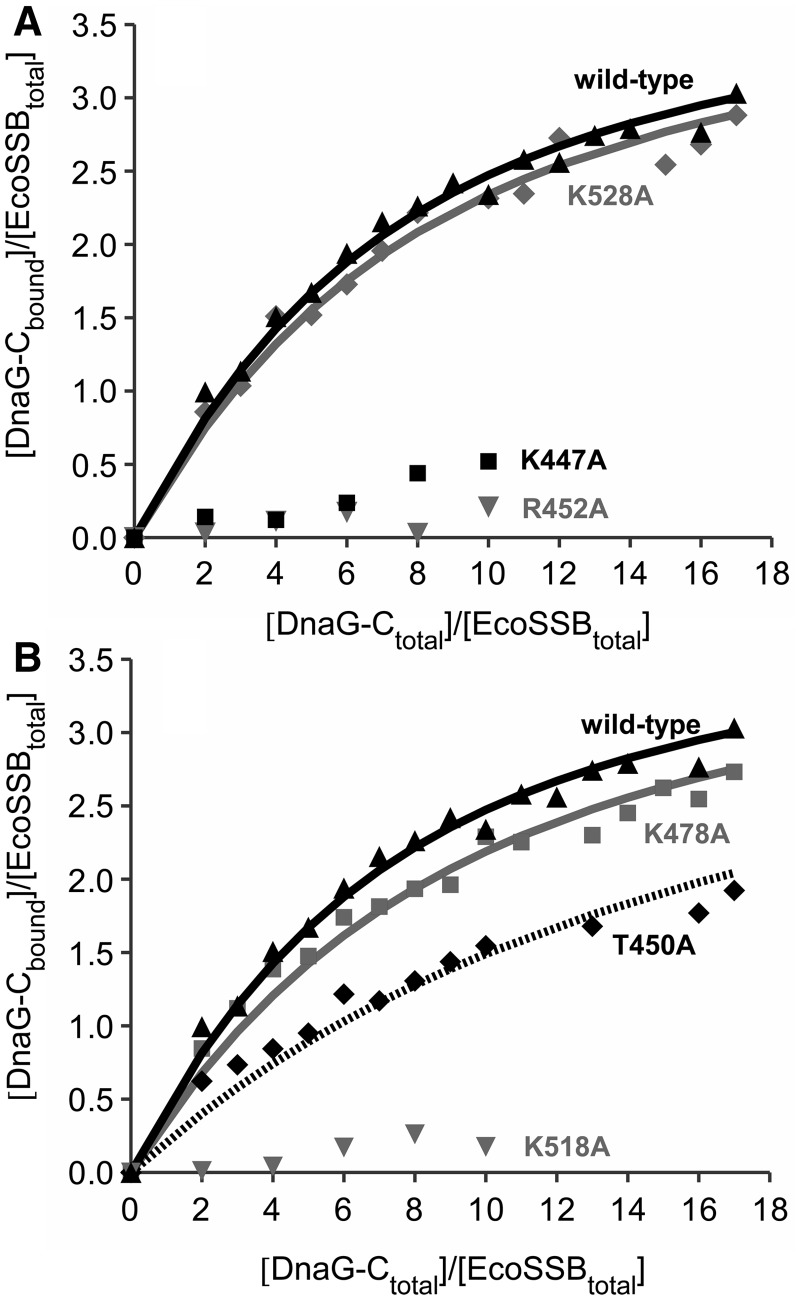
In reaction mixtures of SSB and DnaG-C, only two boundaries could be observed in sedimentation velocity profiles: A slower sedimenting boundary containing free DnaG-C and a faster one containing EcoSSB and all EcoSSB/DnaG-C complexes. When different ratios of EcoSSB and DnaG-C were used, the sedimentation coefficient of this reaction boundary increased with increasing amounts of DnaG-C. Therefore, the reaction must be fast on the timescale of sedimentation (52). As previously described for the SSB/χ interaction (29,46,47,53), integration of the two peaks in the c(s) distribution can be used to determine the concentrations of free and bound SSB-binding partner and subsequently the binding parameters. Fitting a model of four identical independent binding sites for DnaG-C on an EcoSSB tetramer to the data revealed an association constant (KA) of ∼9 × 104 M−1 (Figure 3). An overview of all binding constants determined by analytical ultracentrifugation can be found in Supplementary Table S1.
Figure 3.
Interaction of DnaG-C mutants with SSB. EcoSSB (2.5 µM) was mixed with different amounts of DnaG-C variants in low salt buffer and analysed in sedimentation velocity experiments in an analytical ultracentrifuge. Lines represent theoretical binding isotherms calculated for a simple interaction model of one EcoSSB tetramer binding four DnaG-C molecules with the association constants given below. For comparison, a binding isotherm of the interaction of EcoSSB and DnaG-C wild-type (black triangles) with KA = 8.6 × 104 M−1 is shown in both parts of the figure (solid black line). (A) Replacing K528 by alanine (diamonds) did not significantly change the affinity of DnaG-C to SSB (solid grey line: KA = 7.4 × 104 M−1). Exchanging K447 to alanine (squares), however, had a strong effect on the SSB/DnaG-C interaction. Moreover, the R452A mutation dramatically lowered the binding affinity to SSB (grey triangles). (B) DnaG-C K478A (squares) behaved similarly to wild-type protein (solid grey line: KA = 6.2 × 104 M−1), whereas an exchange of K518 to alanine dramatically decreased the binding affinity to SSB (grey triangles). The T450A mutation (diamonds) lowered the affinity by a factor of three (dashed line: KA = 2.8 × 104 M−1).
DnaG-C interaction with a C-terminal peptide of SSB characterized by ITC
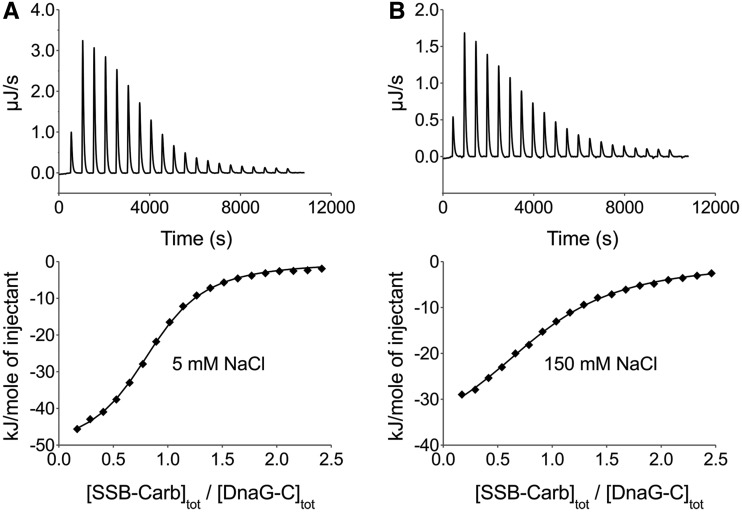
To check whether the C-terminal region of SSB is sufficient for interaction with DnaG-C and to test the salt dependency of the interaction, binding of DnaG-C to a peptide comprising the last nine amino acids of SSB (SSB-Carb) was investigated by ITC experiments. First, DnaG-C was titrated with SSB-Carb peptide in low-salt buffer (5 mM NaCl) as used in analytical ultracentrifugation, but omitting DTT. Fitting a model of n identical independent binding sites for SSB-Carb on DnaG-C to the data yielded a stoichiometry of about one and a binding constant of 2.4 × 105 M−1(Figure 4A). These results are comparable with the values obtained for the binding of DnaG-C to full-length EcoSSB protein (see Figure 3). Therefore, as observed for other SSB-interaction partners (26), it is solely the highly conserved C-terminus of EcoSSB that is responsible for the interaction with DnaG-C. The fact that binding of the peptide occurs with a somewhat higher affinity than binding to full-length protein indicates that the DNA-binding domain of EcoSSB inhibits interaction with the C-terminal region. A similar inhibition was observed for the interaction of both the χ subunit of DNA polymerase III and the replication restart protein PriA with EcoSSB (54). This effect could be overcome by binding SSB to ssDNA, indicating that in absence of ssDNA, the DNA-binding domain interacts with the negatively charged C-terminal region of SSB. Consistently, for χ, a 20-fold increase in SSB-binding affinity could be observed in presence of ssDNA under low salt conditions (18).
Figure 4.
Interaction of DnaG-C with the SSB-Carb peptide as investigated by ITC at different salt concentrations. Upper panels show the raw data obtained for 20 automatic injections of SSB-Carb peptide into the sample cell containing DnaG-C. Lower panels give the integrated heat responses per injection, after correction for heats of dilution obtained by titrating SSB-Carb into sample buffer, normalized to the moles of injected peptide. (A) In a buffer containing 5 mM NaCl, 1.04 mM SSB-Carb were titrated into a solution of 46 µM DnaG-C. (B) In a buffer containing 150 mM NaCl, 1.08 mM SSB-Carb were titrated into a solution of 46 µM DnaG-C. Solid lines represent the best fit with a model of n identical independent binding sites for SSB-Carb on DnaG-C with (A) n = 0.84, KA = 2.4 × 105 M−1, ΔH = −52.0 kJ/mol and (B) n = 0.89, KA = 7.4 × 104 M−1, ΔH = −41.3 kJ/mol. Note that raw data of the Nano ITC calorimeter show a positive sign if heat production occurs during the reaction.
Because the highly conserved C-terminus of EcoSSB contains three aspartate residues and we could chemically cross-link EcoSSB to DnaG and DnaG-C by EDC (see Supplementary Figures S1 and S2), we wanted to test whether the ionic strength influences the DnaG-C/SSB-Carb interaction. At 150 mM NaCl, ITC analysis of this interaction revealed a 1:1 stoichiometry and a binding constant of ∼7 × 104 M−1(Figure 4B). Therefore, an increase in salt concentration from 5 to 150 mM NaCl decreases the affinity only by a factor of about three. This indicates that in the case of DnaG-C, the interaction with the amphipathic C-terminus of EcoSSB is dominated by hydrophobic interactions and hydrogen bonds rather than by electrostatic interactions.
Site-directed mutagenesis identifies lysines on DnaG-C involved in the interaction with SSB
Protein–protein cross-linking, analytical ultracentrifugation and ITC experiments have pinpointed the SSB-binding site to lysine residues on the surface of DnaG-C. This domain of DnaG contains six lysine residues, two of which (K580 and K581) are located in the helix hairpin at the ultimate C-terminus that is required for binding to DnaB helicase (5,9,34). Because it seemed unlikely that these two residues were also implicated in the interaction with SSB, we focused on the remaining four lysines, K447, K478, K518 and K528, and replaced them by alanine using site-directed mutagenesis.
When the mutants were analysed for their interaction with SSB by sedimentation velocity experiments, two of them, K478A and K528A, showed binding affinities similar to DnaG-C wild-type (Figure 3). In the case of the K447A and K518A mutants, however, the interaction with SSB was so severely impaired that no binding parameters could be obtained (Figure 3). This indicates that both K447 and K518 of DnaG-C are involved in the SSB/primase interaction.
A highly conserved pocket on the surface of DnaG-C takes part in the interaction with SSB
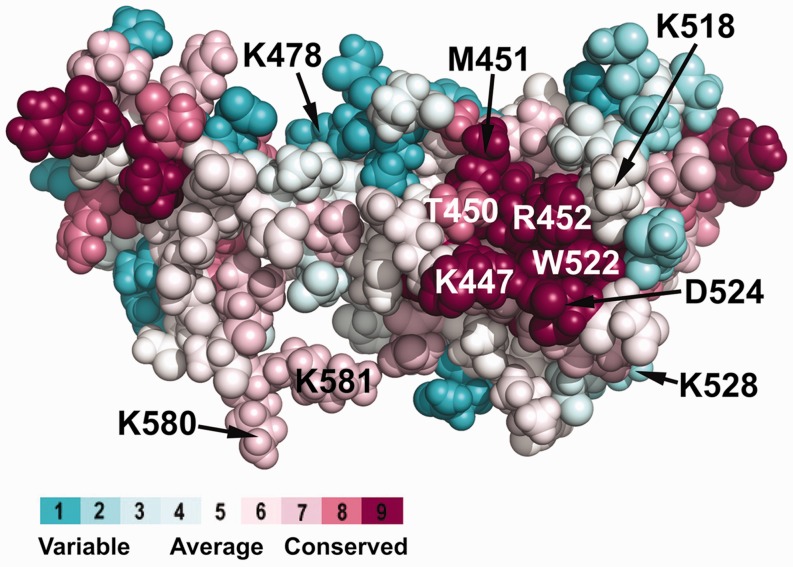
From the 3D structure of DnaG-C (33), it is known that residues K447 and K518 identified by us to be involved in the interaction with SSB are located rather close to each other on the surface of the protein. To identify a possible SSB-binding surface on DnaG-C, a ConSurf analysis (55) was performed that projected an alignment of ∼70 bacterial DnaG proteins on the known structure of E. coli DnaG-C (Figure 5). It revealed that K447 and K518 together with R452 surround a highly conserved hydrophobic pocket (containing T450, M451, I455, L519 and W522) on the surface of the N-terminal subdomain of the protein. The highly conserved C-terminus of SSB is amphipathic, habouring three acidic and three hydrophobic residues (DDDIPF). Therefore, the basic residues on the surface of DnaG-C could interact with the aspartate residues and/or the C-terminal carboxyl group, whereas the hydrophobic pocket could accommodate the proline and phenylalanine residues at the ultimate C-terminus of SSB. A similar array of hydrophobic amino acids with adjacent basic residues forms the other four characterized SSB-binding sites on exonuclease I, RecQ, RecO and the χ subunit of DNA polymerase III, respectively (29,35,56–58).
Figure 5.
Highly conserved basic and hydrophobic amino acid residues are clustered on the surface of DnaG-C. Using the ConSurf server (55), an alignment of ∼70 bacterial DnaG proteins was projected on the known 3D structure of E. coli DnaG-C [pdb code: 2HAJ (33)]. On the right hand side of the structure, a cluster of highly conserved amino acid residues located on the surface of the N-terminal subdomain of DnaG-C can be observed. Similar to the other characterized SSB-binding sites, it consists of a hydrophobic patch (containing T450, M451 and W522), surrounded by a stretch of basic amino acids (K447, K518 and R452). The colour legend shows the conservation scores of the respective residues. Note that the C-terminal lysine residues K580 and K581 involved in interaction of DnaG-C with DnaB helicase (34) are also labelled.
To test whether this highly conserved region is involved in the SSB/primase interaction, two additional amino acids of DnaG-C were exchanged, producing the T450A and R452A mutants, respectively. These proteins were checked for their binding to SSB by analytical ultracentrifugation under low salt conditions. As can be seen from Figure 3B, the T450A mutant showed a significant decrease in affinity to SSB (KA = 2.8 × 104 M−1), and in the case of DnaG-C R452A, the interaction with SSB was again so severely reduced that no binding parameters could be obtained (Figure 3A).
These results indicate that the highly conserved region on the surface of the N-terminal subdomain of DnaG-C identified by ConSurf analysis participates in the SSB/primase interaction.
Chemical shifts obtained from NMR experiments confirm SSB-binding site on DnaG-C
To obtain more information about the SSB-binding site on DnaG-C, an interaction study using 1H,15N heteronuclear NMR spectroscopy was performed. First, an assignment of the 1H,15N correlation was obtained for DnaG-C, using a set of six standard triple resonance experiments. Owing to the different buffer conditions, the assignment deposited in the BMRB (accession number 6284) (9) could be used only in the less-crowded regions of the spectrum; most of the resonances had to be reassigned (Supplementary Figure S3). After addition of the SSB-Carb peptide, containing the last nine amino acids of EcoSSB, the assignment was repeated, using two triple resonance experiments. In the end, the resonances of all amino acids except I525 could be assigned in the peptide-bound form of DnaG-C, some peaks were missing in the free form (see below).
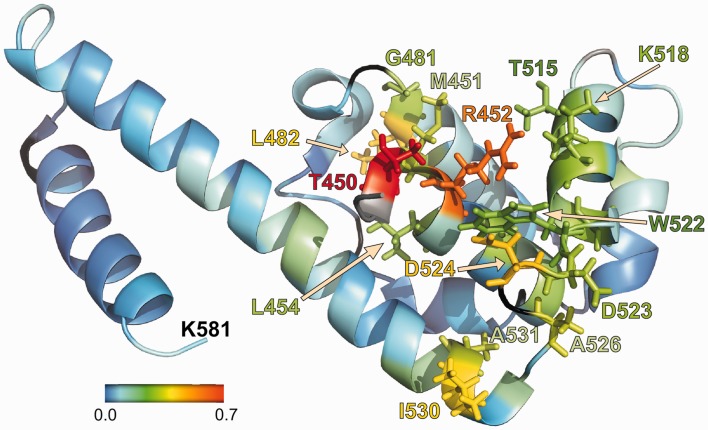
To determine the binding site of the peptide on DnaG-C, a titration was performed, adding increasing amounts of SSB-Carb to the protein and observing the changes in chemical shift (Supplementary Figures S4–S6). The differences in chemical shifts between the free protein and the one with an ∼3-fold molar excess of peptide are shown in Supplementary Table S2 and Supplementary Figure S7. Most changes were complete when a peptide concentration of 0.3 mM (1:1) had been reached. Shift differences are mapped onto the structure of DnaG-C in Figure 6 and are basically restricted to the highly conserved region identified by ConSurf analysis and site-directed mutagenesis (see Figures 3 and 5) that is located in the N-terminal subdomain of the protein (9). The most dramatic shifts were seen in the backbone of T450, which forms part of the hydrophobic pocket, and of R452, which is most probably involved in the binding of acidic residues of the C-terminus of SSB and in the interaction with F177 of SSB (see below). The respective substitution of both residues by alanine decreased the SSB-binding affinity of DnaG-C, with the exchange of R452 abolishing binding nearly completely (see Figure 3A).
Figure 6.
NMR analysis identifies amino acids of DnaG-C involved in binding to the C-terminus of SSB. Changes in chemical shift for the amino proton and nitrogen resonances in the backbone of DnaG-C after addition of SSB-Carb peptide. The shift of the resonances was calculated using the formula shift = sqrt{[Δδ(1H)]2 + [Δδ(15N)/10]2} (where Δδ is the difference in chemical shift in the respective dimensions), and is coloured according to the scheme given in the figure. Amino acids with a shift of >0.2 ppm are shown with side chains using the same colour code. Proline residues and unassigned amino acids are shown in black. Where no shift could be determined (because of peaks missing in the spectrum without peptide, see text), the amino acid is shown in grey. Amino acids 435–446 are missing in the pdb-file 2HAJ (33) and are thus not shown here. Of these residues, however, only L446 did show a significant shift (0.21 ppm).
Apart from the changes in the chemical shifts of the protein backbone, one of the two Trp side chains (W522) showed strong changes in chemical shift on peptide binding (Supplementary Figure S6). Thus, W522, which is involved in the formation of the hydrophobic pocket, is a good candidate for undergoing a stacking interaction with the ultimate phenylalanine residue (F177) of SSB. In addition, several resonances of the arginine side chains (not assigned) and also of some backbone resonances (K447, R448, T449, D511 and D565) became only visible after addition of excess peptide, indicating a rigidification of amino acids that had most likely been in conformational exchange before binding of the peptide.
CONCLUSIONS
During replication of the DNA lagging strand, DnaG primase and the χ subunit of DNA polymerase III compete for binding to SSB to ensure the primer hand-off to the replicative DNA polymerase (22). Whereas the interaction of χ and SSB has been studied thoroughly (29,35), little was known about the interaction of DnaG and SSB. In this work, we locate the binding regions of both proteins to the C-terminal helicase-binding domain of DnaG and the highly conserved C-terminal region of SSB. We identify by ConSurf analysis, site-directed mutagenesis and 1H,15N heteronuclear NMR spectroscopy, a conserved hydrophobic surface pocket in the N-terminal subdomain of DnaG-C as being involved in the binding of the SSB C-terminus. Hydrophobic pockets mediating SSB-binding have also been found on the surface of other SSB-interaction partners, namely χ, exonuclease I, RecO and RecQ (29,35,56–58). In these cases, the ultimate phenylalanine of SSB (F177) is accommodated in the pocket. F177 shows the highest conservation among the C-terminal amino acids of SSB (26) and can be considered invariant. Using NMR analysis of DnaG-C, we could show that not only the chemical shifts of tryptophan 522 backbone change upon addition of SSB-Carb peptide but also those of the side chain. Therefore, W522 is likely to undergo a stacking interaction with F177 of SSB. This stacking interaction could be intensified by a cation-π interaction (59) of F177 with the guanidinium group of R452 of DnaG located above the indole ring of W522 (see Figure 6) in such a way that F177 of SSB could be bound in between. An exchange of this arginine resulted in a dramatic decrease in SSB-binding affinity. In addition and similar to the situation in the other SSB-interaction partners, the SSB-binding pocket of DnaG is surrounded by other basic residues (K447, K518), which are most probably involved in binding of the acidic C-terminal residues and/or the C-terminal carboxyl group of SSB. Substitution of these residues by alanine also resulted in a dramatic decrease in SSB-binding affinity of DnaG-C, making an involvement in hydrogen bond formation probable.
It has been shown that the C-terminal domain of DnaG is also responsible for binding of the replicative helicase DnaB (9). This interaction has been pinpointed to the last eight amino acids in the C-terminal helix hairpin of DnaG-C (5,34). The hydrophobic pocket identified by us, however, is located in the N-terminal subdomain of DnaG-C. Therefore, the two binding regions do not overlap (see Figure 5). Nevertheless, it remains to be tested whether both, helicase and SSB, can simultaneously bind to DnaG primase, or whether the dissociation of the helicase/primase complex is a prerequisite for SSB-binding and thus primer hand-off to the replicative polymerase.
Since we show that the C-terminal domain of DnaG primase is not only responsible for binding to the helicase but also to SSB, it should be considered to rename the ‘helicase-binding domain’ of DnaG ‘protein-interaction domain’.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and 2 and Supplementary Figures 1–7.
FUNDING
Deutsche Forschungsgemeinschaft [CU 35/1-1 to U.C.]. Funding for open access charge: Deutsche Forschungsgemeinschaft.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Lidia Litz for excellent technical assistance, Dr. Andreas Pich for performing mass spectrometry experiments, Drs. Roman Fedorov and Dietmar Manstein for valuable discussions and Dr. Nicholas Dixon, University of Wollongong, for providing pKL1176, the plasmid for DnaG-C expression.
REFERENCES
- 1.LeBowitz JH, McMacken R. The Escherichia coli dnaB replication protein is a DNA helicase. J. Biol. Chem. 1986;261:4738–4748. [PubMed] [Google Scholar]
- 2.Kobori JA, Kornberg A. The Escherichia coli dnaC gene product. III. Properties of the dnaB-dnaC protein complex. J. Biol. Chem. 1982;257:13770–13775. [PubMed] [Google Scholar]
- 3.Kobori JA, Kornberg A. The Escherichia coli dnaC gene product. II. Purification, physical properties, and role in replication. J. Biol. Chem. 1982;257:13763–13769. [PubMed] [Google Scholar]
- 4.Arai K, Kornberg A. Unique primed start of phage phi X174 DNA replication and mobility of the primosome in a direction opposite chain synthesis. Proc. Natl Acad. Sci. USA. 1981;78:69–73. doi: 10.1073/pnas.78.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tougu K, Marians KJ. The interaction between helicase and primase sets the replication fork clock. J. Biol. Chem. 1996;271:21398–21405. doi: 10.1074/jbc.271.35.21398. [DOI] [PubMed] [Google Scholar]
- 6.Yoda K, Yasuda H, Jiang XW, Okazaki T. RNA-primed initiation sites of DNA replication in the origin region of bacteriophage lambda genome. Nucleic Acids Res. 1988;16:6531–6546. doi: 10.1093/nar/16.14.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kitani T, Yoda K, Ogawa T, Okazaki T. Evidence that discontinuous DNA replication in Escherichia coli is primed by approximately 10 to 12 residues of RNA starting with a purine. J. Mol. Biol. 1985;184:45–52. doi: 10.1016/0022-2836(85)90042-7. [DOI] [PubMed] [Google Scholar]
- 8.Tougu K, Peng H, Marians KJ. Identification of a domain of Escherichia coli primase required for functional interaction with the DnaB helicase at the replication fork. J. Biol. Chem. 1994;269:4675–4682. [PubMed] [Google Scholar]
- 9.Oakley AJ, Loscha KV, Schaeffer PM, Liepinsh E, Pintacuda G, Wilce MC, Otting G, Dixon NE. Crystal and solution structures of the helicase-binding domain of Escherichia coli primase. J. Biol. Chem. 2005;280:11495–11504. doi: 10.1074/jbc.M412645200. [DOI] [PubMed] [Google Scholar]
- 10.Okazaki R, Okazaki T, Sakabe K, Sugimoto K. Mechanism of DNA replication possible discontinuity of DNA chain growth. Jpn. J. Med. Sci. Biol. 1967;20:255–260. [PubMed] [Google Scholar]
- 11.Gefter ML, Hirota Y, Kornberg T, Wechsler JA, Barnoux C. Analysis of DNA polymerases II and III in mutants of Escherichia coli thermosensitive for DNA synthesis. Proc. Natl Acad. Sci. USA. 1971;68:3150–3153. doi: 10.1073/pnas.68.12.3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Onrust R, Finkelstein J, Naktinis V, Turner J, Fang L, O'Donnell M. Assembly of a chromosomal replication machine: two DNA polymerases, a clamp loader, and sliding clamps in one holoenzyme particle. I. Organization of the clamp loader. J. Biol. Chem. 1995;270:13348–13357. doi: 10.1074/jbc.270.22.13348. [DOI] [PubMed] [Google Scholar]
- 13.Scheuermann R, Tam S, Burgers PM, Lu C, Echols H. Identification of the epsilon-subunit of Escherichia coli DNA polymerase III holoenzyme as the dnaQ gene product: a fidelity subunit for DNA replication. Proc. Natl Acad. Sci. USA. 1983;80:7085–7089. doi: 10.1073/pnas.80.23.7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stukenberg PT, Studwell-Vaughan PS, O'Donnell M. Mechanism of the sliding beta-clamp of DNA polymerase III holoenzyme. J. Biol. Chem. 1991;266:11328–11334. [PubMed] [Google Scholar]
- 15.Kelman Z, O'Donnell M. DNA polymerase III holoenzyme: structure and function of a chromosomal replicating machine. Annu. Rev. Biochem. 1995;64:171–200. doi: 10.1146/annurev.bi.64.070195.001131. [DOI] [PubMed] [Google Scholar]
- 16.Xiao H, Crombie R, Dong Z, Onrust R, O'Donnell M. DNA polymerase III accessory proteins. III. holC and holD encoding chi and psi. J. Biol. Chem. 1993;268:11773–11778. [PubMed] [Google Scholar]
- 17.Kelman Z, Yuzhakov A, Andjelkovic J, O'Donnell M. Devoted to the lagging strand-the chi subunit of DNA polymerase III holoenzyme contacts SSB to promote processive elongation and sliding clamp assembly. EMBO J. 1998;17:2436–2449. doi: 10.1093/emboj/17.8.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Witte G, Urbanke C, Curth U. DNA polymerase III chi subunit ties single-stranded DNA binding protein to the bacterial replication machinery. Nucleic Acids Res. 2003;31:4434–4440. doi: 10.1093/nar/gkg498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Williams KR, Spicer EK, LoPresti MB, Guggenheimer RA, Chase JW. Limited proteolysis studies on the Escherichia coli single-stranded DNA binding protein. Evidence for a functionally homologous domain in both the Escherichia coli and T4 DNA binding proteins. J. Biol. Chem. 1983;258:3346–3355. [PubMed] [Google Scholar]
- 20.Webster G, Genschel J, Curth U, Urbanke C, Kang C, Hilgenfeld R. A common core for binding single-stranded DNA: structural comparison of the single-stranded DNA-binding proteins (SSB) from E. coli and human mitochondria. FEBS Lett. 1997;411:313–316. doi: 10.1016/s0014-5793(97)00747-3. [DOI] [PubMed] [Google Scholar]
- 21.Raghunathan S, Kozlov AG, Lohman TM, Waksman G. Structure of the DNA binding domain of E. coli SSB bound to ssDNA. Nat. Struct. Biol. 2000;7:648–652. doi: 10.1038/77943. [DOI] [PubMed] [Google Scholar]
- 22.Yuzhakov A, Kelman Z, O'Donnell M. Trading places on DNA–a three-point switch underlies primer handoff from primase to the replicative DNA polymerase. Cell. 1999;96:153–163. doi: 10.1016/s0092-8674(00)80968-x. [DOI] [PubMed] [Google Scholar]
- 23.Sandigursky M, Mendez F, Bases RE, Matsumoto T, Franklin WA. Protein-protein interactions between the Escherichia coli single-stranded DNA-binding protein and exonuclease I. Radiat. Res. 1996;145:619–623. [PubMed] [Google Scholar]
- 24.Genschel J, Curth U, Urbanke C. Interaction of E. coli single-stranded DNA binding protein (SSB) with exonuclease I. The carboxy-terminus of SSB is the recognition site for the nuclease. Biol. Chem. 2000;381:183–192. doi: 10.1515/BC.2000.025. [DOI] [PubMed] [Google Scholar]
- 25.Shereda RD, Bernstein DA, Keck JL. A central role for SSB in Escherichia coli RecQ DNA helicase function. J. Biol. Chem. 2007;282:19247–19258. doi: 10.1074/jbc.M608011200. [DOI] [PubMed] [Google Scholar]
- 26.Shereda RD, Kozlov AG, Lohman TM, Cox MM, Keck JL. SSB as an organizer/mobilizer of genome maintenance complexes. Crit. Rev. Biochem. Mol. Biol. 2008;43:289–318. doi: 10.1080/10409230802341296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Curth U, Genschel J, Urbanke C, Greipel J. In vitro and in vivo function of the C-terminus of Escherichia coli single-stranded DNA binding protein. Nucleic Acids Res. 1996;24:2706–2711. doi: 10.1093/nar/24.14.2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chase JW, L'Italien JJ, Murphy JB, Spicer EK, Williams KR. Characterization of the Escherichia coli SSB-113 mutant single-stranded DNA-binding protein. Cloning of the gene, DNA and protein sequence analysis, high pressure liquid chromatography peptide mapping, and DNA-binding studies. J. Biol. Chem. 1984;259:805–814. [PubMed] [Google Scholar]
- 29.Naue N, Fedorov R, Pich A, Manstein DJ, Curth U. Site-directed mutagenesis of the chi subunit of DNA polymerase III and single-stranded DNA-binding protein of E. coli reveals key residues for their interaction. Nucleic Acids Res. 2011;39:1398–1407. doi: 10.1093/nar/gkq988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frick DN, Richardson CC. DNA primases. Annu. Rev. Biochem. 2001;70:39–80. doi: 10.1146/annurev.biochem.70.1.39. [DOI] [PubMed] [Google Scholar]
- 31.Pan H, Wigley DB. Structure of the zinc-binding domain of Bacillus stearothermophilus DNA primase. Structure. 2000;8:231–239. doi: 10.1016/s0969-2126(00)00101-5. [DOI] [PubMed] [Google Scholar]
- 32.Keck JL, Roche DD, Lynch AS, Berger JM. Structure of the RNA polymerase domain of E. coli primase. Science. 2000;287:2482–2486. doi: 10.1126/science.287.5462.2482. [DOI] [PubMed] [Google Scholar]
- 33.Su XC, Schaeffer PM, Loscha KV, Gan PH, Dixon NE, Otting G. Monomeric solution structure of the helicase-binding domain of Escherichia coli DnaG primase. FEBS J. 2006;273:4997–5009. doi: 10.1111/j.1742-4658.2006.05495.x. [DOI] [PubMed] [Google Scholar]
- 34.Tougu K, Marians KJ. The extreme C terminus of primase is required for interaction with DnaB at the replication fork. J. Biol. Chem. 1996;271:21391–21397. doi: 10.1074/jbc.271.35.21391. [DOI] [PubMed] [Google Scholar]
- 35.Marceau AH, Bahng S, Massoni SC, George NP, Sandler SJ, Marians KJ, Keck JL. Structure of the SSB-DNA polymerase III interface and its role in DNA replication. EMBO J. 2011;30:4236–4247. doi: 10.1038/emboj.2011.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pace CN, Vajdos F, Fee L, Grimsley G, Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laue T, Shah B, Ridgeway T, Pelletier S. Computer-aided interpretation of analytical sedimentation data for proteins. In: Harding S, Rowe A, Horton J, editors. The Royal Society of Chemistry. Cambridge, UK: 1992. pp. 378–387. [Google Scholar]
- 38.Lohman TM, Overman LB. Two binding modes in Escherichia coli single strand binding protein-single stranded DNA complexes. Modulation by NaCl concentration. J. Biol. Chem. 1985;260:3594–3603. [PubMed] [Google Scholar]
- 39.Loscha K, Oakley AJ, Bancia B, Schaeffer PM, Prosselkov P, Otting G, Wilce MC, Dixon NE. Expression, purification, crystallization, and NMR studies of the helicase interaction domain of Escherichia coli DnaG primase. Protein Expr. Purif. 2004;33:304–310. doi: 10.1016/j.pep.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 40.Zabeau M, Stanley KK. Enhanced expression of cro-beta-galactosidase fusion proteins under the control of the PR promoter of bacteriophage lambda. EMBO J. 1982;1:1217–1224. doi: 10.1002/j.1460-2075.1982.tb00016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bohme HJ, Kopperschlager G, Schulz J, Hofmann E. Affinity chromatography of phosphofructokinase using Cibacron blue F3G-A. J. Chromatogr. 1972;69:209–214. doi: 10.1016/s0021-9673(00)83103-9. [DOI] [PubMed] [Google Scholar]
- 42.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 43.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 44.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dam J, Schuck P. Sedimentation velocity analysis of heterogeneous protein-protein interactions: sedimentation coefficient distributions c(s) and asymptotic boundary profiles from Gilbert-Jenkins theory. Biophys. J. 2005;89:651–666. doi: 10.1529/biophysj.105.059584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Witte G, Fedorov R, Curth U. Biophysical analysis of Thermus aquaticus single-stranded DNA binding protein. Biophys. J. 2008;94:2269–2279. doi: 10.1529/biophysj.107.121533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Naue N, Curth U. Investigation of protein-protein interactions of single-stranded DNA-binding proteins by analytical ultracentrifugation. Methods Mol. Biol. 2012;922:133–149. doi: 10.1007/978-1-62703-032-8_8. [DOI] [PubMed] [Google Scholar]
- 48.Schanda P, Kupce E, Brutscher B. SOFAST-HMQC experiments for recording two-dimensional heteronuclear correlation spectra of proteins within a few seconds. J. Biomol. NMR. 2005;33:199–211. doi: 10.1007/s10858-005-4425-x. [DOI] [PubMed] [Google Scholar]
- 49.Sattler M, Schleucher J, Griesinger C. Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Prog. Nucl. Mag. Reson. Spectrosc. 1999;34:93–158. [Google Scholar]
- 50.Lescop E, Schanda P, Brutscher B. A set of BEST triple-resonance experiments for time-optimized protein resonance assignment. J. Magn. Reson. 2007;187:163–169. doi: 10.1016/j.jmr.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 51.Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, Llinas M, Ulrich EL, Markley JL, Ionides J, Laue ED. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 52.Dam J, Velikovsky CA, Mariuzza RA, Urbanke C, Schuck P. Sedimentation velocity analysis of heterogeneous protein-protein interactions: Lamm equation modeling and sedimentation coefficient distributions c(s) Biophys. J. 2005;89:619–634. doi: 10.1529/biophysj.105.059568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.El Houry Mignan S, Witte G, Naue N, Curth U. Characterization of the chi psi subcomplex of Pseudomonas aeruginosa DNA polymerase III. BMC Mol. Biol. 2011;12:43. doi: 10.1186/1471-2199-12-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kozlov AG, Jezewska MJ, Bujalowski W, Lohman TM. Binding specificity of Escherichia coli single-stranded DNA binding protein for the chi subunit of DNA pol III holoenzyme and PriA helicase. Biochemistry. 2010;49:3555–3566. doi: 10.1021/bi100069s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Landau M, Mayrose I, Rosenberg Y, Glaser F, Martz E, Pupko T, Ben-Tal N. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 2005;33:W299–W302. doi: 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lu D, Keck JL. Structural basis of Escherichia coli single-stranded DNA-binding protein stimulation of exonuclease I. Proc. Natl Acad. Sci. USA. 2008;105:9169–9174. doi: 10.1073/pnas.0800741105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shereda RD, Reiter NJ, Butcher SE, Keck JL. Identification of the SSB binding site on E. coli RecQ reveals a conserved surface for binding SSB's C terminus. J. Mol. Biol. 2009;386:612–625. doi: 10.1016/j.jmb.2008.12.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ryzhikov M, Koroleva O, Postnov D, Tran A, Korolev S. Mechanism of RecO recruitment to DNA by single-stranded DNA binding protein. Nucleic Acids Res. 2011;39:6305–6314. doi: 10.1093/nar/gkr199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Crowley PB, Golovin A. Cation-pi interactions in protein-protein interfaces. Proteins. 2005;59:231–239. doi: 10.1002/prot.20417. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.